ABSTRACT
The replication of plus-strand RNA virus genomes is mediated by virally encoded RNA-dependent RNA polymerases (RdRps). We have investigated the role of the C-proximal region in the RdRp of tomato bushy stunt virus (TBSV) in mediating viral RNA synthesis. TBSV is the prototype species in the genus Tombusvirus, family Tombusviridae, and its RdRp is responsible for replicating the viral genome, transcribing two subgenomic mRNAs, and supporting replication of defective interfering RNAs. Comparative sequence analysis of the RdRps of tombusvirids identified three highly conserved motifs in their C-proximal regions, and these sequences were subsequently targeted for mutational analysis in TBSV. The results revealed that these motifs are important for (i) synthesizing viral genomic RNA and subgenomic mRNAs, (ii) facilitating plus- and/or minus-strand synthesis, and (iii) modulating trans-replication of a defective interfering RNA. These motifs were also found to be conserved in other plant viruses as well as in a fungal and insect virus. The collective findings are discussed in relation to viral RNA synthesis and taxonomy.
IMPORTANCE Little is currently known about the structure and function of the viral polymerases that replicate the genomes of RNA plant viruses. Tombusviruses, the prototype of the tombusvirids, have been used as model plus-strand RNA plant viruses for understanding many of the steps in the infectious process; however, their polymerases remain poorly characterized. To help address this issue, the function of the C-terminal region of the polymerase of a tombusvirus was investigated. Three conserved motifs were identified and targeted for mutational analysis. The results revealed that these polymerase motifs are important for determining what type of viral RNA is produced, facilitating different steps in viral RNA production, and amplifying subgenomic RNA replicons. Accordingly, the C-terminal region of the tombusvirus polymerase is needed for a variety of fundamental activities. Furthermore, as these motifs are also present in distantly related viruses, the significance of these results extends beyond tombusvirids.
INTRODUCTION
Replication of the genomes of plus-strand RNA viruses is mediated by virally encoded RNA-dependent RNA polymerases (RdRps) (1). RdRps are translated shortly after infection and associate with other viral or host proteins to assemble into RNA replication complexes (RCs) (2). These RCs are responsible for synthesizing complementary minus strands of the genomes that are then used as the templates for production of plus-strand progeny genomes (2). In some viruses, these RCs are also responsible for synthesizing viral subgenomic (sg) mRNAs that are required for expression of other viral proteins (3). Viral RdRps of animal viruses have been the focus of many functional studies, and there are atomic structures available for several viruses (4). In contrast, much less is known about the RdRps of plant viruses (5).
The genus Tombusvirus (6) is the prototype member of the family Tombusviridae, which currently contains 11 other genera (7). Tombusviruses are among the best-characterized plus-strand RNA plant viruses (6), and a great deal has been learned about viral RNA synthesis in terms of both the RNA elements required and the protein factors involved (8–12). The prototype tombusvirus is tomato bushy stunt virus (TBSV), which encodes two viral proteins critical for viral RNA synthesis. p33 is an accessory replication protein, and p92, its readthrough product, is the RdRp (13, 14). The N-terminal one-third of p92 is identical to p33, due to its coding and expression strategy (Fig. 1A). This shared portion in the two proteins has been extensively studied and contains regions important for many different functions, including organelle targeting, membrane anchoring, p33-p33 and p33-p92 interactions, RNA binding, etc. (9). The nonoverlapping region of p92 harboring the RdRp activity is less well understood functionally. This segment contains the canonical RdRp amino acid motifs that are highly conserved in RdRps (15). One of these motifs, motif VI, containing the GDD triplet, was confirmed via mutational analysis to be important for efficient genome replication of the tombusvirus artichoke mottled crinkle virus (16). Additionally, two RNA-binding domains, termed RNA-binding region 2 (RBR2) and RBR3, have been defined within this section; RBR2 is located internally, while RBR3 is positioned C proximally (Fig. 1B) (17). The C terminus of p92 also harbors a C-terminal (Ct) motif that is essential for sg mRNA transcription but dispensable for genome replication (18) (Fig. 1B). p92 interacts with p33 and a variety of host factors to form a viral RC that is responsible for mediating various steps in the RNA replication process (9).
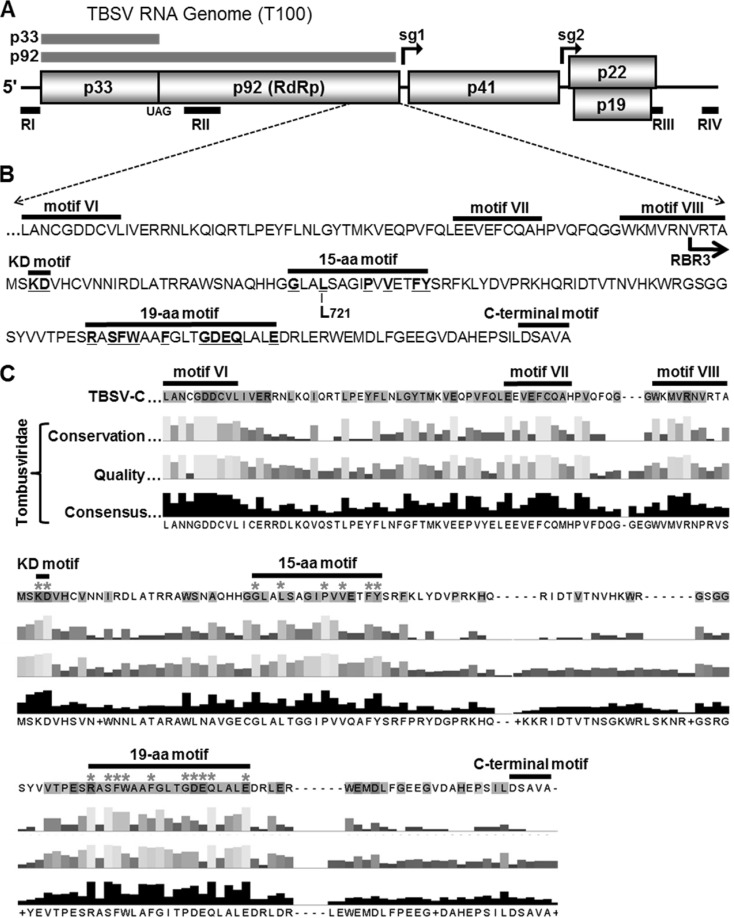
FIG 1.
TBSV genome and C-proximal region of p92. (A) Schematic representation of the TBSV RNA genome with coding regions represented by boxes with the molecular masses (in kDa) of proteins indicated. p33 and its readthrough protein p92 are depicted as gray bars above the genome. The initiation sites of two subgenomic (sg) mRNAs are indicated by black arrows above the genome, while the approximate locations of essential regions RI-RIV present in TBSV DI RNAs are shown below as black bars. (B) C-terminal sequence of TBSV p92. Canonical RdRp motifs (motifs VI to VIII) and the previously reported C-terminal motif are indicated by horizontal bars above the sequence. The locations of the KD, 15-aa, and 19-aa motifs are indicated, with the residues selected for mutational analysis underlined and in bold. The L721 residue, which is important for trans-replication of the DI RNA and the N-terminal border of RNA-binding region 3 (RBR3 with arrow) are also shown. (C) Summary of the alignment of amino acid sequences of RdRps from species in Tombusviridae (see Fig. S1 in the supplemental material for details). A reference TBSV p92 sequence is shown at the top, while conservation, quality, and consensus ratings for the family member comparisons are provided below. Asterisks denote residues that were assessed by mutational analysis.
In addition to directing viral genome replication and the transcription of two sg mRNAs (Fig. 1A), tombusvirus RCs support the replication of two different classes of subviral replicon. Defective interfering (DI) RNAs represent the first type. DI RNAs are deletion mutants of the tombusvirus genome that maintain small segments harboring RNA replication elements (Fig. 1A) (19). These highly truncated replicons are noncoding and thus rely on p33 and p92 provided from their parental (or helper) genome for replication. This type of viral RNA amplification is referred to as trans-replication, because the required viral replication proteins are provided by a different RNA. Tombusvirus satellite RNAs (satRNAs) represent the second class of trans-replicated subviral replicon that requires p33 and p92 provided by a helper tombusvirus genome (20). In contrast to DI RNAs, satRNAs share little sequence identity with their helper genomes; nonetheless, they are recognized and amplified by tombusvirus RCs (21).
In this study, we investigated the roles of motifs in the C-proximal region of TBSV p92 that are conserved in all tombusvirids. Our results point to important roles for these motifs in genome replication, sg mRNA transcription, and DI RNA amplification. Additionally, we show that these motifs are also conserved in other plant, fungal, and insect viruses.
MATERIALS AND METHODS
Sequence analysis.
All RdRp amino acid sequences were obtained from the UniProtKB database (http://www.uniprot.org). Multiple sequence alignments were conducted using ClustalX from the European Bioinformatics Institute's website (http://www.ebi.ac.uk); default values were used for each of the relevant parameters. Sequence accession numbers for the viral proteins used are provided in Fig. S1 in the supplemental material.
A motif search for the 19-amino-acid (aa) motif was performed using the online search engine available from GenomeNet (http://www.genome.jp/). The motif search tool (http://www.genome.jp/tools/motif/MOTIF2.html) was used to query the KEGG VGENES database for the peptide R-x-S-[F or Y or W]-x-x-x-x-x-x-x-x-x-x-Q-x-x-x-E, where x is any amino acid.
Viral constructs.
The full-length infectious TBSV cDNA clone (T100) has been described previously (22), and all replicating genomic mutants were derived from this construct. Nonreplicating mutants were generated from a defective T100 cDNA containing a lethal C-to-G substitution in an essential RNA replication element located in region II of the viral genome (23). The DI RNA and satRNA used (i.e., DI-72 and Sat181) have also been described previously (24, 25). Standard overlapping PCR-based mutagenesis was used to introduce substitutions into the open reading frame of the TBSV RdRp (p92) in both replicating and nonreplicating versions of the TBSV cDNA. All clones were sequenced over the PCR inserts to ensure that only the intended modification was present.
In vitro transcription.
Infectious viral transcripts were generated by in vitro transcription of SmaI-linearized clones as described previously (13). Transcript concentrations were determined using spectrophotometry, and transcript integrity was verified by agarose gel electrophoresis.
Isolation and transfection of protoplasts.
Protoplasts were prepared using 6- to 7-day-old cucumber cotyledons as described previously (24). Isolated protoplasts were transfected using polyethylene glycol-CaCl2 according to the protocol with 5 μg of genomic T100 transcripts and, in coinoculations with subviral replicons, 1 μg of either DI RNA or satRNA (24). Inoculated protoplasts were incubated under constant light for 22 h in a growth chamber at 22°C.
Analysis of viral RNA accumulation.
Total nucleic acids were extracted from protoplasts as described previously (24). For viral RNA analysis, 1.5-μg aliquots of total nucleic acids were separated in 1.4% agarose gels, followed by electrophoretic transfer to nylon membrane. For minus-strand detection, total nucleic acids were treated with glyoxal prior to electrophoresis (26). Northern blot detection of authentic viral and DI RNA plus and minus strands were performed using strand-specific 32P-labeled probes as described previously (26, 27). Radioanalytical quantification of viral RNA accumulation was carried out using a PharosFX Plus Molecular Imager (Bio-Rad) and QuantitiyOne Software (Bio-Rad). Experiments were repeated at least three times and quantified with standard errors.
RESULTS
Identification of conserved motifs in the carboxy-proximal region of RdRps in Tombusviridae.
Sequence comparison of the RdRps of viruses in Tombusviridae revealed the presence of three conserved motifs that were located downstream of the canonical RdRp motifs (i.e., motifs I to VIII) (15) (Fig. 1C; see also Fig. S1 in the supplemental material). The first motif was located immediately C proximal to core motif VIII and was composed of two completely conserved amino acids (K and D); consequently, it was termed the KD motif (Fig. 1B and C). The second motif encompassed 15 residues, several of which were highly conserved, and was termed the 15-amino-acid (15-aa) motif. The third motif, called the 19-aa motif, was located closest to the C terminus and contained a number of invariant residues. Based on the observed conservations, we set out to investigate the possible roles of these newly defined C-proximal motifs in virus reproduction.
Functional analysis of the KD and 15-aa motifs.
To assess the function of residues in the KD and 15-aa motifs, substitutions of selected conserved amino acids were made in an infectious clone of TBSV (Fig. 2A). Wild-type (wt) T100 and mutant TBSV genomes were transfected into protoplasts, and viral genome replication and sg mRNA transcription were monitored via Northern blotting (Fig. 2B). Total RNA was extracted at 22 h postinfection, a time point when viral RNA accumulation in wt infections is subsaturating. Absolute accumulation values of the genomic RNAs were compared, whereas, for sg mRNAs, the ratios of sg mRNA levels to their cognate genomic RNA levels were compared. Replacement of either the K or D residue in the KD motif with alanine (mutants CG1 and CG2) dramatically reduced the levels of viral plus strands, and replacement of both with alanine (mutant CG3) abolished RNA synthesis (Fig. 2B and C). However, for CG1 (K→A), the relative level of plus-strand sg mRNA2 was near wt levels and all minus strands were readily detectable. Similarly, the minus strands for CG2 (D→A) were also detectable, but to a lesser degree. Accordingly, the defects in CG1 and CG2 appeared to affect primarily plus-strand production. In contrast, the double substitution in CG3 (KD→AA) effectively inhibited accumulation of all strands. Transposing the oppositely charged residues in CG4 (KD→DK) also effectively reduced accumulation; however, low levels of minus strands were still evident.
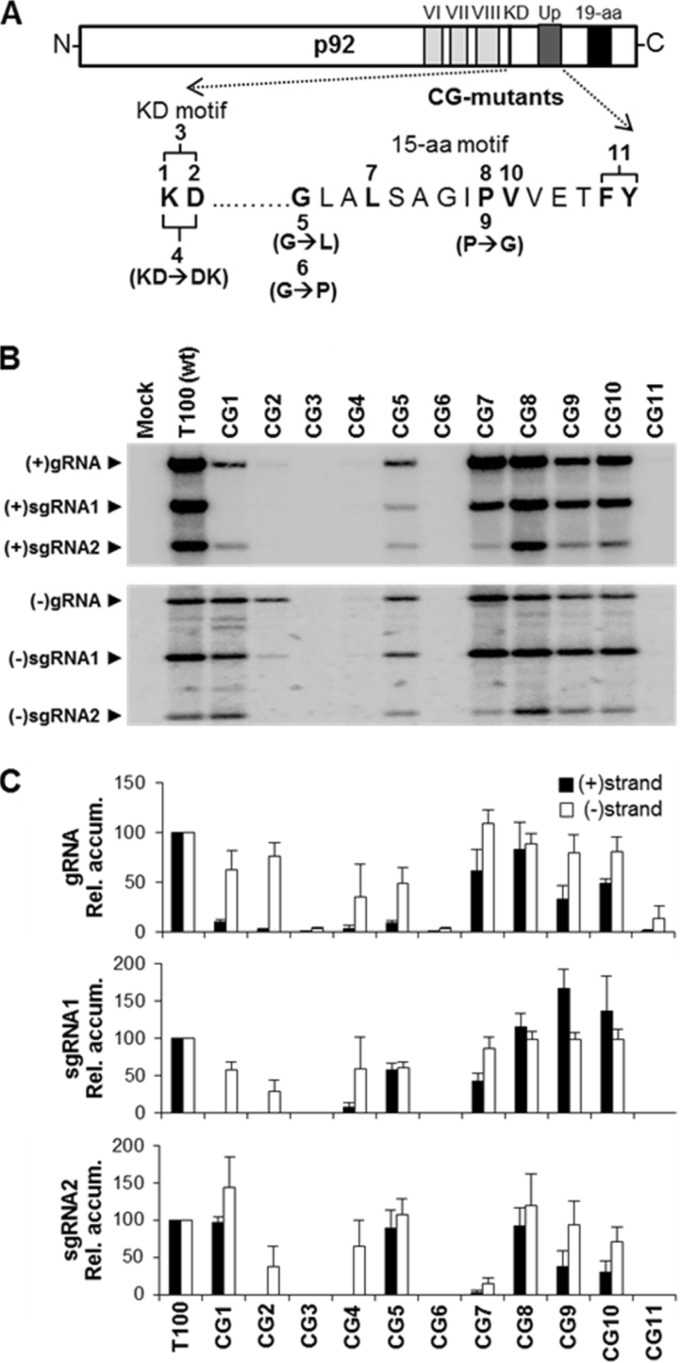
FIG 2.
Mutations in the KD and 15-aa motifs of TBSV p92. (A) A schematic representation of the TBSV p92 protein is shown with the relevant motifs indicated. Residues selected for mutational analysis are indicated in boldface and are numbered according to their corresponding genomic mutant in the CG series. All mutants shown above the sequence contained alanine substitutions, whereas those below contained alternate substitutions, which are specified in parentheses. (B) Northern blot analysis of viral RNA accumulation in protoplasts inoculated with the genomic CG mutants. The identities of the mutants analyzed are provided above each lane, and the positions of subgenomic (sg) and genomic (g) viral RNAs are indicated to the left. The accumulations of plus strands (+) are shown in the upper panel, while those of minus strands (−) appear in the lower panel. (C) Quantification of accumulation levels of plus (black bars) and minus (white bars) sense viral RNAs from CG mutant infections. In the graphs, absolute accumulation values of the genomic RNAs are compared, whereas, for sg mRNAs, the ratios of sg mRNA levels to their cognate genomic RNA levels are compared. Relative accumulation levels are presented with standard errors derived from three independent experiments.
In the 15-aa motif, the N-terminal glycine is conserved in all genera of Tombusviridae, except in dianthoviruses, where this residue is N (see Fig. S1 in the supplemental material). Substitution of the G in TBSV with a bulkier L (CG5) or secondary-structure-disrupting P (CG6) substantially reduced or eliminated viral RNA accumulation, respectively (Fig. 2B and C). However, compared with relative sg mRNA levels, the viral genome for CG5 appeared to be preferentially inhibited at the plus-strand level. The substitution in CG7 (L→A) reduced plus-strand genomic and sg mRNA1 levels to ∼60% and ∼40%, respectively, and sg mRNA2 levels were reduced to near zero for strands of both senses. When the absolutely conserved proline was replaced with A (CG8) or the smaller G (CG9), the A was well tolerated, whereas the G preferentially inhibited the genome and sg mRNA2 at the plus-strand level. Results similar to the last were also observed for CG10, in which the adjacent V was replaced with A. Finally, tandem substitution of the C-proximal aromatic residues in CG11 (FY→AA) abolished viability. Collectively, this mutational analysis revealed that both the KD and 15-aa motifs contain residues important for mediating the accumulation of different viral RNAs and that critical roles are played by the D, G, and tandem FY residues.
Functional analysis of the 19-aa motif.
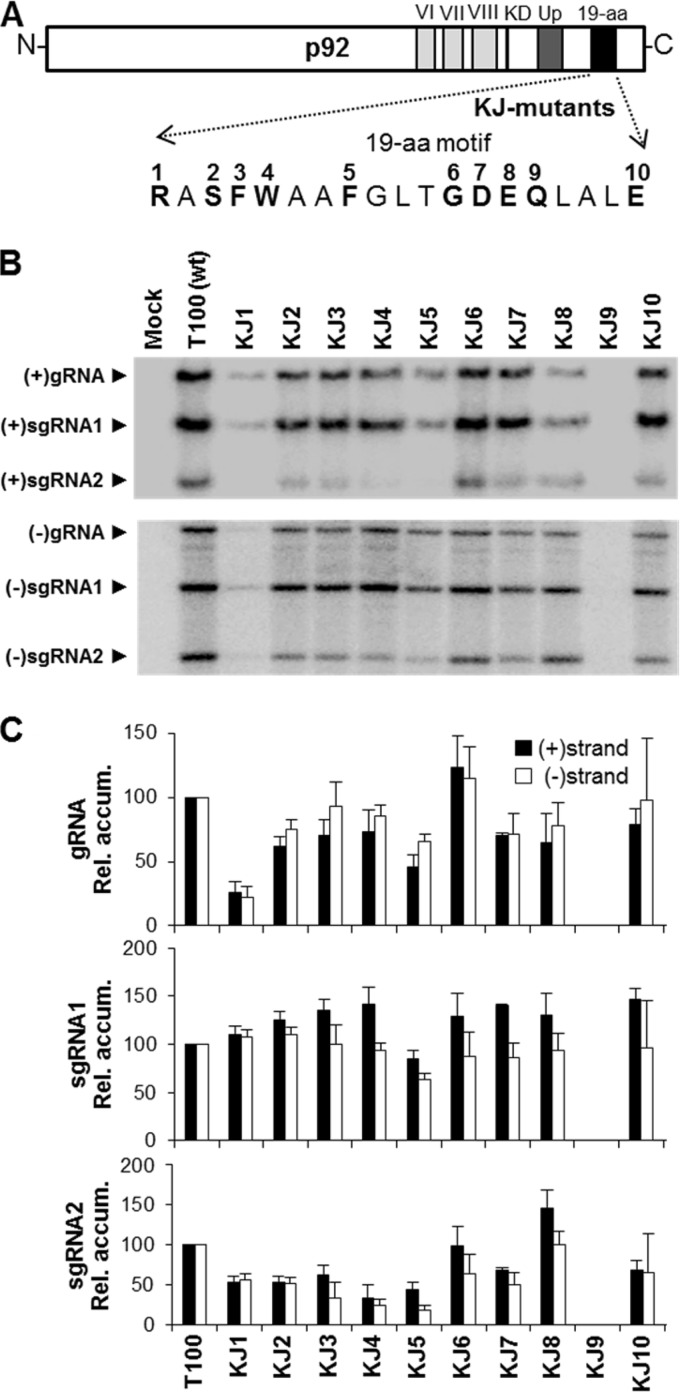
Similar mutational analyses involving substitutions were also performed on selected residues in the 19-aa motif (Fig. 3A). Replacement of the completely conserved R in KJ1 (R→A) preferentially reduced the levels of genome and sg mRNA2 (Fig. 3B and C). Similar effects were also observed by replacing the invariant S in KJ2 (S→A); however, genome levels were less affected. Changing the aromatic residues in KJ3 (F→A), KJ4 (W→A), or KJ5 (F→A) reduced genome levels by approximately one-quarter and sg mRNA2 levels by about one-half. However, in all three cases, the relative levels of sg mRNA1 were similar to that of wt or slightly higher. Sequential replacements of moderately conserved residues in KJ6 (G→A), KJ7 (D→A), and KJ8 (E→A) resulted in near-wt levels of all viral RNAs. Replacement of the completely conserved Q in KJ9 (Q→A) eliminated viral accumulation, while substitution of A for the invariant E in KJ10 (E→A) was well tolerated. Overall, the results of the mutational analysis of the 19-aa domain suggest a central role in sg mRNA2 production, as 7 of the 10 mutants preferentially affected this viral RNA (i.e., KJ1 to KJ5, KJ7, and KJ10). Also, in contrast to expectations based on preservation, replacement of two of the four invariant residues, i.e., S in KJ2 and E in KJ10, had relatively minor effects on virus viability in protoplasts.
FIG 3.
Mutations in the 19-aa motif of TBSV p92. (A) Residues selected for mutational analysis are indicated in boldface and are numbered according to their corresponding genomic mutant in the KJ series. All mutants contained alanine substitutions. (B) Northern blot analysis of viral RNA accumulation in protoplasts inoculated with genomic KJ mutants, as described in the legend to Fig. 2B. (C) Relative accumulation levels of plus- and minus-sense viral RNAs from KJ mutant infections as described in the legend to Fig. 2C.
Comparison of motif functions.
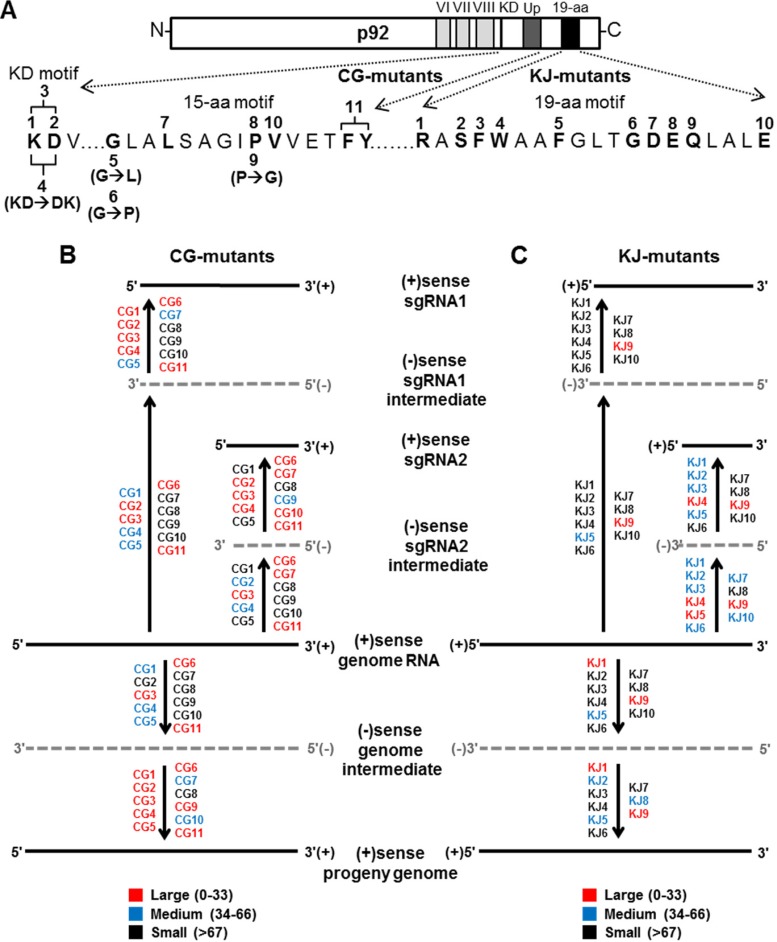
For all three motifs (KD, 15-aa, and 19-aa), defects that either abolished all viral accumulation or preferentially reduced certain viral RNAs were observed. Interestingly, the level of inhibition for many of the mutants in the KD and 15-aa motifs appeared to occur primarily at the plus-strand level (i.e., CG1, CG2, CG4 to CG7, CG9, CG10), because their relative minus-strand levels notably exceeded those of their cognate plus strands. In contrast, the defects from modifications in the 19-aa motif were associated with nearly equivalent levels of reduction in both plus and minus strands, indicative of inhibition mainly at the level of minus-strand production. Together, the results from the analyses of all three motifs in the TBSV RdRp point to fundamental roles for these elements in the production of different viral RNAs at both the plus- and minus-strand levels of synthesis, as summarized in Fig. 4.
FIG 4.
Summary of the impact of TBSV CG and KJ mutants on viral RNA accumulation. (A) All substitutions in the KD, 15-aa, and 19-aa motifs tested in TBSV mutants are indicated. (B, C) Diagrammatic representations of TBSV genome replication and sg mRNA transcription showing the different steps in which mutants affected plus-strand (solid black lines) or minus-strand (dotted gray lines) production. The effects are grouped into three categories: large (red), in which relative accumulation levels were between 0 to 33% of that of wt; medium (blue), levels between 34 to 66% of wt and; small (black), levels greater than 67% of wt.
Viral defects are not due to alterations in genomic cis-acting RNA elements.
The TBSV genome is rich in functional cis-acting RNA elements, many of which reside in coding regions (10–12). Thus, the possibility exists that the defects observed in some mutants were not the result of interference with RdRp activity but instead were due to disruption of important genomic regulatory RNA elements. To investigate this possibility, we employed a trans-replication system in which p33 and p92, expressed from a nonreplicating TBSV genome, mediate amplification of a coinoculated noncoding viral replicon: in this case, a TBSV DI RNA (14). The nonreplicating TBSV genome, nT100, contains a lethal C-to-G substitution in an essential RNA replication element (RII-SL) in the viral genome (23). In this system, the nonreplicating genome acts only as an mRNA and the activity of the encoded replication proteins is determined, independent of genome replication, via monitoring DI RNA amplification. If the relative level of DI RNA replication by a nonreplicating genome containing a certain RdRp mutation is equivalent to or lower than that observed for a replicating genome containing the same mutation, then it can be deduced that the defect is not related to a cis-acting RNA element in the genome. However, if the relative level of the DI RNA exceeds the corresponding relative genome level, then there is a possibility that the defect is related to disruption of a genomic RNA element.
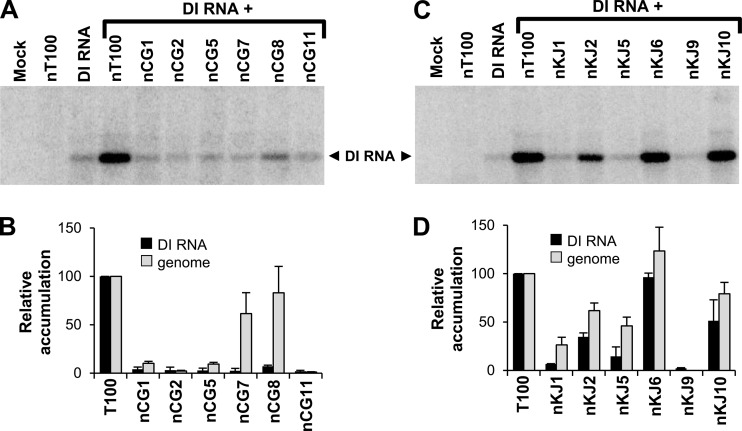
Representative mutations from each of the three motifs were introduced into a nonreplicating TBSV genome (prefixed with “n”) and cotransfected with a DI RNA replicon into protoplasts. The relative levels of DI RNA accumulation were then determined and compared with the relative levels of the genomes of corresponding replicating genomic mutants (Fig. 5). In all cases, the relative DI RNA levels did not exceed those of their corresponding genomic levels, indicating that disruption of genomic RNA elements was not responsible for any of the defective phenotypes (Fig. 5B and D). In most cases, there was relatively good correlation between the two levels; however, for nKJ5, nKJ1, nCG8, and nCG7, the DI RNA levels were substantially lower than their corresponding genome levels, i.e., 3-fold, 4-fold, 12-fold, and 30-fold, respectively (Fig. 5B and D). Thus, for these mutants, it appeared that the modifications in their RdRps significantly decreased their ability to trans-replicate the DI RNA.
FIG 5.
CG and KJ mutations in nonreplicating TBSV genomes affect amplification of a DI RNA. (A, C) Northern blot analysis of DI RNA accumulation in protoplasts coinoculated with nonreplicating genomic CG or KJ mutants and a DI RNA. (B, D) Relative accumulation levels of DI RNA from panels A and C are shown by the black bars and compared with corresponding levels of genomic RNA for replication-competent mutant genomes (gray bars) derived from Fig. 2C and 3C, respectively.
A mutation in the 15-aa motif of the RdRp inhibits viral RNA replication in trans.
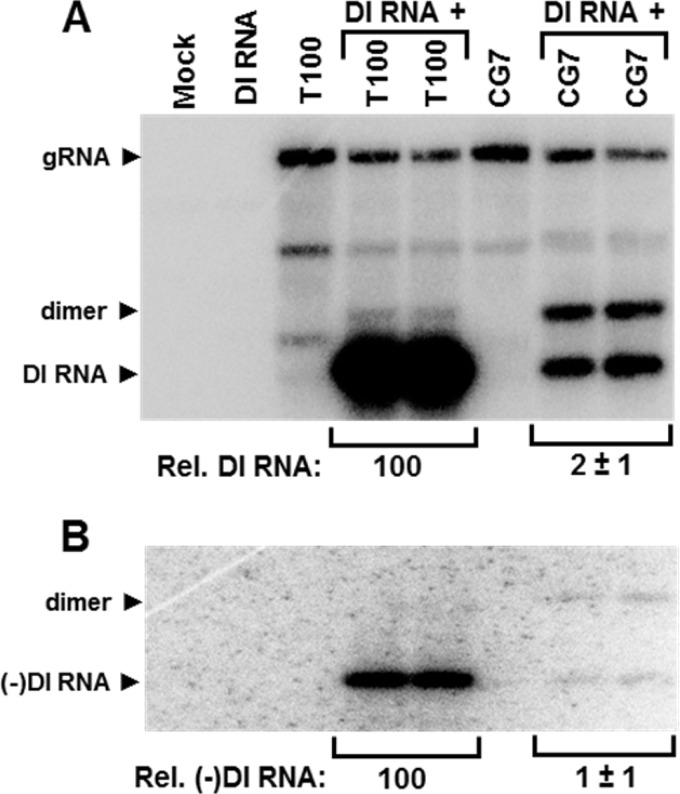
The CG7 mutant, which contained an L→A substitution in the 15-aa motif, was selected for further investigations because it exhibited the largest reduction of trans-replication of the DI RNA (i.e., 30-fold). We first wanted to determine if the trans defect also occurred with a replicating helper genome containing the same substitution. To this end, the replicating genomic CG7 mutant was cotransfected with a DI RNA into plant protoplasts and the level of DI RNA amplification was monitored (Fig. 6). Compared with coinoculations with the wt T100 genome, the relative levels of DI RNA accumulation with CG7 were 50-fold less (Fig. 6A). This value is comparable to the 30-fold difference seen with the nonreplicating nCG7 genome (Fig. 5A and B); therefore, the trans defect was not related to the inability of the helper genome to replicate. Interestingly, the CG7 plus DI RNA coinfection resulted in high levels of accumulation of DI RNA dimer, which was negligible in the wt T100 plus DI RNA coinfection (Fig. 6A). Accordingly, in addition to severely reducing the accumulation level of monomeric DI RNA, the L→A substitution also greatly enhanced the production of the dimeric form. Examination of DI RNA minus strands revealed low levels in the CG7 coinfection, which were comparable to those for the plus strands (Fig. 6B), suggesting a defect at the level of minus-strand production.
FIG 6.
Replication-competent CG7 genomic mutant affects DI RNA amplification. (A) Northern blot analysis of viral RNA accumulation in protoplasts inoculated with the CG7 genomic mutant. Bands corresponding to gRNA, DI RNA, and a DI RNA dimer are labeled on the left. (B) Northern blot analysis of minus-sense DI RNA accumulation from infections whose results are shown in panel A. Relative absolute accumulation levels of DI RNA monomers are provided below each panel.
Importance of L721 in the 15-aa motif for trans-replication of DI RNA and satellite RNA.
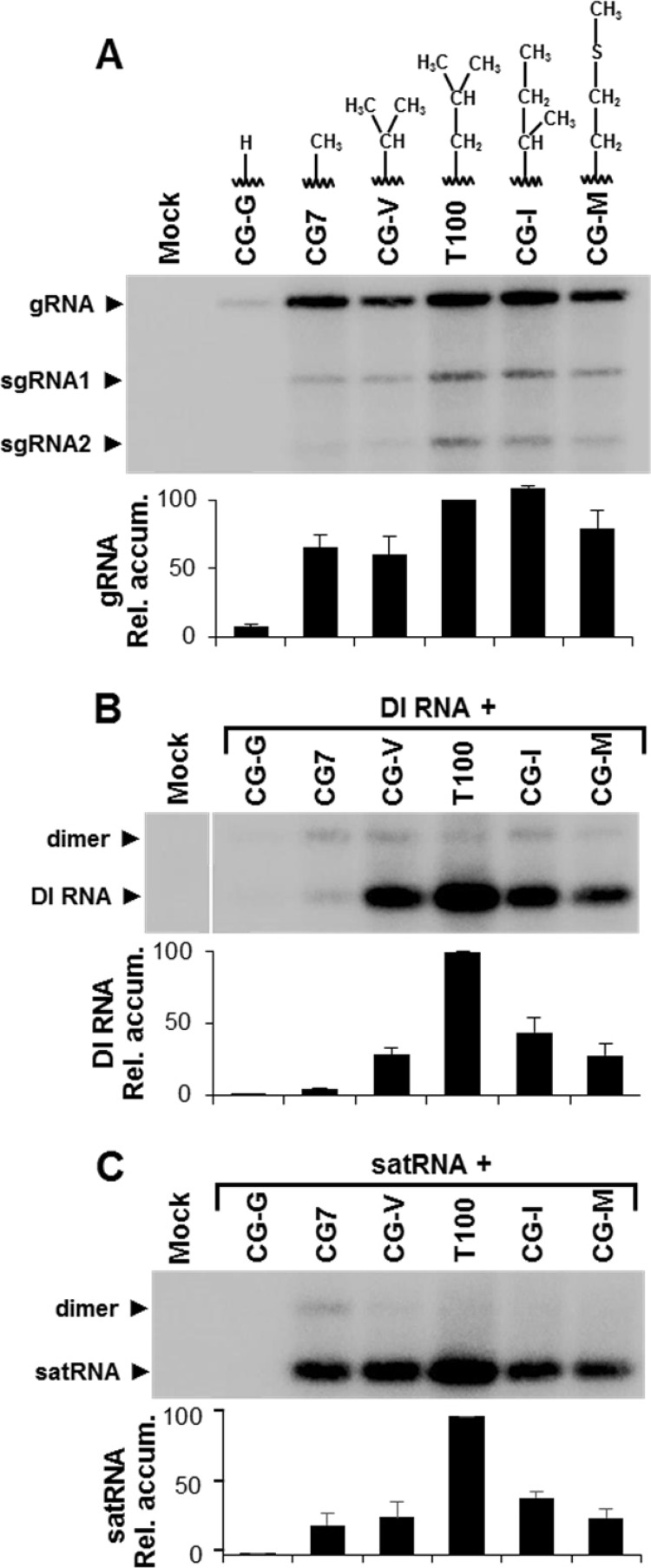
To gain additional insights into the structural requirements for residue 721 of the TBSV RdRp, which is the site of the L→A substitution in CG7, additional replacements were made at this position. In tombusvirids, this residue is occupied most frequently by L (∼68%) and to a lesser extent by I (∼28%) or M (∼4%). Four additional substitutions were made in the wt TBSV genome, which converted L to G (CG-G), V (CG-V), I (CG-I), or M (CG-M) (Fig. 7A). These mutants, as well as wt T100 and CG7, were then tested for replication in protoplasts (Fig. 7A). CG-I exhibited wt levels of viral RNA accumulation, while CG-7, CG-V, and CG-M yielded intermediate levels. The L→G substitution in CG-G was extremely detrimental, resulting in only trace amounts of the genome (Fig. 7A). These results indicate that the L721 position in the RdRp is relatively tolerant of different-sized R groups composed of nonpolar hydrocarbon chains but has a minimum requirement of a methyl moiety.
FIG 7.
Replication-competent TBSV genomic mutants with substitutions at position 721 of p92 affect genome accumulation, as well as DI RNA and satRNA amplification. (A) Northern blot analysis of viral RNA accumulation in protoplasts inoculated with TBSV genomes containing different substitutions of residue 721 of p92, as indicated above each lane. Corresponding genome levels are presented graphically below the blot. (B) Northern blot analysis of protoplasts coinoculated with the mutant genomes shown in panel A and a DI RNA. Relative absolute accumulation levels of DI RNA monomers are provided in the graph. (C) Northern blot analysis of protoplasts coinoculated with the mutant genomes in panel A and a satRNA. Relative absolute accumulation levels of satRNA monomers are provided in the graph.
The same genomes were then used in coinoculations with either DI RNA or SatRNA to determine their effects on trans-replication of these distinct subviral replicons. DI RNA amplification was reduced to between ∼25 and 55% of that of wt for mutants CG-V, CG-I, and CG-M and was undetectable for CG-G (Fig. 7B). A similar accumulation profile was also observed for the satRNA, except for CG-7, where the level was ∼20% for SatRNA (Fig. 7C), compared with ∼4% for DI RNA (Fig. 7B). Thus, with the exception of CG-7, the effects of the RdRp mutations on trans-replication were very similar for the DI RNA and satRNA.
DISCUSSION
Roles of the RdRp carboxy-proximal motifs in virus replication.
Analysis of the conserved motifs in the carboxyl region of the TBSV RdRp revealed important roles for many of the residues therein. These functions were related to (i) the production of specific viral RNAs, (ii) facilitating plus- and/or minus-strand synthesis, and (iii) modulating trans-replication. Previous analysis of the C-proximal region of the TBSV RdRp revealed that a segment encompassing the last ∼131 aa, termed RBR3, possesses RNA-binding activity (17). RBR3 overlaps all three of the newly characterized motifs (Fig. 1B); thus, the function of these motifs may be linked to the ability of the RdRp to interact with viral RNA templates. Additional studies will be necessary to determine if the observed motif-based defects are related to perturbation of the RNA-binding function of RBR3 or are independent of this activity.
Different viral RNAs were affected by modifications in the three motifs, indicating that the C-proximal region of the RdRp is involved in controlling production of both genomic and sg RNAs. For the 19-aa motif, sg mRNA2 was preferentially affected in many mutants (Fig. 3). This motif is located just upstream from the Ct motif, in which the five C-terminal residues are known to be essential for sg mRNA2 transcription but dispensable for genome replication (18). Accordingly, there may be cross talk between the 19-aa and Ct motifs that leads to the observed sg mRNA2 defects. Biased sg mRNA2 reduction was also seen for certain modifications in the 15-aa motif (e.g., CG7 [Fig. 2]); thus, at least three different motifs (19-aa, 15-aa, and Ct) are able to preferentially affect sg mRNA2 levels. Conversely, sg mRNA1 was not inhibited by many of the mutations that reduced sg mRNA2 (Fig. 3), suggesting that although both sg mRNAs are produced via a premature termination mechanism (28, 29), each has specific RdRp requirements. In addition to possessing determinants for transcription, all motifs also contained residues that were absolutely essential for production of all viral RNAs, indicating that these elements govern both specific and general functions related to viral RNA synthesis.
Another interesting finding from this study was that the replication defects resulted from inhibition of either plus- or minus-strand synthesis. Several of the KD and 15-aa motif mutants showed substantial inhibition at the plus-strand level (e.g., CG1, CG2, CG9, and CG10 in Fig. 2). Similar biased suppression of positive strands in protoplast infections has also been observed in genomic mutants of potato virus X (30) and brome mosaic virus RNA3 (31). In contrast, most of the mutations in the 19-aa motif of TBSV were defective at the minus-strand level (Fig. 3). The specificity for a particular strand sense, as well as for production of certain viral RNA species, could be related to perturbation of intrinsic RdRp activity, that is, an effect independent of other proteins. Alternatively, these motifs could mediate RdRp interactions with other proteins that are involved in regulating these properties. Several host proteins have been found to be involved in minus- and/or plus-strand synthesis and are present in complexes with p92 (8, 9, 32). Accordingly, the functional motifs in the C-proximal region of the RdRp could facilitate interactions with such factors. Currently, two host proteins are known to be part of the tombusvirus RC (33, 34) and to interact directly with the unique C-proximal two-thirds in p92 (35). One is eEF1A, and the other is the DBP3/AtRH5 DEAD box RNA helicase (33, 35). The former facilitates minus-strand synthesis (36), while the latter promotes plus-strand production (33). Consequently, the strand-specific phenotypes observed for mutants of the p92 C-proximal motifs could potentially be related to disruption of functional interactions with these host proteins.
Cis- versus trans-replication in tombusviruses.
An interesting property of tombusviruses is their ability to support the replication of different subviral replicons, namely, DI RNAs and satRNAs (20). Normally, in coinfections, TBSV is able to amplify prototypical DI RNAs very efficiently; however, our CG7 mutant was not able to do so (Fig. 6). This DI RNA-resistant mutant also showed a 5-fold reduction in its ability to amplify a satRNA (Fig. 7), indicating a general deficiency in trans-replication of subviral replicons. Previous studies of tombusviruses have identified the N terminus of p33 and an internal region in this protein as determinants that specifically reduce the amplification of satRNAs (37, 38). However, because these regions are present in both the prereadthrough p33 accessory replication protein and readthrough p92, it is unclear in which protein the modifications exerted their effects. Also, these modifications did not alter the ability of the mutant genomes to amplify DI RNAs (38). Thus, our CG7 mutant represents the first DI RNA-resistant phenotype observed in tombusviruses and, to our knowledge, in any plant virus. In addition to inefficiently amplifying the DI RNA, the CG7 mutant also generated approximately equivalent amounts of monomer and dimer forms. Since the dimer is not normally produced at such high relative levels, this modification seems to have also reduced the ability of the RdRp to preferentially select monomer DI RNA templates for replication and/or enhanced its recombination activity that generates the dimer.
DI RNAs can accumulate de novo during serial passage of tombusviruses at a high multiplicity of infection (39). This is believed to be a consequence of recombination or deletion events leading to the formation of the DI RNAs and their subsequent amplification by RdRp supplied in trans by the helper virus (40). Our CG7 mutant may be less disposed to the de novo accumulation of DI RNAs, as any prototypical recombinants generated would not be efficiently amplified. However, alternative DI RNAs, which are compatible with the CG7 mutant, could potentially arise. Genomic mutants defective in DI RNA amplification have been reported for the animal plus-strand RNA virus Sindbis virus (41), as well as the negative-strand RNA virus vesicular stomatitis virus (VSV) (42); however, evidence implicating modifications in the RdRp as the cause of these phenotypes was provided only for VSV (43). In the case of TBSV CG7, both minus and plus strands of the DI RNA were very efficiently inhibited, pointing to a minus-strand defect at the core of the DI RNA resistance (Fig. 6). In contrast, cis-replication of the CG7 genome yielded near-wt levels of minus strands and plus-strand accumulation at ∼60% of that of wt (Fig. 2). This notable difference between trans- and cis-replication illustrates that DI RNAs may not always be accurate surrogate replicons for indirect assessment of genome replication.
Mutant full-length TBSV genomes that do not express either p33 or p92 cannot be complemented for replication by providing the missing viral protein in trans (13; our unpublished data). This indicates that replication of this viral genome is cis-preferential and thus requires replication proteins to be translated directly from the genome that is to be replicated. This type of mechanism was recently confirmed to occur in tobacco mosaic virus, where it was shown that the 126-kDa replication protein, while being translated, binds directly to the 5′ untranslated region (5′ UTR) of its cognate genome (44). For TBSV, its associated DI RNAs and satRNAs have somehow escaped the restriction of cis-replication, which is likely due to the absence, in these replicons, of regulatory genomic RNA elements that control this process. The inhibition of DI RNA replication at the minus-strand level with mutant CG7 implies that the modified RdRp cannot effectively utilize the plus strand as a template for RNA synthesis. In turn, this suggests that the way the RdRp normally engages the full-length genome versus the DI RNA is distinct, in terms of either the RNA-directed assembly of the RC or the initiation of minus-strand RNA synthesis. Additional studies will be required to identify the mechanism responsible for the observed inhibition of trans-replication in CG7.
Prevalence of the carboxy-proximal motifs in tombusvirid-like RNA viruses.
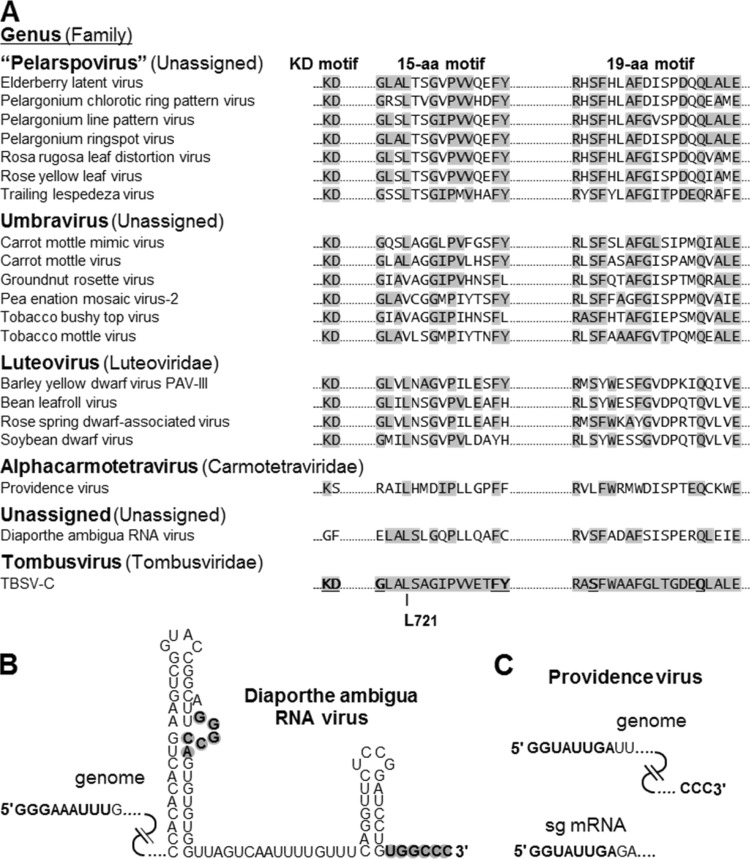
The conservation and confirmed functional importance of the three motifs prompted us to search for additional viruses that may harbor these protein signatures. We found that members of the genus Umbravirus and the provisional genus Pelarspovirus, both of which will likely be assigned to the family Tombusviridae in the future, contained all three motifs (Fig. 8A). Members of the genus Luteovirus, assigned to the family Luteoviridae, also harbored the motifs (Fig. 8A). It has been suggested, based on similarities with tombusvirids, that luteoviruses may be a better fit in the family Tombusviridae (45), and our results lend support to this notion.
FIG 8.
(A) RdRp sequence alignment of the KD, 15-aa, and 19-aa motifs in tombusvirid-like viruses. Amino acids shaded in gray are identical to those in the reference TBSV sequence at the bottom. The L721 residue is indicated, and residues determined to be critical for TBSV viral RNA synthesis are in bold and underlined in the reference sequence. (B) Terminal sequences of the DaRV RNA genome. 5′-terminal nucleotides that are similar to carmovirus RNA replication elements are in bold. For the 3′-terminal sequence, potential RNA secondary structures and a tertiary interaction involving the 3′-end residues (bold and gray shading), which are commonly found in tombusvirids, are presented. (C) Terminal sequences of the PrV RNA genome are shown at the top with nucleotides that are similar to those in tombusvirids in bold. The 3′ end of the genome is not predicted to form any local RNA secondary structures. Below, the 5′-terminal sequence of the sg mRNA, which is identical to the genomic 5′ terminus, is shown in bold. Accession numbers for panel A: elderberry latent virus, AHZ59463.1; Pelargonium chlorotic ring pattern virus, YP_052925.1; Pelargonium line pattern virus, AAU84998.1; Pelargonium ringspot virus, AHZ59473.1; Rosa rugosa leaf distortion virus, AGF70694.1; rose yellow leaf virus, AGF70700.1; trailing lespedeza virus, ADY69093.2; carrot mottle mimic virus, NP_054006.2; carrot mottle virus, YP_002302259.1; groundnut rosette virus, NP_619714.1; pea enation mosaic virus-2, AEM45994.1; tobacco bushy top virus, NP_733848.1; tobacco mottle virus, AAG02571.1; barley yellow dwarf virus PAV-III, AAF62525.1; bean leafroll virus, NP_563609.1; rose spring dwarf-associated virus, YP_001949736.1; soybean dwarf virus, AII15905.1; Providence virus, YP_003620397.1; Diaporthe ambigua RNA virus, AAF22958.1.
Our search also revealed that the carboxy-proximal motifs are conserved, albeit to different degrees, in two other plus-strand RNA viruses, the insect virus Providence virus (PrV; genus Alphacarmotetravirus, family Carmotetraviridae) and a fungal virus, Diaporthe ambigua RNA virus (DaRV; genus and family unassigned) (46, 47). Like all tombusvirids, both of these viruses translate a prereadthrough accessory replication protein and a readthrough RdRp, and sequence similarities between these RdRps and those of tombusvirids have been noted previously (46, 47); however, the relevance of the three motifs investigated here was not addressed. For DaRV, all four invariant residues in the 19-aa motif were maintained (i.e., RxSxxxxxxxxxxxQxxxE), whereas PrV retained all but the S (Fig. 8A). The completely conserved P near the center of the 15-aa motif was also present in both of these viruses (Fig. 8A). Additional positions in these motifs were conserved at levels close to those in the other comparative genera (Fig. 8A), suggesting a similar level of functional importance. In contrast, only the K of the invariant KD motif was present in PrV and neither residue was conserved in DaRV (Fig. 8A). Since the KD residues in TBSV are highly functionally relevant (Fig. 2), these diverged viruses must have relaxed requirements for this particular motif.
As with all tombusvirids, PrV transcribes an sg mRNA and encodes a coat protein that forms icosahedral capsids (46); conversely, DaRV does neither (47). Also of relevance is the observation that terminal nucleotides in both of these RNA genomes conform to those of carmovirus-like tombusvirids, with two to three G's at the 5′ end followed by several A/U's (48), or of most tombusvirids, with three consecutive C's at the 3′ end (Fig. 8B and C) (49). Additionally, DaRV contains typical RNA secondary structures at its 3′ terminus that are found in tombusvirids (i.e., terminal and internal stem-loop structures), including a conserved tertiary interaction between the 3′-terminal sequence and the bulge in an upstream stem (49) (Fig. 8B). For PrV, the 5′-terminal sequence of the sg mRNA is identical to that of the genome, which is another common feature in tombusvirids (10) (Fig. 8C). Therefore, both of these viruses, one an insect virus and the other a fungal virus, contain protein and RNA elements characteristic of the family Tombusviridae, which currently contains only plant viruses. Accordingly, based on the presence of these defining viral features, it may be worth considering both PrV and DaRV for adoption into the family Tombusviridae.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratory for reviewing the manuscript and Baodong Wu for technical assistance.
This research was funded by NSERC, and C.D.G. was supported by an Ontario Graduate Scholarship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03378-14.
REFERENCES
- 1.Te Velthuis AJ. 2014. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci 71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mine A, Okuno T. 2012. Composition of plant virus RNA replicase complexes. Curr Opin Virol 2:669–675. doi: 10.1016/j.coviro.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Sztuba-Solińska J, Stollar V, Bujarski JJ. 2011. Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle. Virology 412:245–255. doi: 10.1016/j.virol.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald SM. 2013. RNA synthetic mechanisms employed by diverse families of RNA viruses. WIREs RNA 4:351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy PD, Pogany J. 2008. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol Biol 451:55–68. doi: 10.1007/978-1-59745-102-4_4. [DOI] [PubMed] [Google Scholar]
- 6.White KA, Nagy PD. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acids Res Mol Biol 78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- 7.Sit TL, Lommel SA. 2010. Tombusviridae, p 1–6 InEncyclopedia of Life Sciences (ELS). JohnWiley & Sons Ltd., Chichester, United Kingdom. doi: 10.1002/9780470015902.a0000756.pub2. [DOI] [Google Scholar]
- 8.Nagy PD, Pogany J, Lin JY. 2014. How yeast can be used as a genetic platform to explore virus-host interactions: from ‘omics’ to functional studies. Trends Microbiol 22:309–316. doi: 10.1016/j.tim.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Nagy PD, Barajas D, Pogany J. 2012. Host factors with regulatory roles in tombusvirus replication. Curr Opin Virol 2:691–698. doi: 10.1016/j.coviro.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Wu B, Grigull J, Ore MO, Morin S, White KA. 2013. Global organization of a positive-strand RNA virus genome. PLoS Pathog 9(5):e1003363. doi: 10.1371/journal.ppat.1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiwan SD, White KA. 2011. Subgenomic mRNA transcription in Tombusviridae. RNA Biol 8:287–294. doi: 10.4161/rna.8.2.15195. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson BL, White KA. 2014. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oster SK, Wu B, White KA. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol 72:5845–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimino PA, Nicholson BL, Wu B, Xu W, White KA. 2011. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog 7(12):e1002423. doi: 10.1371/journal.ppat.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol 28:375–430. [DOI] [PubMed] [Google Scholar]
- 16.Molinari P, Marusic C, Lucioli A, Tavazza R, Tavazza M. 1998. Identification of artichoke mottled crinkle virus (AMCV) proteins required for virus replication: complementation of AMCV p33 and p92 replication-defective mutants. J Gen Virol 79:639–647. [DOI] [PubMed] [Google Scholar]
- 17.Rajendran KS, Nagy PD. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J Virol 77:9244–9258. doi: 10.1128/JVI.77.17.9244-9258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B, White KA. 2007. Uncoupling RNA virus replication from transcription via the polymerase: functional and evolutionary insights. EMBO J 26:5120–5130. doi: 10.1038/sj.emboj.7601931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon AE, Roossinck MJ, Havelda Z. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu Rev Phytopathol 42:415–437. doi: 10.1146/annurev.phyto.42.040803.140402. [DOI] [PubMed] [Google Scholar]
- 21.Rubino L, Pantaleo V, Navarro B, Russo M. 2004. Expression of tombusvirus open reading frames 1 and 2 is sufficient for the replication of defective interfering, but not satellite, RNA. J Gen Virol 85:3115–3122. doi: 10.1099/vir.0.80296-0. [DOI] [PubMed] [Google Scholar]
- 22.Hearne PQ, Knorr DA, Hillman BI, Morris TJ. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177:141–151. [DOI] [PubMed] [Google Scholar]
- 23.Pogany J, White KA, Nagy PD. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol 79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White KA, Morris TJ. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J Virol 68:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chernysheva OA, White KA. 2005. Modular arrangement of viral cis-acting RNA domains in a tombusvirus satellite RNA. Virology 332:640–649. doi: 10.1016/j.virol.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Choi IR, Ostrovsky M, Zhang G, White KA. 2001. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J Biol Chem 276:41761–41768. [DOI] [PubMed] [Google Scholar]
- 27.Ray D, Wu B, White KA. 2003. A second functional RNA domain in the 5′ UTR of the Tomato bushy stunt virus genome: intra- and interdomain interactions mediate viral RNA replication. RNA 9:1232–1245. doi: 10.1261/rna.5630203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HX, White KA. 2004. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J 23:3365–3374. doi: 10.1038/sj.emboj.7600336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi IR, White KA. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J Biol Chem 277:3760–3766. doi: 10.1074/jbc.M109067200. [DOI] [PubMed] [Google Scholar]
- 30.Miller ED, Plante CA, Kim KH, Brown JW, Hemenway C. 1998. Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J Mol Biol 284:591–608. [DOI] [PubMed] [Google Scholar]
- 31.Hema M, Kao CC. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J Virol 78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy PD, Pogany J. 2011. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalev N, Nagy PD. 2014. The expanding functions of cellular helicases: the tombusvirus RNA replication enhancer co-opts the plant eIF4AIII-like AtRH2 and the DDX5-like AtRH5 DEAD-box RNA helicases to promote viral asymmetric RNA replication. PLoS Pathog 10(4):e1004051. doi: 10.1371/journal.ppat.1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J Virol 82:6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Pogany J, Tupman S, Esposito AM, Kinzy TG, Nagy PD. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog 6(11):e1001175. doi: 10.1371/journal.ppat.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Célix A, Burgyán J, Rodríguez-Cerezo E. 1999. Interactions between tombusviruses and satellite RNAs of tomato bushy stunt virus: a defect in sat RNA B1 replication maps to ORF1 of a helper virus. Virology 262:129–138. [DOI] [PubMed] [Google Scholar]
- 38.Rubino L, Russo M. 2012. A single amino acid substitution in the ORF1 of cymbidium ringspot virus determines the accumulation of two satellite RNAs. Virus Res 168:84–87. doi: 10.1016/j.virusres.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Knorr DA, Mullin RH, Hearne PQ, Morris TJ. 1991. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology 181:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng CP, Pogany J, Nagy PD. 2002. Mechanism of DI RNA formation in tombusviruses: dissecting the requirement for primer extension by the tombusvirus RNA dependent RNA polymerase in vitro. Virology 304:460–473. doi: 10.1006/viro.2002.1713. [DOI] [PubMed] [Google Scholar]
- 41.Weiss B, Schlesinger S. 1981. Defective interfering particles of Sindbis virus do not interfere with the homologous virus obtained from persistently infected BHK cells but do interfere with Semliki Forest virus. J Virol 37:840–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DePolo NJ, Giachetti C, Holland JJ. 1987. Continuing coevolution of virus and defective interfering particles and of viral genome sequences during undiluted passages: virus mutants exhibiting nearly complete resistance to formerly dominant defective interfering particles. J Virol 61:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giachetti C, Holland JJ. 1988. Altered replicase specificity is responsible for resistance to defective interfering particle interference of an Sdi- mutant of vesicular stomatitis virus. J Virol 62:3614–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawamura-Nagaya K, Ishibashi K, Huang YP, Miyashita S, Ishikawa M. 2014. Replication protein of tobacco mosaic virus cotranslationally binds the 5′ untranslated region of genomic RNA to enable viral replication. Proc Natl Acad Sci U S A 111(16):E1620–E1628. doi: 10.1073/pnas.1321660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller WA, Liu S, Beckett R. 2002. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol Plant Pathol 3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- 46.Walter CT, Pringle FM, Nakayinga R, de Felipe P, Ryan MD, Ball LA, Dorrington RA. 2010. Genome organization and translation products of Providence virus: insight into a unique tetravirus. J Gen Virol 91:2826–3285. doi: 10.1099/vir.0.023796-0. [DOI] [PubMed] [Google Scholar]
- 47.Preisig O, Moleleki N, Smit WA, Wingfield BD, Wingfield MJ. 2000. A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J Gen Virol 81:3107–3114. [DOI] [PubMed] [Google Scholar]
- 48.Guan H, Carpenter CD, Simon AE. 2000. Analysis of cis-acting sequences involved in plus-strand synthesis of a turnip crinkle virus-associated satellite RNA identifies a new carmovirus replication element. Virology 268:345–354. doi: 10.1006/viro.1999.0153. [DOI] [PubMed] [Google Scholar]
- 49.Na H, White KA. 2006. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology 345:305–316. doi: 10.1016/j.virol.2005.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.