Abstract
High-capacity adenoviral vectors (HCAdVs) are promising tools for gene therapy as well as for genetic engineering. However, one limitation of the HCAdV vector system is the complex, time-consuming, and labor-intensive production process and the following quality control procedure. Since HCAdVs are deleted for all viral coding sequences, a helper virus (HV) is needed in the production process to provide the sequences for all viral proteins in trans. For the purification procedure of HCAdV, cesium chloride density gradient centrifugation is usually performed followed by buffer exchange using dialysis or comparable methods. However, performing these steps is technically difficult, potentially error-prone, and not scalable. Here, we establish a new protocol for small-scale production of HCAdV based on commercially available adenovirus purification systems and a standard method for the quality control of final HCAdV preparations. For titration of final vector preparations, we established a droplet digital polymerase chain reaction (ddPCR) that uses a standard free-end-point PCR in small droplets of defined volume. By using different probes, this method is capable of detecting and quantifying HCAdV and HV in one reaction independent of reference material, rendering this method attractive for accurately comparing viral titers between different laboratories. In summary, we demonstrate that it is possible to produce HCAdV in a small scale of sufficient quality and quantity to perform experiments in cell culture, and we established a reliable protocol for vector titration based on ddPCR. Our method significantly reduces time and required equipment to perform HCAdV production. In the future the ddPCR technology could be advantageous for titration of other viral vectors commonly used in gene therapy.
Introduction
Adenoviral vectors are the most frequently used types of gene therapy vectors in clinical trials.1 Various generations of adenoviral vectors were explored over the past decades and the use of the most advanced version represented by high-capacity adenoviral vectors (HCAdV) is a growing field. HCAdV carry up to 36 kilobases (kb) of foreign DNA allowing efficient transfer of large and complex transgenes. For improved safety they lack all coding sequences for viral proteins, and they can transduce a broad range of different cell types in vitro and in vivo. Moreover, because of the lack of viral coding sequences, the toxicity profile of HCAdV is significantly improved compared with early generation adenoviral vectors.2–4 During the production process of HCAdV, all adenoviral proteins are provided in trans by a helper virus (HV). The packing signal of the HV is flanked by the Cre recombinase DNA recognition sites loxP, and therefore it can be removed during amplification by the HCAdV production cell line that stably expresses Cre recombinase.5 As a result, only the HCAdV genomes with an intact packaging signal are efficiently encapsidated. Another advantage of HCAdV is their stability of transgene expression in vivo.2,3,6,7
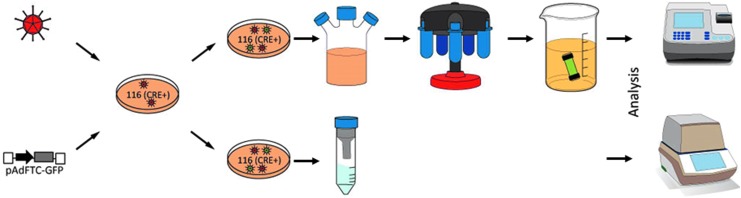
For construction and production of this vector system, a standard protocol is established.8,9 However, with respect to the production procedure and quality control of final HCAdV preparations, challenges remain before broadly exploring this vector system in different applications. One of these challenges is the production of HCAdV virus in a small-scale manner without relying on expansive equipment. The current protocol requires a large amount of tissue culture dishes or the use of a spinner flask providing a final volume of 3 liters of producer cells grown in suspension (Fig. 1). After amplification, the virus is usually purified by two CsCl gradient ultracentrifugation steps and subsequent dialysis. Therefore, these protocols are time-consuming, potentially error-prone, and labor-intensive and may lead to an excess amount of virus that is not needed for experiments performed in vitro.
FIG. 1.
Production strategy of high-capacity adenoviral vectors (HCAdV) using a small-scale or a large-scale procedure. For production of HCAdV the transgene of interest is cloned between the adenoviral 5′ and 3′ inverted terminal repeats (ITRs). Next to the 5′ ITR is an adenoviral packing signal (Ψ) moderating the packaging of the viral genomes. The capsid proteins are provided by a helper virus (HV) in trans that needs to be co-infected during the production process. The packing signal of the HV is flanked by loxP sites, and as the adenoviral producer cell line (116 cells) stably expresses Cre recombinase (CRE+), the packing signal of the HV is excised. After an incubation of 48–72 hr, cells are collected and several freeze–thaw cycles to release the virus particles from the production cell line are performed. Top panel: After amplification, the HCAdV (green) can be purified by two CsCl ultracentrifugation steps and dialyzed in a glycerol-containing storage buffer. Afterward, the purified vector is available in quantities ranging from 1 to 2 ml with an infectious titer ranging from 106 to 109 transducing units (TU)/μl. Bottom panel: The small-scale fast track uses column purification instead of ultracentrifugation, reducing the purification time tremendously. The lysate from infected cells grown in tissue culture dishes is directly loaded onto a column after the cell debris has been cleared and purified within a single step. The final titer of the vector preparation can be either determined by conventional quantitative polymerase chain reaction (qPCR) or by droplet digital PCR (ddPCR).
Here, we aimed at shortening the protocol by implementing commercially available adenovirus purification kits, and therefore bypassing CsCl purification steps for which neither downscaling nor the spinning time can be sufficiently improved (Fig. 1). The usage of small chromatography columns provided by these kits seems to be a valuable alternative for small-scale HCAdV production because the viral particles display sufficient affinity to an anion-exchange column at high salt concentrations wherein most proteins pass the column under the same conditions. By lowering the salt concentration, the viral particles elute because of their negative net charge as a result of their acidic isoelectronic point, which is at neutral pH.10–14 Compared with the standard protocol, the implementation of these systems should shorten the time needed to purify the virus from 2 days to several hours.
The second challenge we wanted to address was the quality control of final vector preparations. Current titration methods are based on optical density, quantitative real-time PCR (qPCR), and slot blot analysis.15–18 Besides the determination of completely assembled viral particles containing viral DNA, which is usually performed by optical density using a photometer or conventional qPCR, a standardized procedure would be advantageous to differentiate between infectious and noninfectious HCAdV and HV contamination levels. The thorough determination of these data is crucial not only because the results have to be evaluated by authorities before conducting clinical trials. Precise measurements of final HCAdV titers are also important because it has been shown in earlier studies that there is a nonlinear dose response in mice, and therefore only a small therapeutic window for HCAdV-based gene therapeutic studies may exist.6,19
Here we developed a new droplet digital PCR (ddPCR) strategy to characterize final adenoviral vector preparations in one single reaction with a special focus on HCAdVs. ddPCR is a method to determine the quantity of DNA by a standard free-end-point method in a high-throughput manner.20 It was used in various approaches, including titration of wild-type viruses in clinically relevant samples.21–24 Within a given sample, a water-in-oil emulsion is formed with droplets of defined volume. The DNA is randomly distributed within these droplets. After endpoint qPCR with hydrolysis probes, the droplets are individually analyzed for the containment of fluorescent dye.20,25,26 This provides the basis for statistical analyses leading to a more reliable result and less influencing factors compared with standard dependent traditional qPCR methods.27
Materials and Methods
Cell lines
For HCAdV production, 116 cells9 (human embryonic kidney-derived HEK293 cells stably expressing Cre-recombinase) were used as a producer cell line. For viral titration experiments, HEK293 and human adenocarcinomic alveolar basal epithelial-derived A549 cells were used. HEK293 and A549 cells were cultured in Dulbecco's modified Eagle's medium (PAN-Biotech). For 116 cells, minimal essential medium (PAN-Biotech) was used. All cell culture medium was supplemented with 10% fetal bovine serum (GE Healthcare) and penicillin/streptomycin (PAN-Biotech) at a concentration of 200 units/ml for penicillin and 0.2 mg/ml streptomycin. During infection with adenovirus, cells were cultured in antibiotics-free medium. During all experiments, cells were incubated at 37°C at a level of 5% CO2 using standard cell culture incubators.
HCAdV vector amplification
The vector HCAdV-pEPito-ΔS/MAR-FRT used for this study was described in our previous publication.28 As HV, a first-generation adenoviral vector lacking early adenovirus genes E1 and E3 containing a packaging signal flanked by loxP sites (AdNG163R-2) was used.9 The preamplification procedure of the HCAdV was performed in accordance to a previously published protocol.8 To compare different purification methodologies, we infected one hundred 150 mm tissue culture dishes at MOIs 10 and 2 for HCAdV and HV, respectively, to assure having a sufficient amount of starting material for the different purification strategies. We used a defined amount of purified virus for amplification of the vector and not cell lysate of infected cells. Two days after infection of 100 tissue culture dishes with purified HCAdV and HV, the cell lysate was harvested for further processing. The rationale for doing that was the fact that we aimed at having the same quality of starting material for all purification strategies. The original titers of the HCAdV and the HV used for infection and amplification were 1.2×108/μl and 2.8×107/μl, respectively.
Virus purification
The purification procedure using the commercially available adenovirus purification systems was performed according to the manufacturer's manual. We explored the Adenovirus Purification Miniprep Kit (Cat. No. VPK-099) provided by Cell Biolabs, the Adeno-X Maxi Purification Kit (Cat. No. 631532) from TaKaRa/Clontech, and the Fast-Trap Virus Purification and Concentration Kit (Cat. No. FTAV00003) provided by Millipore. As a control, a commonly used ultracentrifuge-based purification protocol was used, which was described in our previous publication.8
Physical titer of final vector preparations
To determine the physical titer, also referred to as the OD titer, a 1:20 dilution of the final virus preparation was prepared using a dilution buffer consisting of 10 mM Tris-HCl (pH 7.5), 10 mM EDTA (pH 8.0), and 0.1% SDS. After gently shaking for 30 min at room temperature, the dilution was centrifuged in a microcentrifuge at 15,000 g for 2 min. Subsequently, the absorbance was measured at 260 nm. Because of the relatively low values, four measurements were performed and a mean value was calculated. The physical titer was calculated using the following formula: Physical titer (ml−1)=(absorbance at 260 nm)×(dilution factor)×(1.1×1012)×(36)/(size of HCAdV). In our study the genome length of the vector HCAdV-pEPito-ΔS/MAR-FRT was 33 kb.
Titration experiments
To determine transducing units (TUs) in final vector preparations, 6-well plates with HEK293 and A549 cells at 90% confluency were infected with varying volumes of the final vector preparations using serum-free medium. Three hours after infection, cells were harvested with trypsin (0.05%; PAN-Biotech) and washed with PBS to remove extracellular noninfectious virus particles. Subsequently, total DNA was purified using a conventional protocol based on phenol/chloroform extraction and ethanol precipitation or the DNeasy Blood and Tissue Kit from Qiagen. The quality of the purified DNA was analyzed by visualization using a 1% agarose gel. A qPCR/ddPCR was performed to determine the relation between vector and genomic DNA. Knowing the volume of virus added and the number of cells that were infected initially, we calculated the TUs.
Standard qPCR based on SYBR green to measure HCAdV genomes
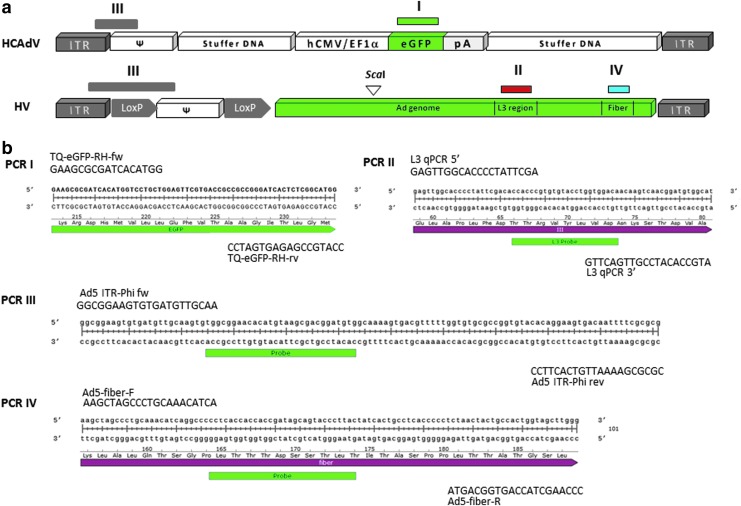
For the standard qPCR protocol specifically detecting the eGFP transgene contained in the vector HCAdV-pEPito-ΔS/MAR-FRT, a protocol recommended by Bio-Rad was applied using the C1000 Touch Thermal Cycler (Fig. 2, PCR I). As PCR parameters, a 5 min denaturation step at 95°C and then 39 rounds of 15 sec denaturation at 95°C, annealing, and extension at 60°C for 60 sec were used. Subsequently, a melting curve was performed using a 0.5°C/sec gradient as quality control. As primers, the following sequences were used: TQ-eGFP-RH-fw, 5′ GAA GCG CGA TCA CAT GGT 3′; TQ-eGFP-RH-rv, 5′CCA TGC CGA GAG TGA TCC 3′. As a standard, the EGFP containing plasmid Pepito was used containing the identical transgene.29
FIG. 2.
Primer and PCR design for HCAdV vectors based on conventional qPCR or ddPCR. (a) To evaluate the final vector preparation in terms of quality and quantity based on qPCR and ddPCR, we used different primer sets binding to different sequences contained in the HCAdV and the HV genomes. Note that only most relevant DNA sequences contained in HCAdV encoding eGFP and the HV are displayed. Primer pair I (green horizontal bar) used for conventional qPCR is binding in the transgene (eGFP) of the HCAdV. Primer pair II (red horizontal bar) used for conventional qPCR is for the detection of a potential harmful HV contamination binding in the L3 region of the hexon that is only present in the genome of the HV. Primer pair III (gray horizontal bar) used for ddPCR detects the transition of the ITR region to the packaging signal Ψ in the HCAdV and the HV, while primer pair IV (blue horizontal bar) detects the fiber gene that is only present in the HV genome. Ψ, packaging signal; hCMV/EF1α, human cytomegalovirus enhancer and elongation factor alpha-1 promoter; loxP, Cre recombinase recognition sites; pA, polyadenylation signal. Restriction enzyme digest (ScaI) was performed before PCR to separate the PCR targets and to increase specificity (triangle). (b) PCR and probe sequences and primer binding sites used for all PCR approaches (I–IV). Note that for PCR setup III only the primer binding sites of the HCAdV are shown. The identical primer set also detects the left arm of the HV that contains a floxed packaging signal as schematically shown in (a).
Conventional probe PCR detecting the HV
As described previously,8 for the detection of the HV a PCR was performed utilizing a fluorescence-labeled probe binding in the L3 region of the HV hexon (Fig. 2, PCR II). Primers and probe used were based on the following sequences: L3 qPCR 5′, 5′ GAG TTG GCA CCC CTA TTC GA 3′; L3 qPCR 3′, 5′ ATG CCA CAT CCG TTG ACT TG 3′; L3 probe FAM (purchased from Eurofins Genomics), 5′ CCA CCC GTG TGT ACC TGG TGG ACA 3′. The PCR was performed using a Bio-Rad C1000 touch Thermal Cycler. The PCR program was designed as follows: 95°C for 3 min for primary denaturation, 95°C for 15 sec, and 60°C for 30 sec. Steps 2 and 3 were repeated 43 times. As standard, a cloned L3 region was used.
Droplet digital PCR
For the simultaneous detection of HCAdV and HV in one reaction, a PCR was performed with a hydrolysis probe and primers binding in the inverted terminal repeat (ITR) and Ψ regions (Fig. 2, PCR III). This PCR detects the left arm of the HCAdV with an unmodified 5′ region and the HV, which, in addition to ITR and packaging signal, contains a loxP site. Primers and probe used had the following sequences: Ad5 ITR-Phi for, 5′-GGC GGA AGT GTG ATG TTG CAA-3′; Ad5 ITR-Phi rev, 5′-CGC GCG AAA ATT GTC ACT TCC T-3′; Ad5 ITR-Phi probe, 5′-Hex CAC ATC CGT CGC TTA CAT GTG TTC CGC CA BHQ-1-3′.
To determine the copy number of HV, a second PCR was performed in a duplex approach with hydrolysis probe and primers binding in the fiber gene (Fig. 2, PCR IV) that is present in the HV genome and wt Ad5 but not in the HCAdV DNA molecule. Sequences of primers and probe are consistent with the official collection of methods according to § 28b of the German Genetic Engineering Act (GenTG),30 containing the following sequences: Ad5-fiber-F, 5′-AAG CTA GCC CTG CAA ACA TCA-3′; Ad5-fiber-R, 5′-CCC AAG CTA CCA GTG GCA GTA-3′; Ad5-fiber-Probe-FAM, 5′-6 FAM CCT CAC CAC CAC CGA TAG CAG TAC CCT TAC BBQ-3′. PCRs were prepared with the required ddPCR Supermix for probes (Cat. No. 186-3010; Bio-Rad) with a final concentration of 400 nM for each primer and 200 nM for each probe, together with 5 units of ScaI and 2 μl sample (0.9–34 ng DNA) to a final volume of 20 μl. Sample DNA concentration was determined with a Quantus Fluorometer (Cat. No. E6150; Promega) and the QuantiFluor dsDNA System (Cat. No. E2670; Promega) according to manufacturer's instructions.
Each reaction was loaded into a sample well of an 8-well disposable cartridge (Cat. No. 186-3006; Bio-Rad) followed by 70 μl of droplet generator oil (Cat. No. 186-3005; Bio-Rad), which was added to the oil wells of the cartridge. Droplets were formed in the QX100 droplet generator (part of Cat. No. 186-3001; Bio-Rad). Droplets were then transferred to a 96-well PCR plate (Cat. No. 951020389), heat-sealed with foil (Cat. No. 181-4040; Bio-Rad) in a PX1 PCR Plate Sealer (Cat. No. 181-4000; Bio-Rad), and amplified with a Bio-Rad C1000 Touch Thermal Cycler (95°C primary denaturation/activation for 10 min, followed by 40 cycles of 94°C for 30 sec and 60°C for 1 min, followed by a final 98°C heat treatment for 10 min). PCRs were analyzed with the QX100 droplet reader (part of Cat. No. 186-3001; Bio-Rad) and data analysis was performed with QuantaSoft software (version 1.5.38.1118; Bio-Rad). Note that no standard is needed for the ddPCR technology.
Results
Amplification and purification of the HCAdV
We amplified the HCAdV HCAd-pEPito-ΔS/MAR-FRT using 116 cells by infecting with an MOI of 10 for the HCAdV and an MOI of 2 for the HV. To produce sufficient amounts of down-stream material for analyses, we amplified the virus in one hundred 15 cm tissue culture dishes. Infection efficiency and amplification was confirmed by eGFP expression during the amplification process (data not shown). After harvesting the crude cell lysate, this virus stock was used for further purification and characterization steps. The crude lysate was analyzed by qPCR using a SYBR-green-based standard protocol measuring eGFP gene copies per cell (Fig. 2, PCR I). Results revealed a titer of 4.07×105 infectious viral particles per μl cell lysate.
Next different virus purification procedures were conducted using the identical crude lysate that were performed by different scientists to eliminate an influence of variations regarding handling of the samples. All kit column-based systems were successful in resulting in functional HCAdV, which was confirmed by eGFP expression in A549 cells (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hgtb).
Establishment of a standard free ddPCR for the detection of HCAdV and HV genomes
We established a ddPCR setup for simultaneous detection of HCAdV and HV genomes displaying a high sensitivity and specify for our targets. The principle of the ddPCR and a comparison to the conventional PCR is schematically displayed in Fig. 3a. Note that, in contrast to conventional qPCR based on SYBR green or labeled probes, no standard curve is required for the ddPCR protocol. To develop a protocol based on the ddPCR technology for titration of adenoviral DNA molecules derived from the adenovirus type 5, we chose two different primer sets. One primer set binds to the adenoviral fiber region and the other primer set binds to the ITR and the adenovirus packing signal Ψ (Fig. 2, PCRs III and IV). For establishment of conditions for the ddPCR approach, we first generated a reference plasmid (pMA-Ad5-ITR/Ψ-fiber) into which the ITR, the packaging signal Ψ, and the fiber region of the adenovirus type 5 were cloned (Supplementary Fig. S2a). To perform ddPCR, a water-in-oil emulsion with droplets of defined volume was formed for each sample. Randomly distributed DNA within these droplets were used to perform endpoint PCR and subsequently individually analyzed for the presence of fluorescent dye. Since the reference target plasmid contained both DNA sequences specifically detected by PCRs amplifying the adenovirus 5′-end and the fiber, each analyzed droplet was positive for both PCR signals and appears in quadrant II of the two-dimensional dot plot (Supplementary Fig. S2b).
FIG. 3.
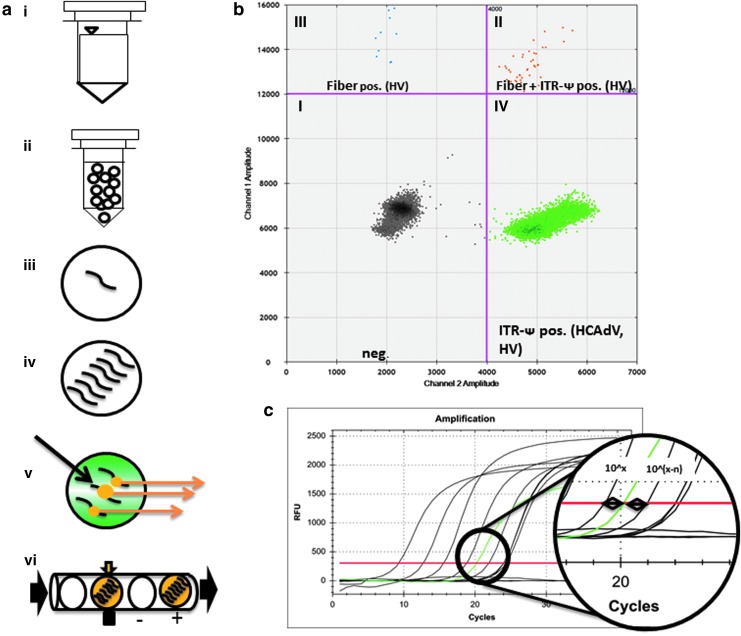
Implementation of ddPCR as new quality control tool for gene therapeutic products. (a) Principle of ddPCR. In sharp contrast to traditional qPCR, ddPCR allows absolute quantification of the desired product instead of relative quantification. (i) For the reaction, standard TaqMan primers and probes are used in a special reaction mix that is capable of forming an emulsion. (ii) After the reaction mix is prepared, the sample is loaded onto a machine, which forms water–in-oil droplets of a defined size, more than 20,000 per sample with a volume of approximately 1 nl. The DNA is equally distributed within these droplets and the sensitivity of the later read out is capable to detect one single copy within one droplet. (iii and iv) A standard end-point PCR is performed. The PCR is independently processed in every droplet. If a droplet contains the desired sequence, the target will be amplified and the reporter dye will be released. (v) If the reporter dye is released during amplification, the droplets will be read as positive, and if no free dye is present, the droplet will be read as negative since there is no fluorescence signal. (vi) The analyses are performed in a capillary in which every single droplet is passing a detector. It is scored as positive or negative with a speed of 1000 droplets/sec. On the basis of the produced I/O code, the protocol was named digital PCR. (b) Representative data analysis for ddPCR. Every droplet is plotted onto a chart relative to the emitted fluorescence signal. Since the system used in this study can read more than one channel for quantification of more than one sequence within a reaction, a plot with an x and y axis is formed. Droplets are then separated in fields containing only one target or both. The x axis displays PCR products formed with primer set III amplifying the ITR-Ψ region (quadrant IV), whereas the y axis (quadrants II and III) represent the fiber PCR. On the basis of the equal size of each droplet and a cutoff for every signal (purple line), it is then mathematically possible to directly calculate the concentration of the target in the sample. The picture shown was analyzed by the Bio-Rad calculation software QuantaSoft. Neg., negative droplets without PCR (quadrant I); Fiber+ITR-Ψ pos., droplets with PCR products for the primer sets III and III (HV) (quadrant II); Fiber pos. (HV), droplets positive for PCR setup IV; ITR-Ψ pos. (HCAdV, HV), droplets positive for PCR setup III (quadrant IV); Ψ, packaging signal. (c) Data analyses using conventional qPCR. During the exponential phase of the amplification, the target interacts with a dye. The resulting fluorescence signal from the sample is measured at every cycle (green curve). If it becomes higher than background (cq value, red line) it is compared with a reference material (black curve) of known concentration in several dilutions, which is also amplified in a different reaction. Compared with ddPCR, the results are depended on the reference material, which leads to a significantly lower sensitivity and more influencing factors compared with the ddPCR.
Next we established a ddPCR protocol to characterize final HCAdV preparations including HV contamination levels. To detect and quantify HCAdV DNA molecules in a given sample, the primer set III (Fig. 2a and b, PCR III) was chosen, which directly amplifies the unmodified adenovirus type 5 5′-end including part of the ITR and the packaging signal of the adenoviral genome. The identical primer pair and probe can be used to amplify ITR region of the HV genome (Fig. 2a, PCR III) that contains a floxed packaging signal. To differentiate between HCAdV and HV genomes in the same PCR, a ddPCR setup was established specifically detecting and quantifying the fiber gene of the adenovirus type 5 genome (Fig. 2, PCR IV). When applying the ddPCR technology for each DNA molecule in one droplet, a PCR product can be obtained. To physically separate the two PCR products (Fig. 2, PCR III and IV) contained in the HV DNA molecule, the PCR sample was digested with ScaI cutting in the HV genome (Fig. 2a). The ScaI restriction enzyme digest was performed to create two distinctive PCRs and to reach similar efficiencies for PCR III and PCR IV. Without ScaI restriction enzyme digest PCR III and PCR IV would otherwise take place on the same DNA molecule for the HV that interferes with the digital PCR strategy.
A representative example of the expected raw data of a final HCAdV preparation is shown in Fig. 3b. The QuantaSoft software allows differentiating between four different fractions of droplets as shown by the four quadrants (Fig. 3b). Negative samples without PCR product are displayed in quadrant I, samples that are positive for the ITR and the packaging signal Ψ derived from HCAdV and HV in quadrant II, samples that are positive for the fiber gene of the HV in quadrant III, and samples that are positive for the ITR attached to the packaging signal Ψ and the fiber gene of the HV in quadrant IV. The latter droplets correlate with PCR samples in which the ScaI digest to physically separate the ITR and packaging signal from the fiber region was incomplete. In contrast to the ddPCR setup, Fig. 3c exemplarily shows data analyses using conventional qPCR. Positive PCR signals above background are measured at each PCR cycle and directly compared with a reference sample (standard curve) with known concentrations of the DNA fragment to be PCR amplified.
In a first attempt we tested our PCR setup for titration of final HCAdV preparations in HEK293 cells, which we usually use for titration of HCAdV vector preparations. Cells were infected with final vector preparations and analyzed 3 hr postinfection. However, after performing the ddPCR we observed a high background signal in HEK293 cells (Supplementary Fig. S3) for the primers covering part of the ITR and the packing signal (Fig. 2, PCR III) in uninfected HEK293 cells. This observation was because of binding of our primers to ITR and packaging signal-derived DNA sequences that are stably integrated into the host genome of HEK293 cells.31,32 Thus, we adapted our protocol and performed the following titration experiments in adenocarcinoma-derived A549 cells in which no adenoviral DNA sequence is stably integrated, which may interfere with our ddPCR data.33,34 With this strategy we could exclude the high background signals in our negative control.
Characterization of the HCAdV preparations obtained by different purification strategies
The physical titer generally measures DNA containing adenoviral virions for all kit preparations and also the CsCl purified virus ranging from 4.8×108 to 2.43×109 optical particles/μl (Fig. 4a). Next we explored our different PCR strategies to determine TUs of HCAdV and HV in final vector preparations. Toward that end we directly compared traditional qPCR and ddPCR. As shown in Fig. 2 the traditional qPCR utilizes one primer set specifically amplifying the eGFP transgene contained in the HCAdV and another primer set detecting part of the adenoviral L3 region (Fig. 2, PCRs I and II).
FIG. 4.
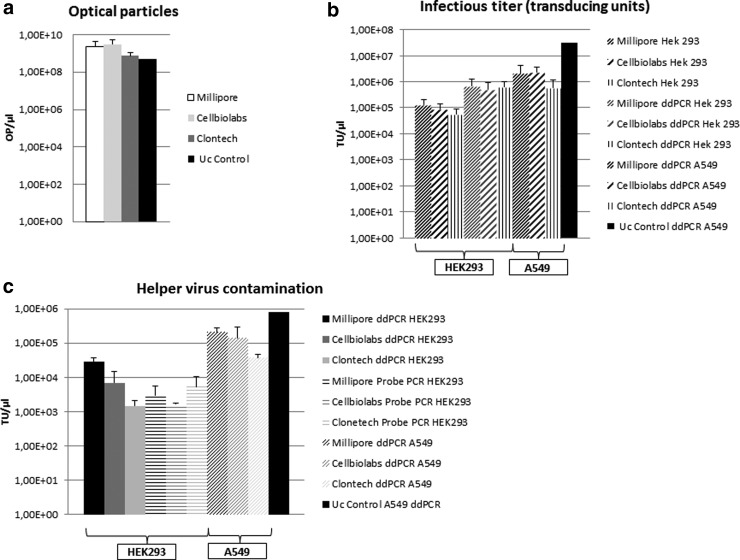
Evaluation of final HCAdV vector preparations. Cell lines in which vector titration was performed (HEK293 cells or A549 cells) and respective companies from which the adenovirus purification kits were obtained are indicated. (a) Physical titers expressed as optical particles measured with a photometer after different purification protocols. A differentiation regarding infectious particles or noninfectious particles is not possible using this approach. OP, optical particles. (b) Measurement of the infectious titer expressed as transducing units (TUs) using primer pair I (qPCR) and III (ddPCR). The diagram shows HCAdV detection in DNA samples collected from different cell lines 3 hr after treatment with the final purified vector preparations using traditional qPCR and ddPCR. For the data shown, different dilutions of the respective purified virus were used and the mean TU was calculated. The cell line in which the titration experiment was performed is indicated. (c) TUs of the HV (primer pair II for qPCR and primer pair IV for ddPCR) within the same preparations. For the ddPCR we acquired both datasets (HCAdV and HV) within one ddPCR run. Uc control, HCAdV purified by ultracentrifugation.
For the titration of the HCAdV we began our experiments using HEK293 cells as a reference cell line because this cell line can be used to quantify TUs of HCAdV based on traditional qPCRs routinely performed in our laboratory. For the kit systems, we had similar infectious titers expressed as TUs for the HCAdV ranging from 5.77×104 to 1.28×105 TU/μl (Fig. 4b). For the digital PCR approach performed in HEK293 cells, we received titers ranging from 3.79×105 to 6.98×105 TU/μl. As expected, this PCR strategy showed a high background signal in uninfected HEK293 cells (see also Supplementary Fig. S2), and therefore this value was subtracted during data analysis. Using A549 cells as a cell line for vector titration, we measured slightly increased TUs compared with the traditional eGFP detecting qPCR and the ddPCR performed in HEK293 cells, ranging from 5.81×105 to 2.26×106 TU/μl (Fig. 4b). This corresponds to a nearly fivefold difference in vector titers for the different column-based purification systems. For the HCAdV preparation purified by the standard protocol using ultracentrifugation (Fig. 1), the infectious titer was significantly higher with 3.13×107 TU/μl (Fig. 4b).
Regarding HV contamination levels in final vector preparations determined in HEK293 cells, we measured lower values for the ddPCR ranging from 1.46×103 to 2.82×104 TU/μl compared with the probe-based qPCR amplifying the L3 region for which we measured between 1.66×103 and 5.40×103 TU/μl (Fig. 4c). The ddPCR performed from DNA isolated from infected A549 cells showed a higher level of HV contamination ranging from 3.74×104 to 2.13×105 TU/μl (Fig. 4c) compared the standard qPCR conducted from total DNA of infected HEK293 cells. For the HCAdV preparation purified by ultracentrifugation, the titer of the HV was 8.2×105 TU/μl (Fig. 4c). The percentage of HV contamination levels within our preparations varied between 0.23% and 9%. The HV contamination levels for the kit-based purification systems were nearly equal in all preparations and varied only with the use of different cell lines (Fig. 4c).
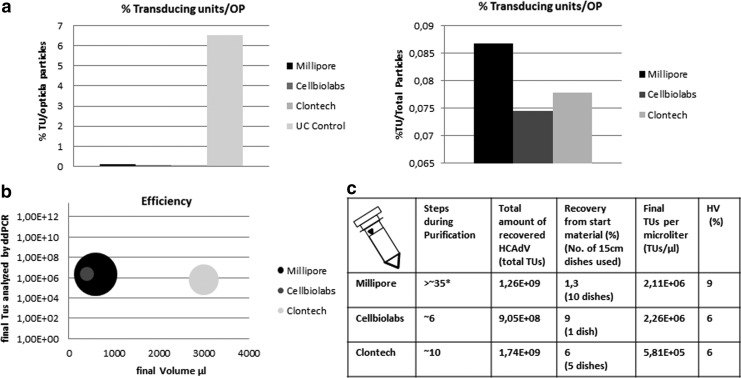
With respect to the ratio between optical units and infectious units, the CsCl preparation was superior compared with the column-purified HCAdV resulting in 6.51% infectious units HCAdV per total numbers of optical particles (Fig. 5a). The ratios for the column-purified HCAdV were significantly lower with values around 0.1% infectious units HCAdV per total optical particles (Fig. 5a). On the basis of the cell surface area used, the respective titer, and the final elution volume, we observed a difference between the different kit systems. The system provided by TaKaRa/Clontech seems to produce higher amounts of virus based on a relatively small growth area. This was in contrast to the systems provided by Cell Biolabs, which use a relatively small cell surface area, resulting in a smaller amount of virus in the final preparation (Fig. 5b). The Millipore system on the other hand relies on a large cell surface area but at least in our hands resulted in a smaller amount of HCAdV in the final vector preparation (Fig. 5b).
FIG. 5.
Column purification and kit performance. (a) Percentage of infectious units of HCAdV contained in final vector preparations compared with total optical particle (OP) numbers. As expected, the ultracentrifuge separates HCAdV lacking a genome from the fully assembled particles, resulting in the highest ratio of infectious units compared with optical particle numbers. The left panel shows all vector preparations, including ultracentrifugation (Uc Control), and the right panel displays only column-based purification procedures. (b) Efficacy of the different kit systems based on cell surface area purified (size of the circle), infectious titer, and final yields. In our hands, the Cellbiolabs system outperformed the other systems since it provided a significant higher amount of virus based on a small amount of starting material. On the basis of quantification of TUs, the systems provided a similar quality of purified virus. (c) Summary and comparison of the different column-based purification systems. The total number of required steps from freezing and thawing the cellular lysate to the final vector preparation is provided. In addition, the total yields of HCAdV expressed as total TUs after the purification procedure for all systems are shown. The third column summarizes the recovery rate of HCAdV from the given amount of starting material expressed as recommended amount of tissue culture dishes that were used for purification. Moreover, the final number of TUs of the HCAdV for each system per microliter and the percentage of HV contamination levels in final vector preparations are summarized for each system. For the vector purified by CsCl gradients, the HV contamination was 2.5%. *The Millipore purification system includes most steps because it makes use of a complex purification device that requires a great number of smaller steps.
Discussion
We successfully implemented a new tool to assess the quality of HCAdV preparations by utilizing a digital ddPCR strategy, which is fast, is easy to handle, and provides more information compared with the most frequently used standard qPCRs. The main new aspect of this new strategy is the possibility to absolutely quantify DNA amounts in a given sample without relying on a standard or a reference sample. This reduces the influencing factors and makes results more reliable and easier to reproduce. The fact that the ddPCR technology is independent of a reference material or a standard also enables direct comparison of viral titers between different preparations and laboratories. A summary of advantages and disadvantages and a direct comparison of the ddPCR technology and the regular qPCR strategy are shown in Supplementary Table S1.
By using different fluorescent probes, we were also able to gather more information about the vector preparations that seem to be indispensable for the use of HCAdV in preclinical and clinical studies. To facilitate measurement of absolute HCAdV titers and HV contamination levels in one sample is of great value. Even though it has been shown that HV-associated toxicity can be neglected in experiments in which a small virus dose is administrated, this may change when injecting a higher dose.35
We used our ddPCR technology to evaluate a small-scale protocol for the production of HCAdV. We could show that, by using column-based commercially available systems, we were able to produce infectious HCAdV in sufficient amounts. As expected, the infectious titers were significantly lower compared with the CsCl-produced virus and also when compared with viruses used in other studies.28,36 Concerning HV contamination levels, we received slightly increased values for the column purification systems compared with other studies9,37 and at the time can only speculate whether this results also in increased toxicity. One reason for this observation could be that using a CsCl gradient-based protocol the HV is, at least to a certain extent, separated away from the HCAdV even though the HV and the HCAdV are similar in size. Concerning the empty particles that are normally produced in excess during the amplification process, the CsCl is capable of efficiently removing these site products. This is not possible for the column-based systems because they can only separate viral particles based on surface properties. This may represent one major disadvantage of column purification-based methods because empty particles may cause unwanted side effects. For instance, incoming viral proteins may cause toxicity to the transduced cell or may even block uptake of the HCAdV.
Clearly, vector recovery and purity are two important issues when using the column-based purification systems and when comparing them to the routinely used CsCl gradient-based systems. We observed an up to fivefold difference in final vector titers when using the column-based systems. However, to really conclude that this corresponds to a significant difference, more column purification needs to be performed and directly compared. Besides the percentage of HV contamination levels in the final HCAdV preparation, which is considered to be one source for cytotoxicity and immunogenicity, the starting material such as the number of initially infected cells may be another source of toxicity. The starting material varies for the different column systems (Fig. 5c) and this may result in different levels of cytotoxicity and/or immunogenicity because of different levels of purity and contaminating levels of cellular proteins in the final vector preparation. However, we would like to point out that, for all purification systems, cell debris were removed by centrifugation before adding to the column.
Although the CsCl gradient seems to be superior compared with other methods with respect to purity and concentration, some features of this technology are also disadvantageous. For instance, it requires complex equipment and large quantities of crude lysate, and since CsCl is toxic to cells, a buffer exchange needs to be performed. This is why we believe that the column purification is an additional solution for proof-of-principle studies in cell culture that can be performed in any laboratory. Using the column procedure, the production of the HCAdV is radically shortened and displays less influencing factors compared with the method based on ultracentrifugation.
In summary, we have shown that HCAdV vectors can be produced in sufficient amounts without using expansive equipment and that final HCAdV vector preparations can be reliably titrated using the ddPCR technology. We would recommend the column-based small-scale protocol for researchers without access to the expensive technical equipment to perform HCAdV purification by CsCl gradients or for researchers who basically need fast access to a small amount of virus to carry out experiments in tissue culture. Furthermore, it is important to point out that the column-based systems are significantly less time-intensive and requires less starting material and media plus plastic ware for conducting cell culture procedures. Regarding ddPCR, we think that, in the future, the ddPCR technology may be also useful to perform quality control of other viral vectors used in gene therapeutic applications.
Supplementary Material
Acknowledgments
We thank Philip Ng (Baylor College, Houston, TX) for providing the helper virus and 116 cells for high-capacity adenovirus production. This work was supported by DFG Grant EH 192/5-1 (A.E.), the UWH Forschungsförderung (E.S. and W.Z), and the PhD program of the University Witten/Herdecke (P.B. and J.D.). J.L. was supported by a stipend from the Chinese Scholarship Council and T.B. by the Else Kröner-Fresenius Foundation.
Author Disclosure Statement
The authors declare that they have no competing financial interests.
References
- 1.Wiley. Gene Therapy Clinical Trials Worldwide, 2014. Available at www.abedia.com/wiley/vectors.php
- 2.Ehrhardt A, Kay MA. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood 2002;99:3923–3930 [DOI] [PubMed] [Google Scholar]
- 3.Schiedner G, Morral N, Parks RJ, et al. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet 1998;18:180–183 [DOI] [PubMed] [Google Scholar]
- 4.Seiler MP, Cerullo V, Lee B. Immune response to helper dependent adenoviral mediated liver gene therapy: challenges and prospects. Curr Gene Ther 2007;7:297–305 [DOI] [PubMed] [Google Scholar]
- 5.Parks RJ, Chen L, Anton M, et al. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA 1996;93:13565–13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hausl MA, Zhang W, Muther N, et al. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther 2010;18:1896–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morral N, O'neal W, Rice K, et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc Natl Acad Sci USA 1999;96:12816–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jager L, Hausl MA, Rauschhuber C, et al. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nat Protoc 2009;4:547–564 [DOI] [PubMed] [Google Scholar]
- 9.Palmer D, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther 2003;8:846–852 [DOI] [PubMed] [Google Scholar]
- 10.Altaras NE, Aunins JG, Evans RK, et al. Production and formulation of adenovirus vectors. Adv Biochem Eng Biotechnol 2005;99:193–260 [DOI] [PubMed] [Google Scholar]
- 11.Blanche F, Cameron B, Barbot A, et al. An improved anion-exchange HPLC method for the detection and purification of adenoviral particles. Gene Ther 2000;7:1055–1062 [DOI] [PubMed] [Google Scholar]
- 12.Burova E, Ioffe E. Chromatographic purification of recombinant adenoviral and adeno-associated viral vectors: methods and implications. Gene Ther 2005;12Suppl 1:S5–S17 [DOI] [PubMed] [Google Scholar]
- 13.Green AP, Huang JJ, Scott MO, et al. A new scalable method for the purification of recombinant adenovirus vectors. Hum Gene Ther 2002;13:1921–1934 [DOI] [PubMed] [Google Scholar]
- 14.Huyghe BG, Liu X, Sutjipto S, et al. Purification of a type 5 recombinant adenovirus encoding human p53 by column chromatography. Hum Gene Ther 1995;6:1403–1416 [DOI] [PubMed] [Google Scholar]
- 15.Crettaz J, Olague C, Vales A, et al. Characterization of high-capacity adenovirus production by the quantitative real-time polymerase chain reaction: a comparative study of different titration methods. J Gene Med 2008;10:1092–1101 [DOI] [PubMed] [Google Scholar]
- 16.Kreppel F, Biermann V, Kochanek S, et al. A DNA-based method to assay total and infectious particle contents and helper virus contamination in high-capacity adenoviral vector preparations. Hum Gene Ther 2002;13:1151–1156 [DOI] [PubMed] [Google Scholar]
- 17.Puntel M, Curtin JF, Zirger JM, et al. Quantification of high-capacity helper-dependent adenoviral vector genomes in vitro and in vivo, using quantitative TaqMan real-time polymerase chain reaction. Hum Gene Ther 2006;17:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas MA, Lichtenstein DL, Krajcsi P, et al. A real-time PCR method to rapidly titer adenovirus stocks. Methods Mol Med 2007;130:185–192 [DOI] [PubMed] [Google Scholar]
- 19.Tao N, Gao GP, Parr M, et al. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol Ther 2001;3:28–35 [DOI] [PubMed] [Google Scholar]
- 20.Mazaika E, Homsy J. Digital droplet PCR: CNV analysis and other applications. Curr Protoc Hum Genet 2014;82:7.24.1–7.24.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunetto GS, Massoud R, Leibovitch EC, et al. Digital droplet PCR (ddPCR) for the precise quantification of human T-lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations. J Neurovirol 2014;20:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden RT, Gu Z, Ingersoll J, et al. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J Clin Microbiol 2013;51:540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibovitch EC, Brunetto GS, Caruso B, et al. Coinfection of human herpesviruses 6A (HHV-6A) and HHV-6B as demonstrated by novel digital droplet PCR assay. PLoS One 2014;9:e92328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strain MC, Lada SM, Luong T, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 2013;8:e55943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 2012;84:1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voigtlander R, Haase R, Muck-Hausl M, et al. A novel adenoviral hybrid-vector system carrying a plasmid replicon for safe and efficient cell and gene therapeutic applications. Mol Ther Nucleic Acids 2013;2:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haase R, Argyros O, Wong SP, et al. pEPito: a significantly improved non-viral episomal expression vector for mammalian cells. BMC Biotechnol 2010;10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beuth. Real-time PCR—nachweis des fiber protein-gens von adenovirus Typ 5. Bundesamt für Verbraucherschutz und Lebensmittelsicherheit, 2013. Available at www.beuth.de/en/technical-rule/bvl-g-1040-1/179717430
- 31.Graham FL, Smiley J, Russell WC, et al. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 1977;36:59–74 [DOI] [PubMed] [Google Scholar]
- 32.Spector DJ. The pattern of integration of viral DNA sequences in the adenovirus 5-transformed human cell line 293. Virology 1983;130:533–538 [DOI] [PubMed] [Google Scholar]
- 33.Chang H, Jackson DG, Kayne PS, et al. Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines. PLoS One 2011;6:e21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieber M, Smith B, Szakal A, et al. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 1976;17:62–70 [DOI] [PubMed] [Google Scholar]
- 35.Muruve DA, Cotter MJ, Zaiss AK, et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J Virol 2004;78:5966–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Muck-Hausl M, Wang J, et al. Integration profile and safety of an adenovirus hybrid-vector utilizing hyperactive sleeping beauty transposase for somatic integration. PLoS One 2013;8:e75344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng P, Beauchamp C, Evelegh C, et al. Development of a FLP/frt system for generating helper-dependent adenoviral vectors. Mol Ther 2001;3:809–815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.