ABSTRACT
The majority of influenza virus-specific antibodies elicited by vaccination or natural infection are effective only against the eliciting or closely related viruses. Rare stem-specific heterosubtypic monoclonal antibodies (hMAbs) can neutralize multiple strains and subtypes by preventing hemagglutinin (HA)-mediated fusion of the viral membrane with the endosomal membrane. The epitopes recognized by these hMAbs are therefore considered promising targets for the development of pan-influenza virus vaccines. Here, we report the isolation of a novel human HA stem-reactive monoclonal antibody, hMAb 1.12, with exceptionally broad neutralizing activity encompassing viruses from 15 distinct HA subtypes. Using MAb 1.12 and two other monoclonal antibodies, we demonstrate that neutralization by hMAbs is virtually irreversible but becomes severely impaired following virus attachment to cells. In contrast, no interference by human anti-influenza virus serum antibodies was found, indicating that apically binding antibodies do not impair access to the membrane-proximal heterosubtypic epitopes. Our findings therefore encourage development of new vaccine concepts aiming at the induction of stem-specific heterosubtypic antibodies, as we provide support for their effectiveness in individuals previously exposed to influenza virus.
IMPORTANCE The influenza A virus hemagglutinin (HA) can easily accommodate changes in its antigenic structures to escape preexisting immunity. This variability restricts the breadth and long-term efficacy of influenza vaccines. Only a few heterosubtypic antibodies (hMAbs), i.e., antibodies that can neutralize more than one subtype of influenza A virus, have been identified. The molecular interactions between these heterosubtypic antibodies and hemagglutinin are well characterized, yet little is known about the functional properties of these antibodies. Using a new, extraordinarily broad hMAb, we show that virus neutralization by hMAbs is virtually irreversible and that efficient neutralization is possible only if stem-specific hMAbs bind to HA before the virus attaches to the cell surface. No interference between strain-specific human serum immunoglobulin and hMAbs was found, indicating that preexisting humoral immunity to influenza virus does not limit the efficacy of stem-reactive heterosubtypic antibodies. This knowledge supports the development of a pan-influenza virus vaccine.
INTRODUCTION
Hemagglutinin (HA), the major surface antigen of influenza A virus, exists in 18 subtypes and is responsible for virus entry into the host cell. Influenza virus vaccines are usually effective against seasonal influenza (1–3), but currently available vaccines elicit antibodies of limited breadth that neutralize only the inoculated and closely related seasonal strains. This strain-specific (or homotypic) nature of the antibody response implies that seasonal vaccines have to be regularly reformulated to reflect antigenic changes acquired by drifting. Furthermore, vaccines have to precisely match the antigenic outfit of the strains predicted to be predominantly circulating and may be ineffective if the prediction fails. Although rather rare, several human heterosubtypic monoclonal antibodies (hMAbs) have been described (4–16) and used to define highly conserved epitopes in the receptor-binding site and in the stem of the influenza virus HA. However, development of a universal influenza virus vaccine against these epitopes has so far been approached unsuccessfully using various strategies (17–22).
To date, it is also not clear whether the membrane-proximal locations of the conserved epitopes bound by broadly neutralizing hMAbs restrict the efficacy of heterosubtypic antibodies if virions are cell associated or if they are saturated with strain-specific, membrane-distally binding serum antibodies. These are likely to represent common conditions under which naturally occurring or elicited heterosubtypic antibodies will encounter the virus in humans.
MATERIALS AND METHODS
Characterization of donor RI13.
Donor RI13, a 30-year-old Caucasian male, was identified in a different study as an individual with an average heterosubtypic antibody response (23). RI13 had been vaccinated six times against influenza A virus prior to blood donation, and cells were harvested prior to the arrival of the swine origin H1N1 virus in 2009.
Isolation and characterization of MAb 1.12.
A phage library was prepared as previously described (24). In brief, frozen peripheral blood mononuclear cells (PBMCs) from donor RI13 were used to purify B cells using anti-CD22-coated magnetically activated cell sorting (MACS) beads (∼1.6 × 106 B cells were isolated). Following total RNA extraction (RNeasy Mini; Qiagen), reverse transcription into cDNA was performed using oligo(dT) primer (Promega) and Superscript II reverse transcriptase (Invitrogen) according to the manufacturers' recommendations. Rearranged variable gene segment families were amplified individually and modified for phage surface expression in 3 subsequent PCRs. The resulting full-length Fab fragments were cloned into the pComb3X phage display vector and used to rescue a phage library with a total of 1.5 × 109 transformants, giving rise to a 3.3 × 1011 phage particles/ml library titer.
This phage display library was enriched for phages binding to biotinylated recombinant trimeric hemagglutinin immobilized on streptavidin-coated magnetic beads (the construction and biochemical characterization of these antigens will be reported elsewhere; the beads were purchased from Promega). Approximately 2.5 × 1012 phage were combined with bead-immobilized hemagglutinin (see below; the final concentration of protein was 50 to 100 nM, as determined for HA dimer) in the first round of selection. A total of 4 rounds of selection were performed with a 50 to 100 nM HA concentration (as determined for HA trimer in rounds 2 to 4) and increasing wash stringency (washing with Tris-buffered saline containing 0.05% Tween 20 [TBST]). Phage clones obtained from the 3rd and 4th rounds were screened for binding to various HAs in enzyme-linked immunosorbent assays (ELISA), and positive clones were sequenced.
A total of 48 clones from the 3rd and 4th rounds were analyzed for binding to recombinantly expressed H2 [A/Japan/305/1957(H2N2)], H3 [A/Moscow/10/1999(H3N2)], and H7 [A/fowl plague/Bratislava/1979(H7N7)] hemagglutinins in ELISA. Remarkably, 43 out of 48 clones showed cross-reactivity to all tested HAs. Sequence analysis revealed that all the selected clones have almost the same heavy chain (HC) derived from the VH1-69 germ line gene joined to the same complementarity-determining region 3 (HCDR3), which contains a stretch of five adjacent tyrosine residues. In contrast, high variability and the use of multiple germ line genes and isotypes have been seen for light chains (LC). We therefore predicted that the heavy chain only is probably responsible for binding to HA, and randomly selected clone 1.12. For expression as an IgG1 molecule, it was cloned into pAbVec and transfected into 293T cells, as described previously (16).
Expression and purification of recombinant HAs.
Recombinant HA was expressed in Sf9 cells using genetically modified baculovirus vectors as described by Ekiert et al. or Stevens et al. (7, 25). In brief, HA open reading frames were modified to contain an insect secretion leader sequence at the N-terminal end and a foldon/trimerization instead of a transmembrane and intracellular domain at the C-terminal end. Secreted recombinant protein was purified on Ni-nitrilotriacetic acid (NTA) columns (GE Healthcare) from cell supernatant harvested 4 days postinfection. To process recombinant HA protein into HA1 and HA2, 10 U of tosyl phenylalanyl chloromethyl ketone-treated trypsin (TPCK) (from bovine pancreas; Sigma-Aldrich) per 1 μg of HA was used for processing at room temperature (RT) for 1 h. Immediately after trypsinization, nonaggregated HA trimers were purified by size exclusion chromatography on a Superdex S-200 gel filtration column (GE Healthcare). All HA proteins used in phage display had an additional cysteine residue introduced at position 158 of HA1 to enable biotinylation (with EZ-link HPDP biotin; Pierce) and immobilization on streptavidin beads in an upside-down orientation.
HA-binding ELISA.
Binding of IgG1 1.12 to various recombinant HAs was assessed by ELISA. To this end, high-binding, half-area plates (Costar) were coated at 4°C with 25 μl/well of 2 to 4 μg/ml HA in phosphate-buffered saline (PBS) overnight. The plates were then blocked with 60 μl of 2% low-fat dry milk in PBS for 1 h at RT. Purified IgG1 was titrated in 0.2% milk-PBS and incubated in a volume of 30 μl on the coated plates at RT for 1 h. Bound IgG1 was detected using a peroxidase-coupled goat anti-human Igκ antiserum (1:5,000; Southern Biotech). Between steps, the plates were washed 4 times with approximately 200 μl of TBST containing 0.1% Tween. The ELISA plates were then developed using Ultra TMB substrate (Pierce) for 5 to 10 min before the reaction was stopped by the addition of 2 N H2SO4. The optical density at 450 nm (OD450) was measured in a PerkinElmer plate reader (EnVision; PerkinElmer Wallac). As a negative control, recombinantly expressed HIV-1 gp120-specific IgG1 b12 was included in all assays.
Competition ELISA.
ELISA plates (half area; high binding; Costar) were coated with 25 μl/well of 2-μg/ml recombinant H1 from A/Puerto Rico/8/1934(H1N1) in PBS at 4°C overnight. The plates were then blocked with 60 μl of 2% milk in PBS at room temperature for 1 h. The blocked plates were incubated in duplicate with 30 μl of serially diluted (in PBS-0.2% milk) human IgG1 1.12 at RT for 1 h and washed 4 times with TBST. The plates were then incubated at RT for 1 h with murine IgG c179 (26) diluted in PBS-0.2% milk at 1 μg/ml and washed 4 times with TBST. Binding of human IgG 1.12 and murine IgG c179 was detected in parallel using either peroxidase-coupled goat anti-human Igκ (Southern Biotech) or rabbit anti-mouse-horseradish peroxidase (HRP) serum (Dako), respectively. The signal was developed as described for HA-binding ELISA.
Neutralization of influenza A viruses.
Titrated IgG 1.12 was mixed with a fixed amount of influenza A virus corresponding to a multiplicity of infection (MOI) of 2 to 3 (∼100,000 PFU/well) in Dulbecco's modified Eagle medium (DMEM) supplemented with 0.2% bovine serum albumin (BSA) and 20 mM HEPES (D/B/H medium). Following incubation at 37°C-5% CO2 for 2 h, the MAb-virus mixture was transferred onto PBS-washed, subconfluent Madin-Darby canine kidney (MDCK) epithelial cells (ATCC CCL-34) growing in 96-well tissue culture plates (Techno Plastic Products [TPP]). To initiate infection, cells were incubated with the antibody-virus mixture at 37°C-5% CO2 for 1 h. The mixture was then removed, and the cells were washed with PBS before they were overlaid with D/B/H medium. After 5 to 7 h (depending on the growth kinetics of the virus isolate) at 37°C-5% CO2, the cells were fixed with methanol, washed, and stained with a fluorescein isothiocyanate (FITC)-labeled anti-NP monoclonal antibody (ATCC; HB-65; 3 μg/ml) diluted in PBS-1% BSA at 4°C overnight. Following 4 washes with PBS, the FITC fluorescence signal was detected at 16 individual positions per well in a multilabel plate reader (PerkinElmer EnVision). For the calculation of the titers, the average values from the 16 fluorescence-measuring points were used to determine the best-fitting Hill curves using Prism 6 software (GraphPad Software). The HIV-1 gp120-specific monoclonal antibody b12 was used as a negative control.
Reversibility of neutralization.
Highly concentrated (ca. 3 × 109 TCID50/ml), sucrose cushion-purified stocks of A/Puerto Rico/8/1934(H1N1) or unpurified A/California/07/2009(H1N1), A/Brisbane/59/2007(H1N1), A/Brisbane/10/2007(H3N2), or A/Chicken/Vietnam/C58/2004(H5N3) virus were diluted 1:5 to 1:30 in 900 μl of D/B/H medium, depending on the titers of the stocks. To obtain a final antibody concentration of 10 μg/ml, 100 μl of the indicated MAb diluted in the same medium was added to the virus. The mixture was then incubated at 37°C-5% CO2 for 2 h before it was combined with 120 μl of 1:10-diluted and PBS-washed MagnaBind amine-derivatized magnetic beads (Pierce) and incubated at 37°C-5% CO2 on a rotator for 1 h. Next, 300 μl of the 1-ml bead-virus-MAb mixture was transferred into 2 individual tubes, with each tube representing one of the test conditions: no dissociation and long-term dissociation. Long-term-dissociation beads were washed 1 time with 400 μl of D/B/H medium and resuspended in 1 ml of D/B/H medium without antibody. In parallel, no-dissociation tubes were filled up to 1 ml with D/B/H medium supplemented with the appropriate MAb (final concentration, 10 μg/ml). The tubes were then incubated at 37°C-5% CO2 on a rotator for 14 h. After this incubation, the supernatant was removed and 100 μl of D/B/H medium supplemented with 10 μg/ml of the indicated MAb was added to the no-dissociation tube. In parallel, the long-term-dissociation samples were washed with D/B/H medium and resuspended in 100 μl of D/B/H medium, and 50 μl of resuspended beads was then transferred to MDCK cells seeded at a density of 2 × 104/well in 96-well plates 1 day before. The plates with cells were incubated at 37°C-5% CO2 for 30 min, placed on an orbital shaker for a few seconds to resuspend the beads, and transferred back to the incubator for an additional 30 min. After this incubation, the beads were again resuspended on an orbital shaker and aspirated, and the cells were washed 1 time with PBS. Further incubation and detection of infectivity were performed as described for the neutralization assay. The HIV-1 gp120-specific antibody b12 was used as a nonneutralizing control.
Mouse protection experiments.
To assess the protective capacity of MAb 1.12, 6- to 8-week-old female C57BL/6 mice were injected intraperitoneally (i.p.) in the lower right abdominal quadrant with 3 or 10 mg/kg of body weight of the indicated antibody in 200 μl PBS 24 h before the animals were anesthetized by i.p. injection with 0.05 mg/kg fentanyl, 5 mg/kg midazolam, 0.5 mg/kg medetomidin. After 5 to 10 min, when surgical tolerance was reached, the mice were weighed and allowed to aspirate 25 μl of A/Puerto Rico/8/1934(H1N1) or mouse-adapted A/Hong Kong/1/68(H3N2) (27) corresponding to 3 50% lethal doses (LD50). The mice were then placed on a heating pad to prevent hypothermia, and virus was allowed to be further aspirated for 10 min before the anesthetics were antagonized by i.p. injection with 2.5 mg/kg atipamezolin, 1.2 mg/kg naloxon, 0.5 mg/kg flumazenil. Weight loss was monitored daily, and mice that had lost more than 20% (first experiment) or 15% (second experiment) of their initial body weight were euthanized and scored as dead.
Neutralization of virus particles attached to cell surfaces.
MDCK cells were seeded at 2 × 104 cells/well 1 day before the experiment. To prevent virus internalization during the subsequent step, the cells were first chilled to 4°C for 15 min and then placed on ice for 15 min before they were washed with ice-cold PBS. To preadsorb virus on the cells, A/Puerto Rico/8/1934(H1N1) or A/California/07/2009(H1N1) virus at an MOI of ∼3 was added to half of the plates and incubated at 4°C for 1.5 h. In parallel, the other half of the MDCK cell plates were prepared using the same procedure but without the addition of virus (virus-in-solution control sample). Both plates were then washed once with ice-cold PBS. MAbs titrated in D/B/H medium were added to the virus-preadsorbed-on-cells plate, while only ice-cold D/B/H medium was added to the virus-in-solution control plate. Both plates were then incubated at 4°C for 2 h. In parallel, A/Puerto Rico/8/1934(H1N1) or A/California/07/2009(H1N1) virus (from the same initial dilution kept at 4°C) corresponding to an MOI of ∼3 was mixed with ice-cold titrated MAbs on a dilution plate and incubated at 4°C for 2 h. Both MDCK cell-containing plates were then washed once with prewarmed PBS. The virus-preadsorbed-on-cells plate was covered with 100 μl of prewarmed D/B/H medium, while the virus-in-solution plate was incubated with the virus and MAb mixture from the titration plate. Infectivity was later detected as described for the neutralization assay. The HIV-1 gp120-specific monoclonal antibody b12 was used as a control.
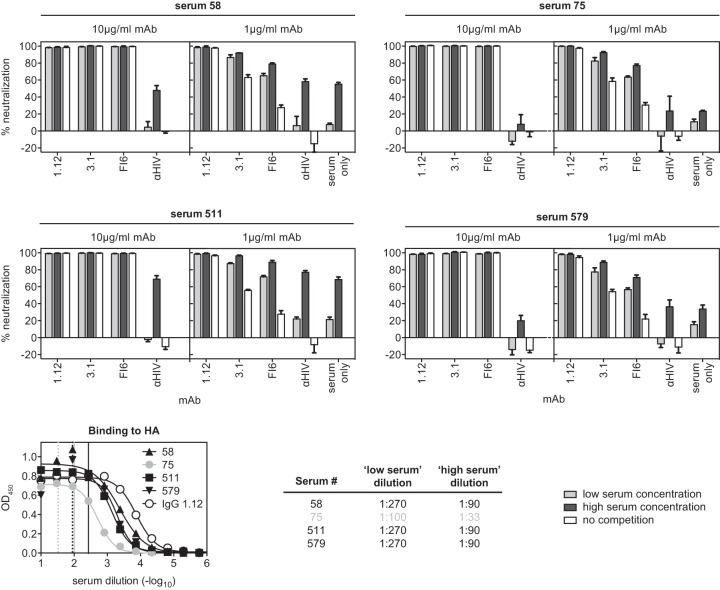
Competition with human sera.
To establish the serum dilutions for the competition assay, the concentrations giving saturating binding were first determined by ELISA on recombinant HA from A/Puerto Rico/8/1934(H1N1). In parallel, the neutralizing activity was determined against the same isolate. Two distinct serum concentrations were chosen: first, the “optimal” dilution, which showed saturating binding but little neutralization, and second, the “neutralizing” dilution, which contained three times as much serum and therefore provided clear yet incomplete virus neutralization (see Fig. 4). To determine potential competition between sera and the monoclonal antibodies, 20 μl of concentrated, sucrose cushion-purified H1N1 A/Puerto Rico/8/1934 virus stock (ca. 3 × 109 TCID50/ml) diluted 1:405 in D/B/H medium was combined with 20 μl of the indicated human serum at the indicated dilution and incubated at 37°C-5% CO2 for 1 h. As a control, no serum or HIV-1-specific MAb b12 was used. Following this incubation, 20 μl of hMAb was added to the virus-serum mixture to obtain a final concentration of 1 or 10 μg/ml. This mixture was then incubated at 37°C-5% CO2 for 1 h to allow binding of the monoclonal antibodies. To determine the residual infectivity after the incubation, 50 μl was transferred to MDCK cells and incubated at 37°C-5% CO2 for 1 h to initiate infection. All subsequent steps were performed as described for staining in neutralization assays.
FIG 4.
Competition with human sera. A/Puerto Rico/8/1934(H1N1) virus was first incubated with sera from the indicated donors for 1 h before the indicated hMAbs were added at a concentration of 10 μg/ml. Residual infectivity of the sample was evaluated and compared to the infectivity of samples without preincubation with human serum. The low serum concentration corresponds to the dilution giving saturated signals in ELISA while having only minor neutralizing activity against the virus. The high serum concentration is three times higher than the low concentration. Representatives of 2 independent, consistent experiments (performed in triplicate) are shown. The error bars indicate standard deviations.
Affinity measurement.
For determination of the affinities of MAbs, a CM5 Biacore chip was covalently coated with goat anti-human Fc (Bethyl Laboratories; A80-104 A) at 0.1 mg/ml to a final density of 1,342 response units (RU) before purified MAb 1.12 or 3.1 at 0.01 mg/ml was captured to a level of 120 RU each. After recording association and dissociation sensograms of recombinant HA concentration series (0.625, 1.25, 2.5, 5, and 10 nM for HA1; 4 and 5 nM and 5, 10, 20, 40, and 80 nM for HA3 and HA12, respectively) at a flow rate of 30 μl/min, the data were fitted to a simple 1:1 binding model (T100 Evaluation Software; Biacore) and the kon (association constant), koff (dissociation constant), and KD (equilibrium dissociation constant) were calculated.
Ethics statements.
All human samples used for this study were obtained from adult volunteers in the context of a broader, accompanying study (EK-17-42) that has been reviewed and approved according to Swiss human subject legislation by the ethics commission of the Canton of Zurich, Switzerland. Written informed consent has been obtained from all participating volunteers.
All animal experiments performed for this study have been reviewed and approved according to the Swiss National Animal Welfare Act under license number 121/210 by the Cantonal Veterinary Office of Zurich, Switzerland.
RESULTS
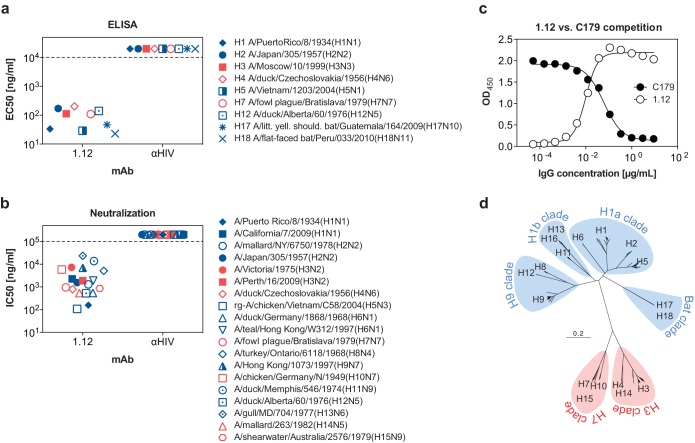
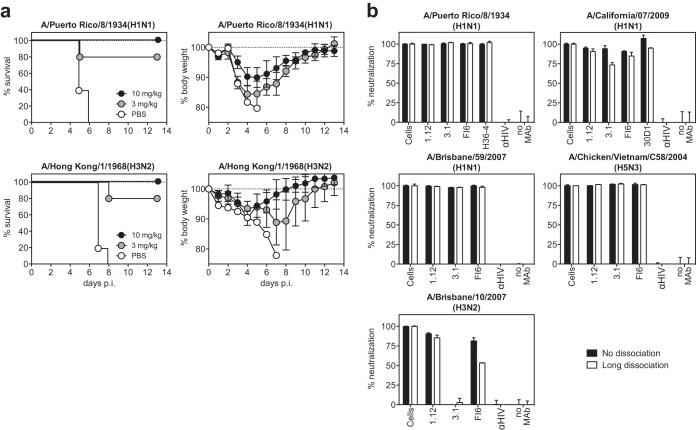
To assess the functional, kinetic, and steric properties of HA stem-reactive antibodies, a test panel of three heterosubtypic MAbs was compiled (hMAbs 1.12, 3.1, and FI6). One of these antibodies, hMAb 1.12, was isolated for this study from a healthy donor using phage display. The second antibody, hMAb 3.1 (16), is genetically related to the well-characterized hMAb FI6 (9) but of more limited breadth (primarily H1a clade). hMAb 1.12 was very remarkable in that it neutralized multiple influenza A virus strains belonging to HA subtypes 1 through 15. Moreover, hMAb 1.12 was found to bind recombinant HA protein from the recently isolated H17 and H18 subtypes (Fig. 1a and b). Binding-competition assays revealed that 1.12 recognizes an epitope overlapping the previously described epitope for the stem-reactive prototype hMAb c179 (Fig. 1c) (5, 28). Like other hMAbs, the in vivo prophylactic protective efficacy of hMAb 1.12 was evaluated in C57BL/6 mice (Fig. 2a). To this end, mice were injected intraperitoneally with 10 or 3 mg/kg of hMAb 1.12 24 h before intranasal infection with 3 LD50 of A/Puerto Rico/8/1934(H1N1) or A/Hong Kong/1/1968(H3N2). The experiment was conducted with 5 mice per group and a 20% weight loss abortion criterion (Fig. 2a) and repeated with 5 mice per group and a more stringent abortion criterion of 15% maximal weight loss [only for A/Puerto Rico/8/1934(H1N1)] (not shown). For A/Puerto Rico/8/1934(H1N1), when data from both experiments were pooled, it was found that at 10 mg/kg, 8 out of 10 animals were protected, whereas at 3 mg/kg, 9 out of 10 mice still survived. In the case of A/Hong Kong/1/1968(H3N2), all mice were protected at 10 mg/kg and 4 out of 5 at the 3-mg/kg dose. The surviving mice did not show apparent signs of morbidity and displayed only moderate weight loss. Interestingly, the time point of antibody application had an influence on protection by MAb 1.12. Intraperitoneal application 3 h before infection conferred the lowest level of protection, whereas intravenous injection 2 h before infection was protective to all animals at both tested doses (15 and 5 mg/kg; 3 mice per group). To further understand the aforementioned differences, mice were injected intravenously with 5 or 15 mg/kg of hMAb 1.12. Serum antibody titers determined after 3 days were found to be a third of the serum antibody concentration measured 1 h after injection (data not shown).
FIG 1.
Specificity of hMAb 1.12. (a) Half-maximal binding concentrations (EC50s) of hMAb 1.12 to recombinantly expressed HA proteins from 9 subtypes. The data represent the means of EC50s obtained in two independent experiments. (b) Half-maximal neutralizing concentrations (IC50s) of MAb 1.12 to a panel of 19 viruses from 15 subtypes. The HIV-1 gp120-specific MAb b12 was used as a negative control in both experiments (αHIV). A representative of at least 2 independent, consistent experiments performed in triplicate is shown. Avian viruses are depicted with open symbols, human isolates with solid symbols, and bat isolates with asterisk-like symbols. Isolates with zoonotic potential are designated with mixed symbols. Isolates from phylogenetic group 1 are depicted in blue and those of phylogenetic group 2 in red. A dashed line is used to indicate the detection limit of the assay. (c) The epitope recognized by hMAb 1.12 was roughly evaluated in a binding-competition ELISA using HA stem-reactive hMAb c179. ELISA plates coated with purified HA from A/Puerto Rico/8/1934(H1N1) were incubated with titrated amounts of hMAb 1.12, washed, and later incubated with a fixed concentration (1 μg/ml) of the murine hMAb c179. Binding of both antibodies was then detected using species-specific secondary antibodies. (d) Phylogenetic tree for all 18 HA subtypes. At total of 1,339 arbitrarily chosen recent nonidentical HA amino acid sequences (from the year 2000 to the present for frequent isolates; from 1985 to the present for rare isolates) were aligned using Muscle 3.8 (34). The tree was built using “neighbor” from the Phylip 3.69 software package (http://evolution.genetics.washington.edu/phylip.html) and illustrated as a rooted tree in FigTree 1.4 (http://tree.bio.ed.ac.uk/software/figtree). Phylogenetic group 1 is indicated in blue and group 2 in red.
FIG 2.
Protection of mice and reversibility of neutralization. (a) Passive immunization of mice. Animals were injected intraperitoneally with the indicated dose of hMAb 1.12 in PBS or with PBS alone (control group) 24 h prior to intranasal infection with a lethal dose of A/Puerto Rico/8/34(H1N1) or mouse-adapted A/Hong Kong/1/1968(H3N2). Mice that dropped below 80 or 85% of their initial body weight were scored as dead and euthanized. For A/Puerto Rico/8/1934(H1N1), pooled data from two separate experiments are shown, each performed with 5 C57BL/6 females per group. One experiment was performed with 15% and one with 20% weight loss as the abortion criteria. For mouse-adapted A/Hong Kong/1/1968(H3N2), only the depicted experiment with a 20% weight loss abortion criterion was performed. (b) Reversibility of neutralization. Three HA stem-reactive antibodies were incubated at a concentration of 10 μg/ml with A/Puerto Rico/8/1934(H1N1), A/California/07/2009(H1N1), A/Brisbane/59/2007(H1N1), A/Brisbane/10/2007(H3N2), or A/Chicken/Vietnam/C58/2004(H5N3) virus (amounts corresponding to an MOI of ∼ 30) before the MAb-virus mixture was captured on magnetic beads. The beads were then processed so that no-dissociation and long-term-dissociation conditions were applied. In the last step, the residual infectivity of each sample was measured on MDCK cells. The HIV-1 gp120-specific MAb b12 was used as a negative control (αHIV). Uninfected and infected cells were included as positive and negative controls, respectively. Representatives of at least 2 consistent experiments (performed in duplicate) are shown. The error bars indicate standard deviations.
The reversibility of heterosubtypic neutralization was then assessed by preincubation of A/Puerto Rico/8/1934(H1N1), A/California/07/2009(H1N1), A/Brisbane/59/2007(H1N1), A/Brisbane/10/2007(H3N2), or A/Chicken/Vietnam/C58/2004(H5N3) virus in solution with 10 μg/ml of one of the hMAbs from our panel or with strain-specific, apically binding MAbs H36-4 (29) and 30D1 (30) (Fig. 2b). The preincubated virus particles were then captured on magnetic beads, and two different dissociation protocols were performed: no dissociation, where captured virus was incubated in the presence of the antibody for 17 h, and long-term dissociation, where antibodies were allowed to dissociate for 14 h before the virus was added to the cells. As depicted in Fig. 2b, even after a prolonged dissociation time of 14 h, virus neutralization was indistinguishable from that of virus permanently incubated in the presence of the antibodies [with the exception A/Brisbane/10/2007(H3N2) neutralization by hMAb FI6, showing partial dissociation of the antibody]. Minor differences seen in A/California/7/2009(H1N1) can largely be attributed to poorer capturing of these virions on beads, and therefore to a poorer signal-to-background ratio, resulting in more noise for the isolate. Nonetheless, it can be concluded that the heterosubtypic epitope is very accessible on free virus particles in solution and that both homo- and heterosubtypic neutralizations are virtually irreversible.
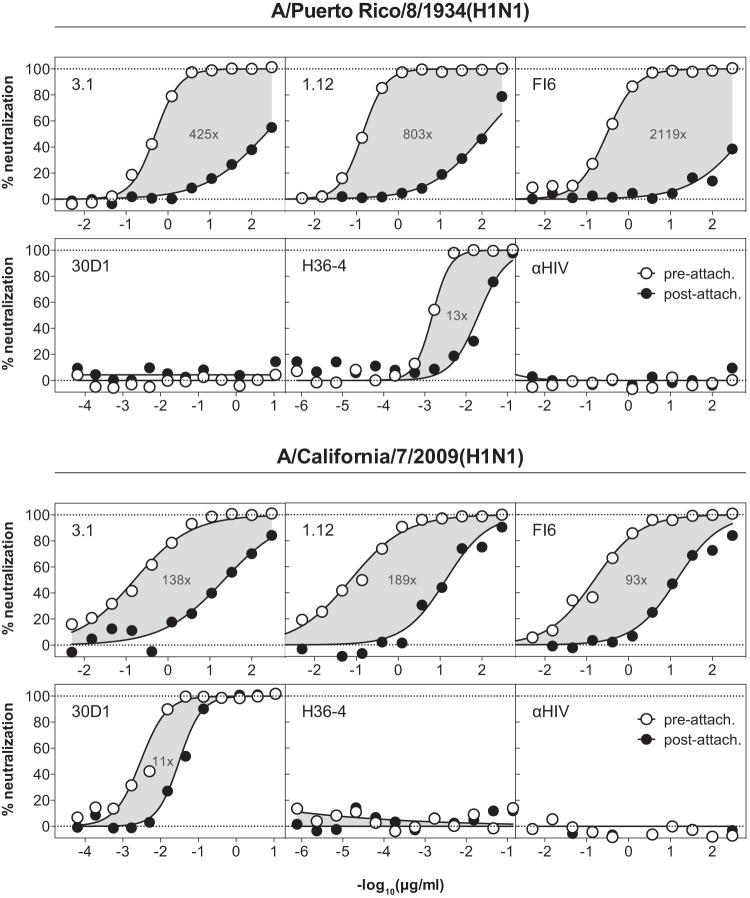
To test whether hMAbs have access to their epitope after virus attachment to the cell surface, we compared the neutralizing activities of hMAbs 1.12, 3.1, and FI6 against both free and cell-attached A/Puerto Rico/8/1934(H1N1) and A/California/07/2009(H1N1). As depicted in Fig. 3, neutralization of cell-attached virus was found to be considerably less (about 2 orders of magnitude or more, depending on the antibody and isolate used) efficient than that of free virions. This was seen for all stem-reactive hMAbs tested, suggesting that these antibodies cannot effectively access their epitopes on hemagglutinin spikes on cell-bound virus. In contrast, the strain-specific and apically binding MAbs 30D1 and H36-4 were both able to neutralize cell-bound virus almost as efficiently as free virus (∼11- to 13-fold differences versus 93- to 2,119-fold in the case of hMAbs).
FIG 3.
Neutralization of virus attached to cell surfaces. A/Puerto Rico/8/1934(H1N1) or A/California/7/2009(H1N1) virus was preadsorbed to MDCK cells at 4°C to avoid virus internalization. The attached viruses were then incubated with titrated amounts of HA stem-reactive (3.1, 1.12, and FI6) or hemagglutination-inhibiting (30D1 and H36-4) MAbs, and residual infectivity was detected. As a control, viruses were mock incubated in cell-free medium before titrated amounts of the MAbs were added. The HIV-1 gp120-specific MAb b12 (αHIV) was used as a nonneutralizing control. Gray areas highlight the differences between the neutralization curves, and numbers indicate the fold difference in the corresponding IC50s. Representatives of at least 2 independent, consistent experiments (performed in duplicate) are shown.
The protective capacity of heterosubtypic antibodies has primarily been determined by passive immunization of seronegative animals. However, since it has been speculated that apically binding strain-specific antibodies may sterically restrict access to the membrane-proximal heterosubtypic epitopes, we tested the neutralizing activities of the antibodies against A/Puerto Rico/8/1934(H1N1) virus particles saturated with human serum antibodies (Fig. 4). To this end, 7 sera that displayed high binding but poor neutralizing titers against this isolate were identified during an independent study (29). For 4 of these sera, we were able to determine concentrations that were saturating but subneutralizing (Fig. 4). Indeed, when virus was incubated with these human sera at the corresponding concentrations (or left mock treated), no reduction in the neutralizing activity resulted from saturation of the virions with human serum antibodies. When both serum and our MAbs were applied at subneutralizing concentrations, additive neutralization was observed (Fig. 4). Thus, strain-specific human serum antibodies do not interfere with neutralization by stem-reactive hMAbs.
DISCUSSION
With this study, we introduced a new hMAb whose breadth is exceeded only by that of hMAb CR9114 (6). Quite interestingly, the two antibodies share common features in that both are encoded by the VH1-69 V gene-joined JH6, and both display an extraordinary stretch of four or five tyrosine residues in HCDR3 of the heavy chain. Although the structure of hMAb 1.12 has not been solved yet, it is safe to assume that, like other VH1-69-encoded antibodies, the central contacts are made by the heavy chain. Indeed, during phage display, many phages that used the 1.12 heavy chain in combination with different light chains were isolated. Unfortunately, and in contrast to hMAb CR9114, hMAb 1.12 does not recognize HA from the influenza B virus genus, but so far, the molecular reasons for this difference remain elusive. In our hands, hMAb FI6, which genetically is more closely related to hMAb 3.1 than to 1.12, struggled to neutralize A/Chicken/Germany/N/1949(H10N7) and to some degree also A/fowl plague/Bratislava/1979(H7N7), but otherwise, it performed comparably in in vitro neutralization assays.
We found that neutralization by hMAbs and strain-specific monoclonal antibodies was virtually irreversible. In the cases of hMAbs 1.12 and 3.1, the slow equilibrium dissociation constant (Table 1) (16) observed in Biacore most likely enables these antibodies to stay attached for a long time. In addition, the dense packing of HA proteins on the virus envelope (31) and the membrane-proximal location of the epitope are likely to further slow down dissociation in that detached antibodies probably reattach rather than diffuse into the supernatant. Induction of conformational changes rendering the HA protein nonfunctional, as postulated for gp120-specific MAbs (32), appears unlikely, as no evidence for antibody-induced conformational changes was found in crystal structures. Strain-specific antibodies, which typically are 1 or 2 orders of magnitude more potent than stem-specific antibodies, were found to neutralize virus virtually irreversibly.
TABLE 1.
Binding avidities for MAb 1.12a
| Isolate | kon (1/mol·s) | koff (1/s) | KD (M) |
|---|---|---|---|
| A/Puerto Rico/8/1934(H1N1) | 4.24 × 105 | ≤5.0 × 10−5 | ≤1.2 × 10−10 |
| A/Moscow/10/1999(H3N2) | 2.95 × 104 | ≤5.0 × 10−5 | ≤1.7 × 10−9 |
| A/Duck/Czechoslovakia/1956(H4N6) | 5.81 × 105 | 2.9 × 10−4 | 5.0 × 10−10 |
| A/Vietnam/1203/2004(H5N1) | 1.06 × 106 | ≤5.0 × 10−5 | ≤4.7 × 10−11 |
| A/Duck/Alberta/60/1976(H12N5) | 1.08 × 105 | 6.6 × 10−5 | 6.1 × 10−10 |
hMAb 1.12 was immobilized on an anti-Fc-coated CM5 chip before binding of soluble recombinant HA was assessed, and binding constants were calculated on a Biacore instrument.
So far, we have no explanation for the poor neutralization of cell-attached virus. Based on the postulated aggregation of an estimated six HA molecules required for fusion (three of which have to undergo a conformational change [33]), we concluded that a cluster of HA molecules is formed whose center is not accessible to hMAbs. Accordingly, three or more HA spikes can still undergo the confirmation change required for infection (33). We believe that the lower impact of prior cell attachment on the neutralizing activity of hemagglutination-inhibiting antibodies originates in the poor affinity of the HA protein for its receptor: apically binding antibodies should have no problem with displacing dissociated receptors if they are sufficiently affine and concentrated. Based on present data, it is not clear why cell-attached A/California/7/2009(H1N1) was less resistant to heterosubtypic neutralization than A/Puerto Rico/8/1934(H1N1). We believe that the extensive adaptation of A/Puerto Rico/8/1934(H1N1) to cell culture probably contributed to this observation.
The densely packed arrangement of the HA and NA spikes on the surfaces of influenza A virions (31), in combination with the membrane-proximal location of the stem epitope, led to speculations that access to these epitopes could be difficult. Moreover, it has been reasoned that this access may even be further restricted if the virions are saturated with preexisting homotypic antibodies that are binding to the strain-specific apical epitopes. If true, this hypothesis could provide an explanation for the poor immunogenicity of the epitope in humans, but it would also call into question the efficacy of passive or active immunization targeting this epitope in humans, as strain-specific antibodies can be found in most individuals. However, we showed that neither concern proved to be relevant in vitro. Accordingly, we do not predict that a universal influenza virus vaccine or therapy based on stem-reactive hMAbs would be impaired by the presence of preexisting strain-specific humoral immunity. However, their action may be limited against cell-associated viruses.
ACKNOWLEDGMENTS
We are grateful to Richard Webby and Scott Kraus from the St. Jude Children's Research Hospital, Memphis, TN, USA; Yves Thomas and Laurent Kaiser from University Hospital of Geneva, Geneva, Switzerland; and Rodney Daniels from the National Institute of Medical Research, London, United Kingdom, for kindly providing viruses from their repositories. We express our gratitude to Silke Stertz, Institute of Medical Virology, University of Zurich, Zurich, Switzerland, and Georg Kochs, Institute of Virology, Freiburg, Germany, for providing reagents. We are grateful to Peter Palese and Adolfo García-Sastre, Icahn School of Medicine at Mt. Sinai Hospital, New York, NY, USA, for providing antibodies H36-4 and 30D1. Last but not least, we are also grateful to Dennis Burton, The Scripps Research Institute, La Jolla, CA, USA, for providing monoclonal antibody b12.
REFERENCES
- 1.Knipe DM, Howley PM. 2007. Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Nichol KL. 2008. Efficacy and effectiveness of influenza vaccination. Vaccine 26(Suppl 4):D17–D22. doi: 10.1016/j.vaccine.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 3.Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 4.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyfus C, Ekiert DC, Wilson IA. 2013. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 87:7149–7154. doi: 10.1128/JVI.02975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 10.Hu W, Chen A, Miao Y, Xia S, Ling Z, Xu K, Wang T, Xu Y, Cui J, Wu H, Hu G, Tian L, Wang L, Shu Y, Ma X, Xu B, Zhang J, Lin X, Bian C, Sun B. 2013. Fully human broadly neutralizing monoclonal antibodies against influenza A viruses generated from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology 435:320–328. doi: 10.1016/j.virol.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, Qin CF, Che XY, Wilson IA. 2013. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyrzucki A, Dreyfus C, Kohler I, Steck M, Wilson IA, Hangartner L. 2014. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J Virol 88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palese P. 2006. Making better influenza virus vaccines? Emerg Infect Dis 12:61–65. doi: 10.3201/eid1201.051043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pica N, Palese P. 2013. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med 64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 19.Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steel J, Lowen AC, Wang T, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1:e00018-10. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, Garcia-Sastre A, Moran TM, Palese P. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A 107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bommakanti G, Lu X, Citron MP, Najar TA, Heidecker GJ, ter Meulen J, Varadarajan R, Liang X. 2012. Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol 86:13434–13444. doi: 10.1128/JVI.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler I, Scherrer AU, Zagordi O, Bianchi M, Wyrzucki A, Steck M, Ledergerber B, Gunthard HF, Hangartner L. 2014. Prevalence and predictors for homo- and heterosubtypic antibodies against influenza A virus. Clin Infect Dis 59:1386–1393. doi: 10.1093/cid/ciu660. [DOI] [PubMed] [Google Scholar]
- 24.Barbas CF III, Burton DR, Scott JK, Silverman GJ. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 25.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 26.Smirnov YA, Lipatov AS, Gitelman AK, Okuno Y, Van Beek R, Osterhaus AD, Claas EC. 1999. An epitope shared by the hemagglutinins of H1, H2, H5, and H6 subtypes of influenza A virus. Acta Virol 43:237–244. [PubMed] [Google Scholar]
- 27.Haller O, Arnheiter H, Lindenmann J. 1979. Natural, genetically determined resistance toward influenza virus in hemopoietic mouse chimeras. Role of mononuclear phagocytes. J Exp Med 150:117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staudt LM, Gerhard W. 1983. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J Exp Med 157:687–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seibert CW, Rahmat S, Krause JC, Eggink D, Albrecht RA, Goff PH, Krammer F, Duty JA, Bouvier NM, Garcia-Sastre A, Palese P. 2013. Recombinant IgA is sufficient to prevent influenza virus transmission in guinea pigs. J Virol 87:7793–7804. doi: 10.1128/JVI.00979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A 103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruprecht CR, Krarup A, Reynell L, Mann AM, Brandenberg OF, Berlinger L, Abela IA, Regoes RR, Gunthard HF, Rusert P, Trkola A. 2011. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J Exp Med 208:439–454. doi: 10.1084/jem.20101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobay MP, Dobay A, Bantang J, Mendoza E. 2011. How many trimers? Modeling influenza virus fusion yields a minimum aggregate size of six trimers, three of which are fusogenic. Mol Biosyst 7:2741–2749. doi: 10.1039/c1mb05060e. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]