Abstract
Herpes simplex virus 1 (HSV-1) genomes are associated with the repressive heterochromatic marks H3K9me2/me3 and H3K27me3 during latency. Previous studies have demonstrated that inhibitors of H3K9me2/me3 histone demethylases reduce the ability of HSV-1 to reactivate from latency. Here we demonstrate that GSK-J4, a specific inhibitor of the H3K27me3 histone demethylases UTX and JMJD3, inhibits HSV-1 reactivation from sensory neurons in vitro. These results indicate that removal of the H3K27me3 mark plays a key role in HSV-1 reactivation.
TEXT
Histone posttranslational modifications, both euchromatic and heterochromatic, are found in association with herpes simplex virus 1 (HSV-1) genes as the genome associates with nucleosomes during latency. The presence of permissive euchromatic marks (H3KAc, H9KAc, and H18K18Ac) in association with the latency-associated transcript (LAT) promoter and 5′ exon during latency is consistent with its transcriptional activity during latency (1–4). Intrinsic to the latent state, however, are several repressive epigenetic marks that are also found in association with lytic genes of the latent HSV-1 genomes, notably, H3K9me2/me3 and H3K27me3, enriched along lytic genes. These marks have been implicated in helping to maintain lytic genes in a repressed state (3, 5, 6).
Presumably, to reactivate from latency, the H3K9me2/me3 and H3K27me3 repressive marks need to be removed by chromatin remodeling enzymes. H3K27me3 is reversibly removed through the action of specific histone demethylases KDM6B/JMJD3 and KDM6A/UTX (7). H3K9me3 can be removed by all members of the JMJD2 family (8), while H3K9me2/me3/1 can be removed by LSD1, KIAA1718, JHDM1F, and JMJD1A (9–12). In this study, we sought to determine whether blocking the removal of H3K27me3 would also block HSV-1 reactivation. This issue was addressed using GSK-J4, a specific inhibitor of both JMJD3 and UTX (13).
Inhibition of JMJD3 and UTX blocks reactivation-induced H3K27me3 demethylation of HSV genomes.
To understand the roles of UTX and JMJD3 in viral reactivation, we used an in vitro model of HSV-1 latency in primary adult murine trigeminal ganglion (TG) neurons (14), where reactivation is stimulated by nerve growth factor (NGF) depletion (15). TGs from 6-week-old outbred Swiss Webster mice were incubated in papain (Worthington, Lakewood, NJ) reconstituted with neurobasal A medium (Gibco; catalog no. 10888-022) followed by Hanks balanced salt solution containing dispase (4 mg/ml) and collagenase (4.7 mg/ml) (Sigma). Mechanically dissociated TGs were purified on an iodixinol gradient (OptiPrep; Sigma). Neuronal cells were counted and plated onto plates precoated with poly–d-lysine and laminin. Cells were maintained in neuralbasal media containing NGF, glial cell line-derived neurotrophic factor, neutrinin, l-glutamine, 2% (vol/vol) B-27 supplement minus AO (Gibco; catalog no. 10889-038), and 1% (vol/vol) penicillin-streptomycin. To suppress actively dividing nonneuronal cells, the media were supplemented with fluorodeoxyuridine for 72 h prior to infection with virus. Three-day-old TG cultures were infected with HSV-1 17syn+ at a multiplicity of infection of 10. The media were replaced after 1 h and cultures maintained for 7 days. Latency was verified by the presence of the LAT and the lack of detectable infectious virus.
It has been demonstrated that the disruption of NGF signaling can induce HSV-1 reactivation and that addition of anti-NGF antibody to explants or cultures can produce robust reactivation (15–17). Therefore, in order to determine the ability of GSK-J4 to block H3K27me3 following NGF-induced reactivation, GSK-J4 (Xcess Biosciences Inc.) was added to latently infected TG neurons, and, 24 h following treatment with the inhibitor, anti-NGF antibody (Abcam; catalog no. ab6198) was added at a working concentration of 2 μg/ml to induce HSV-1 reactivation. Twenty-four hours following α-NGF induction, HSV-1 genomes were analyzed for H3K27me3 by chromatin immunoprecipitation (ChIP). ChIP assays were performed as previously described (1, 3) using anti-H3K27me3 antibody (Millipore; catalog no. 07-449).
Real-time PCRs were performed using Applied Biosystems TaqMan Fast Universal PCR master mix (catalog no. 4352042; Life Technologies, Grand Island, NY, USA) and target-specific primers for immediate-early (ICP4), early (thymidine kinase [TK]), and late (UL20) HSV-1 lytic genes (Table 1). In addition, TaqMan forward and reverse primers and probes were designed for an internal negative control and positive control corresponding to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and Hox A5 cellular genes, respectively (Table 1). For analysis of mRNA transcripts, samples were normalized to the mouse 18S rRNA gene.
TABLE 1.
Real-time PCR primer-probe sets
| Gene target | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| UL20 | CCATCGTCGGCTACTACGTTAC | CGATCCCTCTTGATGTTAACGTACA | CCCGCACCGCCCAC |
| TK | CACGCTACTGCGGGTTTATATAGAC | GGCTCGGGTACGTAGACGATAT | CACCACGCAACTGC |
| Hox A5, upstream | AGCAGCAGGGCCAATTCT | GCTGCCCAAGCCAGCTT | CCCGCGATGCACCC |
| ICP4 CDS | CACGGGCCGCTTCAC | GCGATAGCGCGCGTAGA | CCGACGCGACCTCC |
H3K27me3 ChIP analysis revealed that 50 μM GSK-J4 inhibited the decrease in H3K27me3 that occurred following anti-NGF-induced reactivation in the vehicle-treated group in latently infected neurons (Fig. 1A). In addition, transcripts representing the three classes of HSV-1 lytic genes were all significantly reduced in the presence of GSK-J4 relative to the vehicle-treated control results (Fig. 1B). There was no significant change in the levels of H3K27me3 on both cellular controls examined in cells treated with GSK-J4 compared to vehicle (Fig. 1C). This suggests that the effect seen in Fig. 1A where GSK-J4 treatment caused an increased enrichment of H3K27me on the HSV-1 genomes occurred by virtue of the fact that it blocked the demethylation of H3K27me3 on the reactivating genomes. As shown by examination of 18S RNA levels, there was a significant decrease observed in GSK-J4-treated samples compared to the vehicle-treated sample (Fig. 1D), indicating that GSK-J4 treatment could have been affecting transcription of at least some cellular genes as well. Taken together, these results suggest that during induced reactivation, JMJD3 and UTX actively reduce levels of H3K27me3 demethylation, thereby allowing viral transcription.
FIG 1.
JMJD3/UTX-specific inhibitor GSK-J4 inhibits HSV-1 replication in reactivated TG neurons. (A) Enrichment of H3K27me3 via ChIP analysis of reactivated TG neurons treated with GSK-J4 for HSV-1 genes ICP4, TK, and UL20. Relative quantities of enrichment are represented as bound/bound + unbound. (B) HSV-1 ICP4, TK, and UL20 mRNA levels. Error bars are ± standard deviations (SD). P values were determined using Student's t test (*, P = 0.003; **, P = 0.00001; ***, P < 0.000001). (C and D) Cellular controls for H3K27me3 ChIP (C) and mRNA 18S (D) assays of latently infected TGs stimulated with NGF antibody in the presence of GSK-J4. *, P value < 0.06.
GSK-J4 treatment reduces the production of infectious virus following α-NGF-induced reactivation.
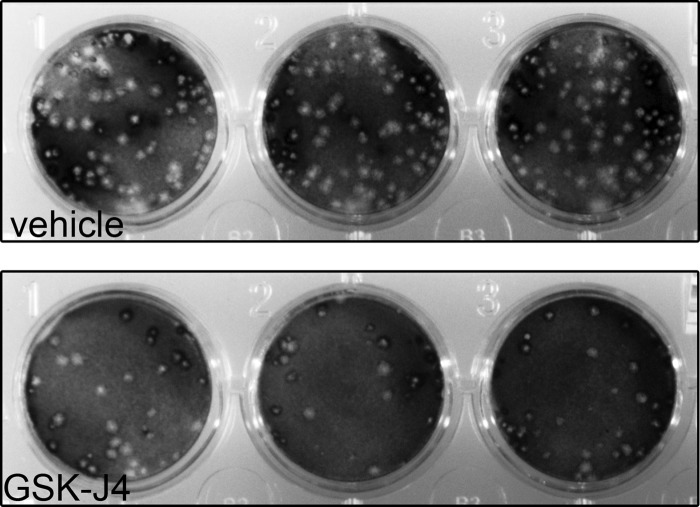
To determine if the ability of GSK-J4 to block JMJD3 and UTX and maintain viral gene repression translated to a block in productive reactivation, we quantified infectious virus particles produced following induced reactivation. Latently infected TG neurons were analyzed 24 h following anti-NGF treatment in the presence or absence of GSK-J4. This analysis indicated that GSK-J4 treatment resulted in a greater than 5-fold reduction in viral yield during reactivation (Fig. 2 and Table 2).
FIG 2.
Plaque assay of infectious HSV-1 particles reactivated from latently infected TG neurons in the presence of JMJD3/UTX-selective inhibitor GSK-J4.
TABLE 2.
Summary of reactivated HSV-1 infectious particles following treatment of latently infected neurons in the presence of JMJD3/UTX-selective inhibitor GSK-J4
| Biological replicate | Vehicle (PFU/ml) | GSK-J4 (PFU/ml)a |
|---|---|---|
| 1 | 517 | 58 |
| 2 | 210 | 82 |
| 3 | 403 | 67 |
| Avg | 377 | 69* |
*, the P value determined with a Student's t test using a two-tailed distribution of vehicle to GSK-J4 is 0.08.
Profiles of HSV-1 epigenomes in latently infected neurons demonstrate the existence of both constitutive and facultative heterochromatic marks (18, 19). It has been demonstrated that H3K9me2/3 demethylases (JMJD2s) and H3K9me1/2 demethylase LSDI reduce HSV-1 reactivation both in vitro and in vivo (20–22). It is difficult to state why inhibitors of the H3K9me2/me3 demethylases did not completely inhibit reactivation fully given issues with penetrance in the tissue and the experimental half-life of the drug. Because it is known that at least as large a proportion of the latent genomes is associated with the H3K27me3-repressive mark, this left open the issue of whether inhibitors of H3K27me3 could also inhibit reactivation by blocking reactivation from HSV-1 genomes that were repressed by this other heterochromatic mark. In summary, the observations presented here indicate that removal of the H3K27me3 mark is required for efficient reactivation of HSV from latency. These results provide new insights into the regulation of the HSV-1 epigenome in latently infected neurons undergoing reactivation and suggest that separate but parallel pathways to reactivation exist based on the need to remove both the H3K9me2/me3 and H3K27me3 heterochromatin marks. Finally, these results suggest that small-molecule inhibition of UTX and JMJD3 histone H3K27me3 demethylases could be a promising strategy for therapeutic intervention for recurrent HSV disease.
ACKNOWLEDGMENT
This work was supported by NIH grant AI48633 (to D.C.B.).
REFERENCES
- 1.Amelio AL, Giordani NV, Kubat NJ, O'Neil JE, Bloom DC. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol 80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubat NJ, Tran RK, McAnany P, Bloom DC. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol 78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cliffe AR, Coen DM, Knipe DM. 2013. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, Nozaki M, Kanai Y, Shinkai Y, Tachibana M. 2013. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 11.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439. [DOI] [PubMed] [Google Scholar]
- 12.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertke AS, Swanson SM, Chen J, Imai Y, Kinchington PR, Margolis TP. 2011. A5-positive primary sensory neurons are nonpermissive for productive infection with herpes simplex virus 1 in vitro. J Virol 85:6669–6677. doi: 10.1128/JVI.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilcox CL, Johnson EM. 1988. Characterization of nerve growth factor-dependent herpes simplex latency in neurons in vitro. J Virol 62:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilcox CL, Smith RL, Freed CR, Johnson EM Jr. 1990. Nerve growth factor-dependence of herpes simplex virus latency in peripheral sympathetic and sensory neurons in vitro. J Neurosci 10:1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 20.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4:e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra165. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]