Abstract
Long-lived post-mitotic cells, such as the majority of human neurons, must respond effectively to ongoing changes in neuronal stimulation or microenvironmental cues through transcriptional and epigenomic regulation of gene expression. The role of epigenomic regulation in neuronal function is of fundamental interest to the neuroscience community, as these types of studies have transformed our understanding of gene regulation in post-mitotic cells. This perspective article highlights many of the resources available to researchers interested in neuroepigenomic investigations and discusses some of the current obstacles and opportunities in neuroepigenomics.
Neuroepigenomics Comes of Age
Epigenetic changes are historically defined as heritable changes that alter transcription but not the underlying DNA sequence. Unlike cells in many other tissues, most neurons in the human brain are post-mitotic (Lacar et al., 2014;Gage and Temple, 2013), with many individual neurons appearing to survive and function for decades. Thus, gene expression and associated synaptic changes are required to effectively respond to altered neuronal inputs, interactions with support cells, or environmental changes (e.g. nutrient levels, drugs of abuse, stress, inflammation, aging, and other microenvironmental triggers). This modulation of neuronal gene expression occurs via transcriptional and epigenomic mechanisms, which are likely to be adapted to accommodate the special requirements of neurons.
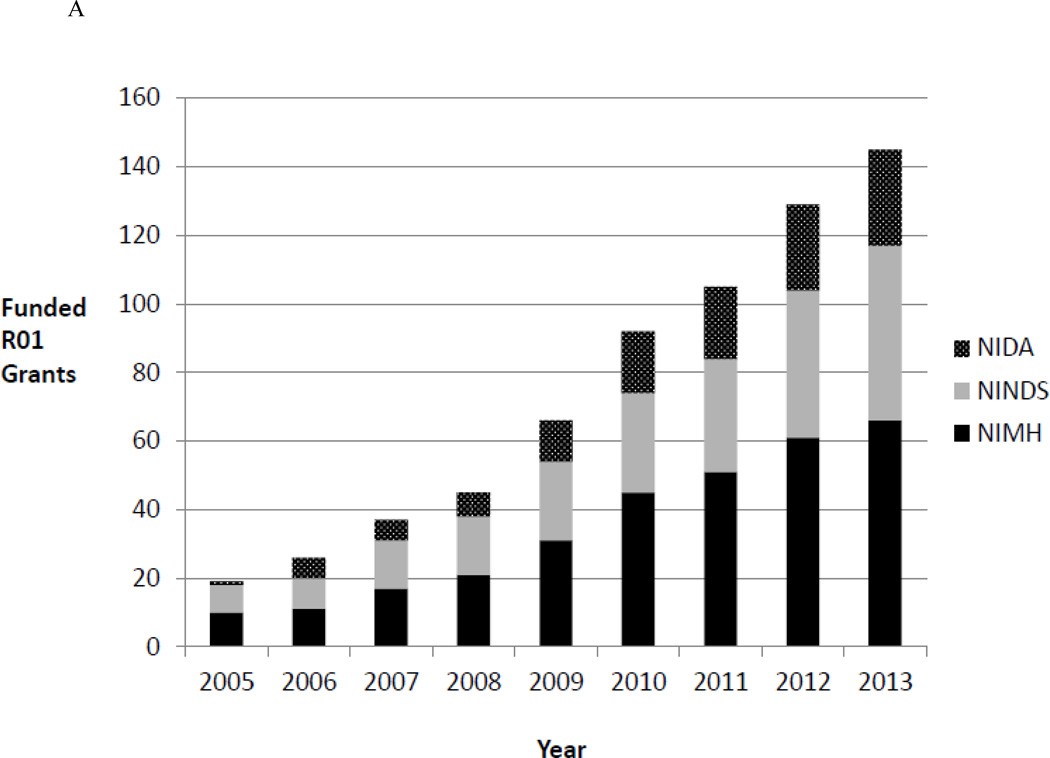
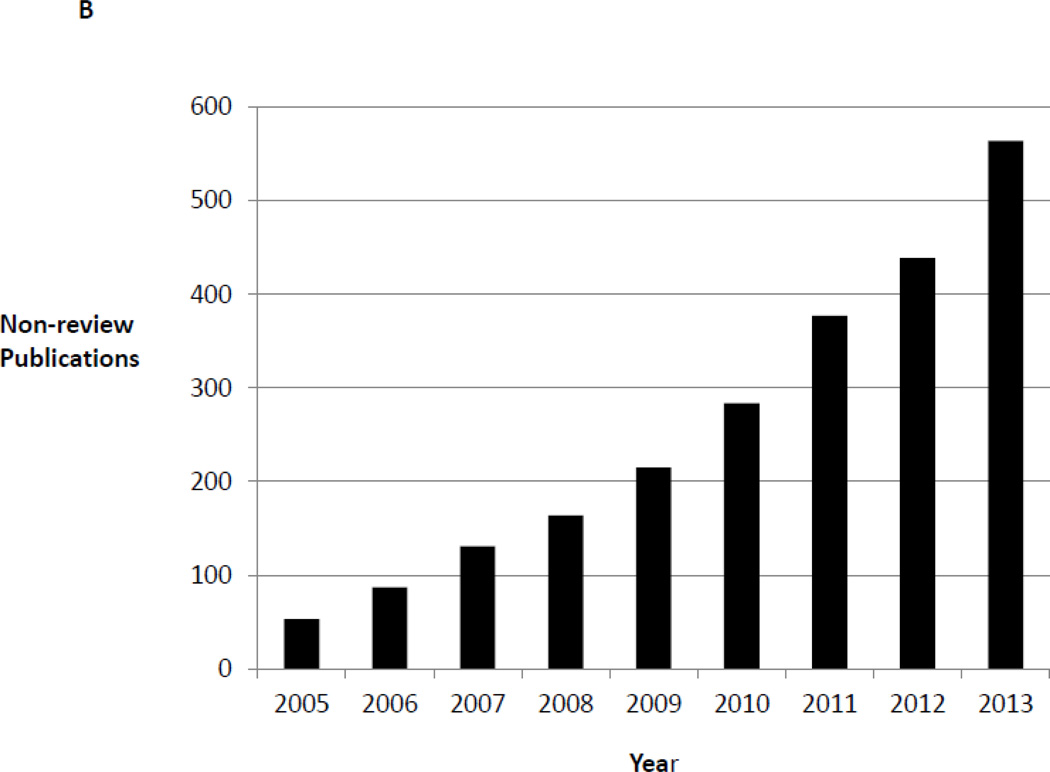
The field of epigenomics has exploded in recent years with improved assays, the generation of genome-wide epigenomic maps from multiple tissues, the identification of a host of epigenetic regulators important in numerous types of cancers, and the potential for the development of novel epigenetic therapies. Does this explosion extend to neuroepigenomics? Figure 1a shows the exponential increase in the number of funded R01 grants related to epigenetics/epigenomics from three neuroscience-focused NIH Institutes, indicating that many researchers are working in this scientific space. Figure 1b shows the increasing number of primary publications on topics that touch upon neuroepigenetics or neuroepigenomics, suggesting that epigenomic questions have captivated the neuroscience community.
Figure 1.
a. Increasing NIH Funded Research in Neuroepigenetics. Figure 1a shows the cumulative number of funded R01 epigenetic/epigenomic grants from 2005–2013 from three neuroscience-focused NIH institutes: National Institute on Drug Abuse (NIDA), National Institute on Mental Health (NIMH), and National Institute of Neurological Disorders and Stroke (NINDS). These data were obtained by searching NIH Reporter (http://projectreporter.nih.gov/reporter.cfm) in June 2014 for funded grants that used the terms epigenetic or epigenomic in their abstract or specific aims.
b. Increasing Numbers of Non-review Publications in Neuroepigenetics. Figure 1b shows the increasing number of non-review publications over time in the area of epigenetics or epigenomics in the nervous system. PubMed (http://www.ncbi.nlm.nih.gov/pubmed) was searched in June 2014 for titles or abstracts that mention epigen (to capture epigenetics or epigenomics) and a nervous system term (nervous system, or neuro or brain). The search was performed to capture only non-review publications.
Forays into neuroepigenetics research have led to a number of groundbreaking discoveries in substance use disorders, brain development, neurodegeneration, intellectual disability, memory, and even in transgenerational inheritance of behavioral phenotypes. Since several reviews have discussed the role of epigenetic regulation in the nervous system, we will briefly highlight a few of the key discoveries below (Sweatt, 2013;Day and Sweatt, 2011;Haggarty and Tsai, 2011a;Ma, 2010;Zocchi and Sassone-Corsi, 2010;Bellet and Sassone-Corsi, 2010;Maze et al., 2011;Maze et al., 2013;Nelson and Monteggia, 2011;Dulac, 2010;Namihira et al., 2008;Pena et al., 2014;Feng and Nestler, 2013;Rahn et al., 2013;Rogers et al., 2011;Bennett et al., 2014). For example, work from Eric Nestler’s laboratory has shown that cocaine exposure leads to defined changes in histone modifications and DNA methylation of neuronal regulators in the nucleus accumbens (Renthal et al., 2007;Renthal et al., 2009;LaPlant et al., 2010;Nestler, 2014). Investigations into autism and intellectual disability disorders indicate that epigenetic regulators (e.g. MECP2, MBD5, JARID1C, DNMT3A, ARID1B) play important roles in these disorders (Moretti and Zoghbi, 2006;Jensen et al., 2005;Tatton-Brown et al., 2014;Tsurusaki et al., 2012;Santen et al., 2012; Talkowski et al., 2012). Several lines of evidence point to an epigenetic basis underlying memory processing. Work from David Sweatt’s laboratory suggests an essential role for epigenetic regulation in memory formation and maintenance (Miller et al., 2008;Miller et al., 2010;Guzman-Karlsson et al., 2014;Day and Sweatt, 2011;Zovkic et al., 2014). Marcelo Wood and colleagues have found Brg1-associated factor (BAF) chromatin remodeling complexes to be necessary for memory and synaptic plasticity (Vogel-Ciernia et al., 2013). Li-Huei Tsai and colleagues have found that histone deacetylase inhibitors can effectively re-establish access to memories after neurodegeneration (Graff et al., 2014; Graff et al., 2012; Rudenko and Tsai, 2014). There is even evidence that certain exposures can lead to intergenerational inheritance of behavioral phenotypes (Szutorisz et al., 2014;Vassoler et al., 2013;Byrnes et al., 2011;Gapp et al., 2014;Dias and Ressler, 2014).
One of the most important epigenetic discoveries in the last several years is the identification of TET-mediated oxidized derivatives of 5-methylcytosine: 5-hydroxycytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC) in mammals (Rudenko and Tsai, 2014;Cheng et al., 2014;Kriaucionis and Heintz, 2009;Tahiliani et al., 2009;Mellen et al., 2012;Ito et al., 2011, Sun et al., 2014). 5hmC is especially abundant in the brain with up to 10-fold higher levels compared to embryonic stem cells and other tissues. hmC modification of DNA, initially discovered in Purkinje cells, is now known to play a critical role in stem cell biology and has emerging roles in other cell types and in nervous system disorders (Rudenko and Tsai, 2014;Cheng et al., 2014;Kriaucionis and Heintz, 2009;Tahiliani et al., 2009;Mellen et al., 2012). For example, analysis in specific brain cell types demonstrates that MeCP2, an epigenetic regulator known for its ability to bind 5mC of inactive gene promoters, binds 5hmC in active gene bodies in Purkinje cells, granule cells and Bergmann glial cells (Mellen et al. 2012). In the brain, this observation is accompanied by the loss of 5mC and an increase in 5hmC in the gene body of active genes. These observations are likely to have important implications in regards to gene expression and brain plasticity.
Tools and technologies for neuroscience research have improved significantly and will continue to improve through projects such as BRAIN (Brain Research through Advancing Innovative Neurotechnologies) http://www.nih.gov/science/brain/2025/index.htm. Neuroepigenomics will no doubt be an important component of many future discoveries in neuroscience. This review focuses on a few of the currently available resources that neuroepigenomics researchers might find useful, including reference epigenome maps, epigenomic assays and imaging tools and recent key discoveries in disease research. We will also discuss several of the current obstacles and opportunities in neuroepigenomics research, including: tools for single cell analysis and epigenomic manipulation, the need for additional brain cell reference epigenome maps, a deeper understanding of the mechanisms of transgenerational epigenetic inheritance, and the further development of epigenetic biomarkers and therapeutics. These obstacles and opportunities will become increasingly important as the field of neuroepigenomics emerges from “adolescence.”
Resources and Tools for Neuroepigenomics
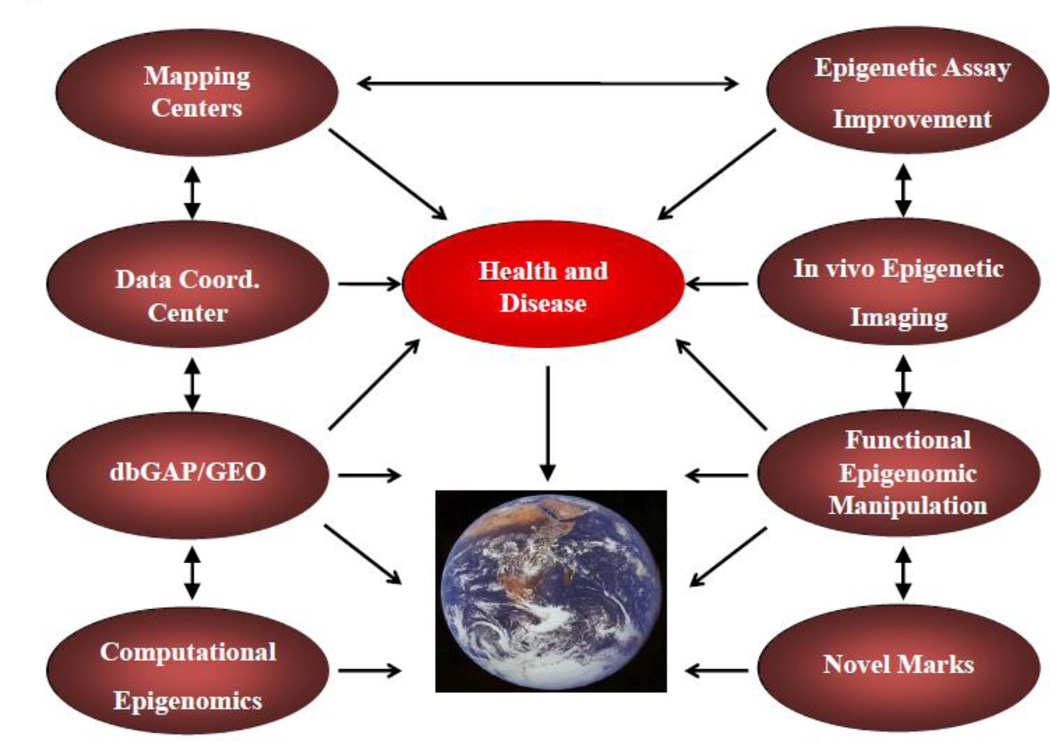
As shown in Figure 2, the Roadmap Epigenomics Program (supported by the NIH Common Fund) consists of multiple components with different functions, including 1. Development of new technologies to improve epigenome-wide assays, advance epigenetic imaging, and enable functional epigenetic manipulation; 2. Identification and characterization of novel epigenetic marks; and 3. Investigation of epigenomic processes underlying human disease (Satterlee et al., in press). Additionally, reference epigenome maps from normal cells and tissues were generated and uniformly processed by the Mapping Consortium and a Data Coordination Center. This data was deposited into NIH databases (Gene Expression Omnibus (GEO) or database of Genotypes and Phenotypes (dbGaP) where it is can be accessed by researchers (Bernstein et al., 2010). Most recently, a Computational Epigenomics component was added to support secondary data analysis studies using reference epigenome mapping data and other user-generated or public datasets to investigate important biological questions or diseases. Overall 83 R01, R21, or RC1 grants were funded through the Roadmap Epigenomics Program. Below we will discuss some of the neuroepigenomic-relevant tools and resources generated by these researchers.
Figure 2. NIH Common Fund (Roadmap) Epigenomics Program Components.
Figure 2 shows the components of the NIH Roadmap Epigenomics Program. The reference epigenome mapping components included Mapping Centers, a Data Coordination Center, and databases where scientists can obtain this information (Gene Expression Omnibus (GEO) and database of Genotypes and Phenotypes (dbGaP). A Computational Epigenomics component was recently funded to support computational investigations into important biological questions or diseases using reference epigenome mapping data. Three technology development initiatives endeavored to improve epigenome-wide assays, improve epigenetic imaging, and enable functional epigenetic manipulation. Two additional components included identification and characterization of novel epigenetic marks as well as investigations into epigenomic processes in human disease.
Reference Epigenome Maps for the Nervous System
In the nucleus, genes are turned on and off via a sophisticated interplay of transcriptional regulators; the consequences of this elaborate dance can be monitored in part through the assay of epigenomic features. The NIH Roadmap Epigenomics Program has generated a comprehensive catalog of epigenome maps for 92 distinct normal human cells and tissues (Bernstein et al., 2010). These maps were anticipated to stimulate a variety of hypothesis-generating studies such as 1. the identification of tissue-specific functional genetic elements, 2. uncovering the breadth of epigenomic plasticity during cellular differentiation, and 3. establishing a normal reference for investigators exploring the effects of environmental or disease on the epigenome (Satterlee, et al., In Press, Bernstein et al., 2010;Pollock et al., 2014).
These reference epigenomes (http://www.roadmapepigenomics.org) are available to the research community and prior publications have outlined how to access and visualize the data (Chadwick, 2012; Satterlee, et al., in Press,). The reference datasets typically include DNA methylation assays, ChIP-seq (chromatin immunoprecipitation followed by sequencing) for a core set of six post-translational histone modifications, and mRNA expression analysis. In some cases, tissues were also assayed for chromatin accessibility using DNAse I hypersensitivity assays. As indicated in Table 1, these assays can be used to help identify gene promoters, tissue-specific enhancers, or actively transcribed or repressed regions of the genome (Rada-Iglesias et al., 2011;Guenther et al., 2007;Kimura, 2013;2012;Barski et al., 2007;Creyghton et al., 2010;Wagner and Carpenter, 2012;Grewal and Jia, 2007; ENCODE Project Consortium, 2012). Table 1 shows the assays used to interrogate a variety of neuroscience-relevant cells and tissues, including: embryonic stem (ES) cells, ES-derived cells including neural progenitor cells, induced pluripotent stem (iPS) cells, post-mortem fetal tissues (brain and spinal cord), and post-mortem adult brain (angular gyrus, anterior caudate, cingulate gyrus, hippocampus, inferior temporal lobe, mid-frontal lobe, and substantia nigra). The Human Epigenome Browser (http://epigenomegateway.wustl.edu/info/) provides an Ensembl-like visualization of this epigenomic data and can even display long range genomic interactions (Zhou et al., 2011;Zhou et al., 2013). The Roadmap Epigenomics Mapping Consortium has also developed a set of experimental protocols, assay standards, and data quality standards/guidelines to aid researchers who wish to perform these types of assays in their own laboratories (http://www.roadmapepigenomics.org/protocols).
Table 1. Selected Neuroepigenomics Data Resources.
This table highlights resources generated by several large scale projects of relevance to neuroepigenomics researchers as of September 2014. Cell or tissue types are shown on the left, while epigenomic modifications, putative functions, and assay type are shown at the top (see text for further details). The upper section of the table describes resources generated by the NIH Roadmap Epigenomics Program (www.roadmapepigenetics.org) (Bernstein et al., 2010). This is followed by data from the International Human Epigenome Consortium (IHEC) (http://ihec-epigenomes.org/research/cell-types) as well as the Encyclopedia of DNA Elements (ENCODE) project (https://www.encodedcc.org) (Bernstein et al., 2012). Please note that the Roadmap Epigenomics Program and ENCODE are both IHEC members, so the IHEC datasets shown in this table were generated by the other IHEC members. Data from the MethylomeDB (http://www.neuroepigenomics.org/methylomedb/), Brain Cloud (http://braincloud.jhmi.edu/), and BrainSpan (http://www.brainspan.org) projects are also shown (Colantuoni et al., 2011; Xin et al., 2012;Miller et al., 2014). A blue square indicates the data is currently available, a green square indicates assays are in progress, while a white square indicates no data is available for a given assay for this cell or tissue type. Abbreviations used are WGBS (whole genome bisulfite sequencing), MeDIP (methylated DNA immunoprecipitation sequencing), MRE (Methylation-sensitive Restriction Enzyme Sequencing), RRBS (reduced representation bisulfite sequencing), TSS (transcription start site), smRNA-seq (small RNA-sequencing), DNase I HS (DNase I hypersensitivity assay), and Methyl-MAPS (Methylation Mapping Analysis by Paired-end Sequencing). Histone modifications (e.g. H3K36me3) were assayed by chromatin immunoprecipitation followed by sequencing.
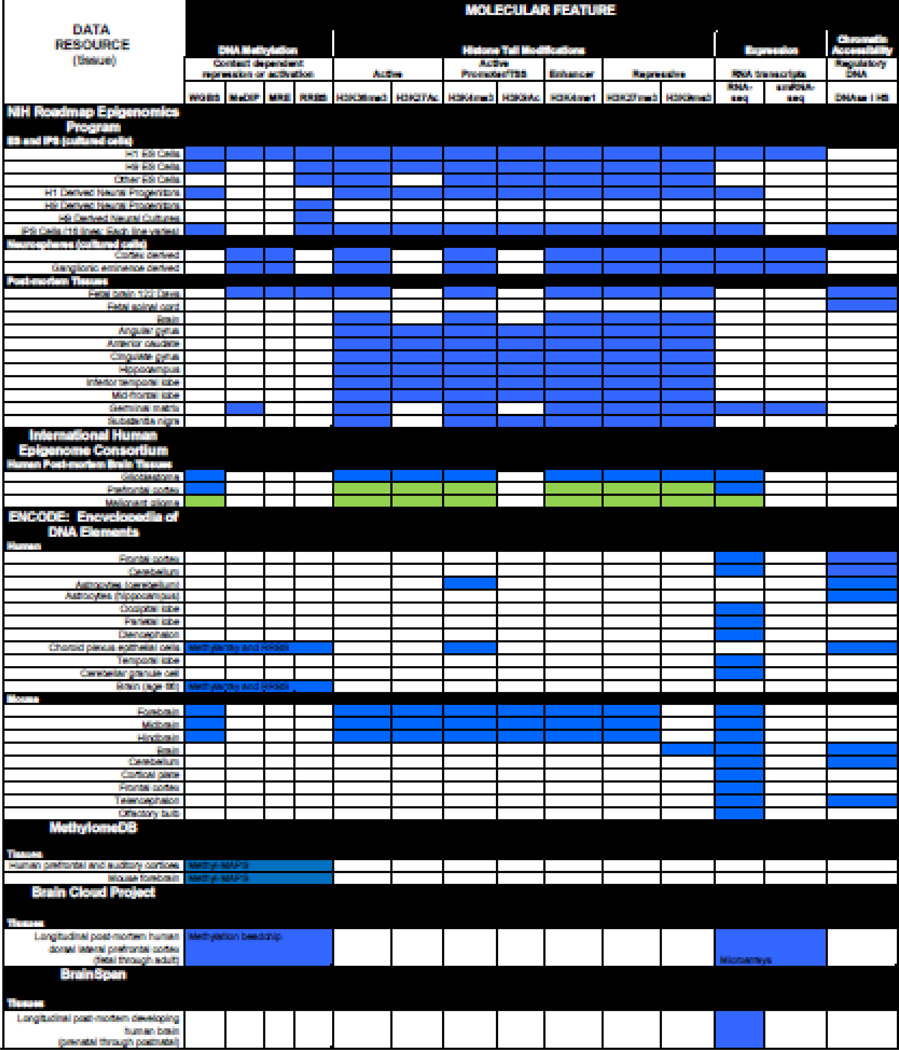 |
Additional datasets relevant for the neuroscience community include IHEC, ENCODE, MethylomeDB, Brain Cloud, and Brainspan. As shown in Table 1, the International Human Epigenome Consortium (IHEC) is generating similar epigenomic data sets for several additional human brain samples including prefrontal cortex, glioblastoma, and malignant glioma (http://ihec-epigenomes.org/outcomes/datasets/). Similarly, the ENCODE project has data for several human post-mortem brain regions (https://www.encodedcc.org)(Bernstein et al., 2012). As a part of the mouse ENCODE project, epigenomic and gene expression assays have been performed on forebrain, midbrain, hindbrain, and a number of other brain regions. MethylomeDB provides genome-wide DNA methylation data for selected mouse and human brain regions (http://www.neuroepigenomics.org/methylomedb/) (Xin et al., 2012). Two projects have collected longitudinal data that includes fetal development. The Brain Cloud project showcases gene expression and DNA methylation data for post-mortem human prefrontal cortices from fetal development through the aging adult (http://braincloud.jhmi.edu/) (Colantuoni et al., 2011), while BrainSpan provides transcriptome data for the developing human brain from pre-natal through post-natal development (http://www.brainspan.org/). See Table 1 for details about these selected resources for neuroepigenomics.
These datasets can be exploited for a variety of scientific investigations. For example, epigenomic data has enhanced the analysis of Genome Wide Association Study (GWAS) data (Ernst et al., 2011;Trynka et al., 2013;Karczewski et al., 2013;Pasquali et al., 2014;Maurano et al., 2012). In one study, researchers found that a significant percentage of disease-associated single nucleotide polymorphisms (SNPs) from GWAS occur in DNAse I hypersensitive sites which are frequently associated with transcription factor binding (Maurano et al., 2012). This combined epigenomic/GWAS analysis can enable researchers to mine GWAS datasets for relevant gene variants that were not statistically nominated using standard analysis methods. Aberrant gene silencing or activation could explain some of the variability in GWAS findings, and underscores the value of integrating epigenomic data with gene expression and genotype information. Remarkably, epigenomic/GWAS analyses can also be useful for predicting the cell types or tissues most likely to be impacted by a human disease phenotype (Maurano et al., 2012). For human brain disorders, a comprehensive set of cell type and brain region specific epigenomic datasets could enhance our ability to identify new gene variants involved in disease and help to corroborate or even predict cell types or brain regions disrupted in human brain disorders.
Epigenomic Assay and Imaging tools
Some of the technologies developed through the NIH Roadmap Epigenomics Program have contributed greatly to our ability to perform epigenetics research. For example the MethylC-seq whole genome bisulfite sequencing (WGBS) assay developed in the Ecker lab can be used to characterize methylomes, which are defined as all of the methylated and non-methylated DNA cytosine residues in a cell type or tissue (Lister et al., 2009). This assay was used to pioneer the exploration of mammalian methylomes and the publication describing it has been cited over 1082 times as of June 2014. These researchers found that a significant fraction of DNA methylation occurred in a non-CG context in human embryonic stem cells and later revealed important DNA methylation differences between human embryonic stem cells and induced pluripotent stem cells (Lister et al., 2009; Lister et al., 2011). MethylC-seq and related methylome assays have been applied to a variety of mammalian cell and tissue types, including the brain, by the Roadmap Mapping Consortium (see Table 1), as well as other researchers (Guo et al., 2014; Hovestadt et al., 2014; Varley et al., 2013). For example, during mammalian brain development 5-methylcytosine and 5-hydroxymethylcytosine undergo profound reconfiguration (Lister et al., 2013). The development or improvement of other epigenomic assays has also significantly enhanced our ability to interrogate the epigenome. For example, padlock probes allow the interrogation of DNA methylation at investigator-selected specific regions of the genome without the need for expensive whole genome sequencing (Deng et al., 2009;Diep et al., 2012), while nanofluidic approaches have been used to investigate the epigenomic state of single molecules (Cipriany et al., 2012; Cipriany et al., 2010).
Epigenomic assays have been improving steadily, however they typically provide measurements for only a single point in time. Our ability to observe chromatin features dynamically and in vivo has been quite limited. Researchers in the Lomvardas and Larabell labs are using soft X-ray tomography (SXT) to investigate chromatin domains in mouse olfactory neurons. Each neuron expresses only one olfactory receptor; the remaining ones are silenced. These studies show that reductions in lamin b receptor levels lead to the aggregation of the silenced olfactory receptors in the nuclear periphery, while the active receptors lie within an active transcriptional zone (Clowney et al., 2012). In the future, SXT could be combined with a fluorescence complementation strategy to enable visualization of epigenetic regulators in vivo. Using this strategy, a fluorescent signal is only observed following interaction of two proteins labeled with partial complementary fluorescent domains. As these and related approaches improve our ability to image chromatin features in vivo (including non-coding RNAs, DNA binding proteins, and modified histones), it is hoped that neuroscientists will be able to use these tools to better investigate how chromatin territories are associated with gene regulation in the nervous system.
A critical consideration for brain researchers is our almost complete inability to obtain brain specimens from living humans for epigenomic analysis. Each mammalian cell-type is believed to exhibit a distinct epigenome, thus interrogation of the brain epigenomes of specific cell types may be essential for disease diagnosis. Some researchers funded through the Roadmap Epigenomic Program have been exploring methods for in vivo imaging of epigenetic enzymes to begin to overcome this obstacle. Specifically these researchers are developing Positron Emission Tomography (PET) radiotracers for Class I and Class III histone deacetylases (HDACs) (Wang et al., 2013;Schroeder et al., 2013;Yeh et al., 2013). Development and pharmacokinetic optimization of these in vivo brain permeable PET ligands that monitor HDAC levels or activity in humans could improve accuracy of disease diagnosis, enable monitoring of the efficacy of epigenetic therapeutics, or enhance our ability to explore the effects of environmental factors such as psychosocial stress or substance abuse.
Disease investigations
Historically, cancer researchers have been the most strenuous pursuers of epigenetic studies. However many scientists have wondered about the potential role of epigenetic regulation in other diseases and chronic conditions including those that impact the nervous system. To encourage work in this area, the Roadmap Epigenomics Program supported research projects that investigated potential epigenetic changes that underlie a number of diseases including autism, Alzheimer’s Disease, schizophrenia, bipolar disorder, and substance use disorders. Some of the publications associated with these investigations can be found at the following website (http://www.roadmapepigenomics.org/publications).
Of particular interest are two recent Epigenome-Wide Association Studies (EWAS) which profile alterations in CpG methylation in post-mortem brain regions of patients with Alzheimer’s Disease (AD). The investigators independently converged on several loci including CpG dinucleotides near ANK1, RPL13, CDH23, and RHBDF2 (De Jager et al., 2014;Lunnon et al., 2014;Lord and Cruchaga, 2014). Interestingly, calculations by one group suggested that the 71 CpG variants they identified explained 28.7% of the variance in neuritic amyloid plaque burden, while all known AD gene variants from GWAS studies explained only 13.9% of the variance (De Jager et al., 2014;Lord and Cruchaga, 2014). Thus epigenomic studies can reveal new candidate loci involved in brain diseases and suggest that DNA methylation may play a role in the onset or progression of AD.
Technology, Tool, and Research Needs for Neuroepigenomics
Although the field of neuroepigenomics has made great strides, it is clear that even greater progress has been hampered by specific obstacles that must be overcome. Briefly we will describe some of the technology needs for neuroepigenomics, such as tools for epigenomic manipulation and robust single cell assays. Similarly, resource and dataset needs in neuroepigenomics include: expanding neuroepigenomic datasets, exploration of human neuroepigenomics using imaging technologies and post-mortem brain resources, as well as the exploration and development of neuroepigenomic surrogates and biomarkers. Finally there are some very exciting opportunities in neuroepigenomics research that should not be overlooked including exploration of the 4-dimensional (4D) structure of neuronal genomes, somatic mosaicism in neuronal cells, environmental epigenomics, investigation of mechanisms of intergenerational inheritance of behavioral phenotypes, and further development of epigenetic neurotherapeutics.
Technology Needs for Neuroepigenomics
Tools for Cell-type Specific, Locus-specific, and Temporal Manipulation of Neuronal Epigenomes
In the nervous system, optogenetic and chemogenetic strategies have been instrumental in enabling neuroscientists to explore questions regarding neuronal function (Wess et al., 2013;Dong et al., 2010;Chow et al., 2012;Fenno et al., 2011). However long term changes in neuronal function are associated with concomitant changes in gene expression via transcription factor and epigenomic regulation. Our ability to investigate long term gene expression changes in the nervous system via manipulation of the epigenome and the associated expression of genes has lagged in comparison; most strategies employ small molecule modulators (e.g. HDAC inhibitors) or RNA silencing methods. These approaches provide limited temporal control and impact many cell types and genomic loci. The ability to conduct spatiotemporal manipulation in vivo would enable researchers to probe the effects of locus-specific changes to the epigenome in neuronal or glial cells in a reversible manner. Some researchers are already making important strides in this direction. For example, Feng Zhang and colleagues have developed first generation genetic tools called LITES (light-inducible transcriptional effectors) that enable researchers to optically control transcriptional and epigenetic states (Konermann et al., 2013).
In order to address this critical technology gap, the Roadmap Epigenomics Program is supporting the development of a variety of robust tools and technologies in this area. These include manipulating the epigenome at specific loci using genome editing technologies (e.g. TALE, CRISPR), temporally regulating the epigenome via opto-epigenetic or chemo-epigenetic strategies, and exploiting available genetic tools to achieve cell-type specificity in key model organisms. It will be of great interest to see what fundamental questions in neuronal gene regulation can be answered using these platforms.
Single Cell Analyses
During normal growth and differentiation, cells must tightly control if, when, and where gene expression occurs. Epigenetic marks are critical in ensuring that the correct gene expression patterns are maintained and transmitted to the next generation of cells. Each type of cell displays a distinct epigenomic profile that reflects its past experiences and developmental potential (Gifford et al., 2013;Zhu et al., 2013). This epigenetic profile is read by the transcriptional machinery, creating a unique gene expression signature. Current technologies permit epigenetic marks to be studied in extracts from large populations of cells, yet epigenetic gene regulation occurs within single living cells (Wills et al., 2013). Distinct microenvironments within cellular niches likely influence the molecular and cellular phenotypes of different cell types. Induced pluripotent stem (iPS) cells provide a striking example of this cellular diversity, since only some of the precursor cells have the necessary plasticity to de-differentiate into a pluripotent state (Smith et al., 2010). This ability to transform from a differentiated state to an iPS cell may reflect the individual epigenomes of these cells. Unfortunately current technologies do not permit analysis of a given epigenomic modification in a single cell at a global scale. This challenge is especially acute in learning and memory studies, where epigenomic changes may occur in response to neural activity (Zovkic et al., 2013). Although improvements have allowed glimpses into the DNA methylation and gene expression profiles at the single cell level, more work must be done to enable assay of histone modifications and other chromatin features at the single cell level (Hayashi-Takanaka et al., 2011;Shalek et al., 2014;Patel et al., 2014; Smallwood et al., 2014;Iourov et al., 2012). There is great hope that the Common Fund Single Cell Analysis program (https://commonfund.nih.gov/Singlecell) will facilitate the development of platforms with the capability of studying the epigenomes and gene expression profiles of individual cells.
Resource and Dataset Needs for Neuroepigenomics
Expanded Neuroepigenomic Datasets
Although the Roadmap Epigenomics Program and other projects have generated valuable epigenomic datasets, systematic efforts to apply molecular phenotyping strategies to the nervous system are lacking. A comprehensive atlas of molecular phenotypes that spans a wide variety of brain regions, brain cell types, and developmental stages for both human and mouse is crucial for understanding the molecular etiology and ontology of neurological diseases. Key molecular phenotypes should encompass chromatin features (e.g. histone or DNA modifications), transcription factor binding sites, and mRNA/ncRNA expression whenever possible. The histone variant H2A.Z was recently shown to play a role in memory consolidation suggesting that histone variants would be an important molecular feature to assay (Zovkic et al., 2014). Secondary molecular phenotypes that could be assayed include modified RNAs, circular RNAs, and higher order chromatin structure. Ideally, these molecular phenotypes will be connected whenever possible to brain cell morphology, connectivity, and electrophysiological measures. In addition to helping elucidate the ontology of disease, molecular phenotypes will aid in the interpretation of GWAS and other datasets that investigate psychiatric diseases, as well as help to delineate candidate cell-type specific molecular targets for the development of small molecule therapeutics.
Human Brain Disease Epigenomics: Post-mortem and Imaging Resources
Investigation of the human brain epigenome is a necessary step to understanding long term changes in gene regulation and gene expression that may be associated with neurodevelopmental or neuropsychiatric disorders. It is also important to determine the extent to which the molecular pathways that regulate brain phenotypes in rodent and non-human primate models are recapitulated in humans. For studies exploring the mechanisms of disease processes, it is of critical importance to study the epigenomic changes that occur in the tissue and cell types specifically associated with that disease. Yet unlike diseases involving blood or skin, epigenomic assay of the human brain is particularly problematic. It is uncommon to obtain fresh surgical specimens and, even when available, these brain samples are typically associated with a preexisting disease state.
Post-mortem human studies are therefore essential for investigating some epigenomic changes associated with brain diseases. However, alterations occurring at or near the time of death (e.g. changes in oxygen levels or brain pH) can negatively impact the stability and levels of molecular brain phenotypes. Minimization of the interval between death and brain collection is essential for maximal preservation of molecular phenotypes (Birdsill et al., 2011). In general, DNA methylation marks can be fairly well preserved in post-mortem brain samples and mRNA profiling has been performed successfully (Ernst et al., 2008;Twine et al., 2011;Kang et al., 2011;Colantuoni et al., 2011). However, histone modifications and other chromatin features may be less stable, and some classes or subsets of RNAs may have differential sensitivity to the post-mortem interval (Barrachina et al., 2012). Animal studies have been used to examine alterations in mRNA expression associated with the interval between post-dissection and tissue preservation. Depending on the length of the interval, clear gene expression signatures emerge both temporally and functionally (e.g. hypoxia inducible genes, heat shock proteins, stress-response genes etc.) (Sanoudou et al., 2004;Zhao et al., 2006; Durrenberger et al., 2010; Catts et al.,2005;Trotter et al., 2002). To better address the extent to which fresh and post-mortem human tissues differ, freshly resected normal or diseased tissue with minimal time to preservation can be compared to more widely available post-mortem human brain tissue. Comparisons of gene expression profiles for these two conditions are currently underway for human non-brain tissue in the Common Fund Genes, Tissue, and Expression (GTEx) program (http://www.gtexportal.org/home/). One confounding issue is the lack of appropriate controls with human post-mortem research due to unique genotypes and environmental exposures. However post-mortem brain studies have been successful for helping understand human brain function and disorders such as autism and Alzheimer’s disease (Twine et al., 2011;Kang et al., 2011;Colantuoni et al., 2011; Mill et al., 2008; Voineagu et al., 2011; Davies et al., 2012; De Jager et al., 2014;Lunnon et al., 2014).
An alternative possibility for exploring the human brain epigenome is the use of in vivo imaging approaches. As described earlier, PET ligands can be used to measure the amount and activity of certain HDACs. Although this does not reveal epigenomic information at the single gene level or for an individual cell type, these approaches may have value for disease diagnosis and measurement of therapeutic efficacy. The development of in vivo molecular imaging approaches to monitor a greater variety of epigenomic readers, writers, and erasers as well as approaches that enable more refined measurement of epigenomic features would revolutionize neuroscience research.
Neuroepigenomic Surrogates and Biomarkers
Given our inability to obtain human brain samples from living patients, another approach of great potential clinical utility is to identify robust peripheral indicators that closely reflect both the phenotypic and epigenomic changes identified in disease-relevant brain cells. These peripheral indicators could include accessible cell types such as blood cells, skin, buccal samples, olfactory epithelia or even body fluids. Both animal and human studies could be used to advance our knowledge in this area. Animal studies would enable experimental control of genotype and environment and would provide a more detailed understanding of how epigenomic events in the brain are correlated with molecular phenotypes in more accessible cell types. Parallel human experiments could be carried out using post-mortem brain and peripheral tissues from the same donor. The Common Fund Genes, Tissue, Expression (GTEx) program (http://www.gtexportal.org/home/) will in part address the latter scientific question, since one goal of this program is to capture genotype and RNA-seq information for 50 tissues from each individual donor. When the GTEx program is completed, it is expected that data for 900 individual donors will be available. Epigenomic and other molecular phenotype assays will be added to the GTEx datasets for some tissues, which may help to identify the surrogate tissues most salient for brain investigations (Lonsdale, et al., 2013). However, a surrogate tissue strategy may not yield useful biomarkers for all neurological disorders. A recent Alzheimer’s Disease EWAS study reports that the DNA methylation state of surrogate tissues (cerebellum and whole blood) does not recapitulate disease relevant DNA methylation differences in brain tissues impacted by AD (superior temporal gyrus and prefrontal cortex) (Lunnon et al., 2014).
Body fluids are another potential source for generation of informative biomarkers. Extracellular vesicles and proteins associated with extracellular RNAs, exRNAs, appear to move through the body and act in ways analogous to the endocrine system (Christianson et al., 2014;Lai and Breakefield, 2012;Yang et al., 2011). The best current candidates to test for exRNA content include cerebral spinal fluid (CSF) and blood serum, although other bodily fluids could provide additional informative biomarkers (Saman et al., 2012;Revenfeld et al., 2014). Studies aimed at generating methods to purify extracellular vesicles derived solely from brain could, theoretically, prove an exceptionally useful source of biomarkers for brain disorders.
Additional Opportunities in Neuroepigenomic Research
The Neuronal Genome: 4-Dimensional Structure
Evidence from imaging, as well as genome conformation assays (e.g. Hi-C, ChIA-PET), indicates that cellular genomes have complex and dynamic three-dimensional structures (Lieberman-Aiden et al., 2009;Fullwood et al., 2009;Clowney et al., 2012;Mitchell et al., 2014). Although our knowledge of the structure of neuronal or glial genomes is poorly understood, recent studies demonstrate that olfactory neurons display a complex genomic architecture that differs between olfactory neuron types depending upon the gene expression status of individual olfactory receptors (Clowney et al., 2012). Furthermore ATP-dependent chromatin remodeling proteins such as BAF53b have neurodevelopmental, synaptic plasticity, and memory functions, suggesting that genome conformation plays an important functional role in the nervous system (Yoo et al., 2009;Staahl and Crabtree, 2013;Vogel-Ciernia et al., 2013). However a systematic investigation of the 4D structure of the genome conformation of distinct neuronal cell types is needed to understand the extent of the relationship between genome conformation and neuronal function. The recently launched Common Fund 4D-Nucleome program will begin to address some of the scientific questions in this area (http://commonfund.nih.gov/4Dnucleome/index). Studies aimed at gaining a deeper understanding of how transcription factor binding, epigenomic modifications, and 4D nuclear architecture correlate with gene expression would yield important insights into the complexities and dynamics of gene regulation in normal tissues and during disease processes.
Brain Somatic Mosaicism and Epigenomic Regulation
It is often assumed that the genomes of all of our cells are identical. However, this is not always the case, as enucleated red blood cells, haploid germ cells, and immune cells are genomically distinct from the majority of the cells in the rest of the body. Each haploid germ cell contains a distinct genome, while every B-cell and T-cell undergoes a unique recombination event that generates a repertoire of immune cells that are poised to attack different types of antigens (Alt and Baltimore, 1982; Roth et al., 1992). It is becoming increasingly clear that as cells in the nervous system differentiate, they may undergo genomic rearrangements or acquire copy number or other structural variations, which ultimately can lead to significant levels of somatic mosaicism in the brain. For example Jerold Chun and colleagues demonstrated that distinct cells from the nervous system differ dramatically in their complement of chromosomes (Rehen et al., 2005;Bushman and Chun, 2013; Westra et al., 2010). Furthermore, L1 retrotransposons can become active, jumping into different chromosomal locations within the brain (Singer et al., 2010;Muotri et al., 2010;Erwin et al., 2014; Reilly et al., 2013). In some organisms, epigenomic regulation modulates transposon activity (Lorenz et al., 2012;Creasey et al., 2014). Interestingly MeCP2, which is mutated in patients with Rett Syndrome (an autism spectrum disorder), can regulate L1 transposition (Amir et al., 1999;Muotri et al., 2010). MeCP2 is known for its role in regulating epigenetic processes; it binds 5mC residues, as well as the oxidized derivative, 5hmC. 5hmC levels are higher in the brain than any other tissue (Kriaucionis and Heintz 2009), and it would be intriguing to understand the potential roles that 5mC and 5hmC may play in retrotransposon activation.
In addition to somatic mosaicism on the genomic level, females display partial somatic mosaicism on an epigenetic level due to X-chromosome inactivation. X-inactivation is apparently random and leads to clustering of daughter cells; some will inactivate the maternal X chromosome, while others will inactivate the paternal X chromosome. DNA methylation, histone code changes and expression of ncRNAs mediate this process (Gendrel and Heard 2014). X-inactivation can contribute to disease phenotypes in Rett Syndrome (Braunschweig et al. 2004) and other neurobehavioral disorders (Lasalle and Yasui 2009), where the males are often more highly affected than the females. In addition, regions subject to genomic imprinting also display localized somatic mosaicism at the epigenetic level, as the epigenotypes on the maternal alleles differ from the paternal alleles. Mistakes in this process lead to several imprinted disorders, including Prader-Willi Syndrome and Angelman Syndrome (Horsthemke and Wagstaff, 2008;Reis et al. 1994). Individuals affected by Prader-Willi Syndrome exhibit specific behavioral phenotypes that include hyperphagia and obsessive compulsive disorder (Saitoh et al. 1997), while those affected by Angelman Syndrome display severe developmental delay, an easily excitable personality with an inappropriately happy affect, profound movement and balance deficits, as well as seizures (Lossie et al. 2001). One important area for future research is to explore the extent to which somatic mosaicism, at the genomic and epigenomic levels, occurs in the brain and whether or not it underlies neurodevelopmental, neuropsychiatric, or substance abuse disorders.
Environmental Epigenomics Investigations
Environmental exposures such as prenatal environment, early childhood or adult trauma, psychiatric medications, or exposure to substances of abuse are associated with epigenomic alterations in particular brain cell types (Zhang et al., 2013; Satterlee, 2013; Pena et al., 2014;Nestler, 2014;McGowan et al., 2009; Miller et al., 2010;Rutten and Mill, 2009;Toffoli et al., 2014). Although several studies have documented these changes, they have not been systematically investigated. In most cases it is unclear how long these presumed environmental epimutations perdure and what molecular pathways are involved in maintaining or reversing these changes (Robison and Nestler, 2011;Zhang et al., 2013;Guerrero-Bosagna et al., 2013;Berger et al., 2009). One approach to address these questions would be to perform tightly controlled, systematic experiments in which genetically identical animals are quantitatively exposed to environmental stressors, such as psychosocial stress or substances of abuse and then phenotyped for a suite of epigenomic brain features at different time points. These studies would determine the long-term plasticity and persistence (days, weeks, months) of brain epigenome changes and enable researchers to begin to functionally characterize the biological processes involved in formation, maintenance, or erasure of brain epigenome features resulting from environmental exposures.
Mechanisms of Intergenerational Inheritance
There is evidence that exposure to certain chemical toxins, social environments, or nutrient levels can, in some cases, lead to persistence of particular phenotypes in subsequent generations. These phenotypes appear to be transmitted without an apparent DNA mutation and can be transmitted even when subsequent generations have not been exposed to the environmental factor (Youngson and Whitelaw, 2008;Skinner et al., 2008;Champagne, 2008;Weaver et al., 2004;Crews et al., 2012;Carone et al., 2010). For example, endocrine disruptor (bisphenol A) exposure can lead to behavioral effects on social recognition in subsequent generations (Rissman and Adli, 2014). Work from Chris Pierce and colleagues showed that male rats that self-administered cocaine had sons, but not daughters, that were resistant to the acquisition of cocaine self-administration and that this phenotype was correlated with Bdnf promoter Histone-3 acetylation in sperm from cocaine-exposed fathers (Vassoler et al., 2013;Vassoler and Sadri-Vakili, 2014). Exposure to tetrahydrocannabinol (THC), morphine, nicotine, or methamphetamine also appears to impact phenotypes in subsequent generations (e.g. heroin seeking, anxiety), although the mechanisms by which this occurs remain to be elucidated (Rehan et al., 2013;Zhu et al., 2014;Itzhak et al., 2014;Szutorisz et al., 2014;Byrnes et al., 2011). Chronic stress exposure can lead to increased anxiety and other behavioral phenotypes in subsequent generations (Saavedra-Rodriguez and Feig, 2013), while early life trauma in mice impacts metabolic and behavioral phenotypes in the next generation and may be transmitted through sperm RNAs (Gapp et al., 2014). There has even been a report that a parental olfactory experience (fear conditioning paired with the odorant acetophenone) is associated with increased DNA methylation of the acetophenone odorant receptor in sperm. The resulting progeny exhibited increased sensitivity to acetophenone (Dias and Ressler, 2014).
Future studies aimed at validating claims of intergenerational inheritance are critical to ensure that any reported findings are robust and reproducible in different laboratories. It will also be crucial to identify the molecular mechanisms by which changes or risk factors are transmitted to and manifested in subsequent generations. It is often hypothesized that germline transmission of epigenetic features accounts for the persistence of some phenotypes over multiple generations, so it will be important to investigate the association of germline epigenetic modifications (and any RNAs that might influence these modifications) with offspring phenotype. It will also be critical to explore alternate hypotheses, including neurobehavioral or societal transmission or transmission through infectious agents such as viruses or the parental microbiome (Youngson and Whitelaw, 2008). Despite these caveats, this area of research has great relevance to disease prevention, since knowledge that a particular environmental exposure could lead to phenotypes in subsequent generations would have profound public health implications.
Epigenetic Neurotherapeutics
If a disease is caused by the presence of a particular gene variant or mutation, then gene therapy approaches may ultimately be necessary to correct the disorder. However epigenetic changes are inherently more plastic than DNA-based mutations and thus should be more readily impacted by small molecule therapeutics (Haberland et al., 2009;Haggarty and Tsai, 2011b). We will briefly touch on two areas of epigenetic neurotherapeutic investigation: histone deacetylase (HDAC) inhibitors and histone acetylation readers such as BRD4. HDAC inhibitors have been used to treat T-cell lymphoma as well as bipolar disorder, migraines, and seizures (Sharma et al., 2010;Mack, 2006;Bialer and Yagen, 2007). In animal models, HDACis can positively influence depression as well as cognitive defects in an Alzheimer’s Disease model (Kilgore et al., 2010;Fischer et al., 2007b). For substance abuse, prolonged inhibition of Class I HDACs with MS-275 blocks cocaine locomotor sensitization (Kennedy et al., 2013). The HDAC inhibitor sodium butyrate has been shown by Marcelo Wood and colleagues to facilitate extinction of cocaine-associated cues (Malvaez et al., 2010), suggesting that HDACis or perhaps other epigenetic therapeutics could be particularly efficacious if used in concert with behavioral therapies.
One limitation of HDACis is that they are pleiotropic and can impact many different cells and genetic loci. It is known that certain epigenetic “reader” proteins can bind to a subset of histone modifications. Thus researchers targeted BRD4, which can bind to a subset of acetylated lysines, in an attempt to generate therapeutics with more specificity than HDACis (Dey et al., 2003). JQ1, a small molecule inhibitor of BRD4, has promise as a potential treatment for acute myeloid leukemia (AML) (Zuber et al., 2011; Filippakopoulos et al., 2010). Researchers are also beginning to target histone methylation enzymes as well as other epigenetic “readers, writers, and erasers” to treat a variety of diseases including nervous system disorders (Grant, 2009;Hamada et al., 2010;Fiskus et al., 2009). It would be of great value to systematically develop small molecule modulators of epigenetic readers, writers, and erasers to 1. serve as chemical probes to investigate the functions of these enzymes, 2. for development into ligands for in vivo imaging studies, and 3. as potential lead compounds for future therapeutic development (Arrowsmith et al., 2012). Some work in this area is currently being pursued by the Structural Genomics Consortium http://www.thesgc.org/epigenetics.
Summary
As the field of neuroepigenomics matures it is likely to produce revolutionary new insights into the regulation of gene expression in neurodevelopmental and neuroplastic processes in cells that can persist for decades. It will also likely yield new and perhaps paradigm-shifting opportunities for the diagnosis, treatment and prevention of diseases of the nervous system. In this review we described a few of the tools and technologies available to neuroepigenomics researchers currently, including reference epigenome maps for nervous system cells and tissues, improved epigenomic assays, improved ways to monitor epigenetic enzymes and changes in vivo, and a deeper understanding of epigenetic mechanisms in nervous system disorders.
We have also delineated some of the obstacles and opportunities in this area including: improved tools for manipulating epigenetic processes, additional reference epigenomes for the nervous system, improved technologies for single cell analyses, validation of animal epigenetic studies using human post-mortem tissue, investigation into the potential of surrogate tissues and body fluids as biomarkers, and exploration of the 4-D chromatin structure of nervous system-relevant cell types. Another important area for future work is to better understand how acute or chronic environmental exposures (e.g. early life stress, drugs of abuse, environmental toxins, diet, inflammation, aging) impact the brain epigenome both somatically (including the somatic genome) as well as in subsequent generations.
There are three additional gaps and opportunities in neuroepigenomics that should be mentioned briefly. The first is that the computational needs for neuroepigenomic research are challenging, and it will be important to develop user-friendly computational tools to enable researchers to mine and exploit epigenomic information effectively. Secondly, the typical graduate or post-doctoral training program for neuroscientists differs greatly from that of epigenomicists. The development of explicit training programs in neuroepigenomics would help researchers be able to move more seamlessly between these two scientific “worlds” and better train the next generation of neuroepigenomicists. Finally as neuroepigenomic tools and resources improve, it is essential that individual researchers continue to initiate cutting edge explorations into the epigenetic mechanisms of nervous system disorders. Without an understanding of these mechanisms at a deep level, it will be impossible to improve the prevention, diagnosis, and treatment of nervous system disorders due to epigenetic dysregulation.
Acknowledgements
The authors would like to thank the members of the NIH Epigenomics Work Group, as well as the epigenetics researchers in the NIH Roadmap Epigenomics Program and beyond that have built a strong scientific foundation for this work over many years and are moving this field forward. The views expressed in this article are solely those of the authors and may not necessarily reflect those of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrea Beckel-Mitchener, Email: amitchen@mail.nih.gov.
Roger Little, Email: alittle@nida.nih.gov.
Dena Procaccini, Email: dena.procaccini@nih.gov.
Joni L. Rutter, Email: jrutter@nida.nih.gov.
Amy C. Lossie, Email: amy.lossie@nih.gov.
References
- Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Barrachina M, Moreno J, Villar-Menendez I, Juves S, Ferrer I. Histone tail acetylation in brain occurs in an unpredictable fashion after death. Cell Tissue Bank. 2012;13:597–606. doi: 10.1007/s10561-011-9278-9. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci. 2010;123:3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Yu L, Yang J, Srivastava GP, Aubin C, De Jager PL. Epigenomics of Alzheimer's disease. Transl Res. 2014 doi: 10.1016/j.trsl.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialer M, Yagen B. Valproic Acid: second generation. Neurotherapeutics. 2007;4:130–137. doi: 10.1016/j.nurt.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12:311–318. doi: 10.1007/s10561-010-9210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig D, Simcox T, Samaco RC, LaSalle JM. X-Chromosome inactivation ratios affect wild-type MeCP2 expression within mosaic Rett syndrome and Mecp2−/+ mouse brain. Hum Mol Genet. 2004;13:1275–1286. doi: 10.1093/hmg/ddh142. [DOI] [PubMed] [Google Scholar]
- Bushman DM, Chun J. The genomically mosaic brain: aneuploidy and more in neural diversity and disease. Semin Cell Dev Biol. 2013;24:357–369. doi: 10.1016/j.semcdb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes JJ, Babb JA, Scanlan VF, Byrnes EM. Adolescent opioid exposure in female rats: transgenerational effects on morphine analgesia and anxiety-like behavior in adult offspring. Behav Brain Res. 2011;218:200–205. doi: 10.1016/j.bbr.2010.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Catts SV, Fernandez HR, Taylor JM, Coulson EJ, Lutze-Mann LH. A microarray study of post-mortem mRNA degradation in mouse brain tissue. Brain Res Mol Brain Res. 2005;138:164–177. doi: 10.1016/j.molbrainres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol. 2008;29:386–397. doi: 10.1016/j.yfrne.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Bernstein A, Chen D, Jin P. 5-Hydroxymethylcytosine: A new player in brain disorders? Exp Neurol. 2014 doi: 10.1016/j.expneurol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Han X, Boyden ES. Genetically encoded molecular tools for light-driven silencing of targeted neurons. Prog Brain Res. 2012;196:49–61. doi: 10.1016/B978-0-444-59426-6.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson HC, Svensson KJ, Belting M. Exosome and microvesicle mediated phene transfer in mammalian cells. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Cipriany BR, Murphy PJ, Hagarman JA, Cerf A, Latulippe D, Levy SL, Benitez JJ, Tan CP, Topolancik J, Soloway PD, Craighead HG. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc Natl Acad Sci U S A. 2012;109:8477–8482. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriany BR, Zhao R, Murphy PJ, Levy SL, Tan CP, Craighead HG, Soloway PD. Single molecule epigenetic analysis in a nanofluidic channel. Anal Chem. 2010;82:2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, Van EF, Regulski M, Meyers BC, Martienssen RA. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508:411–415. doi: 10.1038/nature13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci U S A. 2012;109:9143–9148. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep D, Plongthongkum N, Gore A, Fung HL, Shoemaker R, Zhang K. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9:270–272. doi: 10.1038/nmeth.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Allen JA, Farrell M, Roth BL. A chemical-genetic approach for precise spatio-temporal control of cellular signaling. Mol Biosyst. 2010;6:1376–1380. doi: 10.1039/c002568m. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger PF, Fernando S, Kashefi SN, Ferrer I, Hauw JJ, Seilhean D, Smith C, Walker R, Al-Sarraj S, Troakes C, Palkovits M, Kasztner M, Huitinga I, Arzberger T, Dexter DT, Kretzschmar H, Reynolds R. Effects of antemortem and postmortem variables on human brain mRNA quality: a Brain Net Europe study. J Neuropathol Exp Neurol. 2010;69:70–81. doi: 10.1097/NEN.0b013e3181c7e32f. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: In vitro and post-mortem brain studies. J Neurosci Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. Epigenetic mechanisms of drug addiction. Curr Opin Neurobiol. 2013;23:521–528. doi: 10.1016/j.conb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007b;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, Balusu R, Koul S, Atadja P, Marquez VE, Bhalla KN. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Noncoding RNAs and Epigenetic Mechanisms During X-Chromosome Inactivation. Annu Rev Cell Dev Biol. 2014 doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, Holson E, Xu J, Sun J, Neve RL, Mach RH, Haggarty SJ, Tsai LH. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. Targeting histone demethylases in cancer therapy. Clin Cancer Res. 2009;15:7111–7113. doi: 10.1158/1078-0432.CCR-09-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Savenkova M, Haque MM, Nilsson E, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of altered Sertoli cell transcriptome and epigenome: molecular etiology of male infertility. PLoS One. 2013;8:e59922. doi: 10.1371/journal.pone.0059922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, Zhu H, Chang Q, Gao Y, Ming GL, Song H. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Karlsson MC, Meadows JP, Gavin CF, Hablitz JJ, Sweatt JD. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Tsai LH. Probing the role of HDACs and mechanisms of chromatin-mediated neuroplasticity. Neurobiol Learn Mem. 2011a;96:41–52. doi: 10.1016/j.nlm.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Tsai LH. Probing the role of HDACs and mechanisms of chromatin-mediated neuroplasticity. Neurobiol Learn Mem. 2011b;96:41–52. doi: 10.1016/j.nlm.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Suzuki T, Mino K, Koseki K, Oehme F, Flamme I, Ozasa H, Itoh Y, Ogasawara D, Komaarashi H, Kato A, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors. J Med Chem. 2010;53:5629–5638. doi: 10.1021/jm1003655. [DOI] [PubMed] [Google Scholar]
- Hovestadt V, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, Tachibana M, Shinkai Y, Kurumizaka H, Nozaki N, Kimura H. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39:6475–6488. doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, Yurov YB. Single cell genomics of the brain: focus on neuronal diversity and neuropsychiatric diseases. Curr Genomics. 2012;13:477–488. doi: 10.2174/138920212802510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak Y, Ergui I, Young JI. Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.7. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, van EH, Kleefstra T, Hamel B, Moraine C, Gecz J, Turner G, Reinhardt R, Kalscheuer VM, Ropers HH, Lenzner S. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Dudley JT, Kukurba KR, Chen R, Butte AJ, Montgomery SB, Snyder M. Systematic functional regulatory assessment of disease-associated variants. Proc Natl Acad Sci U S A. 2013;110:9607–9612. doi: 10.1073/pnas.1219099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han MH, Bassel-Duby R, Olson EN, Nestler EJ. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–445. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Parylak SL, Vadodaria KC, Sarkar A, Gage FH. Increasing the resolution of the adult neurogenesis picture. F1000Prime Rep. 2014;6:8. doi: 10.12703/P6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Breakefield XO. Role of exosomes/microvesicles in the nervous system and use in emerging therapies. Front Physiol. 2012;3:228. doi: 10.3389/fphys.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J, Cruchaga C. The epigenetic landscape of Alzheimer's disease. Nat Neurosci. 2014;17:1138–1140. doi: 10.1038/nn.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz DR, Mikheyeva IV, Johansen P, Meyer L, Berg A, Grewal SI, Cam HP. CENP-B cooperates with Set1 in bidirectional transcriptional silencing and genome organization of retrotransposons. Mol Cell Biol. 2012;32:4215–4225. doi: 10.1128/MCB.00395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, Theriaque D, Hutson A, Nicholls RD, Zori RT, Williams CA, Driscoll DJ. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease. Nat Neurosci. 2014;17:1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. Epigenetic modifications: significance in drug addiction and treatment. Epigenomics. 2010;2:183–186. doi: 10.2217/epi.10.15. [DOI] [PubMed] [Google Scholar]
- Mack GS. Epigenetic cancer therapy makes headway. J Natl Cancer Inst. 2006;98:1443–1444. doi: 10.1093/jnci/djj447. [DOI] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108 doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Noh KM, Allis CD. Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacology. 2013;38:3–22. 3035–3040. doi: 10.1038/npp.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AC, Bharadwaj R, Whittle C, Krueger W, Mirnics K, Hurd Y, Rasmussen T, Akbarian S. The genome in three dimensions: a new frontier in human brain research. Biol Psychiatry. 2014;75:961–969. doi: 10.1016/j.biopsych.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363:2099–2109. doi: 10.1098/rstb.2008.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: effects on behavior and synaptic transmission. Neurobiol Learn Mem. 2011;96:53–60. doi: 10.1016/j.nlm.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76(Pt B):259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena CJ, Bagot RC, Labonte B, Nestler EJ. Epigenetic Signaling in Psychiatric Disorders. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock JD, Wu DY, Satterlee JS. Molecular neuroanatomy: a generation of progress. Trends Neurosci. 2014;37:106–123. doi: 10.1016/j.tins.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn EJ, Guzman-Karlsson MC, David SJ. Cellular, molecular, and epigenetic mechanisms in non-associative conditioning: implications for pain and memory. Neurobiol Learn Mem. 2013;105:133–150. doi: 10.1016/j.nlm.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan VK, Liu J, Sakurai R, Torday JS. Perinatal nicotine-induced transgenerational asthma. Am J Physiol Lung Cell Mol Physiol. 2013;305:L501–L507. doi: 10.1152/ajplung.00078.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A, Dittrich B, Greger V, Buiting K, Lalande M, Gillessen-Kaesbach G, Anvret M, Horsthemke B. Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am J Hum Genet. 1994;54:741–747. [PMC free article] [PubMed] [Google Scholar]
- Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, III, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, III, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Revenfeld AL, Baek R, Nielsen MH, Stensballe A, Varming K, Jorgensen M. Diagnostic and Prognostic Potential of Extracellular Vesicles in Peripheral Blood. Clin Ther. 2014;36:830–846. doi: 10.1016/j.clinthera.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Rissman EF, Adli M. Transgenerational Epigenetic Inheritance: Focus on Endocrine Disrupting Compounds. Endocrinologyen. 2014;2014:1123. doi: 10.1210/en.2014-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD. The epigenetics of Alzheimer's disease--additional considerations. Neurobiol Aging. 2011;32:1196–1197. doi: 10.1016/j.neurobiolaging.2011.02.025. [DOI] [PubMed] [Google Scholar]
- Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- Rudenko A, Tsai LH. Epigenetic modifications in the nervous system and their impact upon cognitive impairments. Neuropharmacology. 2014;80:70–82. doi: 10.1016/j.neuropharm.2014.01.043. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr Bull. 2009;35:1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra-Rodriguez L, Feig LA. Chronic social instability induces anxiety and defective social interactions across generations. Biol Psychiatry. 2013;73:44–53. doi: 10.1016/j.biopsych.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Buiting K, Cassidy SB, Conroy JM, Driscoll DJ, Gabriel JM, Gillessen-Kaesbach G, Glenn CC, Greenswag LR, Horsthemke B, Kondo I, Kuwajima K, Niikawa N, Rogan PK, Schwartz S, Seip J, Williams CA, Nicholls RD. Clinical spectrum and molecular diagnosis of Angelman and Prader-Willi syndrome patients with an imprinting mutation. Am J Med Genet. 1997;68:195–206. [PubMed] [Google Scholar]
- Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NC, Hall GF. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287:3842–3849. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanoudou D, Kang PB, Haslett JN, Han M, Kunkel LM, Beggs AH. Transcriptional profile of postmortem skeletal muscle. Physiol Genomics. 2004;16:222–228. doi: 10.1152/physiolgenomics.00137.2003. [DOI] [PubMed] [Google Scholar]
- Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EA, Hilhorst-Hofstee Y, Wessels MW, den Hollander NS, Ruivenkamp CA, van Ommen GJ, Breuning MH, den Dunnen JT, van HA, Kriek M. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Satterlee JS, Beckel-Mitchener A, McAllister K, Procaccini DC, Rutter JL, Tyson FL, Helbling-Chadwick L. Mukesh Verma., editor. Community Resources and Technologies Developed through the NIH Roadmap Epigenomics Program. Cancer Epigenetics: Risk Assessment, Diagnosis, Treatment, and Prognosis. 2015 doi: 10.1007/978-1-4939-1804-1_2. In press. [DOI] [PubMed] [Google Scholar]
- Satterlee JS. Randy Jirtle, Fred Tyson., editors. Book chapter “Epigenomic and Non-coding RNA Regulation in Addictive Processes”. Environmental Epigenomics in Health and Disease Vol 2 Epigenetics and Complex Diseases. 2013 [Google Scholar]
- Schroeder FA, Chonde DB, Riley MM, Moseley CK, Granda ML, Wilson CM, Wagner FF, Zhang YL, Gale J, Holson EB, Haggarty SJ, Hooker JM. FDG-PET imaging reveals local brain glucose utilization is altered by class I histone deacetylase inhibitors. Neurosci Lett. 2013;550:119–124. doi: 10.1016/j.neulet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;509:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008;3:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Crabtree GR. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr Opin Neurobiol. 2013;23:903–913. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Guan M, Li X. 5-hydroxymethylcytosine-mediated DNA demethylation in stem cells and development. Stem Cells Dev. 2014;23:923–930. doi: 10.1089/scd.2013.0428. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, Ren Y, Miller ML, Blitzer RD, Hurd YL. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology. 2014;39:1315–1323. doi: 10.1038/npp.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffoli LV, Rodrigues GM, Jr, Oliveira JF, Silva AS, Moreira EG, Pelosi GG, Gomes MV. Maternal exposure to fluoxetine during gestation and lactation affects the DNA methylation programming of rat's offspring: modulation by folic acid supplementation. Behav Brain Res. 2014;265:142–147. doi: 10.1016/j.bbr.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Trotter SA, Brill LB, Bennett JP., Jr Stability of gene expression in postmortem brain revealed by cDNA gene array analysis. Brain Res. 2002;942:120–123. doi: 10.1016/s0006-8993(02)02644-6. [DOI] [PubMed] [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PLoS One. 2011;6:e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Sadri-Vakili G. Mechanisms of transgenerational inheritance of addictive-like behaviors. Neuroscience. 2014;264:198–206. doi: 10.1016/j.neuroscience.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16:42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang YL, Hennig K, Gale JP, Hong Y, Cha A, Riley M, Wagner F, Haggarty SJ, Holson E, Hooker J. Class I HDAC imaging using [(3)H]CI-994 autoradiography. Epigenetics. 2013;8:756–764. doi: 10.4161/epi.25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wess J, Nakajima K, Jain S. Novel designer receptors to probe GPCR signaling and physiology. Trends Pharmacol Sci. 2013;34:385–392. doi: 10.1016/j.tips.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S, Chun J. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills QF, Livak KJ, Tipping AJ, Enver T, Goldson AJ, Sexton DW, Holmes C. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- Xin Y, Chanrion B, O'Donnell AH, Milekic M, Costa R, Ge Y, Haghighi FG. MethylomeDB: a database of DNA methylation profiles of the brain. Nucleic Acids Res. 2012;40:D1245–D1249. doi: 10.1093/nar/gkr1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh HH, et al. Imaging epigenetic regulation by histone deacetylases in the brain using PET/MRI with (1)(8)F-FAHA. Neuroimage. 2013;64:630–639. doi: 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Labonte B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology. 2013;38:111–123. doi: 10.1038/npp.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Zhu BL, Ishikawa T, Quan L, Li DR, Maeda H. Real-time RT-PCR quantitative assays and postmortem degradation profiles of erythropoietin, vascular endothelial growth factor and hypoxia-inducible factor 1 alpha mRNA transcripts in forensic autopsy materials. Leg Med (Tokyo) 2006;8:132–136. doi: 10.1016/j.legalmed.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Zhou X, Lowdon RF, Li D, Lawson HA, Madden PA, Costello JF, Wang T. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PA, Costello JF, Wang T. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lee KP, Spencer TJ, Biederman J, Bhide PG. Transgenerational transmission of hyperactivity in a mouse model of ADHD. J Neurosci. 2014;34:2768–2773. doi: 10.1523/JNEUROSCI.4402-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol. 2010;20:432–440. doi: 10.1016/j.conb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature. 2014 doi: 10.1038/nature13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]