ABSTRACT
The alpha interferon (IFN-α)-inducible restriction factor myxovirus B (MxB) blocks HIV-1 infection after reverse transcription but prior to integration. MxB binds to the HIV-1 core, which is composed of capsid protein, and this interaction leads to inhibition of the uncoating process of HIV-1. Previous studies suggested that HIV-1 restriction by MxB requires binding to capsid. This work tests the hypothesis that MxB oligomerization is important for the ability of MxB to bind to the HIV-1 core. For this purpose, we modeled the structure of MxB using the published tertiary structure of MxA. The modeled structure of MxB guided our mutagenic studies and led to the discovery of several MxB variants that lose the capacity to oligomerize. In agreement with our hypothesis, MxB variants that lost the oligomerization capacity also lost the ability to bind to the HIV-1 core. MxB variants deficient for oligomerization were not able to block HIV-1 infection. Overall, our work showed that oligomerization is required for the ability of MxB to bind to the HIV-1 core and block HIV-1 infection.
IMPORTANCE MxB is a novel restriction factor that blocks infection of HIV-1. MxB is inducible by IFN-α, particularly in T cells. The current work studies the oligomerization determinants of MxB and carefully explores the contribution of oligomerization to capsid binding and restriction. This work takes advantage of the current structure of MxA and models the structure of MxB, which is used to guide structure-function studies. This work leads to the conclusion that MxB oligomerization is important for HIV-1 capsid binding and restriction.
INTRODUCTION
The myxovirus resistance proteins represent a family of interferon-inducible factors with a wide range of antiviral activities (1, 2). The myxovirus B (MxB) gene was originally cloned from a human glioblastoma cell line treated with alpha interferon (IFN-α) (3, 4). MxB as well as the related protein MxA belongs to the dynamin-like family of proteins, which have diverse functions ranging from vesicle transport to antiviral activity (1, 5–10). The most-studied dynamin-like protein that exhibits antiviral activity is MxA (1, 2). Contrary to the role of MxB, the antiviral role of MxA has been extensively studied for viruses, including influenza virus (1, 11–14), tick-born thogotovirus (15), African swine fever virus (16), hepatitis B virus (17), and La Crosse virus (18, 19). The antiviral activity of the long form of MxB was recently described (8, 20–22); these investigations led to the discovery that the IFN-α-inducible protein MxB blocks HIV-1 infection.
Genetic evidence suggested that HIV-1 capsid is the determinant for the ability of MxB to block HIV-1 infection (8, 21, 22). In agreement with these findings, we have recently demonstrated that MxB binds to the HIV-1 capsid and prevents the uncoating process of HIV-1 (23). In addition, the work of others and our work showed that the 90 N-terminal amino acids of MxB are important for its ability to bind capsid and restrict infection (23–25). The use of MxB large deletion variants showed that oligomerization is important for the ability of MxB to block HIV-1 infection (23). This work tested the hypothesis that the oligomerization of MxB provides the necessary avidity to the 90 N-terminal amino acids of MxB to bind capsid. For this purpose, we took advantage of the ample oligomerization studies performed with the MxA protein.
MxB, like MxA, is composed of a GTPase domain connected by a bundle-signaling element (BSE) to the stalk domain (Fig. 1A). Extensive structural studies have demonstrated that the stalk domain of MxA is important for its oligomerization (11). In addition, the stalk region of MxA is a tetramer in solution (11). Interestingly, the crystallization of the stalk region of MxA revealed an oligomerization pattern that requires the interaction of different interfaces (11). Mutagenesis of these interfaces revealed two classes of mutations: (i) disruption of the MxA tetramer with the predominant formation of dimers (interface 1) and (ii) disruption of the MxA tetramer with the predominant formation of monomers (interface 2). Interestingly, a loss of MxA oligomerization correlates with a loss of antiviral function (11). This work explores the role of equivalent mutations in the ability of MxB to oligomerize, bind capsid, and restrict HIV-1 infection. Our findings suggest that oligomerization of MxB is required for capsid binding and restriction. We also explored whether the previously described putative leucine zipper domain plays a role in MxB oligomerization, capsid binding, and restriction of HIV-1 infection.
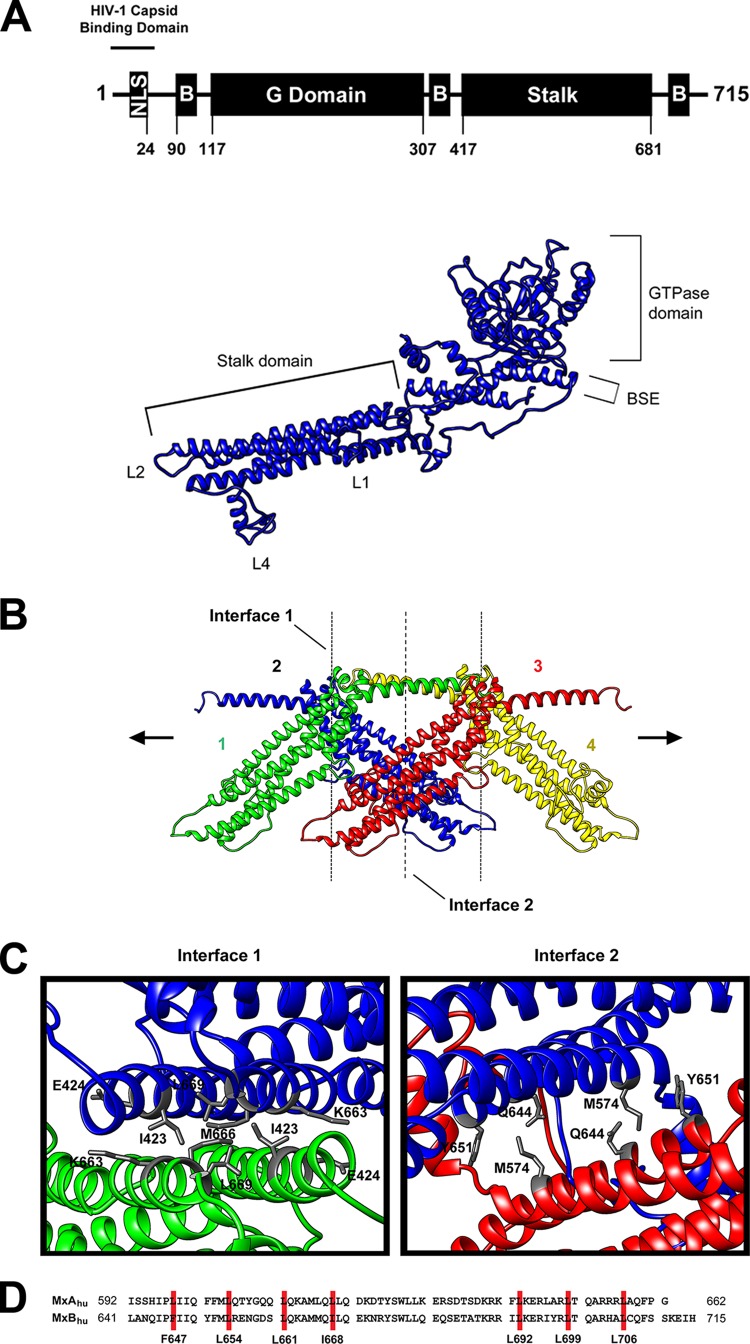
FIG 1.
Predicted structure and oligomerization of MxB. (A) Predicted structural model of human MxB. The wild-type human MxB protein is depicted. The numbers of the amino acid residues at the boundaries of the MxB domains are indicated. The structure of MxB was modeled using the published tertiary structure of MxA (PDB accession number 3SZR) using I-Tasser and Chimera (version 1.9) software. The nuclear localization signal (NLS), stalk domain, GTPase domain (G Domain), and bundle-signaling element (B) are indicated. (B) Predicted oligomerization interactions between MxB monomers. The MxB monomer was modeled into the dynamin 1 higher-order structure formation. (C) Top-down view of interface 1 and interface 2 of the predicted MxB structure. (D) Sequence alignment of human MxA (MxAhu) and human MxB (MxBhu) showing the putative leucine zipper domain in red.
MATERIALS AND METHODS
MxB oligomerization assay.
Approximately 107 human 293T cells were cotransfected with plasmids expressing MxB variants tagged with FLAG and hemagglutinin (HA). After 24 h, cells were lysed in 0.5 ml of whole-cell extract (WCE) buffer (50 mM Tris [pH 8.0], 280 mM NaCl, 0.5% Igepal, 10% glycerol, 5 mM MgCl2, 50 μg/ml ethidium bromide, 50 U/ml nuclease [Benzonase; Roche]). Lysates were centrifuged at 14,000 rpm for 1 h at 4°C. Postspin lysates were then precleared using protein A-agarose (Sigma) for 1 h at 4°C; a small aliquot of each of these lysates was stored for use as the input. Precleared lysates containing the tagged proteins were incubated with anti-FLAG-agarose beads (Sigma) for 2 h at 4°C. Anti-FLAG-agarose beads were washed three times in WCE buffer, and immune complexes were eluted using 200 μg of FLAG tripeptide/ml in WCE buffer. The eluted samples were separated by SDS-PAGE and analyzed by Western blotting using either anti-HA or anti-FLAG antibodies (Sigma).
EGS cross-linking experiments with MxB variants.
Human 293T cells (107) were transfected with plasmids expressing wild-type MxA, wild-type MxB, or mutant MxB tagged with FLAG. At 24 h after transfection, cells were resuspended in 2 ml of lysis buffer (0.5% NP-40 in 1× phosphate-buffered saline [PBS]). Lysates were centrifuged at 15,000 rpm for 1 h at 4°C. Supernatants (120 μl) were collected and incubated with 40 μl of sulfo-ethylene glycol bis(succinimidyl succinate) (sulfo-EGS; Thermo Scientific) at different final concentrations for 30 min. Ten microliters of 1 M Tris-HCl (pH 7.5) was then added to the lysate-EGS mixture to stop the cross-linking reaction. The mixtures were then incubated with 40 μl of 6× SDS-PAGE loading buffer for 30 min at 37°C. Samples were separated by SDS-PAGE and analyzed by Western blotting using anti-FLAG antibodies (Sigma).
Binding of MxB variants to in vitro-assembled HIV-1 CA-NC complexes.
293T cells were transfected with plasmids expressing wild-type or mutant MxB proteins. At 48 h after transfection, cell lysates were prepared as follows: previously washed cells were resuspended in hypotonic lysis buffer (10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]). The cell suspension was frozen, thawed, and incubated on ice for 10 min. Afterwards, the lysate was centrifuged at maximum speed in a refrigerated Eppendorf microcentrifuge (∼14,000 × g) for 5 min. The supernatant was supplemented with 1/10 volume of 10× PBS and then used in the binding assay. In some cases, samples containing the MxB variants were diluted with extracts prepared in parallel from untransfected cells. To test binding, 5 μl of capsid-nucleocapsid (CA-NC) particles assembled in vitro was incubated with 200 μl of cell lysate at room temperature for 1 h (26, 27). A fraction of this mixture was stored (input). The mixture was spun through a 70% sucrose cushion (70% sucrose, 1× PBS, 0.5 mM DTT) at 100,000 × g in an SW55 rotor (Beckman) for 1 h at 4°C. After centrifugation, the supernatant was carefully removed and the pellet was resuspended in 1× SDS-PAGE loading buffer (pellet). The level of MxB proteins was determined by Western blotting with an anti-HA or anti-FLAG antibody as described above. The level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting using anti-p24 CA antibodies.
Creation of cells stably expressing wild-type and mutant MxB proteins.
Retroviral vectors encoding wild-type or mutant human MxB proteins were created using the LPCX vector. The MxB proteins contained a FLAG epitope tag at the C terminus. Recombinant viruses were produced in 293T cells by cotransfecting the LPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid expresses the vesicular stomatitis virus (VSV) G envelope glycoprotein, allowing efficient entry into a wide range of vertebrate cells. Cf2Th canine thymocytes were transduced and selected in puromycin (Sigma).
Infection with viruses expressing GFP.
Recombinant HIV-1 expressing green fluorescent protein (GFP) was prepared as described previously (28). Recombinant viruses were pseudotyped with the VSV G glycoprotein. For infections, 3 × 104 Cf2Th cells stably expressing the different MxB variants were seeded in 24-well plates and incubated at 37°C with virus for 24 h. Cells were washed and returned to the culture for 48 h. Subsequently, GFP-positive cells were analyzed using a flow cytometer (Becton Dickinson).
RESULTS
Oligomerization determinants of MxB.
We have previously demonstrated that the ability of MxB to oligomerize is important for HIV-1 restriction by the use of large deletions (23). Particularly, we demonstrated that deletion of the ∼100 C-terminal amino acids of MxB impaired its ability to oligomerize and restrict HIV-1 infection (23). To explore the contribution of MxB oligomerization to function, we modeled the structure of MxB using the published tertiary structure of MxA using the I-Tasser online protein structure and function prediction software (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (11). Using the modeled structure of MxB (Fig. 1A) and the mutagenesis information previously obtained for MxA (11), we performed a structure-guided mutagenesis of MxB. For this purpose, we modeled the two interfaces between one monomer of MxB and a second monomer based on the higher-order assembly described for MxA (11) (Fig. 1B and C). As shown in Fig. 1B and C, we targeted residues on interfaces 1 and 2 of MxB (Fig. 1B and C). On interface 1, we targeted residues I423, E424, K663, M666, and L669 (Fig. 1C and Table 1); mutations of the equivalent MxA residues resulted in disruption of the MxA tetramer with the predominant formation of dimers (11). On interface 2, we targeted M574, Q644, and Y651 (Fig. 1C and Table 1); the equivalent mutations in MxA resulted in the disruption of the tetramer with the predominant formation of monomers (11). Mutagenic analysis of the residues in these interfaces will reveal whether MxB exhibits interactions similar to those of MxA in order to form oligomers.
TABLE 1.
MxB variant phenotypese
| Region | MxA |
MxB |
|||||
|---|---|---|---|---|---|---|---|
| Variant | Tetramer oligomerization phenotype | Variant | Phenotype |
||||
| Oligomerization by IPa | Oligomerization cross-linkingb | Binding to CA-NC tubes ± SDc | % restriction of HIV-1d | ||||
| WT | WT | Yes | Yes | 1.00 ± 0 | 100 | ||
| Interface 1 | I376D | 1 | I423D | Yes | Yes | 0.95 ± 0.07 | ND |
| D377K | 1 | E424R | Yes | Yes | 0.99 ± 0.29 | 92 ± 11 | |
| K614D | 1 | K663D | Yes | Yes | 1.04 ± 0.08 | 80 ± 13 | |
| L617D | 1 | M666D | Yes | Yes | 1.03 ± 0.11 | ND | |
| L620D | 1 | L669D | Yes | Yes | 1.14 ± 0.03 | 52 ± 10 | |
| Interface 2 | H595D | 2 | Q644D | No | No | 0.32 ± 0.01 | 0 |
| M527D | 2 | M574D | No | No | 0.14 ± 0.10 | 0 | |
| F602D | 2 | Y651D | No | No | 0.11 ± 0.09 | ND | |
| M527D/F602D | 2 | M574D/Y651D | No | ND | 0.09 ± 0.06 | 0 | |
| Putative leucine zipper | L598K | ND | F647K | Yes | ND | ND | ND |
| L605K | ND | L654K | Yes | ND | ND | ND | |
| L612K | ND | L661K | No | No | 0.15 ± 0.05 | 0 | |
| L619K | ND | I668K | Yes | ND | ND | ND | |
| L643K | ND | L692K | Yes | ND | ND | ND | |
| L650K | ND | L699K | Yes | No | 0.87 ± 0.27 | 99 ± 1 | |
| L657K | ND | L706K | Yes | No | 1.06 ± 0.30 | 90 ± 5 | |
Wild-type and mutant MxB-FLAG proteins were assayed for their association with wild-type MxB-HA using immunoprecipitation (IP), as described in Materials and Methods.
Wild-type and mutant MxB-FLAG proteins were assayed for oligomerization by chemical cross-linking using EGS, as described in Materials and Methods.
Binding to the HIV-1 capsid complexes was determined for each MxB variant, as described in Materials and Methods. The binding is expressed as the amount of bound fraction relative to input, where the value for the wild type is equal to 1.0, and the means ± standard deviations from three experiments are shown.
Restriction was measured by infecting cells expressing the indicated MxB variant with HIV-1–GFP. After 48 h, the percentage of GFP-positive (infected) cells was determined by flow cytometry. Restriction is expressed as the percentage of HIV-1 restriction by the MxB variants relative to HIV-1 restriction by wild-type MxB, which was considered to be 100%. The means ± standard deviations from three independent experiments are shown.
WT, wild type; 1, loss of tetramerization resulting in the predominant formation of dimers; 2, loss of tetramerization resulting in the predominant formation of monomers; Yes, oligomerization was similar to that observed for the wild-type MxB protein; No, oligomerization was not observed; ND, not determined.
Previous investigations suggested that MxA exhibits a putative leucine zipper domain important for oligomerization (29); however, these experiments were performed by deletion of large segments of MxA and not by the use of single point mutations. In agreement with the hypothesis that MxB contains a putative leucine zipper domain similar to that of MxA (Fig. 1D), we have shown that disruption of the putative leucine zipper domain of MxB by using the variant L661K completely abolishes the ability of MxB to oligomerize (23). To assess whether residues in the putative leucine zipper domain of MxB contributes to oligomerization, this work targeted the seven residues corresponding to the putative leucine zipper domain (Table 1): F647, L654, L661, I668, L692, L699, and L706. Mutagenic analysis of these residues will establish their contribution to MxB oligomerization.
Contribution of interface 1, interface 2, and the putative leucine zipper domain to MxB oligomerization.
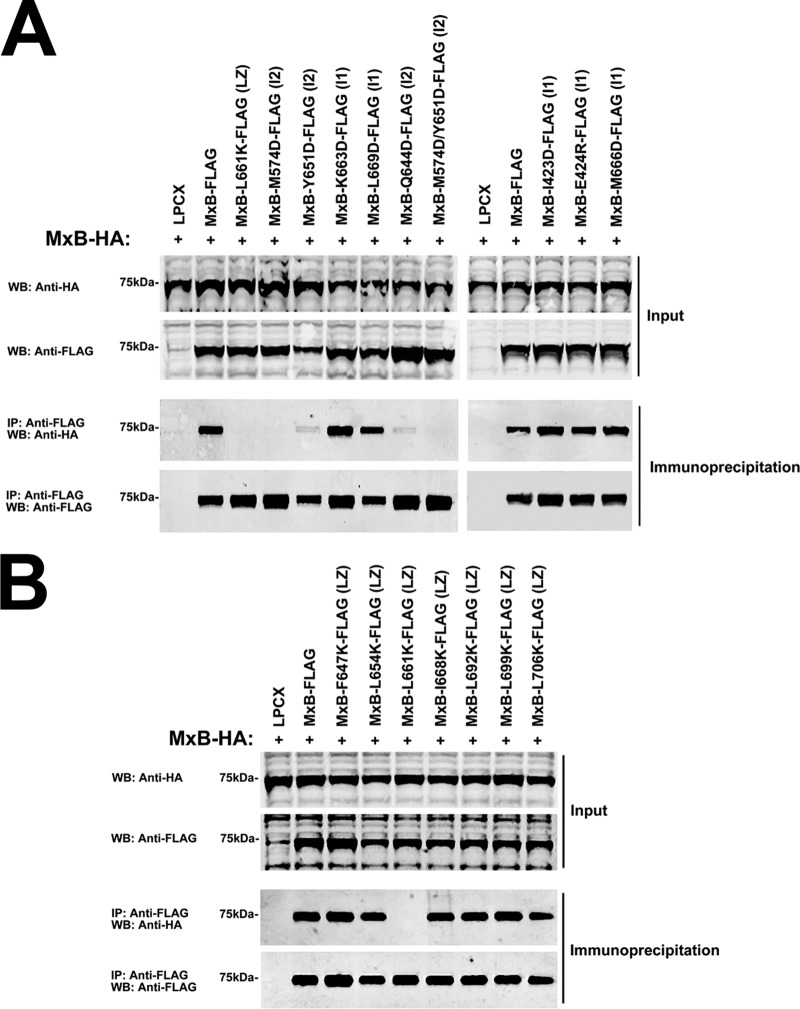
To understand the role of interfaces 1 and 2 in the ability of MxB to form oligomers (Fig. 1C), we created a set of MxB variants in which individual residues of interfaces 1 and 2 were changed, as it has been done for MxA (Table 1) (11). Subsequently, we tested the ability of the different MxB variants to form oligomers by using our previously described MxB oligomerization assay in mammalian cells (30). For this purpose, we evaluated the biochemical ability of wild-type and mutant MxB proteins tagged with a FLAG epitope to immunoprecipitate an MxB protein tagged with an HA epitope in mammalian cells (Fig. 2A). As previously shown, MxB-FLAG interacts with MxB-HA (Fig. 2A). The ability of MxB to oligomerize was not affected in MxB variants with disruptions of interface 1 residues (Fig. 2A and Table 1). Tetramerization was disrupted in MxA variants with mutations equivalent to those in the MxB variants, resulting in the predominant formation of MxA dimers (11) (Table 1). On the contrary, every MxB variant with a disruption of interface 2 lost the ability to oligomerize (Fig. 2A and Table 1). Interestingly, tetramerization was disrupted in the MxA variants with mutations equivalent to those in MxB variants, resulting in the predominant formation of MxA monomers (11) (Table 1).
FIG 2.
Contribution of interface 1, interface 2, and putative leucine zipper residues to MxB oligomerization. (A) The ability of MxB variants with mutations in interface 1 and interface 2 to oligomerize was tested in human 293T cells. For this purpose, 293T cells were transfected with plasmids expressing wild-type MxB-HA and MxB-FLAG mutants. Cells were lysed at 24 h posttransfection and analyzed by Western blotting using anti-HA and anti-FLAG antibodies (Input). Lysates were immunoprecipitated using anti-FLAG agarose beads, and immunoprecipitates were eluted using 3× FLAG peptide. Elutions were analyzed by Western blotting using anti-HA and anti-FLAG antibodies (Immunoprecipitation). Representative results from three independent experiments are shown. (B) The ability of MxB putative leucine zipper variants to oligomerize was tested as described in the legend to panel A. Experiments were repeated at least three times, and representative results are shown. WB, Western blotting; IP, immunoprecipitation; LZ, putative leucine zipper; I1, interface 1; I2, interface 2.
Next, we tested whether the putative leucine zipper on MxB (Fig. 1D), predicted by sequence homology to MxA, plays a role in the ability of MxB to oligomerize. As shown in Fig. 2B, none of the tested MxB variants, with the exception of the MxB L661K variant, lost the oligomerization ability. These results show that most of the residues in the putative leucine zipper domain are not involved in the ability of MxB to oligomerize.
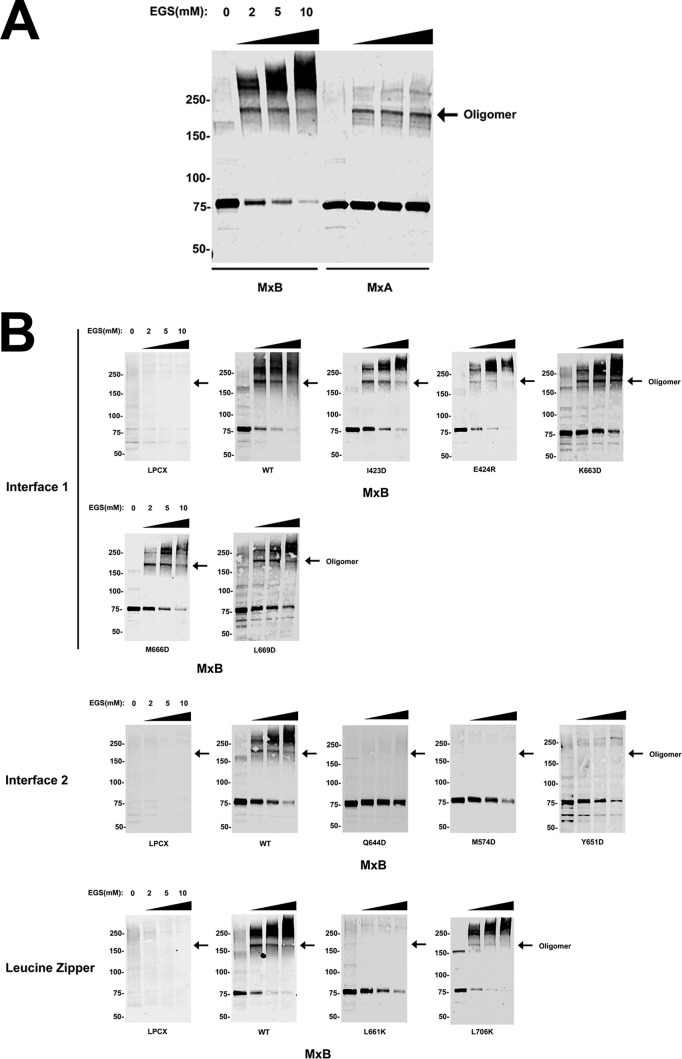
To confirm our oligomerization observations using a different approach, we expressed MxB variants in mammalian cells and performed chemical cross-linking experiments using ethylene glycol bis(succinimidyl succinate) (EGS), as described previously (31). As shown on Fig. 3A, the use of different concentrations of EGS on MxB or MxA resulted in cross-linking of the proteins. In the case of MxA, a fraction of the protein formed oligomers of ∼150 to 250 kDa by chemical cross-linking, as it has been previously shown (29). In addition to forming similar oligomers of ∼150 to 250 kDa, MxB also assembled into complexes that migrated above 250 kDa, which suggests that MxB forms higher-order structures (Fig. 3A). Next, we tested the ability of the different MxB variants to oligomerize by EGS cross-linking. In agreement with the results of our immunoprecipitation experiments, all MxB variants with mutations in interface 1 showed the ability to oligomerize (Fig. 3B). Similarly, all MxB variants with mutations in interface 2 lost their ability to oligomerize (Fig. 3B). We also confirmed by chemical cross-linking the results of the immunoprecipitation experiments performed on the MxB variants with mutations in the putative leucine zipper domain (Fig. 3B).
FIG 3.
Oligomerization of MxB variants by chemical cross-linking. (A) Human 293T cells were transfected with plasmids expressing FLAG-tagged human MxB or MxA. Cells were lysed at 24 h posttransfection. Lysates were incubated with EGS at the indicated concentrations and analyzed by Western blotting using anti-FLAG antibodies. (B) Similarly, the oligomerization ability of FLAG-tagged MxB variants with mutations in interface 1, interface 2, and the leucine zipper was measured by cross-linking experiments. Arrows, the oligomeric form of the indicated protein; WT, wild type. Numbers on the left are molecular masses (in kilodaltons).
Overall, these experiments demonstrated that an intact interface 2 is required for MxB oligomerization, which has also been shown for MxA (11). Although residue L661 is not in close proximity to interface 2, its disruption resulted in the loss of oligomerization, suggesting that this amino acid might be stabilizing a tertiary structure in the MxB monomer which is required for oligomerization.
Contribution of oligomerization to the ability of MxB to bind HIV-1 capsid.
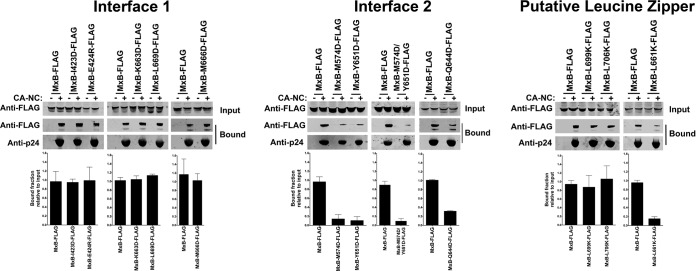
We have previously demonstrated that HIV-1 restriction by MxB requires the ability of MxB to bind to the HIV-1 capsid (23). To understand the role of oligomerization in the ability of MxB to bind capsid, we tested the ability of interface 1, interface 2, and putative leucine zipper MxB variants to bind in vitro-assembled HIV-1 CA-NC complexes (Fig. 4 and Table 1) as described previously (26). MxB variants with mutations in interface 1 bound in vitro-assembled HIV-1 CA-NC complexes as strongly as the wild-type MxB (Fig. 4 and Table 1). In contrast, MxB variants with mutations in interface 2 lost the ability to bind in vitro-assembled HIV-1 CA-NC complexes (Fig. 4 and Table 1). These results are in agreement with our hypothesis that MxB binding to the HIV-1 capsid requires oligomerization of MxB. In addition, we tested the ability of some MxB variants with mutations in the putative leucine zipper domain of MxB to bind capsid and showed that only the MxB L661K variant lost the ability to bind capsid (Fig. 4 and Table 1), and it also lost the ability to oligomerize (Fig. 2 and 3). These experiments showed that MxB variants that lost the oligomerization ability bound poorly to the HIV-1 capsid, suggesting that oligomerization is providing the avidity for MxB to interact with the HIV-1 capsid. Alternatively, oligomerization of MxB might be also creating the binding site that interacts with the capsid. Overall, our results suggest that oligomerization is important for the ability of MxB to bind to the HIV-1 capsid.
FIG 4.
Contribution of interface 1, interface 2, and putative leucine zipper residues to the ability of MxB to bind to in vitro-assembled HIV-1 CA-NC complexes. The ability of MxB variants to bind in vitro-assembled HIV-1 CA-NC complexes was measured. 293T cells were transfected with plasmids expressing FLAG-tagged wild-type or mutant MxB proteins. At 24 h posttransfection, the cells were lysed. Subsequently, the lysates were incubated at room temperature for 1 h with in vitro-assembled HIV-1 CA-NC complexes. The mixtures were applied to a 70% (wt/vol) sucrose cushion and centrifuged. Input, the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were analyzed by Western blotting using anti-FLAG antibodies. The pellet from the 70% sucrose cushion (Bound) was analyzed by Western blotting using anti-FLAG or anti-p24 antibodies. Similar results were obtained in three independent experiments, and the standard deviation for the bound fraction relative to the results for the input is shown.
Contribution of oligomerization to the ability of MxB to block HIV-1 infection.
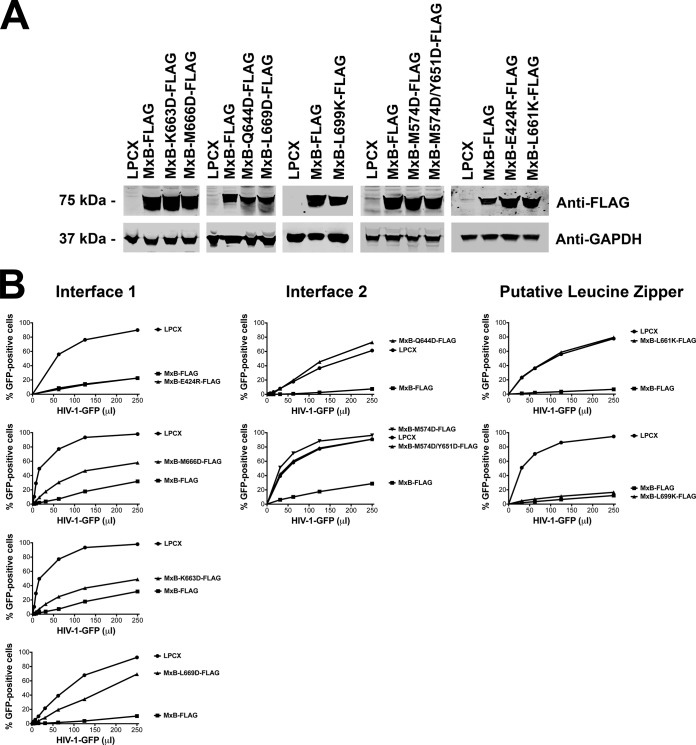
To understand the contribution of oligomerization and capsid binding to the ability of MxB to block HIV-1 infection, we tested the ability of MxB variants that lost oligomerization and capsid-binding abilities to block HIV-1 infection. To this end, we stably expressed MxB variants in Cf2Th cells using the pLPCX vector system (Fig. 5A). We show only the results for cell lines where the expression of the mutant was equal to or greater than that of wild-type MxB since our preliminary observations indicated that the restriction of MxB is sensitive to expression. As shown in Fig. 5B, MxB variants that lost oligomerization and capsid-binding abilities also lost the ability to block HIV-1 infection. On the contrary, MxB variants that did not disrupt oligomerization and capsid binding were not affected in their ability to block HIV-1 infection. As previously shown, the MxB L661K variant also lost its ability to oligomerize, bind capsid, and block HIV-1 infection (23). The results of these experiments suggest that oligomerization is necessary for the ability of MxB to block HIV-1 infection.
FIG 5.
Residues on interface 2 are important for HIV-1 restriction by MxB. (A) Wild-type and mutant MxB proteins were stably expressed in Cf2Th cells. Levels of expression in whole-cell extracts were measured by Western blotting using anti-FLAG antibodies. The protein load was assayed by Western blotting using anti-GAPDH (anti- glyceraldehyde-3-phosphate dehydrogenase) antibodies. (B) Cf2Th cells stably expressing wild-type and mutant MxB proteins were challenged with increasing amounts of HIV-1–GFP. At 48 h postinfection, the percentage of GFP-positive cells was measured by flow cytometry. As a control, Cf2Th cells stably transduced with the empty vector LPCX were challenged with HIV-1. Similar results were obtained in three independent experiments, and representative results are shown.
DISCUSSION
The ability of restriction factors to interact with the HIV-1 core requires oligomerization. For example, oligomerization is essential for the ability of rhesus monkey TRIM5α proteins to bind to the HIV-1 core. In the case of TRIM5α proteins, oligomerization provides the necessary avidity for the SPRY domain to interact with capsid (32–36). In agreement with this, oligomerization of the restriction factor MxA is important for its ability to block viral infection (37). Because we have previously demonstrated that MxB binds to the HIV-1 core (23), this work analyzed the contribution of oligomerization to the ability of MxB to bind to the HIV-1 capsid and restrict HIV-1 infection.
This work took advantage of the results of structure-function studies performed on the MxB-related protein MxA (11, 37). To study the oligomerization determinants of MxB, we modeled the primary sequence of MxB into the previously solved structure of MxA (37) and used this structure to guide our structure-function studies. This work allowed the discovery of MxB variants that lose their ability to oligomerize, as measured by immunoprecipitation and chemical cross-linking (MxB variants with mutations in interface 2; Fig. 1). Interestingly, equivalent mutations in MxA disrupted tetramerization, resulting in the predominant formation of monomers. In contrast, the use of equivalent mutations from MxA that disrupted tetramerization, resulting in the predominant formation of dimers, did not have an effect on the ability of MxB to oligomerize (MxB variants with mutations in interface 1; Fig. 1). The findings of these experiments pointed out the determinants used by MxB to oligomerize in human cells, as it has been previously shown for MxA (11).
The oligomerization ability of MxA is essential for it to work as an antiviral factor. For example, oligomerization permits the direct interaction of MxA with the hepatitis B virus core (17). Interestingly, our results suggested that oligomerization of MxB is required to bind to the HIV-1 capsid. This suggests the possibility that MxB might be forming an array of proteins on the surface of the HIV-1 core, as shown for rhesus monkey TRIM5α (38, 39). The possibility that MxB, like rhesus monkey TRIM5α, forms an array of proteins on the surface of the HIV-1 core is interesting, since the effect on uncoating by these two different restriction factors is opposite; MxB prevents uncoating, while rhesus monkey TRIM5α accelerates uncoating (23, 40, 41). Overall, our results are in agreement with a model in which oligomerization provides the avidity for MxB to interact with capsid. Alternatively, MxB oligomerization might be necessary for the formation of a capsid-binding site. Future investigations will determine whether MxB forms a protein array on the surface of the HIV-1 core.
While the manuscript was under review, Fribourgh et al. reported findings similar to ours (42): (i) the modeled MxB structure described here is very similar to the reported structure, and (ii) oligomerization of MxB is important for restriction.
ACKNOWLEDGMENTS
We thank the NIH/AIDS Repository Program for providing valuable reagents, such as antibodies and drugs.
NIH grants R01 AI087390, R21 AI102824, and R56 AI108432 to F.D.-G funded this work. C.B. acknowledges support from National Institutes of Health grant T32 AI07501.
REFERENCES
- 1.Mitchell PS, Emerman M, Malik HS. 2013. An evolutionary perspective on the broad antiviral specificity of MxA. Curr Opin Microbiol 16:493–499. doi: 10.1016/j.mib.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haller O, Kochs G. 2011. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res 31:79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 3.Melén K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I. 1996. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J Biol Chem 271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- 4.Aebi M, Fäh J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol 9:5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haller O. 2013. Dynamins are forever: MxB inhibits HIV-1. Cell Host Microbe 14:371–373. doi: 10.1016/j.chom.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. 2010. Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J Biol Chem 285:28419–28424. doi: 10.1074/jbc.R110.145839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faelber K, Gao S, Held M, Posor Y, Haucke V, Noé F, Daumke O. 2013. Oligomerization of dynamin superfamily proteins in health and disease. Prog Mol Biol Transl Sci 117:411–443. doi: 10.1016/B978-0-12-386931-9.00015-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. 2013. The interferon-inducible MxB protein inhibits HIV-1 Infection. Cell Host Microbe 14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Morlot S, Roux A. 2013. Mechanics of dynamin-mediated membrane fission. Annu Rev Biophys 42:629–649. doi: 10.1146/annurev-biophys-050511-102247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNew JA, Sondermann H, Lee T, Stern M, Brandizzi F. 2013. GTP-dependent membrane fusion. Annu Rev Cell Dev Biol 29:529–550. doi: 10.1146/annurev-cellbio-101512-122328. [DOI] [PubMed] [Google Scholar]
- 11.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. 2010. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 12.Pavlovic J, Zürcher T, Haller O, Staeheli P. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64:3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao H, Killip MJ, Staeheli P, Randall RE, Jackson D. 2013. The human interferon-induced MxA protein inhibits early stages of influenza A virus infection by retaining the incoming viral genome in the cytoplasm. J Virol 87:13053–13058. doi: 10.1128/JVI.02220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matzinger SR, Carroll TD, Dutra JC, Ma Z-M, Miller CJ. 2013. Myxovirus resistance gene A (MxA) expression suppresses influenza A virus replication in alpha interferon-treated primate cells. J Virol 87:1150–1158. doi: 10.1128/JVI.02271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J Virol 69:3904–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Netherton CL, Simpson J, Haller O, Wileman TE, Takamatsu H-H, Monaghan P, Taylor G. 2009. Inhibition of a large double-stranded DNA virus by MxA protein. J Virol 83:2310–2320. doi: 10.1128/JVI.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li N, Zhang L, Chen L, Feng W, Xu Y, Chen F, Liu X, Chen Z, Liu W. 2012. MxA inhibits hepatitis B virus replication by interaction with hepatitis B core antigen. Hepatology 56:803–811. doi: 10.1002/hep.25608. [DOI] [PubMed] [Google Scholar]
- 18.Kochs G, Janzen C, Hohenberg H, Haller O. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc Natl Acad Sci U S A 99:3153–3158. doi: 10.1073/pnas.052430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reichelt M, Stertz S, Krijnse-Locker J, Haller O, Kochs G. 2004. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic 5:772–784. doi: 10.1111/j.1600-0854.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- 20.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, Hué S, Barclay WS, Schulz R, Malim MH. 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nuñez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/PREACCEPT-6453674081373986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goujon C, Moncorgé O, Bauby H, Doyle T, Barclay WS, Malim MH. 2014. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J Virol 88:9017–9026. doi: 10.1128/JVI.01269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, Strouvelle VP, Zang TM, Willett BJ, Boutell C, Bieniasz PD, Wilson SJ. 2014. Host and viral determinants of Mx2 antiretroviral activity. J Virol 88:7738–7752. doi: 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Brandariz-Nuñez A, Fricke T, Ivanov DN, Sarnak Z, Diaz-Griffero F. 2014. Binding of the rhesus TRIM5α PRYSPRY domain to capsid is necessary but not sufficient for HIV-1 restriction. Virology 448:217–228. doi: 10.1016/j.virol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Griffero F, Perron M, McGee-Estrada K, Hanna R, Maillard PV, Trono D, Sodroski J. 2008. A human TRIM5α B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology 378:233–242. doi: 10.1016/j.virol.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melén K, Ronni T, Broni B, Krug RM, von Bonsdorff CH, Julkunen I. 1992. Interferon-induced Mx proteins form oligomers and contain a putative leucine zipper. J Biol Chem 267:25898–25907. [PubMed] [Google Scholar]
- 30.Fricke T, Brandariz-Nunez A, Wang X, Smith AB, Diaz-Griffero F. 2013. Human cytosolic extracts stabilize the HIV-1 core. J Virol 87:10587–10597. doi: 10.1128/JVI.01705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz-Griffero F, Kar A, Perron M, Xiang S-H, Javanbakht H, Li X, Sodroski J. 2007. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol 81:10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. 2008. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5 protein. J Virol 82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. 2008. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol 82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. 2006. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Biris N, Tomashevski A, Bhattacharya A, Diaz-Griffero F, Ivanov DN. 2013. Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core. J Mol Biol 425:5032–5044. doi: 10.1016/j.jmb.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Griffero F. 2011. Caging the beast: TRIM5α binding to the HIV-1 core. Viruses 3:423–428. doi: 10.3390/v3050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao S, von der Malsburg A, Dick A, Faelber K, Schröder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein A reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. 2009. A B-box 2 surface patch important for TRIM5 self-association, capsid binding avidity, and retrovirus restriction. J Virol 83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci U S A 108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J. 2007. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology 369:400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fribourgh JL, Nguyen HC, Matreyek KA, Alvarez FJD, Summers BJ, Dewdney TG, Aiken C, Zhang P, Engelman A, Xiong Y. 2014. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16:627–638. doi: 10.1016/j.chom.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]