Abstract
Autoinflammatory disease and hyperinflammatory syndromes represent a growing number of diseases associated with inappropriately controlled inflammation in multiple organs. Systemic inflammation commonly results from dysregulated activation of innate immune cells, and therapeutic targeting of the IL-1β pathway has been used to ameliorate some of these diseases. Some hyperinflammatory syndromes, however, such as hemophagocytic lymphohistiocytosis and the newly classified proteasome disability syndromes, are refractory to such treatments, suggesting that other factors or environmental stressors may be contributing. In comparing two cytokine reporter mouse strains, we identify IFN-γ as a mediator of systemic autoinflammatory disease. Chronically elevated levels of IFN-γ resulted in progressive multiorgan inflammation and two copies of the mutant allele resulted in increased mortality accompanied by myeloproliferative disease. Disease was alleviated by genetic deletion of T-bet. These studies raise the possibility that therapeutics targeting the IFN-γ pathway might be effective in hyperinflammatory conditions refractory to IL-1β–targeted therapies.
Introduction
Systemic autoinflammatory diseases are diseases involving abnormal inflammation in multiple tissues, and they can present periodically throughout life (1). These disorders comprise a spectrum of inflammatory conditions and can be both mono- and polygenic in nature (1, 2). Unlike autoimmune diseases, which are commonly associated with breaks in self-tolerance by adaptive immune cells, autoinflammatory diseases are associated with aberrant activation of the innate immune system. Several of the mutations associated with autoinflammatory disorders occur in the IL-1β pathway (3). IL-1β is a proinflammatory cytokine and can induce tissue damage when levels reach a critical threshold (4). As such, therapeutics that target IL-1β or antagonize the IL-1β receptor have been effective in the treatment of a number of autoinflammatory diseases (5, 6). There remain a number of autoinflammatory conditions, however, that induce overt inflammation and excessive innate immune cell activation that are refractory to anti–IL-1β treatment (6). Whether targeting other proinflammatory cytokines might afford effective therapy is unknown, but models for these inflammatory syndromes remain lacking.
IFN-γ is a proinflammatory cytokine that has been implicated as a mediator in at least two different types of autoinflammatory conditions. One type is characterized by hemophagocytic lymphohistiocytosis (HLH) and macrophage activation syndrome (MAS). HLH and MAS are pathologic inflammatory disorders associated with defects in NK and CD8+ CTL function (7–9). Although the triggers for disease progression are not fully defined, inflammatory cytokines contribute to the pathology (10–13). Lymphocytic choriomeningitis virus or murine CMV infection of mice with mutations in known HLH-relevant genes develops symptoms resembling HLH-like autoinflammatory disease, and they have led to the suggestion that IFN-γ from CD8+ T cells may contribute to the pathogenesis of the disease (14–16). Similarly, repeated dosing of IFN-γ or TLR ligands known to induce IFN-γ corroborated the idea that cytokines were not only associated with disease progression but might be causative for some of the symptoms of the disease (17, 18).
The second class of autoinflammatory diseases refractory to IL-1β but characterized by elevated IFN-γ levels is the proteasome disability syndromes (PDS). PDS represent rare autoinflammatory conditions with mutations in the immunoproteasome (19–21). Within this group three syndromes have been described: chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE); joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced childhood onset lipodystrophy (JMP); and Nakajo–Nishimura syndrome (6, 22). Both CANDLE and JMP present with high levels of serum IFN-γ and cells exhibit a gene signature consistent with IFN-γ–mediated activation (20, 21). Thus, IFN-γ likely plays an important role in PDS pathogenesis. However, the triggers for and sources of IFN-γ in CANDLE and JMP remain undefined.
In this study, we describe two strains of IFN-γ reporter mice generated by targeting an IRES/yellow fluorescent protein (YFP) reporter cassette downstream of the endogenous ifng gene. The targeting constructs differed only in the 3′ untranslated region of the IFN-γ mRNA, with one strain using a polyA bovine growth hormone (BGH) sequence and one preserving the endogenous ifng polyA sequence. Because the reporters were targeted into the endogenous IFN-γ locus, cells from the mice facilitated an accurate assessment of IFN-γ expression and regulation as compared with wild-type mice. Mice containing IFN-γ with the foreign BGH polyA tail developed an overabundance of IFN-γ–secreting cells and elevated serum IFN-γ, resulting in tissue pathology, mortality, and morbidity consistent with autoinflammatory disease. Additionally, mice with two mutant IFN-γ alleles developed myeloproliferative disease, thus revealing a link between inflammatory cytokines and the development of myelodysplastic syndromes. This finding is consistent with recent studies showing that IFN-γ released during infection can regulate myelopoiesis (23–25). Importantly, disease was completely alleviated by retargeting the original gene locus with a construct that re-established use of the endogenous ifng polyA sequence. Establishing genetic deficiency of T-bet also reversed the inflammatory hallmarks, which, in mice, consisted of many of the diagnostic criteria for HLH/MAS and PDS syndromes in humans. These studies suggest that there is a critical threshold of IFN-γ that, when achieved either locally in tissues or systemically, drives pathology consistent with autoinflammatory pathology. As such, these mice constitute important models to study the role of IFN-γ in the pathogenesis of autoinflammatory disease.
Materials and Methods
Mice
YFP-enhanced transcript for IFN-γ (Yeti) and IFN-γ reporter with endogenous polyA tail (Great) mice have been described and were bred 10 generations onto BALB/c and C57BL/6 backgrounds for these experiments (26, 27). In brief, Yeti and Great mice were generated by introducing an IRES–enhanced YFP (eYFP) construct after the stop codon of IFN-γ by homologous recombination, which leads to transcription of a bicistronic IFN-γ/IRES/eYFP mRNA and translation of both IFN-γ and eYFP from the same mRNA. This allows analysis of IFN-γ–competent cells in vivo by detection of YFP expression without the need for restimulation. Yeti mice differ from Great mice in that a bovine growth hormone polyA sequence was placed downstream of the YFP sequence to stabilize the mRNA. Mice were maintained in the University of California San Francisco or Duke University specific pathogen-free animal facility in accordance with institutional guidelines. Animals for these studies were used in accordance with Institutional Animal Care and Use Committee, Laboratory Animal Resource Center, and Division of Laboratory Animal Resources protocols established at the University of California San Francisco and Duke University.
Infections
Leishmania major strain WHOM/IR/-/173 was prepared and injected as 0.5–1 × 106 metacyclic promastigotes in the hind footpad of mice. Listeria monocytogenes strain 10403S strain (DP-L4056) was provided by Dr. D. Portnoy (University of California Berkeley). L. monocytogenes was grown to log phase and CFU were assayed by plating on blood agar. Mice were injected with 5 × 105 CFU (LD50 = 1 × 105) (28).
Flow cytometric analysis
Single-cell suspensions were prepared and labeled with Abs as listed: for IFN-γ reporter detection and phenotyping in Yeti and Great mice, YFP+ cells were analyzed for expression of CD4, CD8, CD3, and NK1.1. Granulocytes, monocytes, and neutrophils were labeled with Abs as listed: CD4, CD11c, Ly6G, CD11b, Ly6C, Ly6B (7/4), Gr1, MHC class II (MHC-II) (I-Ab), and CD40. Samples were analyzed on a LSR II or FACSCanto II (BD Biosciences). Where designated, a dump channel of PerCP-Cy5.5–labeled Abs was used to reduce nonspecific staining. Live lymphocytes were gated by DAPI exclusion, size, and granularity based on forward and side scatter. For intracellular cytokine staining, live/dead exclusion was performed and cells were fixed, permeabilized, and stained for intracellular IFN-γ as described (29, 30).
In vitro CD4 T cell differentiation
Cells were isolated from the lymph nodes of Great, Yeti, or wild-type animals, and CD4+ T cells were enriched by magnetic purification according to manufacturer’s instructions (CD4 negative selection, Miltenyi Biotec). Purified CD4+ T cells were incubated with irradiated APCs obtained from TCR-α–deficient mice to eliminate T cell contamination. Th1 cultures were performed as described (31).
ELISA
To detect total IFN-γ from serum of infected mice, 96-well plates were coated with purified anti–IFN-γ (R4-6A2, anti–IFN-γ) and incubated with 4- to 512-fold dilutions of serum. Bound IFN-γ was detected using biotinylated anti–IFN-γ (XMG1.2, biotin–anti-IFN-γ) followed by streptavidin-HRP and substrate o-phenylenediamine. Samples were prepared in duplicate and concentrations were determined by comparing to IFN-γ standards using an ELISA plate reader (Delta Soft). Plasma ferritin (1:40 dilution) and fibrinogen (1:10,000 dilution) were detected from duplicate samples by ELISA according to the manufacturer’s instructions (Immunology Consultants Laboratory, Newberg, OR).
Real-time quantitative PCR
Cells were harvested and RNA was purified from in vitro cultures at the indicated time points. Prior to reverse transcription, RNA was treated with DNAse (DNase I kit, Invitrogen) to remove contaminating genomic DNA. cDNA was generated according to the manufacturer’s instructions using SuperScript III first-strand synthesis (Invitrogen) with random hexamer primers. Real-time quantitative PCR reaction was performed in triplicate using 1:1 and 1:10 dilutions of cDNA as template. Transcripts were quantified by Power SYBR Green (Applied Biosystems) incorporation using a StepOnePlus real-time PCR system (Applied Biosystems) and plotted relative to expression of GAPDH or β-actin. Real-time primers were designed in exon-1 (5′-GCTTTGCAGCTCTTCCTCATGG-3′) and exon-3 (5′-TGCATCCTTTTTCGCCTTGC-3′) to ensure that only mRNA generated from the reporter allele would be amplified as the second defective allele contained a 2 kb neomycin cassette in the second exon, which introduced a termination codon after the first 30 aa of the mature protein (32). Using this approach, only the reporter allele generated a product at the expected size of 312 nt.
Results
Homozygous Yeti IFN-γ reporter mice fail to thrive
The first IFN-γ reporter mouse strain, designated Yeti, consists of a targeted insertion of an IRES, eYFP, and BGH polyA tail sequence, all inserted 3′ to the endogenous ifng gene (33). All known regulatory sequences in the endogenous locus are otherwise intact. The BGH polyA tail was used to stabilize the eYFP transcript, thus increasing sensitivity of the reporter, but also resulted in replacement of the endogenous polyA tail of the bicistronically expressed IFN-γ mRNA.
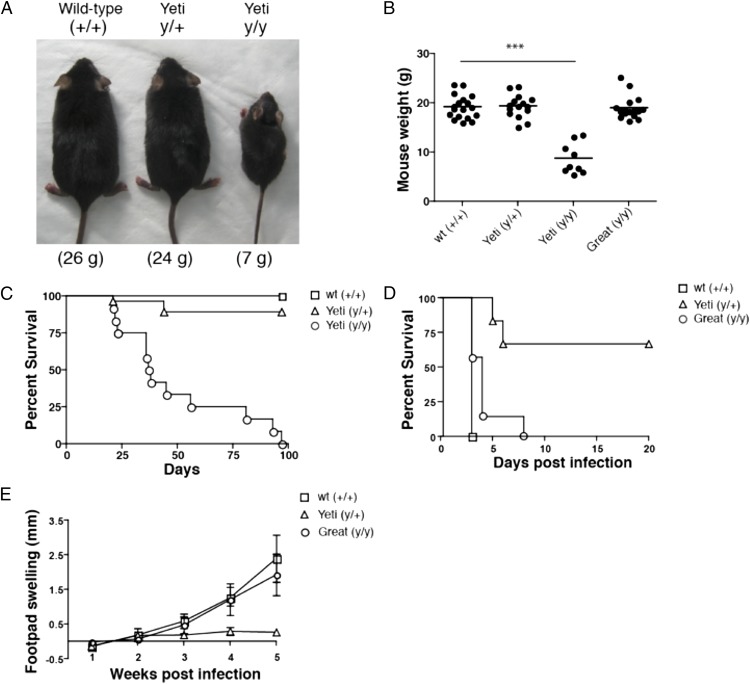
Although heterozygous Yeti C57BL/6 mice grew normally, homozygous mice containing two Yeti alleles failed to thrive as compared with their wild-type littermates. By 5–6 wk of age, heterozygous and wild-type mice, which did not differ, weighed 2- to 3-fold more than homozygous Yeti mice (Fig. 1A, 1B). Runting in homozygous Yeti mice was associated with increased mortality and 75% of homozygous mice died between 6 and 8 wk of age (Fig. 1C).
FIGURE 1.
Mouse weight, survival and response to infection. (A) Representative 6-wk-old wild-type (+/+), Yeti heterozygous (y/+), and Yeti homozygous (y/y) littermates. (B) Weights of 6- to 8-wk-old female mice. Graph represents a composite of weights taken from multiple litters. Each point represents an individual mouse weight with line denoting average weight of each genotype (n = 9–18 mice/group; ***p < 0.0001, two-tailed t test). (C) The percentage survival of wild-type (□), heterozygous (△), and homozygous (○) Yeti mice by days after birth (n = 9–27 mice/group). Graph is a composite of multiple litters. (D) The percent survival of wild-type (□), heterozygous Yeti (△), and homozygous Great (○) days after Listeria infection (n = 6–8 mice/group). Graph is representative of two independent experiments. (E) Footpad swelling in wild-type (□), heterozygous Yeti (△), and homozygous Great (○) BALB/c mice infected with L. major. Number represents size of infected footpad minus uninfected contralateral footpad (millimeters ± SD; n = 5 mice/group).
Heterozygous Yeti mice are resistant to infection
Despite the dramatic differences in weight gain and survival in homozygous Yeti mice, there was little effect on growth or mortality of heterozygous littermates (Fig. 1A–C). To assess whether heterozygous Yeti mice had functionally important elevations in IFN-γ, we infected wild-type and heterozygous Yeti littermates with a lethal dose of L. monocytogenes, a facultative intracellular bacterium whose control depends on IFN-γ (34, 35). Whereas wild-type littermates succumbed to infection within 3 d (Fig. 1D), heterozygous Yeti mice all lived >3 d and most survived for the entire 3 wk when the experiment was terminated (Fig. 1D).
We next crossed Yeti mice for 10 generations onto the BALB/c background and infected heterozygous Yeti BALB/c mice and littermates with L. major, an intracellular protozoan whose control depends on IFN-γ but that normally induces an aberrant and fatal Th2 response in BALB/c mice (36). Although littermate BALB/c mice showed progressive infection after footpad injection (Fig. 1E), heterozygous Yeti BALB/c mice were protected from disease, with the footpad swelling where the parasites were injected, never reaching the values observed in wild-type animals (Fig. 1E). Thus, despite the normal weight and survival in heterozygous Yeti mice, these mice demonstrated evidence for increased IFN-γ responses in comparison with wild-type littermates on two different genetic backgrounds.
Great IFN-γ reporter mice mount wild-type responses to infection
To show definitively that the aberrant phenotypes of the Yeti reporter mice were caused by introduction of the BGH polyA sequence, we generated a second targeted mouse strain that differed only by insertion of the IRES/eYFP element in the same 3′ region of the endogenous ifng gene but without the BGH polyA sequence, thus leaving the endogenous polyA sequence intact to regulate expression and/or translation. This second mouse reporter strain is designated Great. In contrast to Yeti mice, homozygous Great mice gained weight and grew normally as compared with wild-type mice (Fig. 1B). Additionally, Great mice demonstrated susceptibility to Listeria and L. major that was comparable to wild-type animals when assessed on the C57BL/6 and BALB/c backgrounds, respectively (Fig. 1D, 1E). In sum, retargeting the IFN-γ gene to reestablish use of the endogenous polyA element relieved the excessive IFN-γ phenotype of the Yeti strain, including the failure to thrive and the aberrant response to infectious challenges. Thus, alterations of IFN-γ mediated by the BGH polyA sequence underlie the phenotypes in both the heterozygote and homozygote Yeti strains as revealed by the normal phenotypes in the retargeted Great strains.
Yeti mice have increased IFN-γ production
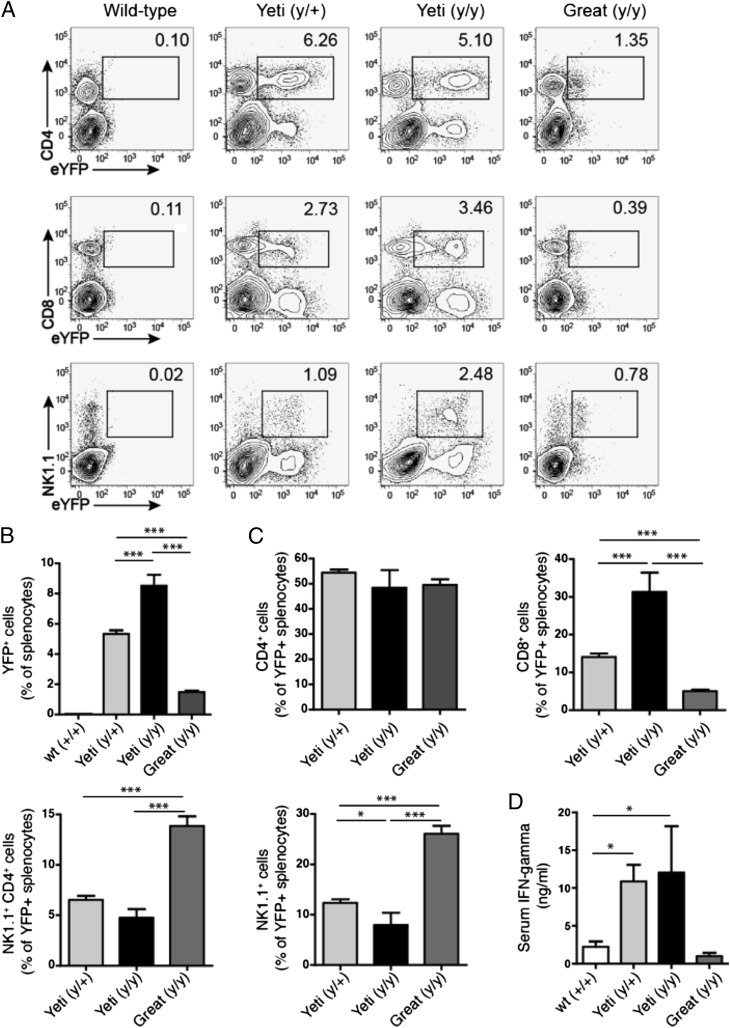
Protective immunity to Listeria and Leishmania requires IFN-γ, as mice deficient in this cytokine succumb to infection by these organisms. The enhanced resistance of Yeti mice suggested that these mice were capable of generating more IFN-γ than were wild-type animals. Indeed, both homozygous and heterozygous Yeti mice showed enhanced reporter activity in subsets of spleen cells as compared with wild-type and Great mice. Thus, ∼5 and 3% of eYFP+ splenocytes in unchallenged Yeti mice were CD4+ and CD8+ T cells, respectively (Fig. 2A). NK1.1+ cells were also marked as IFN-γ–producing cells and comprised ∼1 and 2.5% of total splenocytes in heterozygous and homozygous Yeti mice, respectively (Fig. 2A). In comparison, Great mice had reduced levels of eYFP+ cells at rest in each of these cell subsets (Fig. 2A). Heterozygous and homozygous Yeti mice had 5- and 8-fold more IFN-γ–producing cells among total spleen cells, respectively, than did Great mice (Fig. 2B).
FIGURE 2.
IFN-γ reporter expression and serum cytokine levels. (A) Reporter expression in CD4+, CD8+, NK1.1+ splenocytes from wild-type, Great, and Yeti reporter mice at rest. Plots gated on single-cell suspensions of live (DAPI−) splenocytes. The numbers in each gate represent the percentage of eYFP+ cells among total splenocytes. Plots are representative of two independent experiments (n = 5–25 mice total). (B) The graphs represent eYFP reporter–positive cells as a percentage of total splenocytes (mean percentage ± SEM). Graphs represent data points combined from at least two independent experiments (n = 5–25/group; ***p < 0.0001, two-tailed t test). (C) The graph represents eYFP expression of CD4+, CD8+, NK1.1+ cells as a percentage of total eYFP+ splenocytes in wild-type, Yeti, and Great mice (percentage ± SEM). Data points combined from at least two independent experiments (n = 5-25 mice/group; *p < 0.01, ***p < 0.0001, two-tailed t test). (D) The graph represents serum IFN-γ in wild-type, Yeti, and Great mice (ng/ml ± SEM). Data points combined from 10 independent experiments (n = 9–55 mice/group; *p < 0.01, two-tailed t test).
In an effort to better understand the contribution of specific cell types to the elevated levels of IFN-γ, we assessed the percentages of eYFP+ CD4+, CD8+, NKT, and NK cells among total IFN-γ–producing cells. Although IFN-γ–producing CD4+ T cells represented the same percentage of total reporter-positive splenocytes in Yeti and Great mice, CD8+ T cells represented a significantly higher percentage in Yeti mice. In contrast, NK and NKT cells represented the most prevalent IFN-γ–producing subsets in Great mice (Fig. 2C). As constitutive IFN-γ mRNA expression has been demonstrated in these innate lymphocytes but not in naive T cells (26), Great mice appear to regulate the cytokine in a manner similar to wild-type mice.
To test whether the increased percentage of eYFP+ reporter cells in Yeti mice resulted in increased IFN-γ protein levels, we measured concentrations of IFN-γ in the serum of Great, Yeti, and wild-type animals by ELISA. Supporting the reporter expression data, serum from heterozygous and homozygous Yeti mice contained >10-fold more IFN-γ than serum collected from wild-type and Great mice, which did not differ (Fig. 2D). In sum, retargeting the ifng locus relieved the elevated IFN-γ levels and the aberrant growth and infectious phenotypes of the Yeti mice, thus directly implicating dysregulation of IFN-γ in these processes.
Enhanced IFN-γ protein and mRNA in cells from Yeti mice
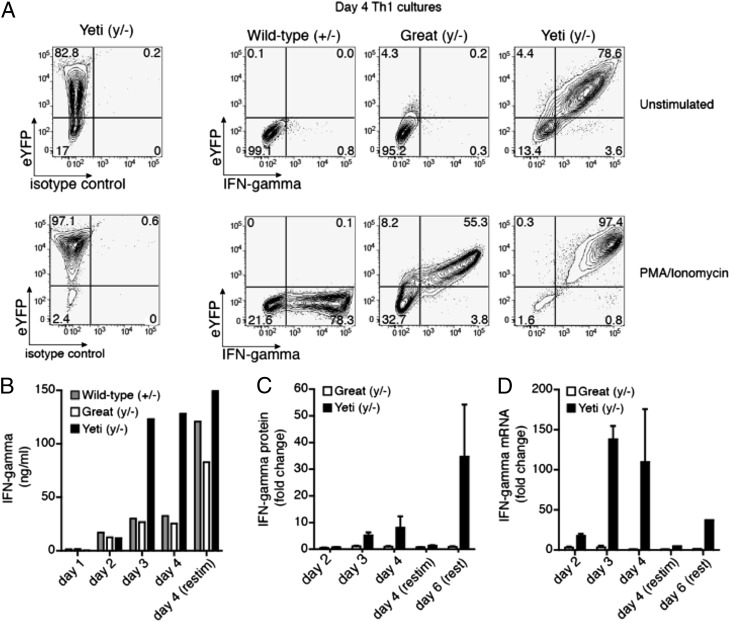
As Yeti and Great reporter constructs differ only in their 3′ untranslated regions (UTR), we hypothesized that the discrepancies in serum levels of cytokine resulted from an increased abundance of IFN-γ mRNA transcripts. To test this hypothesis, we crossed Yeti, Great, and wild-type mice to IFN-γ–deficient mice to assess IFN-γ mRNA expression and cytokine production from a single reporter or wild-type allele. Confining IFN-γ mRNA expression to a single allele in the various heterozygous mice allowed us to directly compare the expression of intracellular IFN-γ mRNA and protein from each reporter strain without the caveats associated with contributions from the wild-type allele. CD4+ T cells from each of the mice were cultured under Th1 conditions for 4 d and assessed for their ability to generate eYFP and intracellular IFN-γ spontaneously or after a second stimulation with PMA and ionomycin. After 4 d of culture, <1% of resting wild-type and Great CD4+ T cells stained positive for intracellular IFN-γ, and only 4.3% of Great CD4+ T cells continued to express the eYFP reporter (Fig. 3A). After restimulation with PMA/ionomycin, 60–78% of Great and wild-type CD4+ T cells stained positive for intracellular IFN-γ (Fig. 3A). All of the cytokine-positive CD4+ T cells from Great mice coexpressed intracellular IFN-γ and eYFP, thus validating that the Great construct sustains endogenous IFN-γ at levels similar to the wild-type IFN-γ allele (as the IFN-γ is present only on the targeted allele). Although <1% of Great Th1 cells continued to produce intracellular IFN-γ by 4 d after primary stimulation, fully 78% of Yeti Th1 cells continued to express intracellular IFN-γ even before the secondary stimulation. After restimulation with PMA/ionomycin, >97% of cells from the Yeti/– mice were positive for IFN-γ. Thus, Yeti Th1 cells continue to produce substantial amounts of cytokine even at times during the culture period when Great and wild-type cells have ceased producing IFN-γ.
FIGURE 3.
Yeti mice but not Great mice have increased IFN-γ protein and mRNA in Th1 cultures. Sorted naive CD4+ T cells cultured with irradiated APCs and soluble anti-CD3 and anti-CD28 for 4 d under Th1 conditions. Wild-type, Yeti, and Great mice were crossed to IFN-γ–deficient mice to restrict reporter analysis to one allele. Thus, each mouse has one functional and one deleted IFN-γ allele (wild-type, +/−; Yeti, y/−; Great, y/−) (A) Cells were either left unstimulated (top panel) or restimulated (bottom panel) with PMA/ionomycin. Plots are gated on live (Live/Dead violet−) CD4+ T cells, and the number in each quadrant represents the percentage of cells with eYFP or intracellular anti–IFN-γ expression. Contour plots are representative of three independent experiments (n = duplicate wells/experiment). (B) The graph represents the amount of IFN-γ secreted into the supernatant of Th1 cultures from wild-type (gray bars), Yeti (black bars), and Great (white bars) CD4+ T cells at the indicated time points (ng/ml). Wells were restimulated as indicated with PMA/ionomycin for 6 h on day 4 of culture. Data are representative of three independent experiments. (C) The graph represents the relative change in secreted IFN-γ from Yeti (black bars) and Great (white) CD4+ T cells compared with wild-type cells. Data are representative of 3 independent experiments. (D) The graph represents IFN-γ mRNA transcripts from Yeti (filled bars) and Great (open bars) CD4+ T cells plotted as fold change over wild-type levels. Relative mRNA was normalized to the housekeeping gene GAPDH. Data are representative of three independent experiments.
To confirm that the prolonged intracellular production of IFN-γ by cells from Yeti mice led to increased amounts of the secreted cytokine, the concentrations of IFN-γ in Th1 culture supernatants were measured. During the first 2 d, CD4+ T cells from Yeti mice secreted levels of IFN-γ that were similar to those of wild-type and Great CD4+ T cells (Fig. 3B, 3C). However, by days 3 and 4, Yeti cultures contained 4-fold more IFN-γ than did wild-type or Great cultures. Restimulation on day 4 led to an accumulation of secreted IFN-γ in wild-type cultures similar to that generated by Yeti CD4+ T cells, suggesting that the observed differences were not a result of an outgrowth or preferential survival of Yeti CD4+ T cells. Of note, the differences in cytokine production between Great and Yeti CD4+ T cells also occurred when cultures were rested for an additional 2 d after secondary stimulation with PMA/ionomycin. Under these conditions, Yeti cultures contained 35-fold more cytokine when normalized to wild-type cultures as compared with Great cultures (Fig. 3C). This differential increase in cytokine expression among Great and Yeti Th1 cells was also apparent at the mRNA level, as Yeti cultures contained >100-fold more IFN-γ transcript than did Great cell cultures relative to wild-type (Fig. 3D). After restimulation with PMA/ionomycin, mRNA levels in wild-type and Great cultures rose such that transcripts were equivalent among all three culture conditions. Thus, IFN-γ expression is initiated in the appropriate cells and under appropriate conditions in these knock-in reporter mice but cytokine expression is maintained aberrantly in the Yeti strain containing the BGH polyA sequence. The finding that wild-type and Great cells generate equal levels of IFN-γ mRNA as do Yeti cell cultures after restimulation suggests that the increased IFN-γ mRNA observed prior to restimulation in Yeti Th1 cells is due to accumulation of transcript over time rather than an intrinsic ability of Yeti cells to generate more mRNA. The mechanism by which increased stability of the Yeti transcript leads to increased IFN-γ mRNA accumulation remains to be fully explored, but the elimination of the UAUUA-rich 3′UTR of the endogenous IFN-γ mRNA, which is known to destabilize cytokine mRNA, is the likely explanation and is consistent with elimination of the phenotype by the retargeted Great allele (37–40).
Yeti mice exhibit overt inflammation consistent with autoinflammatory disease
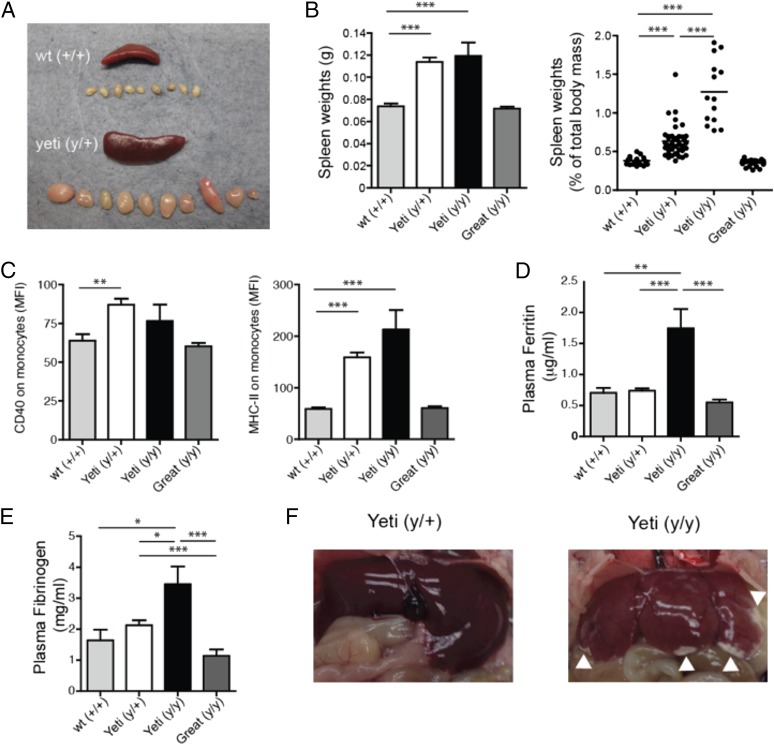
IFN-γ is a proinflammatory cytokine and, consistent with persistent inflammation, even Yeti heterozygous mice had spontaneous splenomegaly and lymphadenopathy (Fig. 4A). This was even more evident in homozygous Yeti mice when the weight of the spleen is presented as a percentage of the total body mass of the mouse (Fig. 4B). This method takes into account the reduced body size of homozygous Yeti mice. Of note, homozygous Great mice were comparable to wild-type animals in comparing spleen weight and lymph node size, demonstrating that these mice do not experience the inflammatory processes that occur in the Yeti mice.
FIGURE 4.
Splenomegaly, lymphadenopathy, and monocyte activation in resting Yeti mice with features of autoinflammatory disease. (A) Representative sizes of the spleen and peripheral lymph nodes in 6-wk-old wild-type and heterozygous Yeti mice. (B) Graphs represents total spleen weight (left panel; grams ± SEM) or as a percentage of total body mass (right panel; percentage ± SEM) in 5- to 6-wk-old wild-type (gray bars), heterozygous Yeti (white bars), homozygous Yeti (black bars), and Great (dark gray bars) mice. Graphs are a composite of mice from at least 10 experiments (n = 15–77 mice/group; ***p < 0.0001, two-tailed t test). (C) Graphs represent the mean fluorescence intensity (MFI) of CD40 (left panel) or I-Ab (right panel) expression on CD11b+GR1int monocytes (MFI ± SEM) isolated from the spleen of wild-type (gray bars), heterozygous Yeti (white bars), homozygous Yeti (black bars), and Great (dark gray bars) mice. Graphs are a composite of mice from at least seven experiments (n = 9–44 mice/group; **p < 0.001, ***p < 0.0001, two-tailed t test). (D) Graph represents plasma ferritin levels in 5- to 6-wk-old wild-type (gray bars), heterozygous Yeti (white bars), homozygous Yeti (black bars), and Great (dark gray bars) mice (μg/ml ± SEM). Data points are combined from four independent experiments (n = 5–25 mice/group; **p < 0.001, ***p < 0.0001, two-tailed t test). (E) Graph represents plasma fibrinogen levels in 5- to 6-wk-old wild-type (gray bars), heterozygous Yeti (white bars), homozygous Yeti (black bars), and Great (dark gray bars) mice (μg/ml ± SEM). Data points are combined from four independent experiments (n = 5–25 mice/group; *p < 0.01, ***p < 0.0001, two-tailed t test). (F) Representative livers from heterozygous and homozygous Yeti mice. Arrowheads denote areas of pathology consistent with necrosis, ischemia, and fibrosis.
IFN-γ induces CD40 and MHC-II on myeloid cells, and these molecules are commonly used as markers of activated APCs. Splenic monocytes from both heterozygous and homozygous 5- to 6-wk-old Yeti mice spontaneously expressed elevated CD40 and MHC-II on their cell surface as compared with wild-type animals (Fig. 4C), consistent with an ongoing activated state. In contrast, Great mice showed no significant differences in monocyte activation as compared with wild-type mice. Taken together, these data demonstrate a direct relationship between prolonged exposure to high levels of IFN-γ and induction of a persistent activated state on myeloid cells in mice.
As implied by their names, HLH/MAS are syndromes associated with increased macrophage and monocyte activation, splenomegaly, and prolonged fever. Although not considered causative or diagnostic, these syndromes often present with a secondary cytokine storm that resembles the high IFN-γ levels established in Yeti mice. Indeed, the sustained IFN-γ signature in these mice was also consistent with symptoms seen in CANDLE and JMP. The similar inflammatory phenotype observed among HLH/MAS and PDS patients and Yeti mice prompted additional investigations of these mice as a potential model for these autoinflammatory diseases. Serum ferritin in both mice and humans is normally <0.3 μg/ml (41). Consistent with HLH-like disease in humans, where serum ferritin levels >0.5 μg/ml comprise one of the eight diagnostic criteria, homozygous Yeti mice had consistently elevated ferritin levels (1.5–2.0 μg/ml) (Fig. 4D) (42).
Although hypofibrinogenemia is one of the diagnostic criteria for HLH disease, fibrinogen levels were increased in homozygous Yeti mice (Fig. 4E) (42). Recent studies suggest that fewer than 38–46% of patients diagnosed with secondary HLH present with hypofibrinogenemia (43, 44). Homozygous Yeti mice also showed pathologic evidence of liver damage, likely a result of poor blood perfusion and ischemia (Fig. 4F). Thus, IFN-γ likely contributes directly to some aspects of the pathology associated with HLH or MAS, but perhaps not to all markers for the disease.
Homozygous Yeti mice show features of myeloproliferative disease
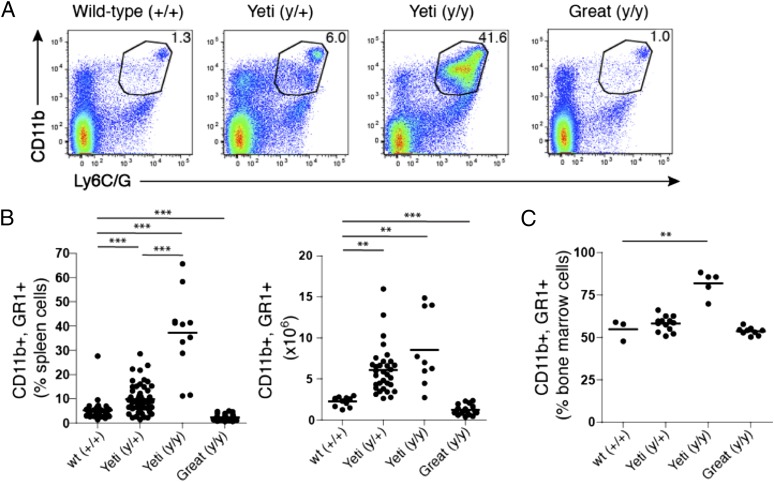
To assess whether the increase in splenic volume in Yeti mice was due to an outgrowth of specific subsets of cells, we assessed the cellular composition of the spleen and bone marrow. Although lymphocytes such as B and T cells were similar in all strains of mice, cells marked by the coexpression of CD11b and Ly6C/G (GR1) varied substantially. Yeti heterozygous mice showed a range of myeloid cell composition in the spleen that was 5–10% higher than that observed in their wild-type and Great counterparts (Fig. 5A, 5B). Furthermore, Yeti mice exhibited a pronounced increase in CD11b+GR1+ cells with 40% of the splenocytes coexpressing these markers; this number reached 65% in homozygous Yeti mice (Fig. 5A, 5B). The total number of granulocytes was also significantly increased in Yeti mice (Fig. 5B). The observed increase in cellularity was not confined to the spleen because CD11b+GR1+ myeloid cells were also increased in the bone marrow (Fig. 5C).
FIGURE 5.
Myeloproliferation in Yeti mice. (A) Plots represent the percentage of CD11b+GR1+ populations in the spleen of the indicated mice. Plots are gated on single-cell suspensions of live (DAPI−) spleen cells. The numbers in each gate represent the percentage of GR1+CD11b+ cells among total splenocytes. Plots are representative of at least four independent experiments. (B) The graphs represent the percentage and total number of CD11b+GR1+ cells among total spleen cells from the indicated mice. Graphs represent data compiled from at least five independent experiments (percentage or number of total spleen cells ± SEM; n = 11–52 mice/group; **p < 0.001, ***p < 0.0001, two-tailed t test). (C) The graph represents the percentage of CD11b+GR1+ cells in total bone marrow of the indicated mice. Graphs represent data compiled from at least five independent experiments (percentage of total spleen cells ± SEM; n = 11–52 mice/group; **p < 0.002, two-tailed t test).
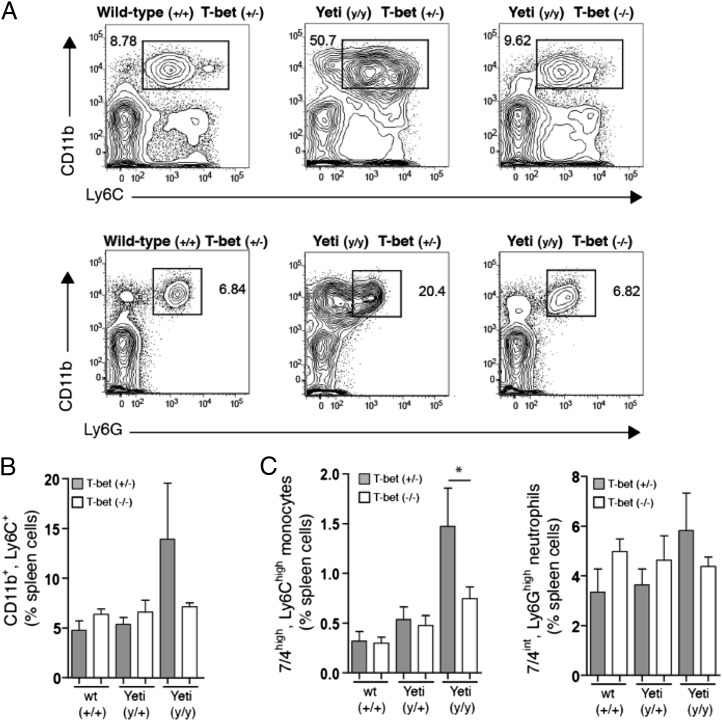
Because IFN-γ production and its downstream effects are dependent to a large extent on the transcription factor T-bet, we assessed whether the observed increase in GR1+CD11b+ cells in Yeti mice was T-bet–dependent. Although no difference in CD11b+Ly6C+ cells were seen in the bone marrow and spleen of T-bet–deficient and T-bet–sufficient wild-type animals, T-bet deficiency in homozygous Yeti mice restored the percentage of CD11b+Ly6C+ cells to wild-type levels (Fig. 6A, 6B). Because CD11b+Ly6C+ cells make up a broad family of immune cells, including neutrophils, monocytes, macrophages, and certain dendritic cell subsets, we further differentiated these cells using neutrophil- and monocyte-specific markers. Ly6G–7/4high monocytes were significantly increased in homozygous Yeti animals as compared with wild-type littermates (Fig. 6C). This increase in monocytes was abrogated in T-bet–deficient Yeti animals. The most abundant population of CD11b+Ly6C− cells in Yeti mice was Ly6G+ neutrophils, which represented 25% of all splenocytes (Fig. 6A). Ly6G+7/4int neutrophils were also increased in homozygous Yeti mice as compared with wild-type animals, but T-bet deficiency affected this population to a lesser extent than did that observed with monocytes (Fig. 6C). These results suggest that the increased disease severity observed between heterozygous and homozygous Yeti mice is associated with a proliferative disorder that predominantly affects myeloid cells.
FIGURE 6.
Myeloproliferation in Yeti mice is dependent on T-bet. (A) Plots are gated on single-cell suspensions of live (DAPI−) spleen cells. The numbers in each gate represent the percentage of CD11bhighLy6C+ granulocytes (top panel) and CD11b+Ly6G+ neutrophils among total splenocytes. Plots are derived from littermates of the indicated genotype and are representative of eight independent experiments. (B) The graphs represent the percentage of CD11b+Ly6C+ cells of total spleen in T-bet–sufficient (+/−, filled bars) or T-bet–deficient (−/−, open bars) mice. Data were compiled from eight independent experiments (percentage ± SEM; n = 3–13 mice/group). (C) The graphs represent the percentage of 7/4highLy6Chigh monocytes and 7/4intLy6G+ neutrophils among total splenocytes from T-bet–sufficient (+/−, filled bars) or T-bet–deficient (−/−, open bars) mice. Data were compiled from eight independent experiments (percentage ± SEM; n = 3–13 mice/group; *p < 0.05, two-tailed t test).
IFN-γ–mediated autoinflammatory disease is ameliorated in T-bet–deficient Yeti mice
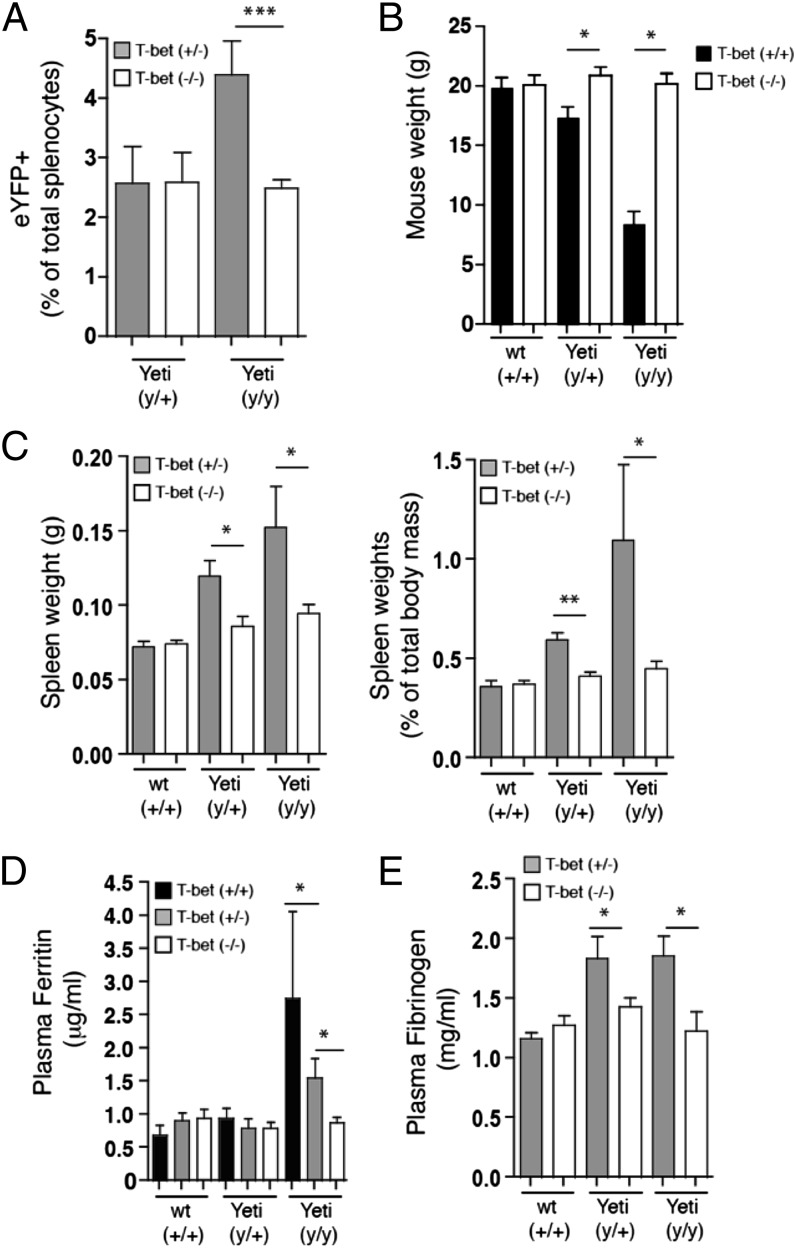
Because T-bet deficiency significantly reduced neutrophil and monocyte numbers in the spleen and bone marrow of Yeti homozygous mice, it suggested that T-bet might influence additional aspects of the disease. Consistent with this hypothesis, T-bet–deficient Yeti mice had reduced numbers of IFN-γ reporter–positive cells in the spleen as compared with T-bet–sufficient Yeti mice (Fig. 7A). Furthermore, there was no decrease in survival among homozygous Yeti mice on the T-bet–deficient background, which also restored normal weight gain, demonstrating that disease severity was greatly attenuated when IFN-γ production was diminished (Fig. 7B). Inflammatory disease manifestations were also significantly reduced in T-bet–deficient Yeti mice, and spleen weights and plasma ferritin levels were comparable to wild-type animals (Fig. 7C, 7D). Elevated fibrinogen levels also returned to baseline in T-bet–deficient Yeti animals (Fig. 7E).
FIGURE 7.
Hallmarks of hyperinflammatory disease and associated morbidity in Yeti mice are dependent on T-bet. (A) Graph represents eYFP+ cells as a percentage of total splenocytes from T-bet–sufficient (filled bars) or T-bet–deficient (open bars) mice. Data represent mice compiled from nine independent experiments (percentage ± SEM; n = 6–13 mice/group; ***p < 0.0001, two-tailed t test). (B) Graph represents weight of 5- to 6-wk-old T-bet–sufficient (+/+, filled bars) or T-bet–deficient (−/−, open bars) mice. Data were compiled from mice taken from four independent experiments (weight ± SEM; n = 3–23 mice/group; *p < 0.02, two-tailed t test). (C) Graphs represents total spleen mass (left panel; grams ± SEM) or spleen weight as a percentage of total body mass (right panel; percentage ± SEM) in 6- to 10-wk-old T-bet–sufficient (filled bars) or T-bet–deficient (open bars) mice. Data were compiled from mice from eight independent experiments (n = 3–16 mice/group; *p < 0.02, **p < 0.001, two-tailed t test). (D) Graph represents plasma ferritin levels in 5- to 10-wk-old T-bet–sufficient (black bars) or T-bet–deficient (white bars) mice. Data points represent mice and are combined from 11 independent experiments (μg/ml ± SEM; n = 3–18 mice/group; *p < 0.02, two-tailed t test). (E) Graph represents plasma fibrinogen levels in 5- to 10-wk-old T-bet–sufficient (filled bars) or T-bet–deficient (open bars) mice. Data points represent mice combined from seven independent experiments (mg/ml ± SEM; n = 3–9 mice/group; *p < 0.05, two-tailed t test).
Discussion
These studies demonstrate that IFN-γ can itself mediate the development of autoinflammatory-like syndromes in mice. High levels of IFN-γ were sufficient to induce many of the disease-associated hallmarks associated with autoinflammatory syndromes, including some of those associated with HLH. The direct role of IFN- γ was demonstrated by retargeting the gene locus to alleviate the syndrome and independently by crossing animals to a T-bet–deficient background to establish that loss of this major regulator of IFN-γ was sufficient to abrogate the autoinflammatory disease. Taken together, these studies demonstrate that cytokines themselves can be a substantial factor contributing to disease progression, as well as to the morbidity and mortality associated with autoinflammatory disease. Furthermore, these results highlight the need to consider proinflammatory cytokines such as IFN-γ as mediators of disease progression, such that anti-cytokine therapies might potentially ameliorate tissue and organ dysfunction. As suggested by these findings, targeting IFN-γ might be helpful in diseases refractory to IL-1β– and TNF-targeted therapies.
Despite these observations, questions remain regarding the role of IFN-γ in autoinflammatory disease. First, what are the relevant cellular sources of cytokine in these diseases? It is well established that IFN-γ from CD8+ T cells is essential for the development of virally induced HLH (14–16). However, a viral trigger is not found in all HLH patients, suggesting that other IFN-γ–producing cell types may be important in disease progression in some circumstances. Thus, it remains possible that other IFN-γ–producing cells, other than CD8+ T cells, are contributing to disease in various autoinflammatory conditions. In support of this, CD8+ T cells, NK cells, and other lymphocytes were deemed unnecessary for CpG-mediated mouse models of HLH (17). The sources of the elevated IFN-γ in PDS syndromes remain even less clear, and, given the lack of mouse systems available to study PDS, inferences from YETI mice may be important in establishing models useful for understanding the role of cytokines in PDS-like disease.
Comparisons between the Yeti and Great mice establish directly that alteration of mRNA at the 3′UTR of the IFN-γ mRNA is sufficient to cause disease. This suggests that it may not be a specific cell that is responsible for disease morbidity, but rather the total level of IFN-γ. This was evident given the different disease manifestations observed in comparing heterozygous and homozygous Yeti mice, where the presence of the second Yeti allele in homozygous animals resulted in significantly higher morbidity and mortality. Given our inability to identify any difference among the subsets of IFN-γ–producing cells in the homozyogous as compared with heterozygous mice, it would suggest that the levels of IFN-γ rather than any particular cell type are responsible for disease progression. Thus, a threshold of IFN-γ becomes established before significant deleterious effects occur. As such, complete inhibition of IFN-γ may not be necessary to prevent clinical HLH or PDS. Maintaining cytokine levels below a requisite threshold may allow the host to avoid the subsequent multiorgan involvement resulting from excessive inflammation. Although our study supports the idea that systemic cytokines can induce disease, the similar serum levels observed between homozygous and heterozygous Yeti mice would indicate that the increase in morbidity could be a result of the summation of localized effects of cytokine reaching a distinct threshold within tissues. Indeed, we speculate that total tissue IFN-γ–producing cells reach pathologic levels in homozygous Yeti mice, and this will be an important area for future study. It may also be reasonable to consider whether mutations in the 3′UTR of IFN-γ are associated with inflammatory syndromes accompanying hematologic malignancies in humans. Intriguingly, mutations in the 3′UTR of TNF, another inflammatory cytokine, results in organ-specific pathology in the joints (arthritis) and bowel (colitis) in mice, and in contrast to the pathology established by the similar mutation in the IFN-γ 3′UTR as described in this study, suggesting that distinct inflammatory cytokines will induce distinct syndromes when dysregulated (39). Of relevance to our findings, the studies in the mice with TNF mutations set the groundwork for the successful use of anti-TNF therapies in human arthritis and colitis syndromes.
Finally, our studies suggest a link between the severity of autoinflammatory disease and the development of myeloproliferative disease. IFN-γ is known to skew hematopoiesis toward production of monocytes (45). Consistent with this, homozygous Yeti mice show marked increases in monocytes in both the bone marrow and spleen. The excessive hematopoiesis observed in Yeti mice is interesting given that some patients with certain leukemic malignancies can develop concurrent HLH or MAS (46–50). Although it is thought that HLH and MAS are byproducts associated with the leukemia, our results suggest that the relationship between HLH-like disorders and myeloproliferative syndromes may be more complicated. Of note, gain-of-function mutations in JAK2, a critical component of IFN-γ receptor signal transduction, have been associated with myeloproliferative neoplasms and leukemias (51–55). Thus, increased IFN-γ levels leading to chronic receptor signaling in Yeti mice may phenocopy aspects of JAK/STAT-mediated myeloproliferation. As a result, JAK/STAT inhibitors may represent additional therapeutic candidates in certain autoinflammatory disorders and autoinflammatory-associated leukemias.
In summary, Yeti and Great mice represent new tools to study the role of IFN-γ and IFN-γ–producing cells in inflammatory disorders, cancer, or infectious disease. Because these mice contain fluorescent reporters under regulation of the endogenous IFN-γ locus, they facilitate direct visualization of cell sources of this important cytokine. Importantly, the Great mice do not appear to suffer from off-target effects and accurately delineate IFN-γ in situ, whereas Yeti mice offer a spontaneous model to study states of persistently elevated IFN-γ. As such, these mice provide a unique opportunity to explore the role of IFN-γ in the development of autoinflammatory diseases, and they should prove useful in both understanding disease pathogenesis as well as in identifying novel therapeutic targets.
Acknowledgments
We thank the University of California San Francisco transgenic facility for injection services, A. Reinhardt (University of Nebraska Medical Center), C. Lowell, and A. Molosky for review and comments, and N. Flores and Z. Wang for technical expertise.
This work was supported by National Institute of Allergy and Infectious Diseases Grants AI026918, AI030663, and AI119944, the Howard Hughes Medical Institute, and the Sandler Asthma Basic Research Center at the University of California San Francisco.
Abbreviations used in this article:
- BGH
- bovine growth hormone
- CANDLE
- chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature
- eYFP
- enhanced YFP
- Great
- IFN-γ reporter with endogenous polyA tail
- HLH
- hemophagocytic lymphohistiocytosis
- JMP
- joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced childhood onset lipodystrophy
- MAS
- macrophage activation syndrome
- MHC-II
- MHC class II
- PDS
- proteasome disability syndrome
- UTR
- untranslated region
- Yeti
- yellow fluorescent protein–enhanced transcript for IFN-γ
- YFP
- yellow fluorescent protein.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Masters S. L., Simon A., Aksentijevich I., Kastner D. L. 2009. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 27: 621–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGonagle D., McDermott M. F. 2006. A proposed classification of the immunological diseases. PLoS Med. 3: e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinarello C. A. 2011. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117: 3720–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinarello C. A. 2009. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27: 519–550. [DOI] [PubMed] [Google Scholar]
- 5.Federici S., Martini A., Gattorno M. 2013. The central role of anti-IL-1 blockade in the treatment of monogenic and multi-factorial autoinflammatory diseases. Front. Immunol. 4: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldbach-Mansky R. 2012. Immunology in clinic review series; focus on autoinflammatory diseases: update on monogenic autoinflammatory diseases: the role of interleukin (IL)-1 and an emerging role for cytokines beyond IL-1. Clin. Exp. Immunol. 167: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imashuku S. 1997. Differential diagnosis of hemophagocytic syndrome: underlying disorders and selection of the most effective treatment. Int. J. Hematol. 66: 135–151. [DOI] [PubMed] [Google Scholar]
- 8.Stark B., Cohen I. J., Pecht M., Umiel T., Apte R. N., Friedman E., Levin S., Vogel R., Schlesinger M., Zaizov R. 1987. Immunologic dysregulation in a patient with familial hemophagocytic lymphohistiocytosis. Cancer 60: 2629–2636. [DOI] [PubMed] [Google Scholar]
- 9.Schneider E. M., Lorenz I., Müller-Rosenberger M., Steinbach G., Kron M., Janka-Schaub G. E. 2002. Hemophagocytic lymphohistiocytosis is associated with deficiencies of cellular cytolysis but normal expression of transcripts relevant to killer-cell-induced apoptosis. Blood 100: 2891–2898. [DOI] [PubMed] [Google Scholar]
- 10.Billiau A. D., Roskams T., Van Damme-Lombaerts R., Matthys P., Wouters C. 2005. Macrophage activation syndrome: characteristic findings on liver biopsy illustrating the key role of activated, IFN-γ-producing lymphocytes and IL-6- and TNF-α-producing macrophages. Blood 105: 1648–1651. [DOI] [PubMed] [Google Scholar]
- 11.Xu X. J., Tang Y. M., Song H., Yang S. L., Xu W. Q., Zhao N., Shi S. W., Shen H. P., Mao J. Q., Zhang L. Y., Pan B. H. 2012. Diagnostic accuracy of a specific cytokine pattern in hemophagocytic lymphohistiocytosis in children. J. Pediatr. 160: 984–990.e1. [DOI] [PubMed] [Google Scholar]
- 12.Imashuku S., Hibi S., Fujiwara F., Todo S. 1996. Hyper-interleukin (IL)-6-naemia in haemophagocytic lymphohistiocytosis. Br. J. Haematol. 93: 803–807. [DOI] [PubMed] [Google Scholar]
- 13.Imashuku S., Hibi S., Fujiwara F., Ikushima S., Todo S. 1994. Haemophagocytic lymphohistiocytosis, interferon-gamma-naemia and Epstein-Barr virus involvement. Br. J. Haematol. 88: 656–658. [DOI] [PubMed] [Google Scholar]
- 14.Jordan M. B., Hildeman D., Kappler J., Marrack P. 2004. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood 104: 735–743. [DOI] [PubMed] [Google Scholar]
- 15.Pachlopnik Schmid J., Ho C. H., Chrétien F., Lefebvre J. M., Pivert G., Kosco-Vilbois M., Ferlin W., Geissmann F., Fischer A., de Saint Basile G. 2009. Neutralization of IFNγ defeats haemophagocytosis in LCMV-infected perforin- and Rab27a-deficient mice. EMBO Mol. Med. 1: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crozat K., Hoebe K., Ugolini S., Hong N. A., Janssen E., Rutschmann S., Mudd S., Sovath S., Vivier E., Beutler B. 2007. Jinx, an MCMV susceptibility phenotype caused by disruption of Unc13d: a mouse model of type 3 familial hemophagocytic lymphohistiocytosis. J. Exp. Med. 204: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens E. M., Canna S. W., Slade K., Rao S., Kreiger P. A., Paessler M., Kambayashi T., Koretzky G. A. 2011. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J. Clin. Invest. 121: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoller E. E., Lykens J. E., Terrell C. E., Aliberti J., Filipovich A. H., Henson P. M., Jordan M. B. 2011. Hemophagocytosis causes a consumptive anemia of inflammation. J. Exp. Med. 208: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arima K., Kinoshita A., Mishima H., Kanazawa N., Kaneko T., Mizushima T., Ichinose K., Nakamura H., Tsujino A., Kawakami A., et al. 2011. Proteasome assembly defect due to a proteasome subunit β type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. USA 108: 14914–14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Ramot Y., Torrelo A., Paller A. S., Si N., Babay S., Kim P. W., Sheikh A., Lee C. C., Chen Y., et al. 2012. Mutations in proteasome subunit β type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 64: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal A. K., Xing C., DeMartino G. N., Mizrachi D., Hernandez M. D., Sousa A. B., Martínez de Villarreal L., dos Santos H. G., Garg A. 2010. PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am. J. Hum. Genet. 87: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanazawa N. 2012. Nakajo-Nishimura syndrome: an autoinflammatory disorder showing pernio-like rashes and progressive partial lipodystrophy. Allergol. Int. 61: 197–206. [DOI] [PubMed] [Google Scholar]
- 23.Schürch C., Riether C., Amrein M. A., Ochsenbein A. F. 2013. Cytotoxic T cells induce proliferation of chronic myeloid leukemia stem cells by secreting interferon-γ. J. Exp. Med. 210: 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belyaev N. N., Biró J., Langhorne J., Potocnik A. J. 2013. Extramedullary myelopoiesis in malaria depends on mobilization of myeloid-restricted progenitors by IFN-γ induced chemokines. PLoS Pathog. 9: e1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNamara K. C., Oduro K., Martin O., Jones D. D., McLaughlin M., Choi K., Borjesson D. L., Winslow G. M. 2011. Infection-induced myelopoiesis during intracellular bacterial infection is critically dependent upon IFN-γ signaling. J. Immunol. 186: 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stetson D. B., Mohrs M., Reinhardt R. L., Baron J. L., Wang Z. E., Gapin L., Kronenberg M., Locksley R. M. 2003. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinhardt R. L., Liang H. E., Locksley R. M. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brockstedt D. G., Giedlin M. A., Leong M. L., Bahjat K. S., Gao Y., Luckett W., Liu W., Cook D. N., Portnoy D. A., Dubensky T. W., Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 101: 13832–13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhardt R. L., Khoruts A., Merica R., Zell T., Jenkins M. K. 2001. Visualizing the generation of memory CD4 T cells in the whole body. Nature 410: 101–105. [DOI] [PubMed] [Google Scholar]
- 30.Khoruts A., Mondino A., Pape K. A., Reiner S. L., Jenkins M. K. 1998. A natural immunological adjuvant enhances T cell clonal expansion through a CD28-dependent, interleukin (IL)-2-independent mechanism. J. Exp. Med. 187: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price A. E., Reinhardt R. L., Liang H. E., Locksley R. M. 2012. Marking and quantifying IL-17A-producing cells in vivo. PLoS ONE 7: e39750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalton D. K., Pitts-Meek S., Keshav S., Figari I. S., Bradley A., Stewart T. A. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259: 1739–1742. [DOI] [PubMed] [Google Scholar]
- 33.Stetson D. B., Mohrs M., Mallet-Designe V., Teyton L., Locksley R. M. 2002. Rapid expansion and IL-4 expression by Leishmania-specific naive helper T cells in vivo. Immunity 17: 191–200. [DOI] [PubMed] [Google Scholar]
- 34.Buchmeier N. A., Schreiber R. D. 1985. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc. Natl. Acad. Sci. USA 82: 7404–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang S., Hendriks W., Althage A., Hemmi S., Bluethmann H., Kamijo R., Vilcek J., Zinkernagel R. M., Aguet M. 1993. Immune response in mice that lack the interferon-gamma receptor. Science 259: 1742–1745. [DOI] [PubMed] [Google Scholar]
- 36.Reiner S. L., Locksley R. M. 1995. The regulation of immunity to Leishmania major. Annu. Rev. Immunol. 13: 151–177. [DOI] [PubMed] [Google Scholar]
- 37.Peppel K., Vinci J. M., Baglioni C. 1991. The AU-rich sequences in the 3′ untranslated region mediate the increased turnover of interferon mRNA induced by glucocorticoids. J. Exp. Med. 173: 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C. H., Curtis J. D., Maggi L. B., Jr., Faubert B., Villarino A. V., O'Sullivan D., Huang S. C., van der Windt G. J., Blagih J., Qiu J., et al. 2013. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153: 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398. [DOI] [PubMed] [Google Scholar]
- 40.Ogilvie R. L., Sternjohn J. R., Rattenbacher B., Vlasova I. A., Williams D. A., Hau H. H., Blackshear P. J., Bohjanen P. R. 2009. Tristetraprolin mediates interferon-γ mRNA decay. J. Biol. Chem. 284: 11216–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee T. S., Shiao M. S., Pan C. C., Chau L. Y. 1999. Iron-deficient diet reduces atherosclerotic lesions in apoE-deficient mice. Circulation 99: 1222–1229. [DOI] [PubMed] [Google Scholar]
- 42.Filipovich A., McClain K., Grom A. 2010. Histiocytic disorders: recent insights into pathophysiology and practical guidelines. Biol. Blood Marrow Transplant. 16(1, Suppl.): S82–S89. [DOI] [PubMed] [Google Scholar]
- 43.Park H. S., Kim D. Y., Lee J. H., Lee J. H., Kim S. D., Park Y. H., Lee J. S., Kim B. Y., Jeon M., Kang Y. A., et al. 2012. Clinical features of adult patients with secondary hemophagocytic lymphohistiocytosis from causes other than lymphoma: an analysis of treatment outcome and prognostic factors. Ann. Hematol. 91: 897–904. [DOI] [PubMed] [Google Scholar]
- 44.Lehmberg K., Pink I., Eulenburg C., Beutel K., Maul-Pavicic A., Janka G. 2013. Differentiating macrophage activation syndrome in systemic juvenile idiopathic arthritis from other forms of hemophagocytic lymphohistiocytosis. J. Pediatr. 162: 1245–1251. [DOI] [PubMed] [Google Scholar]
- 45.Breen F. N., Hume D. A., Weidemann M. J. 1991. Interactions among granulocyte-macrophage colony-stimulating factor, macrophage colony-stimulating factor, and IFN-gamma lead to enhanced proliferation of murine macrophage progenitor cells. J. Immunol. 147: 1542–1547. [PubMed] [Google Scholar]
- 46.Devecioglu O., Anak S., Atay D., Aktan P., Devecioglu E., Ozalp B., Saribeyoglu E. 2009. Pediatric acute lymphoblastic leukemia complicated by secondary hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer 53: 491–492. [DOI] [PubMed] [Google Scholar]
- 47.O’Brien M. M., Lee-Kim Y., George T. I., McClain K. L., Twist C. J., Jeng M. 2008. Precursor B-cell acute lymphoblastic leukemia presenting with hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer 50: 381–383. [DOI] [PubMed] [Google Scholar]
- 48.Arachchillage D. R., Carr T. F., Kerr B., Hawkins K., Kelsey A., Judge M., Wynn R. F. 2010. Juvenile myelomonocytic leukemia presenting with features of neonatal hemophagocytic lymphohistiocytosis and cutaneous juvenile xanthogranulomata and successfully treated with allogeneic hemopoietic stem cell transplant. J. Pediatr. Hematol. Oncol. 32: 152–155. [DOI] [PubMed] [Google Scholar]
- 49.Shin H. T., Harris M. B., Orlow S. J. 2004. Juvenile myelomonocytic leukemia presenting with features of hemophagocytic lymphohistiocytosis in association with neurofibromatosis and juvenile xanthogranulomas. J. Pediatr. Hematol. Oncol. 26: 591–595. [DOI] [PubMed] [Google Scholar]
- 50.Unal S., Cetin M., Kutlay N. Y., Elmas S. A., Gumruk F., Tukun A., Tuncer M., Gurgey A. 2010. Hemophagocytosis associated with leukemia: a striking association with juvenile myelomonocytic leukemia. Ann. Hematol. 89: 359–364. [DOI] [PubMed] [Google Scholar]
- 51.Baxter E. J., Scott L. M., Campbell P. J., East C., Fourouclas N., Swanton S., Vassiliou G. S., Bench A. J., Boyd E. M., Curtin N., et al. Cancer Genome Project 2005. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365: 1054–1061. [DOI] [PubMed] [Google Scholar]
- 52.Jones A. V., Kreil S., Zoi K., Waghorn K., Curtis C., Zhang L., Score J., Seear R., Chase A. J., Grand F. H., et al. 2005. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 106: 2162–2168. [DOI] [PubMed] [Google Scholar]
- 53.Kralovics R., Teo S. S., Buser A. S., Brutsche M., Tiedt R., Tichelli A., Passamonti F., Pietra D., Cazzola M., Skoda R. C. 2005. Altered gene expression in myeloproliferative disorders correlates with activation of signaling by the V617F mutation of Jak2. Blood 106: 3374–3376. [DOI] [PubMed] [Google Scholar]
- 54.Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C. 2005. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352: 1779–1790. [DOI] [PubMed] [Google Scholar]
- 55.James C., Ugo V., Le Couédic J. P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., et al. 2005. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434: 1144–1148. [DOI] [PubMed] [Google Scholar]