ABSTRACT
Worldwide, approximately 160 million people are chronically infected with hepatitis C virus (HCV), seven distinct genotypes of which are discriminated. The hallmarks of HCV are its genetic variability and the divergent courses of hepatitis C progression in patients. We assessed whether intragenotypic HCV variations would differentially trigger host innate immunity. To this end, we stimulated human primary plasmacytoid dendritic cells (pDC) with crude preparations of different cell culture-derived genotype 2a HCV variants. Parental Japanese fulminant hepatitis C virus (JFH1) did not induce interferon alpha (IFN-α), whereas the intragenotypic chimera Jc1 triggered massive IFN-α responses. Purified Jc1 retained full infectivity but no longer induced IFN-α. Coculture of pDC with HCV-infected hepatoma cells retrieved the capacity to induce IFN-α, whereas Jc1-infected cells triggered stronger responses than JFH1-infected cells. Since the infectivity of virus particles did not seem to affect pDC activation, we next tested Jc1 mutants that were arrested at different stages of particle assembly. These experiments revealed that efficient assembly and core protein envelopment were critically needed to trigger IFN-α. Of note, sequences within domain 2 of the core that vitally affect virus assembly also crucially influenced the IFN-α responses of pDC. These data showed that viral determinants shaped host innate IFN-α responses to HCV.
IMPORTANCE Although pegylated IFN-α plus ribavirin currently is the standard of care for the treatment of chronic hepatitis C virus infection, not much is known about the relevance of early interferon responses in the pathogenesis of hepatitis C virus infection. Here, we addressed whether intragenotypic variations of hepatitis C virus would account for differential induction of type I interferon responses mounted by primary blood-derived plasmacytoid dendritic cells. Surprisingly, a chimeric genotype 2a virus carrying the nonstructural genes of Japanese fulminant hepatitis C virus (JFH1) induced massive type I interferon responses, whereas the original genotype 2a JFH1 strain did not. Our detailed analyses revealed that, not the virus infectivity, but rather, the efficiency of virus assembly and core protein envelopment critically determined the magnitude of interferon responses. To our knowledge, this is the first example of hepatitis C virus-associated genetic variations that determine the magnitude of innate host responses.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection, currently affecting approximately 160 million people worldwide (1), is one of the major causes of hepatitis, liver cirrhosis, and hepatocellular cancer (2). Combination treatment with pegylated interferon alpha IFN-α) and ribavirin has been the standard of care (3), and only recently, directly acting antivirals were licensed and triple therapy has further improved treatment options (4). HCV is a single-stranded positive-sense RNA virus belonging to the genus Hepacivirus of the family Flaviviridae and is subdivided into seven major genotypes (5, 6). The immunological processes associated with the establishment of chronic HCV infection are only partially understood. The role of endogenously induced type I IFN responses and the molecular mechanism of IFN induction remain incompletely defined. Although the NS3-4A protease complex inhibits IFN signaling (7–9), it was recently demonstrated that in liver biopsy specimens from chronically infected individuals, numerous IFN-stimulated genes (ISG) were induced (10, 11).
In various viral infections, plasmacytoid dendritic cells (pDC) are major type I IFN producers (12, 13). Constitutive IRF7 expression allows pDC to mount rapid cytokine responses upon endosomal Toll-like receptor 7 (TLR7) or TLR9 triggering. The role of pDC in HCV infection is not yet fully resolved (14). A recent study revealed the abundant presence of pDC in the livers of chronic HCV patients (15), whereas diverse findings have been published about reduced numbers and impaired function of pDC under such conditions (16–20). In in vitro stimulation experiments, human pDC produced large amounts of type I IFN upon direct cell-to-cell contact with HCV-infected hepatoma cells, whereas cell-free virus did not induce IFN-α (21). Of note, IFN-α induction was TLR7 dependent and required viral RNA replication, but no virion formation, in the stimulating cells (21).
In the present study, we analyzed cytokine responses mounted by primary human pDC that were stimulated with cell culture-derived Japanese fulminant hepatitis C virus (JFH1) or the intragenotypic chimera Jc1, both isolates assigned to genotype 2a. As published before, we found that cell-free JFH1 preparations did not stimulate pDC to mount IFN-α responses. Nevertheless, we observed that crude preparations of cell-free Jc1 did induce massive IFN-α responses. Coculture experiments with infected hepatoma cells and pDC revealed that JFH1-infected cells triggered reduced IFN-α responses compared with Jc1-infected cells. The analysis of additionally shuffled chimeras indicated that determinants within domain 2 of the core protein critically affected the magnitude of IFN-α responses. Furthermore, complete viral assembly was needed, whereas the infectivity of the released particles was not critical, indicating that variations of viral determinants that affect particle formation influence the magnitude of the innate immune response.
MATERIALS AND METHODS
Cell culture, cell lines, and antibodies.
All cell lines were grown in Dulbecco's modified minimal essential medium (DMEM) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum. For virus production, cells were cultivated in serum-free medium (adenovirus expression medium [AEM]).
Plasmids and virus production.
The plasmids pFK-JFH1 (22) and pFK-Jc1 (23) have been described recently. Mixed chimeras were created by PCR-based cloning strategies. Sequences are available upon request. The detailed virological properties of these HCV chimeras have been described previously (24). Constructs of the core domain chimeras have also been described previously (25). For preparation of virus stocks, Huh7.5 hepatoma cells were electroporated with 10 μg in vitro-transcribed viral RNA (26). After 4 h, the medium was changed to serum-free AEM. After 48 h and 72 h of incubation, the cell-free supernatant was filtered through 0.45-μm filters and ultrafiltered through Amicon centrifugal filters (Millipore). Partial purification of the virus was performed either by heparin column (27) or by ultracentrifugation in a Sorvall Ultra WX80 centrifuge at 4°C and 24,000 rpm in a TH-641 wing-out rotor for 4 h, with or without a 20% sucrose cushion. Virus titers were determined as previously described (28).
Vesicular stomatitis virus M2 (VSV-M2) is a natural variant of the VSV wild-type strain HR (29) and is routinely propagated on BHK-21 cells and titrated on Vero cells. Stocks were prepared as culture supernatants 72 h after infection at a low multiplicity of infection (MOI) (0.01).
pDC isolation.
pDC were isolated from buffy coats of healthy donors by Ficoll density centrifugation and magnetically activated cell sorting (MACS) separation (Diamond Plasmacytoid Dendritic Cell Isolation Kit; Miltenyi Biotec). All buffy coats were obtained from the Blutbank Springe, which is a blood transfusion service where all donors gave their written informed consent that their blood can also be used for scientific purposes. Isolated pDC were cultivated in serum-free DC medium (CellGenix) enriched with 10 ng/ml interleukin 3 (IL-3) (CellGenix) in a 96-well plate at a density of 2 × 105 cells/well at a final volume of 200 μl.
pDC stimulation and determination of IFN-α in cell culture supernatant.
pDC were stimulated with various stimuli for 18 h before cell-free supernatant was collected and analyzed for cytokines by an enzyme-linked immunosorbent assay (ELISA) method (eBioscience), as indicated by the manufacturer.
Flow cytometry analysis (FACS) of pDC.
To analyze surface marker expression, cells were washed in fluorescence-activated cell sorter (FACS) buffer and incubated for 20 min at 4°C with fluorescence-labeled antibodies directed against CD303/BDCA-2 (Miltenyi Biotec); and immunoglobulin-like transcript 7 (ILT-7) (eBioscience). After washing with FACS buffer, the cells were analyzed by flow cytometry (LSR II; BD).
Coculture experiments.
Highly HCV-permissive Huh7.5 (30) or Lunet N (31) cells were electroporated with 10 μg HCV RNA; seeded in 96-well plates at a density of 1 × 105 or 6.6 ×104, respectively; and incubated for 24 h. Then, the cells were washed with phosphate-buffered saline (PBS) before pDC were coincubated with the cells for 18 h.
Immunofluorescence.
Electroporated Lunet cells were seeded onto glass coverslips in 24-well plates. The cells were fixed 24 h postinfection with 500 μl of PBS supplemented with 3% (wt/vol) paraformaldehyde for 10 min at room temperature. Subsequently, the cells were washed three times, permeabilized with 0.5% Triton X-100 in PBS, and washed three times with PBS prior to antibody staining. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) (Life Technologies GmbH) for 1 min.
Statistical analysis.
Statistical analyses were done using GraphPad Prism 5. The one-tailed nonparametric Mann-Whitney test was used for the analysis of differences between groups with unmatched pair values, and the one-tailed nonparametric Wilcoxon signed-rank test was used for the analysis of differences between groups with matched pair values. For a grouped analysis, a Kruskal-Wallis test with Dunn's multiple-comparison test was used.
RESULTS
pDC mount IFN-α responses upon stimulation with crude preparations of HCV strain Jc1, but not strain JFH1.
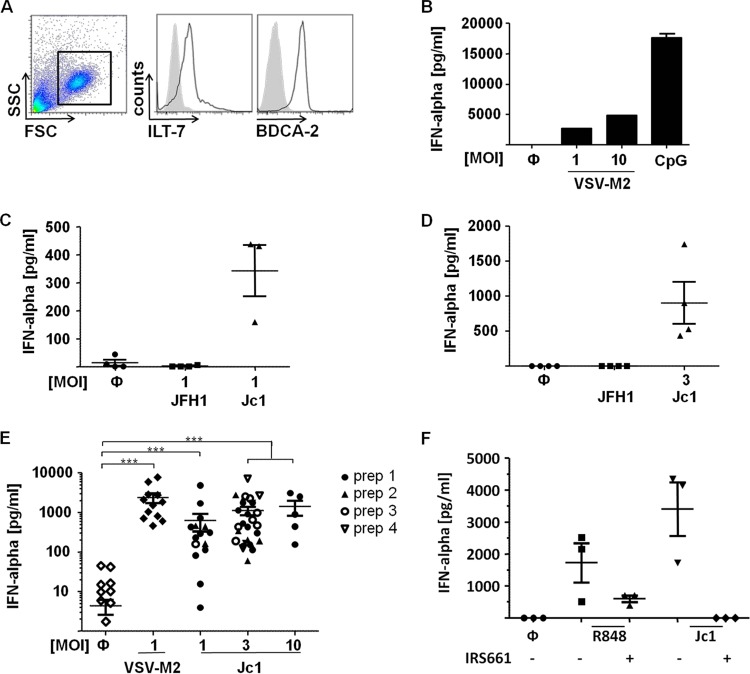
To study HCV-induced cytokine responses of primary human pDC, peripheral blood mononuclear cells (PBMC) were isolated from buffy coats of healthy donors, and pDC were MACS enriched. pDC preparations typically showed a purity of more than 98%, as determined by FACS analysis (Fig. 1A). To ensure their functionality, pDC were stimulated with VSV-M2 (29, 32) or with the TLR9 agonist CpG ODN 2216, which resulted in substantial IFN-α induction (Fig. 1B). For pDC stimulation with HCV, the cell culture-derived HCV strains JFH1 and Jc1 were used. Jc1 is a genotype 2a intragenotypic virus chimera consisting of J6CF- and JFH1-derived genome portions fused at a junction site within the NS2 protein (23). Compared with JFH1, the J6CF-derived core and the p7 protein allow Jc1 to yield enhanced virus titers in cell culture (23–25).
FIG 1.
pDC stimulated with crude preparations of HCV strain Jc1, but not HCV strain JFH1, mount IFN-α responses. (A) MACS-purified pDC, as used throughout this study, were stained with BDCA-2- and ILT-7-specific antibodies and FACS analyzed. FSC, forward scatter; SSC, side scatter. Live cells were gated (boxed area), and the expression of ILT-7 and BDCA-2 (black line) was plotted against unstained samples (shaded curve). (B) pDC isolated from one donor were stimulated with VSV-M2 at the indicated MOI, with CpG (1 μM) or left untreated (Ф). After 18 h of incubation, IFN-α was determined in the cell-free supernatant by an ELISA method. The error bars indicate standard errors of the mean (SEM). (C) pDC were stimulated with crude preparations of HCV strain JFH1 or Jc1 at an MOI of 1 and analyzed as for panel B. (D) pDC were stimulated with crude preparations of JFH1 or Jc1 preparations normalized for equal viral RNA contents, which were equivalent to Jc1 at an MOI of 3. After 18 h of incubation, IFN-α was assessed in the supernatant by an ELISA method. Experiments with pDC isolated from at least three different donors are shown (pDC, n = 3) in panels C and D. (E) pDC were stimulated with VSV-M2 or four different crude preparations of Jc1 at the indicated MOI. After 18 h of incubation, IFN-α was assessed in the supernatant by an ELISA method. pDC were from 5 to 35 donors. ***, P < 0.0001 (Kruskal-Wallis test with Dunn's multiple-comparison test). (F) pDC were pretreated with the TLR7 inhibitor IRS661 (0.175 μM) for 30 min before stimulation with either the TLR7 stimulus R848 (0.5 μg/ml) or Jc1 at an MOI of 3. After 18 h, IFN-α production was assessed in the supernatant by an ELISA method (pDC, n = 3).
As described previously by others (21), treatment of pDC with crude preparations of JFH1 did not induce IFN-α responses of pDC. On the other hand, Jc1 induced massive IFN-α production, irrespective of whether crude Jc1 preparations were normalized for equal infectivity or for similar viral RNA contents (Fig. 1C and D). Repetition of this experiment with pDC isolated from a total of 35 different donors and viruses derived from four different Jc1 preparations confirmed the stable IFN-α induction by Jc1 (Fig. 1E). Notably, upon Jc1 stimulation at MOIs of 1 and 3, dose-dependent IFN-α induction was observed, whereas stimulation at an MOI of 10 did not further increase overall IFN-α production, implying a bell-shaped dose-dependent activation curve (Fig. 1E). The observation that crude preparations of JFH1 did not induce IFN-α whereas Jc1 did was surprising, because JFH1 and Jc1 are assigned to genotype 2a, share identical sequences in all nonstructural genes, and show only modest sequence differences within the structural genes. Interestingly, stimulation with neither Jc1 nor VSV-M2 induced the production of other cytokines, such as IL-6, IL-12, IL-10, IL-4, and IL-1β (data not shown). We next addressed whether direct stimulation with crude virus preparations induced pDC via TLR7, as previously shown for the coculture of HCV-infected cells together with pDC (21). Therefore, pDC were pretreated with the TLR7 inhibitor IRS661 before stimulation with Jc1 or the TLR7 agonist R848. The IRS661-mediated inhibition of both responses indicated that, indeed, Jc1 stimulation of pDC was conferred by TLR7 triggering (Fig. 1F). Thus, the above-described experiments indicated that viral determinants influenced interferon responses mounted by pDC.
Partially purified Jc1 does not induce IFN-α responses.
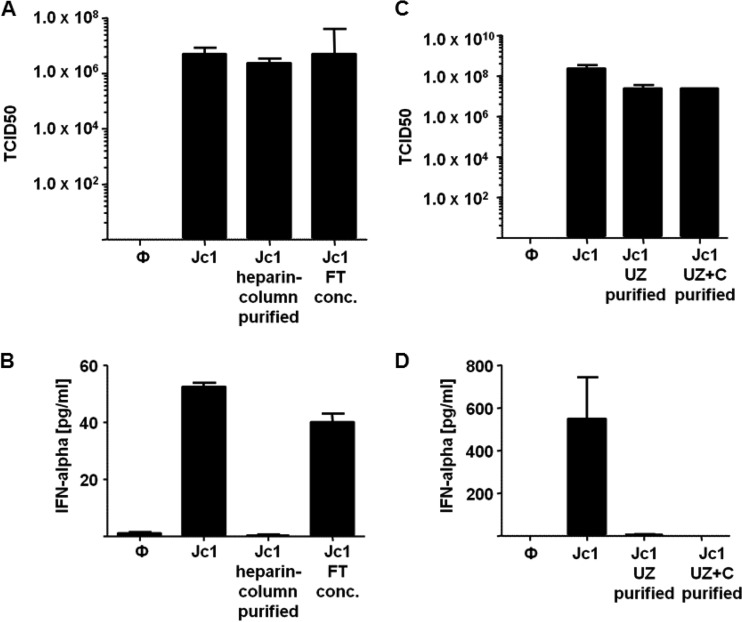
To more closely examine the mechanism of IFN-α induction by Jc1, we partially purified the virus particles. To this end, crude preparations of Jc1 were subjected to heparin column purification (27). Virus eluted from the column and the flowthrough still containing virus particles were adjusted to an MOI of 1. Note that under our experimental conditions, approximately 60% of infectious particles bound to the heparin column, whereas the remaining virus passed through. Nevertheless, virus eluted from the column showed infectivity similar to that of the virus before purification (Fig. 2A). Upon stimulation of pDC with partially purified Jc1 eluted from the column, no IFN-α responses were induced, while the original crude preparation and the virus-containing flowthrough did induce responses (Fig. 2B). Thus, infectious virus alone was not able to stimulate IFN-α responses of pDC. Therefore, we hypothesized that some small components derived from HCV-infected hepatoma cells that pass through 0.45-μm filters during preparation of crude virus were required, either alone or in conjunction with infectious virus, to trigger IFN-α responses.
FIG 2.
pDC stimulated with partially purified HCV strain Jc1 do not mount IFN-α responses. (A) The 50% tissue culture infective doses (TCID50) of a crude Jc1 preparation, partially heparin column-purified Jc1, and the concentrated flowthrough (FT conc.). (B) pDC were stimulated with crude preparations of Jc1 (Jc1), partially heparin column-purified Jc1, or the flowthrough of the heparin column purification of Jc1 at an MOI of 1. After 18 h, IFN-α production was assessed in the supernatant by an ELISA method (pDC, n = 3). (C) TCID50 of a crude Jc1 preparation, ultracentrifuged Jc1 (Jc1 UZ), and Jc1 ultracentrifuged through a sucrose cushion (Jc1 UZ+C). (D) pDC were stimulated with crude preparations of Jc1 or Jc1 that was collected after ultracentrifugation with or without a sucrose cushion (pDC, n ≥ 4). The error bars indicate SEM.
To further study the requirements of IFN-α induction by HCV preparations, Jc1 purified by ultracentrifugation with or without a sucrose cushion was tested. Although the partially purified virus recovered after ultracentrifugation showed infectivity comparable to that of the crude virus preparation (Fig. 2C), virus partially purified by either ultracentrifugation method did not induce IFN-α responses, whereas crude Jc1 preparations did (Fig. 2D). In conclusion, infectious virus alone was not sufficient to trigger IFN-α responses of pDC.
Jc1- and JFH1-infected hepatoma cells induce pDC to mount IFN-α responses.
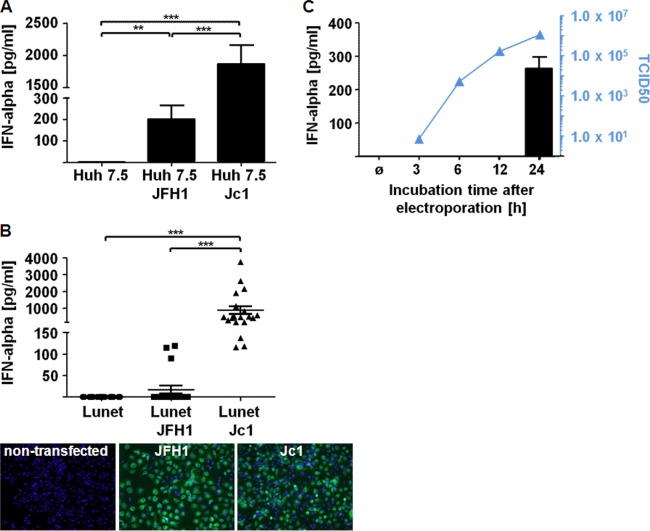
To identify viral determinants that crucially contribute to pDC stimulation, we performed coculture experiments with infected hepatoma cells and pDC. To this end, Huh7.5 hepatoma cells were electroporated with viral RNA and then incubated for 24 h, and finally, the transfectants were coincubated with pDC for an additional 18 h. As observed previously (21), under such conditions, JFH1-infected cells triggered pDC to mount IFN-α responses (Fig. 3A). Under similar conditions, Jc1-infected Huh7.5 cells also induced IFN-α, while IFN-α responses induced by Jc1-infected cells were more abundant than those induced by JFH1-infected cells (Fig. 3A). Notably, hepatoma cells subjected to electroporation with HCV RNA were not stimulated to express IFN-α themselves (data not shown). As Jc1 is more readily produced by transfected Huh7.5 cells than JFH1 and newly formed virus may reinfect Huh7.5 cells, which would further enhance the amount of infected cells, in a subsequent experiment, Lunet N cells, which are CD81 deficient and thus cannot be reinfected by HCV, were used (31). Indeed, upon transfection of Lunet N cells with JFH1 or Jc1 RNA and subsequent incubation for 24 h, similar numbers of infected cells were detected in immunofluorescence analysis (Fig. 3B). Eighteen hours after coculture of pDC with Jc1- or JFH1-infected Lunet N cells, Jc1-infected cells triggered IFN-α responses, which were again higher than those induced by JFH1-infected cells (Fig. 3B). These experiments indicated that differences in the structural genes of Jc1 and JFH1 influenced the capacity of infected cells (or crude virus preparations) to stimulate pDC. To study after how long an incubation time transfected cells acquired the potential to induce IFN responses, transfectants were incubated for 3, 6, 12, and 24 h, and then pDC were added for another 18 h. Of note, after 12 h of incubation, the supernatant of transfected Lunet N cells already contained approximately 90% of the virus levels found after 24 h of incubation (Fig. 3C). Interestingly, transfectants incubated for 24 h induced massive IFN-α responses, whereas transfectants incubated for shorter periods did not (Fig. 3C). Incubation for up to 72 h did not further increase the IFN-α levels (data not shown). These results indicated that cell-derived components triggering IFN-α responses of pDC appeared later than infectious virus in the supernatant of infected cells.
FIG 3.
pDC cocultured with hepatoma cells transfected with Jc1 or JFH1 mount IFN-α responses. (A) Huh7.5 cells were transfected with JFH1 or Jc1 RNA and incubated for 24 h. Transfected hepatoma cells were cocultured with pDC for 18 h, and IFN-α was determined in the supernatant by an ELISA method (pDC, n = 4). ***, P = 0.0002; **, P = 0.0072 (Wilcoxon test). (B) Lunet N cells, which are CD81 deficient and thus cannot be infected by HCV, were transfected with JFH1 or Jc1 RNA and incubated for 24 h. Shown is immunofluorescence analysis of JFH1- or Jc1-transfected Lunet N cells stained for NS3. Transfectants were cocultured with pDC for 18 h, and IFN-α was assessed in the supernatant by an ELISA method (pDC, n = 19). ***, P < 0.0001 (Wilcoxon test). (C) Lunet N cells were transfected 3 h, 6 h, 12 h, and 24 h prior to coculture with pDC for 18 h. The TCID50 was determined as a measure of the amount of virus produced during the 18 h of coincubation (pDC, n = 3). The error bars indicate SEM.
Strain-specific core domain 2 sequences of HCV affect the magnitude of pDC-mediated IFN-α responses.
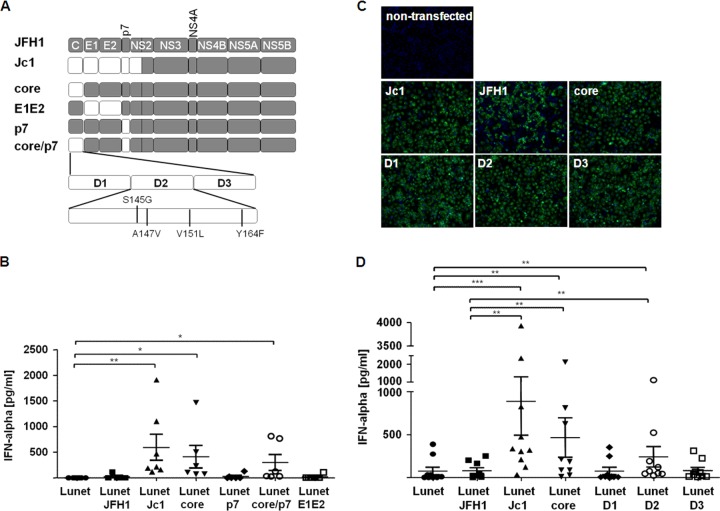
To map viral determinants that account for differential stimulation of pDC by JFH1 and Jc1, we next used a set of JFH1 chimeras that carry various portions of the J6CF-derived coding region instead of the cognate JFH1-derived genome segment (Fig. 4A). These chimeras include constructs encoding either core or p7 of J6CF, alone or together, or E1 and E2 of J6CF, as recently described (24). Interestingly, Lunet N cells electroporated with constructs encoding J6CF-derived core induced significantly higher IFN-α responses than viruses containing JFH1-derived core, irrespective of the origin of the other structural genes (Fig. 4B), indicating that core sequences played a crucial role.
FIG 4.
Determinants within domain 2 of core critically affect the induction of IFN-α responses by pDC. (A) Lunet N cells were transfected with Jc1, JFH1, or JFH1 chimeras encoding different portions of the J6CF isolate, as indicated. In the schematic overview of the constructs used, J6CF-derived genome segments are depicted in white, whereas JFH1-derived genes are shown in gray. (B) After 24 h of incubation, cells transfected with the construct depicted in panel A were cocultured with pDC for 18 h, and IFN-α was assessed in the cell culture supernatant by an ELISA method (pDC, n ≥ 5). **, P = 0.0078; *, P = 0.0156 (Wilcoxon test). (C) Immunofluorescence analysis of Lunet N cells transfected with JFH1, Jc1, or the different core chimeras stained for NS3, as shown in panel A. (D) Lunet N cells were transfected with chimeras containing one of the three domains of the J6-derived core protein. Then, the transfectants were cocultured with pDC for 18 h, and IFN-α was assessed in the supernatant by an ELISA method (pDC, n = 9). ***, P = 0.001; **, P ≤ 0.0039 (Wilcoxon test). The error bars indicate SEM.
To further elucidate which part of the core protein determined the magnitude of IFN-α responses, we made use of core domain chimeras containing only one of the three core domains from the J6CF clone within the JFH1 backbone (25). To ensure that similar numbers of cells transfected with the different virus chimeras were used in the stimulation experiments, NS3 protein-positive cells were monitored by immunofluorescence staining (Fig. 4C). After cocultivation of pDC with Lunet N cells electroporated with the indicated constructs, the J6CF-derived domain 2 of core was sufficient to induce elevated IFN-α responses (Fig. 4D). Interestingly, cells expressing chimeras with either the whole core domain or only domain 2 of core deleted did not trigger pDC to mount IFN-α responses (data not shown).
Notably, the D2 portion of core is important for lipid droplet association and efficient virus production. In fact, residues differing between J6CF and JFH1 within this domain (Fig. 4A) are responsible for increased virus production of HCV genomes (25). Thus, the studies with J6CF-JFH1 chimeras indicated that those viral determinants that facilitate efficient virus particle formation (particularly J6CF-derived core domain 2 [25]) also conferred induction of enhanced IFN-α responses. Therefore, viral determinants within the D2 domain of core critically affected pDC stimulation.
Efficient HCV assembly and core protein membrane envelopment, but not highly infectious virus particles themselves, are critically required for efficient induction of IFN production in pDC.
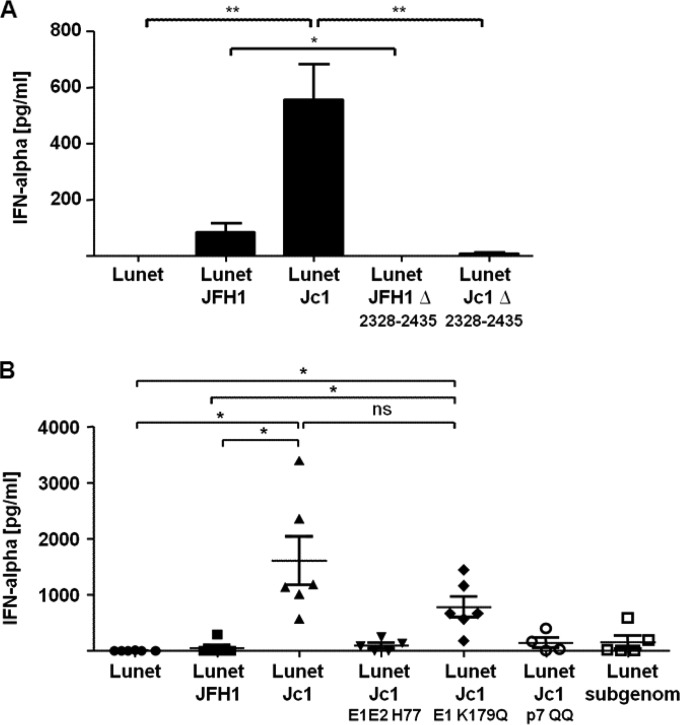
Given the above-mentioned correlations and considering that cell-free, partially purified, highly infectious HCV did not efficiently stimulate IFN-α production, we intended to study whether efficient assembly of viral progeny is sufficient to trigger IFN responses or whether, in addition, release of infectious virus is needed. To this end, we cocultured Lunet N cells transfected with derivatives of Jc1 and JFH1 genomes carrying a large in-frame deletion within domain 3 of the NS5A protein. This deletion abrogates virus assembly at an early stage by disrupting the proper association of core and NS5A with lipid droplets (33). Interestingly, in the Jc1 and JFH1 context, deletion of NS5A domain III strongly decreased induction of IFN-α (Fig. 5A). Since this deletion did not reduce HCV RNA replication and viral protein expression (33), we concluded that assembly of infectious HCV and/or specific properties of NS5A were required for IFN-α triggering.
FIG 5.
The efficiency of HCV assembly affects the induction of IFN-α responses by pDC. (A) Lunet N cells transfected with Jc1, JFH1, or the mutant RNAs carrying an in-frame deletion of NS5A domain III (Jc1Δ2328-2435 and JFH1Δ2328-2435 [33]) were incubated for 24 h. Subsequently, the transfectants were cocultured with pDC for 18 h, and IFN-α release was assessed in the supernatant by an ELISA method (pDC, n = 4). **, P = 0.0078; *, P = 0.037 (Wilcoxon test). The error bars indicate SEM. (B) Lunet N cells transfected with a subgenomic JFH1 replicon or with full-length constructs were incubated for 24 h. Subsequently, the transfectants were cocultured with pDC for 18 h. Then, the IFN-α content of the cell-free supernatant was determined by an ELISA method (pDC, n ≥ 4). *, P = 0.0156; ns, not significant (Wilcoxon test). The error bars indicate SEM.
Assembly of infectious HCV progeny proceeds through several distinct steps, including (i) trafficking of core protein to lipid droplets, (ii) recruitment of NS5A and viral RNA to lipid droplets, (iii) capsid envelopment, and, ultimately, (iv) release of infectious progeny. To determine whether complete HCV morphogenesis, including production and release of infectious progeny, was needed to stimulate pDC, we used a set of Jc1-derived viral mutants that are arrested at distinct steps of the HCV assembly pathway. As a reference for basic stimulation of pDC in the absence of HCV assembly, we used cells transfected with a subgenomic HCV replicon encoding solely NS3 to NS5B proteins. In agreement with a previous study (21), coculture of pDC with replicon-transfected Lunet N cells triggered IFN-α release, albeit only at moderate levels (Fig. 5B). Notably, transfection of the full-length JFH1 genome did not increase IFN-α production of pDC, indicating again that the ability to produce infectious progeny did not increase pDC responsiveness to HCV. As expected, Jc1-transfected cells triggered approximately 10-fold-higher IFN release, which was reduced when a viral genome carrying an inactivating mutation of p7 was transfected or when a Jc1 variant that encodes H77 genotype 1a-derived E1-E2 proteins instead of the cognate J6CF-derived proteins was used (Fig. 5B). Since both inactivation of p7 and expression of genotype 1a glycoproteins in the context of Jc1 prevented membrane envelopment of HCV core protein (24, 34), we concluded that efficient envelopment of HCV core protein was necessary for robust stimulation of pDC. Strikingly, Jc1/E1-K179Q, which carried a mutation disturbing E1-E2 heterodimerization (34, 35), triggered IFN-α release only approximately 3-fold less efficiently than wild-type Jc1 and approximately 10-fold more powerfully than JFH1 (Fig. 4E). As this E1 mutation reduces the infectivity of intracellular and extracellular virus by approximately 100-fold but does not grossly affect core protein envelopment and virus release (34), we concluded that production of infectious virus was dispensable for eliciting high-level IFN-α responses. Collectively, these results indicated that specific features of J6CF-derived core protein, which are critical for efficient virus production of the Jc1 chimera, but not high infectivity of virus particles, are important to trigger enhanced IFN responses. Moreover, only when virus assembly proceeded beyond the step of core protein envelopment was IFN production triggered efficiently. In summary, this showed that the effective assembly, but not the infectivity, of the viral particles determined the magnitude of IFN responses mounted by pDC.
DISCUSSION
In this study, we report that the cell culture-derived HCV strains JFH1 and Jc1, both consisting entirely of genotype 2a-derived viral sequences, display highly divergent properties regarding the induction of IFN-α responses of human primary pDC, i.e., Jc1 induced IFN-α responses and JFH1 did not. This observation suggested that differences between the viral strains profoundly influenced the magnitude of innate immune responses mounted by pDC. While cell-free JFH1 virus preparations, as described previously, did not induce any IFN-α responses of pDC, crude, but cell-free, culture supernatants containing infectious Jc1 virus particles readily triggered pDC to mount abundant IFN-α responses. Since Jc1 differs from JFH1 only in the viral structural genes (core, E1, and E2), as well as p7 and NS2, these findings initially suggested that different features of the Jc1-derived virions might induce enhanced IFN-α responses. However, partial purification of Jc1 particles by either heparin-mediated affinity chromatography or ultracentrifugation rendered Jc1 unable to stimulate IFN-α release. Notably, the purification procedures did not affect viral infectivity. Therefore, we consider it unlikely that virus particle features differing between Jc1 and JFH1 were determining the magnitude of the IFN response triggered by culture supernatants containing viral particles.
Notably, our work suggests that viral determinants substantially affect the induction of IFN-α responses of pDC. This conclusion is based on our observation that, both in stimulation assays with cell-free virus and in the context of coculture experiments on HCV-transfected cells with pDC, IFN-α responses were much more robust for Jc1-derived preparations than with JFH1. Given that the cell-free virus preparations were normalized for equal concentrations of viral RNA or infectivity, and since the cocultured cells carried comparable abundances of viral RNA and protein, we consider it unlikely that divergent abundances of viral components were responsible for this observation.
The influence of the structural genes on the magnitude of IFN-α responses could be clearly shown by using the chimeras that contained only one of the structural genes of the J6CF isolate inserted into the JFH1 backbone. In this system, we found that the insertion of J6CF-derived core sequences into a JFH1 backbone significantly increased the induction of IFN-α. This enhanced IFN-α induction capacity was correlated with an improved assembly efficiency of the core chimera. Whereas JFH1 produced 100-fold-reduced titers compared with Jc1, the insertion of the J6CF-derived core into the JFH1 backbone increased the viral production yield by 50-fold (24). This enhanced assembly efficiency was achieved by the exchange of only domain 2 sequences by Shavinskaya et al. (25). Of note, we found that the D2 JFH1 chimera also induced significantly enhanced IFN-α responses compared with JFH1. To prove the assumption that virus assembly and IFN-α induction are correlated, we used an assembly-deficient NS5AΔD3 mutant that did not trigger pDC to express IFN-α, irrespective of whether the deletion was included in the context of JFH1 or Jc1. To further dissect the impact of the assembly process on the induction of IFN-α responses, we made use of virus variants with mutations that arrest HCV assembly at different stages. The viral mutants Jc1 p7 QQ and Jc1 E1E2 H77 were shown to have a defect in assembly at, or prior to, core protein envelopment (24, 34). When we transfected Lunet cells with these constructs and cocultured them with pDC, we could not detect any IFN-α induction, arguing that the assembly process needed to proceed beyond the stage of capsid envelopment to trigger high-level IFN-α responses. Moreover, we utilized a mutant with a defect in E1E2 heterodimerization (Jc1 E1 K179Q) that has been reported to produce 100-fold fewer infectious particles than Jc1, whereas the release of virions was essentially undisturbed, as evidenced by almost normal release of viral core protein into the culture fluid (34). Cells transfected with this chimera were able to induce IFN-α responses of pDC. These results suggested that the efficient assembly of particles was needed for the induction of IFN-α but that the amount and/or infectiousness of the particles did not play a key role.
Altogether, our observations indicate that core protein features provided by domain 2 are not sufficient to trigger abundant IFN-α release and that completion of membrane envelopment is needed to fully activate pDC. Assuming this, it is conceivable that only viral protein complexes competent in virus envelopment and involving core, E1-E2, and p7 recruit the cellular machinery transferring viral RNA into nascent exosomes with maximal efficiency. This notion is further supported by experiments by Dreux et al. which showed that inhibitors of exosome production, as well as knockdown of host factors involved in production of such secreted membrane vesicles, ablated stimulation of pDC by HCV (36). The observation that viral-strain-specific differences have a pronounced impact on HCV assembly (24, 25) and at the same time influence the induction of an IFN-α response by pDC provides an appropriate setting to define the molecular pathways shared by virus assembly and exosome formation. Moreover, it will be interesting to explore to what extent viral differences with regard to core sequences, assembly efficiencies, and stimulation of IFN-α release from pDC influence the natural course of hepatitis C.
ACKNOWLEDGMENTS
We are grateful to Takaji Wakita for JFH1, to Jens Bukh for the J6CF strain, to Ralf Bartenschlager for the core chimeras and NS5A deletion mutants, and to Charles Rice for the NS5A-specific antibody 9E10 and the Huh7.5 cells. We give special thanks to Theresa Frenz for critically reading the manuscript and to the DRK-Blutspendedienst NSTOB Springe for providing buffy coats.
We declare that we have no competing financial interests.
This work was supported by the Hannover Biomedical Research School (HBRS), the Center for Infection Biology (ZIB), and Topics 1 and 2 of the Programme Infection Research of the Helmholtz Centre for Infection Research. U.K. and T.P. were funded by the Helmholtz-Alberta Initiative, Infectious Diseases Research (HAI-IDR SO-073). This work was also supported by the SFB900 (project A6) and by grant PI734/2-1, all provided by the Deutsche Forschungsgemeinschaft (DFG) and the i-Med Initiative to T.P. and U.K. E.S. was supported by the DFG (STE 1954/1-1). S.P. was supported by a stipend from the international research training group 1273 (IRTG 1273) provided by the DFG.
TWINCORE, Centre for Experimental and Clinical Infection Research, is a joint venture between the Hanover Medical School and the Helmholtz Centre for Infection Research in Braunschweig.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Freeman AJ, Dore GJ, Law MG, Thorpe M, Von Overbeck J, Lloyd AR, Marinos G, Kaldor JM. 2001. Estimating progression to cirrhosis in chronic hepatitis C virus infection. Hepatology 34:809–816. doi: 10.1053/jhep.2001.27831. [DOI] [PubMed] [Google Scholar]
- 3.Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, Shianna KV, Tanaka Y, Thomas DL, Booth DR, Goldstein DB. 2011. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology 53:336–345. doi: 10.1002/hep.24052. [DOI] [PubMed] [Google Scholar]
- 4.Heim MH. 2013. 25 years of interferon-based treatment of chronic hepatitis C: an epoch coming to an end. Nat Rev Immunol 13:535–542. doi: 10.1038/nri3463. [DOI] [PubMed] [Google Scholar]
- 5.Nakano T, Lau GM, Sugiyama M, Mizokami M. 2012. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int 32:339–345. doi: 10.1111/j.1478-3231.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- 6.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 8.Cheng G, Zhong J, Chisari FV. 2006. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci U S A 103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bigger CB, Brasky KM, Lanford RE. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol 75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A 99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barchet W, Cella M, Colonna M. 2005. Plasmacytoid dendritic cells—virus experts of innate immunity. Semin Immunol 17:253–261. doi: 10.1016/j.smim.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. 2011. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol 29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulsenheimer A, Gerlach JT, Jung MC, Gruener N, Wachtler M, Backmund M, Santantonio T, Schraut W, Heeg MH, Schirren CA, Zachoval R, Pape GR, Diepolder HM. 2005. Plasmacytoid dendritic cells in acute and chronic hepatitis C virus infection. Hepatology 41:643–651. doi: 10.1002/hep.20592. [DOI] [PubMed] [Google Scholar]
- 15.Lau DT, Fish PM, Sinha M, Owen DM, Lemon SM, Gale M Jr. 2008. Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology 47:799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- 16.Kanto T, Inoue M, Miyatake H, Sato A, Sakakibara M, Yakushijin T, Oki C, Itose I, Hiramatsu N, Takehara T, Kasahara A, Hayashi N. 2004. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J Infect Dis 190:1919–1926. doi: 10.1086/425425. [DOI] [PubMed] [Google Scholar]
- 17.Dolganiuc A, Chang S, Kodys K, Mandrekar P, Bakis G, Cormier M, Szabo G. 2006. Hepatitis C virus (HCV) core protein-induced, monocyte-mediated mechanisms of reduced IFN-alpha and plasmacytoid dendritic cell loss in chronic HCV infection. J Immunol 177:6758–6768. doi: 10.4049/jimmunol.177.10.6758. [DOI] [PubMed] [Google Scholar]
- 18.Wertheimer AM, Bakke A, Rosen HR. 2004. Direct enumeration and functional assessment of circulating dendritic cells in patients with liver disease. Hepatology 40:335–345. doi: 10.1002/hep.20306. [DOI] [PubMed] [Google Scholar]
- 19.Longman RS, Talal AH, Jacobson IM, Rice CM, Albert ML. 2005. Normal functional capacity in circulating myeloid and plasmacytoid dendritic cells in patients with chronic hepatitis C. J Infect Dis 192:497–503. doi: 10.1086/431523. [DOI] [PubMed] [Google Scholar]
- 20.Shiina M, Rehermann B. 2008. Cell culture-produced hepatitis C virus impairs plasmacytoid dendritic cell function. Hepatology 47:385–395. doi: 10.1002/hep.21996. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. 2010. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A 107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A 103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinmann E, Doerrbecker J, Friesland M, Riebesehl N, Ginkel C, Hillung J, Gentzsch J, Lauber C, Brown R, Frentzen A, Pietschmann T. 2013. Characterization of hepatitis C virus intra- and intergenotypic chimeras reveals a role of the glycoproteins in virus envelopment. J Virol 87:13297–13306. doi: 10.1128/JVI.01708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 26.Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J Virol 82:7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zahn A, Allain JP. 2005. Hepatitis C virus and hepatitis B virus bind to heparin: purification of largely IgG-free virions from infected plasma by heparin chromatography. J Gen Virol 86:677–685. doi: 10.1099/vir.0.80614-0. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 29.Waibler Z, Detje CN, Bell JC, Kalinke U. 2007. Matrix protein mediated shutdown of host cell metabolism limits vesicular stomatitis virus-induced interferon-alpha responses to plasmacytoid dendritic cells. Immunobiology 212:887–894. doi: 10.1016/j.imbio.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, Pietschmann T. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog 6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed M, Mitchell LM, Puckett S, Brzoza-Lewis KL, Lyles DS, Hiltbold EM. 2009. Vesicular stomatitis virus M protein mutant stimulates maturation of Toll-like receptor 7 (TLR7)-positive dendritic cells through TLR-dependent and -independent mechanisms. J Virol 83:2962–2975. doi: 10.1128/JVI.02030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentzsch J, Brohm C, Steinmann E, Friesland M, Menzel N, Vieyres G, Perin PM, Frentzen A, Kaderali L, Pietschmann T. 2013. Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathog 9:e1003355. doi: 10.1371/journal.ppat.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. 2012. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]