ABSTRACT
Assembly-activating protein (AAP) of adeno-associated virus serotype 2 (AAV2) is a nucleolar-localizing protein that plays a critical role in transporting the viral capsid VP3 protein to the nucleolus for assembly. Here, we identify and characterize AAV2 AAP (AAP2) nuclear (NLS) and nucleolar (NoLS) localization signals near the carboxy-terminal region of AAP2 (amino acid positions 144 to 184) (AAP2144–184). This region contains five basic-amino-acid-rich (BR) clusters, KSKRSRR (AAP2BR1), RRR (AAP2BR2), RFR (AAP2BR3), RSTSSR (AAP2BR4), and RRIK (AAP2BR5), from the amino terminus to the carboxy terminus. We created 30 AAP2BR mutants by arginine/lysine-to-alanine mutagenesis or deletion of AAP2BRs and 8 and 1 green fluorescent protein (GFP)-AAP2BR and β-galactosidase–AAP2BR fusion proteins, respectively, and analyzed their intracellular localization in HeLa cells by immunofluorescence microscopy. The results showed that AAP2144–184 has redundant multipartite NLSs and that any combinations of 4 AAP2BRs, but not 3 or less, can constitute a functional NLS-NoLS; AAP2BR1 and AAP2BR2 play the most influential role for nuclear localization, but either one of the two AAP2BRs is dispensable if all 4 of the other AAP2BRs are present, resulting in 3 different, overlapping NLS motifs; and the NoLS is shared redundantly among the five AAP2BRs and functions in a context-dependent manner. AAP2BR mutations not only resulted in aberrant intracellular localization, but also attenuated AAP2 protein expression to various degrees, and both of these abnormalities have a significant negative impact on capsid production. Thus, this study reveals the organization of the intermingling NLSs and NoLSs in AAP2 and provides insights into their functional roles in capsid assembly.
IMPORTANCE Adeno-associated virus (AAV) has become a popular and successful vector for in vivo gene therapy; however, its biology has yet to be fully understood. In this regard, the recent discovery of the assembly-activating protein (AAP), a nonstructural, nucleolar-localizing AAV protein essential for viral capsid assembly, has provided us a new opportunity to better understand the fundamental processes required for virion formation. Here, we identify clusters of basic amino acids in the carboxy terminus of AAP from AAV serotype 2 (AAV2) that act as nuclear and nucleolar localization signals. We also demonstrate their importance in maintaining AAP expression levels and efficient production of viral capsids. Insights into the functions of AAP can elucidate the requirements and process for AAV capsid assembly, which may lead to improved vector production for use in gene therapy. This study also contributes to the growing body of work on nuclear and nucleolar localization signals.
INTRODUCTION
Adeno-associated virus (AAV) is a small, single-stranded DNA virus from the parvovirus family that has become a successful vector for gene delivery. The recent achievements in the field of AAV vector research have called attention to the incompletely understood life cycle of the virus. The AAV genome comprises two genes, rep and cap, which encode the nonstructural Rep proteins and the structural VP proteins, respectively. The AAV virion is composed of 60 subunits comprising the three VP proteins, VP1, VP2, and VP3, encoded by a single open reading frame (ORF) in the cap gene. Recently, a nonstructural viral protein encoded by an alternative ORF within the cap gene was identified and termed assembly-activating protein (AAP) for its indispensable role in capsid formation (1–3). The AAP (AAP2) from AAV serotype 2 (AAV2) is a nucleolar-localizing protein that binds to VP proteins through interacting domains in the amino (N) terminus of AAP2 (1), transports the VP proteins to the nucleolus, and promotes capsid assembly (3). Therefore, AAP2 is expected to have both a nuclear localization signal (NLS) and a nucleolar localization signal (NoLS) within its protein sequence. However, such organelle-targeting sequences in AAP2 remain to be identified and characterized.
The most common mechanism for targeting a protein to the nucleus is by an NLS that is recognized by one of the nuclear import proteins, termed importins, which are part of the large family of transport proteins known as karyopherins (4). Classical NLSs can be either monopartite, such as the PKKKRKV sequence in simian virus 40 (SV40) large T antigen (5), or bipartite, such as the KRPAATKKAGQAKKKK sequence in nucleophosmin (6, 7). These classical NLSs are bound by the adaptor protein importin-α, which is then bound by importin-β, forming a heterotrimeric complex consisting of the two importin proteins and the cargo protein. Importin-β mediates nuclear entry of the heterotrimer through the nuclear pores by its increasing affinity for nucleoporins along the inside of the nuclear pore complex (8). If the cargo protein also contains an NoLS, it can then be targeted to the nucleolus through charge-based interactions (9) or interactions with nucleolar proteins (10, 11), although the specific requirements defining nucleolar localization are not as well understood as those for nuclear import.
As AAP2 is able to localize to the nucleolus (3), we hypothesized that it would contain both an NLS and an NoLS responsible for this intracellular localization and that these signals would be critical to its function in capsid assembly. Because a protein region rich in basic amino acid residues is a hallmark of NLSs and NoLSs, we tested our hypothesis on the carboxy (C)-terminal region of AAP2, amino acid positions 144 to 184 (AAP2144–184), where there are five basic-amino-acid-rich (BR) clusters. By fusing green fluorescent protein (GFP) or the β-galactosidase protein with an AAP2 protein segment of interest and by creating a series of arginine/lysine-to-alanine mutations or deletions in AAP2144–184, we were able to identify NLSs and NoLSs and elucidate their redundant and overlapping nature. Mutations in this NLS- or NoLS-containing region resulted in not only aberrant intracellular localization, but also substantial reduction in AAP2 expression and capsid production, showing the multifaceted functional importance of the NLS-NoLS in AAP2.
MATERIALS AND METHODS
Plasmid construction.
pCMV-FLAG-cmAAP2 is a plasmid expressing a codon-modified (cm) version of the wild-type AAP2 with an N-terminal FLAG tag under the control of the human cytomegalovirus (CMV) immediate-early gene enhancer/promoter (12). This FLAG-tagged AAP2 is translated from the ATG start codon. The codon-modified AAP2 ORF was utilized to maximize expression in human cells and to prevent recombination between the AAV2-RepVP3 viral genome (12) and AAP2 plasmid DNA through the homologous sequence during AAV production in human embryonic kidney (HEK 293) cells. pCMV-FLAG-cmAAP2 mutant plasmids were created by site-directed mutagenesis. pCMV-GFP is an enhanced GFP (eGFP)-expressing plasmid under the control of the same CMV enhancer/promoter as that in pCMV-FLAG-cmAAP2 and was used to construct plasmids expressing GFP fused with the AAP2144–184 peptide at its C terminus. pAAV2-RepVP3 is a plasmid that expresses all the Rep proteins and the VP3 protein from AAV2 but does not express VP1, VP2, or AAP2 (12). An adenovirus helper plasmid, pHelper, was purchased from Agilent. pCMV-AAV2VP3 is a plasmid expressing the AAV2 VP3 protein under the same CMV enhancer/promoter (12). pAAV-CMV-lacZ is a plasmid containing an alcohol dehydrogenase (Adh)-lacZ fusion transgene under the control of the CMV enhancer/promoter (13). This plasmid expresses cytosolic β-galactosidase in cells transfected with the plasmid. To construct the pAAV-CMV-lacZ-AAP2144–184 plasmid expressing β-galactosidase fused with the AAP2144–184 peptide, the peptide-coding nucleotide sequence was introduced at the N terminus between the start and second codons of the Adh-lacZ gene ORF in the plasmid pAAV-CMV-lacZ. pCMV-FLAG-cmAAP2-GFP is a plasmid expressing the wild-type full-length AAP2 fused with a FLAG tag and GFP at the N terminus and C terminus, respectively. This protein configuration allows simultaneous detection of the wild-type AAP2 in cells using three different approaches: anti-FLAG antibody immunostaining, direct detection of GFP fluorescence, and anti-GFP antibody immunostaining.
Cells.
HEK 293 cells (AAV293) were purchased from Stratagene. The HeLa human cervical cancer cell line was obtained from the American Type Culture Collection (ATCC). HEK 293 cells and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS), l-glutamine, and penicillin-streptomycin.
Immunofluorescence microscopy and data analysis.
HeLa cells were seeded on coverslips in 12-well plates and transfected with plasmid DNA using polyethyleneimine (PEI). Forty-eight hours after transfection, the cells were fixed with 4% paraformaldehyde at room temperature, permeabilized with 0.2% Tween 20, blocked with 8% bovine serum albumin (BSA), and stained with mouse monoclonal anti-FLAG M2 antibody (F1804; Sigma-Aldrich, St. Louis, MO) and rabbit polyclonal anti-nucleostemin antibody (AB5689 [Millipore, Billerica, MA] or sc-67012 [Santa Cruz Biotechnology, Dallas, TX]), followed by DAPI (4′,6-diamidino-2-phenylindole), Alexa Fluor 488-AffiniPure goat anti-mouse IgG antibody (115-545-166; Jackson ImmunoResearch, West Grove, PA), and Cy3-AffiniPure goat anti-rabbit IgG antibody (111-165-144; Jackson ImmunoResearch). For imaging of AAP2 and VP proteins together, the cells were stained with rat monoclonal anti-DYKDDDDK (FLAG) antibody (NBP1-06712; Novus Biological, Littleton, CO), mouse monoclonal anti-AAV VP1/VP2/VP3 antibody (B1) (03-61058; American Research Products, Inc., Waltham, MA), and rabbit polyclonal anti-nucleostemin antibody (sc-67012; Santa Cruz Biotechnology), followed by DAPI, Alexa Fluor 488-AffiniPure goat anti-mouse IgG antibody (115-545-166; Jackson ImmunoResearch), Cy3-AffiniPure donkey anti-rat IgG antibody (712-165-153; Jackson ImmunoResearch), and Alexa Fluor 647-AffiniPure goat anti-rabbit IgG antibody (111-605-144; Jackson ImmunoResearch). For a subcellular-localization analysis of β-galactosidase, the cells were treated in the same manner as for the double immunostaining of AAP2 and nucleostemin described above, except that mouse monoclonal anti-β-galactosidase antibody (B0271; Sigma-Aldrich) was used in place of mouse monoclonal anti-FLAG M2 antibody. For antigen retrieval by protease treatment, transfected HeLa cells were fixed with 4% paraformaldehyde at 48 h posttransfection, permeabilized with 0.2% Tween 20, incubated with trypsin (2.5 μg/ml) at 37°C for 10 min, treated with 1 mM phenylmethylsulfonyl fluoride (PMSF) at room temperature for 1 min, and immunostained using mouse monoclonal anti-eGFP antibody (F56-6A1.2.3; Thermo Scientific, Waltham, MA) and rat monoclonal anti-DYKDDDDK (FLAG) antibody (NBP1-06712; Novus Biological, Littleton, CO), followed by Alexa Fluor 647-AffiniPure donkey anti-mouse IgG antibody (A-31571; Invitrogen, Grand Island, NY) and Cy3-AffiniPure donkey anti-rat IgG antibody (712-165-153; Jackson ImmunoResearch). The cells were imaged on a Zeiss LMS 710 laser scanning confocal microscope. In this study, we defined NLS (+) proteins as those that showed exclusive nuclear accumulation, NoLS (+) proteins as those that showed obvious nucleolar enrichment, and nucleolar exclusion (Ex) proteins as those that showed clear exclusion from the nucleolus.
The functional role of each AAP2BR in nuclear and nucleolar localization was evaluated in the following manner. The following mutants harboring a pair of AAP2BR mutations showed impaired nuclear trafficking: AAP2mt12, -mt17, -mt21, -mt22, and -mt27. In these 5 mutants, when correction of one AAP2BR mutation back to the wild-type amino acid sequence resulted in an NLS (+) phenotype, we interpreted it as showing that the AAP2BR that was corrected has an NLS role. Likewise, we focused on the nucleolar-excluded mutants AAP2mt17, -mt22, and -mt27 harboring a pair of AAP2BR mutations to assess the NoLS role of each AAP2BR. In these 3 mutants, when correction of one AAP2BR mutation back to the wild-type amino acid sequence resulted in an NoLS (+) phenotype, we interpreted it as showing that the AAP2BR that was corrected has an NoLS role. The NLS roles of a given pair of AAP2BRs were also assessed statistically as described below.
X-Gal staining.
HEK 293 cells were seeded on a 6-well plate and transfected with 2 μg of either pAAV-CMV-lacZ or pAAV-CMV-lacZ-AAP2144–184 plasmid DNA. Twenty-four hours after transfection, the cells were fixed with 2% formaldehyde-0.2% glutaraldehyde in phosphate-buffered saline (PBS) for 5 min, washed with PBS, and stained with 5 mM FeK4(CN)6–5 mM FeK3(CN)6–2 mM MgCl2–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (1 mg/ml) in PBS at 37°C for 1 h.
AAV production and an ELISA specific for intact AAV2 particles.
AAV2 VP3-only viral particles were produced by transcomplementation, where VP3 and AAP2 (the wild type or mutant) were expressed in HEK 293 cells from two separate plasmids (12). In brief, HEK 293 cells were transfected with pAAV2-RepVP3, pCMV-FLAG-cmAAP2 (the wild type or mutant), and pHelper using PEI in 6-cm dishes. Forty-eight hours after transfection, the cells were washed in PBS and resuspended in 100 μl of HEPES-buffered saline (pH 7.4). The cells were lysed by three freeze-thaw cycles using a dry-ice–ethanol bath, and the supernatants were collected. The resulting cell lysates containing viral particles were then subjected to an A20 antibody-based intact AAV2 capsid-specific enzyme-linked immunosorbent assay (ELISA) using the AAV2 Titration ELISA Kit (Progen, Heidelberg, Germany) according to the manufacturer's instructions.
Western blot analysis.
HEK 293 cells were transfected with either the wild-type or mutant pCMV-FLAG-cmAAP2 plasmid DNA. Forty-eight hours posttransfection, the HEK 293 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Complete Mini; Roche, Indianapolis, IN). Protein concentrations in the cell lysates were determined with a DC Protein Assay Kit (Bio-Rad, Hercules, CA). The same amount of total cell lysates (60 or 80 μg per lane) was separated on an 8% SDS-PAGE gel, transferred onto a polyvinylidene difluoride (PVDF) membrane, and reacted with mouse monoclonal anti-FLAG M2 antibody and monoclonal mouse anti-α-tubulin antibody (sc-32293; Santa Cruz Biotechnology), followed by goat polyclonal anti-mouse IgG antibody (sc-2055; Santa Cruz Biotechnology) conjugated to horseradish peroxidase. The signals on the blots were visualized and quantified using the FluorChem M system (ProteinSimple, Santa Clara, CA). The data were collected from a biologically duplicated set of experiments.
Statistical analyses.
Differences in AAV particle production yields were statistically assessed by the two-tailed Welch t test. An unconditional exact test was used to statistically evaluate the association between the presence or absence of a given combination of 2 intact AAP2BRs and the presence or absence of an NLS using a 2-by-2 contingency table. For the purpose of this statistical analysis, the NLS (+) and NLS (−) mutants among AAP2mt10 to -mt27 were defined as those that showed exclusively nuclear accumulation and those that did not belong to the NLS (+) category, respectively. There were 9 NLS (+) and 9 NLS (−) AAP2BR mutants. The null hypothesis is that there is no association between AAV2BRs and NLSs. Since we analyzed only 18 combinations out of all 25 possible combinations of AAP2BR mutations that leave at least 2 intact AAP2BRs (10 combinations of 2 intact BRs with 3 mutated BRs, 10 combinations of 3 intact BRs with 2 mutated BRs, and 5 combinations of 4 intact BRs with 1 mutated BR), a range of P values are given in which all the possible outcomes from the 7 AAP2BR mutants that were not assessed are taken into account. The P values that would be obtained if all 25 AAP2BR mutants were analyzed are expressed as P25. The P values obtained from the 18 AAP2BR mutants are expressed as P18.
RESULTS
The C terminus of AAP2 contains both an NLS and an NoLS.
AAP2 has an amino acid stretch rich in basic amino acid residues, AAP2144–184, near the C terminus (Fig. 1). This region harbors a KRSR sequence that matches the classical monopartite NLS motif, K(K/R)X(K/R) (14), and a putative NoLS, RRIK (15, 16), and shares features characteristic of NoLSs, including a high proportion of basic amino acids and proximity to the C terminus (17). It is also possible that these two basic amino acid clusters might serve as a bipartite NLS with a long linker sequence (14, 18, 19). Therefore, we hypothesized that AAP2144–184 contains both an NLS and an NoLS. To test this hypothesis, we transfected HeLa cells with the pCMV-GFP-AAP2144–184 plasmid expressing the GFP fused with the AAP2144–184 peptide at its C terminus or with pCMV-GFP, the control parental GFP plasmid devoid of the AAP2144–184 peptide. Forty-eight hours posttransfection, the control GFP was observed diffusely throughout the cytoplasm and nucleoplasm, while the GFP-AAP2144–184 fusion protein was localized predominantly to the nucleolus (Fig. 2A and B). Since GFP can diffuse into the nucleus without an NLS due to its small size (27 kDa), we also investigated whether the AAP2144–184 peptide could target a large cytoplasmic protein, β-galactosidase (∼120 kDa), to the nucleolus. Both immunofluorescence microscopic and cytochemical analyses demonstrated strong nucleolar enrichment of the otherwise predominantly cytosolic β-galactosidase when the enzyme was fused with the AAP2144–184 peptide (Fig. 2C and D). Addition of the canonical NLS derived from SV40 large T antigen (PKKKRKV) at the N terminus of β-galactosidase was not sufficient for nucleolar targeting, showing a nucleolar-exclusion pattern, although the protein accumulated in the nucleus (data not shown). These observations strongly supported our hypothesis that the AAP2144–184 region contains both an NLS and an NoLS.
FIG 1.
Amino acid sequences of the wild type (wt) and mutants (mt) of AAP2 near the C terminus and their roles as an NLS and/or NoLS. The sequence of amino acid positions from 144 to 184 in the wild-type AAP2 is shown at the top, followed by the sequences of a total of 30 AAP2 mutants with arginine/lysine-to-alanine substitutions (mt1 to mt27) or deletions (mt28 to mt30). The 5 BR clusters are indicated by lines above the wild-type sequence. The red and light-blue letters show basic amino acids and alanine mutations, respectively. The blue underlines indicate deletions. The dots in the sequences indicate residues that are the same as those of the wild type. The presence of a fully functional NLS (N) and/or NoLS (No), determined by immunofluorescence microscopy (Fig. 3), is indicated on the right. A plus in the N column indicates that the protein is observed exclusively in the nucleus regardless of its subnuclear localization. A plus in the No column indicates that the protein is strongly associated with the nucleolus. The mutants that showed a nucleolar-exclusion pattern are indicated by Ex.
FIG 2.
Intracellular localization of GFP or bacterial β-galactosidase fused with the AAP2144–184 region at the C or N terminus, respectively. HeLa or HEK 293 cells were transiently transfected with plasmid pCMV-GFP-AAP2144–184 (A), pCMV-GFP (B), pAAV-CMV-lacZ-AAP2144–184 (C), or pAAV-CMV-lacZ (D), using PEI. pCMV-GFP-AAP2144–184 and pAAV-CMV-lacZ-AAP2144–184 are plasmids expressing the GFP-AAP2144–184 fusion protein and the β-galactosidase–AAP2144–184 fusion protein, respectively, under the control of the CMV promoter. For HeLa cells, the signals were detected by immunofluorescence microscopy using corresponding antibodies, except for GFP, for which direct fluorescence was imaged. For HEK 293 cells, X-Gal staining was performed. Scale bars, 20 μm.
FLAG-tagged wild-type AAP2 and nucleostemin colocalize in the nucleus and nuclear bodies.
It has been established that AU1-tagged AAP2 and fibrillarin, an endogenous nucleolar protein, colocalize in the nucleolus in HeLa cells (3). We first investigated whether the N-terminally FLAG-tagged AAP2 we used for this study also localizes in the nucleolus as expected. To this end, HeLa cells were transfected with pCMV-FLAG-AAP2 and immunostained with anti-FLAG and anti-nucleostemin antibodies. The FLAG-tagged wild-type AAP2 was found to localize exclusively to the nucleus with significant nucleolar enrichment (Fig. 3A). The AAP2 signals were also found tightly associated with nucleostemin-positive nuclear bodies (Fig. 3A). The AAP2 signals in the nucleolus detected by immunostaining were stronger in the periphery than in the center, exhibiting a ring-shaped pattern with a central hollow. As discussed below, this nucleolar staining pattern is most likely an artifact caused by overexpression of a nucleolar-localizing protein (20). After trypsin treatment of the fixed cells, FLAG staining was visible throughout the nucleolus (Fig. 4). This observation confirmed that the FLAG peptide fused with AAP2 at the N terminus does not interfere with its intracellular localization. The functional integrity of the FLAG-tagged AAP2 had been confirmed previously (12).
FIG 3.
Intracellular localization of various AAP2 mutants with arginine/lysine-to-alanine substitutions or deletions within the BR clusters, AAP2BR1 to AAP2BR5. HeLa cells were transiently transfected with a plasmid expressing the wild-type or a mutant AAP2 with an N-terminal FLAG tag. The cells were fixed 48 h posttransfection, immunostained with anti-FLAG antibody (green) and anti-nucleostemin antibody (red), and counterstained with DAPI (blue), before being imaged on a Zeiss LSM 710 confocal microscope with a 100× objective. Representative cell images are shown for the wild type and each mutant. (A) Wild-type AAP. (B) AAP2mt10, showing a staining pattern representing all the AAP2BR1 mutants, AAP2mt1 to -mt10 (the data for AAP2mt1 to -mt9 are not shown). (C to V) AAP2mt11 to -mt30. Formation of multiple nuclear bodies containing both AAP2 and nucleostemin was evident in many cells expressing the wild-type AAP2 (A) and the AAP2 mutants showing an intracellular localization pattern similar to that of the wild type (B, C, F, G, T, and U). The identity and the role of these speckled structures have yet to be determined. The 5-digit numbers in the bottom right corner of each FLAG-AAP2 panel indicate mutations introduced in each AAV2BR in each mutant (1, wild type; 0, alanine mutation; -, deletion). For example, 01110 (mt12) indicates BR1−/BR2+/BR3+/BR4+/BR5−. Scale bar, 20 μm.
FIG 4.
Immunofluorescence microscopic analysis of a FLAG-tagged AAP2 fused with GFP in HeLa cells with or without proteinase treatment. (A to C) HeLa cells were transiently transfected with pCMV-FLAG-cmAAP2-GFP expressing the wild-type full-length AAP2 fused with a FLAG tag and GFP. Intracellular localization of the fusion protein was analyzed with or without trypsin treatment by anti-FLAG antibody immunostaining (A), direct detection of GFP fluorescence (B), and anti-GFP antibody immunostaining (C) under a fluorescence microscope. (D) Merged images. Scale bar, 20 μm.
Basic amino acid clusters in the C terminus of AAP2 constitute redundant multipartite NLSs and NoLSs.
The C-terminal region AAP2144–184, where we found that the AAP2 NLS and NoLS reside, contains five BR clusters separated by 3 to 7 amino acids: KSKRSRR (AAP2BR1), RRR (AAP2BR2), RFR (AAP2BR3), RSTSSR (AAP2BR4), and RRIK (AAP2BR5), from the N terminus to the C terminus (Fig. 1). To dissect the functional roles of these basic amino acids in the nuclear and nucleolar localization of AAP2, we constructed plasmids expressing a series of mutant AAP2 proteins by site-directed mutagenesis using the pCMV-FLAG-cmAAP2 plasmid (Fig. 1). Each mutant AAP2 was expressed in HeLa cells by plasmid DNA transfection and analyzed for its intracellular localization by immunofluorescence microscopy (Fig. 3).
We first created AAP2 mutants 1 to 12 (Fig. 1, mt1 to mt12) to investigate the roles of AAP2BR1 and AAP2BR5, which carry the classical monopartite NLS motif, KRSR, and the putative NoLS motif, RRIK, respectively (basic amino acid residues in the motifs are underlined). Removal of positive charges from AAP2BR1 (AAP2mt1 to -mt10) or from AAP2BR5 (AAP2mt11) did not abolish the ability of mutant AAP2s to translocate to the nucleus with strong nucleolar enrichment and robust accumulation in the nucleostemin-positive nuclear bodies (Fig. 3B and C). Removal of positive charges from AAP2BR1 and AAP2BR5 in conjunction (AAP2mt12), however, resulted in substantial, if not complete, nuclear exclusion and nucleolar exclusion (Fig. 3D). This first set of experiments indicated that there are at least two NLSs that involve either AAP2BR1 or AAP2BR5 within AAP2144–184. We then created an additional set of 18 AAP2 mutants, 13 to 30 (Fig. 1, mt13 to mt30), to further investigate the role of each AAP2BR in nuclear and nucleolar localization. In these mutants, the positive charge in one or more AAP2BRs was diminished by alanine substitutions or deletions in various combinations. The 7 AAP2 mutants that had only 1 mutant AAP2BR and 4 intact AAP2BRs (mt10, mt11, mt13, mt14, mt15, mt28, and mt29) were all found exclusively in the nucleus with obviously enhanced association with the nucleolus (Fig. 3B, C, E to G, T, and U). In keeping with this observation, GFP-AAP2144–184 fusion mutants that had four intact AAP2BRs were observed exclusively in the nucleus, with strong enrichment in the nucleolus (Fig. 5A to C). All 13 mutants that had 3 or fewer intact AAP2BRs exhibited attenuated or complete loss of nucleolar association, and among them, 9 (mt16, mt17, mt20, mt21, mt22, mt24, mt25, mt27, and mt30) showed various degrees of impaired nuclear transport and cytoplasmic retention (Fig. 3D and H to V). AAP2 is a small protein that can passively cross the nuclear envelope through the nuclear pore complexes. The cytoplasmic signals that are stronger than the nuclear signals in AAP2mt12, -mt20, and -mt24, therefore, might indicate the presence of a nuclear export signal (NES) in AAP2. However, we have not observed any nuclear-cytoplasmic shuttling of the AAP2 protein during the course of AAV2 particle production in HEK 293 cells by plasmid transfection, where AAP2 consistently remains located in the nucleolus or nucleostemin-positive nuclear bodies (L. F. Earley and H. Nakai, unpublished observation). In addition, three different NES prediction programs (21–23) failed to identify any potential CRM1-binding NES in the AAP2 amino acid sequence. Thus, the above-mentioned observations are most likely due primarily to various degrees of impairment of nuclear import caused by different mutations, although the involvement of an unidentified NES cannot be totally ruled out. Taken together, these observations indicate that the NLS and NoLS in AAP2 are redundant and that various combinations of 4 AAP2BRs out of 5 AAP2BRs within the AAP2144–184 region are able to constitute a functional multipartite NLS-NoLS.
FIG 5.
Microscopic assessment of the abilities of the AAP2BRs to target GFP to the nucleus and the nucleolus. The method used was the same as that for Fig. 2. (A to F) GFP-AAP2144–184mt13 to -mt26 are GFPs fused with the 41-amino-acid-long AAP144–184 region containing their corresponding AAP2BR mutations. (G and H) GFP-AAP2144-150 and GFP-AAP2144-156 are GFPs fused with AAP2BR1 only (7 amino acids) and AAP2BR1-AAP2BR2 only (13 amino acids), respectively. Scale bar, 20 μm.
Copresence of AAP2BR1 and AAP2BR2 is an important but not absolute requirement for the AAP2 NLS.
The immunofluorescence microscopy analysis described above revealed that all 10 AAP2BR mutants that exhibited impaired nuclear translocation and various degrees of cytoplasmic retention had either or both AAP2BR1 and AAP2BR2 mutations, and all 10 combinatorial mutations involving AAP2BR1 or AAP2BR2 impaired nuclear transport of AAP2 to various degrees. A statistical comparison of various combinations of two intact AAP2BRs and the presence of a fully functional NLS in the AAP2 mutants revealed that the combination of AAP2BR1 and AAP2BR2 is strongly associated with nuclear localization (Table 1). The GFP fusion experiment also demonstrated that AAP2BR1 by itself is not sufficient for active nuclear transport and that copresence of AAP2BR1 and AAP2BR2 is required for nuclear accumulation (Fig. 5D to H). When we assessed the NLS role of each AAP2BR based on its ability to restore a fully functional NLS to AAP2 mutants (see Materials and Methods), we found that all 5 AAP2BRs can contribute to the facilitation of nuclear localization of AAP2. These observations indicate that, although all 5 AAP2BRs have an NLS role, the presence of AAP2BR1 and AAP2BR2 in conjunction plays the most influential role in nuclear targeting. Due to the redundant nature of the AAP2 NLS, either one of these two AAP2BRs is dispensable if all 4 of the other AAP2BRs are present, as evidenced by the nuclear localization of AAP2mt10 and -mt13.
TABLE 1.
Associations between various combinations of two AAP2BRs and the presence of an NLS in AAP2144–184
| AAP2BR | Association with [no./total (%)]a: |
|||
|---|---|---|---|---|
| AAP2BR1 | AAP2BR2 | AAP2BR3 | AAP2BR4 | |
| AAP2BR2 | 7/7 (100) (P18 = 0.002; P25 = 0.001–0.017) | |||
| AAP2BR3 | 4/7 (57) (P18 = 1.000; P25 = 0.186–1.000) | 4/5 (80)b (P18 = 0.211; P25 = 0.014–1.000) | ||
| AAP2BR4 | 4/7 (57) (P18 = 1.000; P25 = 0.186–1.000) | 4/5 (80)b (P18 = 0.211; P25 = 0.014–1.000) | 3/6 (50) (P18 = 1.000; P25 = 0.317–1.000) | |
| AAP2BR5 | 4/7 (57) (P18 = 1.000; P25 = 0.186–1.000) | 4/4 (100)b (P18 = 0.031; P25 = 0.002–1.000) | 3/5 (60) (P18 = 0.696; P25 = 0.113–1.000) | 3/5 (60) (P18 = 0.696; P25 = 0.113–1.000) |
Eighteen AAP2 mutants (AAP2mt10 to -mt27) were categorized into two groups according to the presence or absence of an NLS as defined in the legend to Fig. 1. There were 9 NLS (+) and 9 NLS (−) AAP2 mutants. An unconditional exact test was performed to statistically evaluate the association between the presence or absence of a given combination of 2 AAP2BRs and the presence or absence of an NLS. The null hypothesis is that there is no association between AAV2BRs and NLSs. Two types of P values (P18 and P25) are provided, as described in Materials and Methods. Shown are the frequencies of NLS (+) AAP2 mutants among all the mutants that carry a given combination of two different AAP2BRs.
The statistical analysis for the combination does not give a conclusive result due to the wide range of P25 values across the statistically significant cutoff value of 0.05.
All the AAP2BRs can contribute to nucleolar localization and function in a context-dependent manner.
The NoLS role of each AAP2BR was investigated based on the functional restoration of nucleolar-excluded AAP2 mutants, as detailed in Materials and Methods. This analysis revealed an obvious NoLS role in AAP2BR1, -2, -4, and -5. For example, the NoLS role of AAP2BR2 could be demonstrated by the observation that, when the mutated AAP2BR2 was corrected back to the wild-type sequence in the nucleolar-excluded AAP2mt17 (Fig. 3I), nucleolar retention was restored, as evidenced with AAP2mt11 (Fig. 3C). A comparison between AAP2mt19, showing modest nucleolar accumulation, and AAP2mt26, showing nucleolar exclusion, which differ only by the presence or absence of AAP2BR3, revealed that AAP2BR3 also functions as a weak NoLS (Fig. 3K and R). These results indicate that effective nucleolar accumulation is a consequence of the cooperation of the redundant NoLSs residing in the five AAP2BRs in a given combination of AAP2BRs. Interestingly, AAP2mt26 (AAP2BR1+/BR2+/BR3−/BR4−/BR5−, where basic amino acids in the BRs are retained [plus] or mutated to alanine [minus]) exhibited nearly complete nuclear localization with nucleolar exclusion (Fig. 3R), while GFP-AAP2144–184mt26, carrying AAP2144–184 with the same AAP2BR mutations, and GFP-AAP2144-156, in which GFP was fused with a 13-mer peptide containing only AAP2BR1 and AAP2BR2, showed nuclear and nucleolar accumulation (Fig. 5F and H). The same discrepancy in the pattern of nucleolar accumulation was observed in AAP2mt22 (Fig. 3N) and GFP-AAP2144–184mt22 (Fig. 5E). It has previously been shown that adding a nonapeptide containing 9 basic amino acids to the C terminus of GFP allows nucleolar accumulation of the protein, not through a specific NoLS motif, but presumably through nonspecific, electrostatic interactions with negatively charged nucleolar components (9). There are 8 basic amino acids within the AAP2BR1-AAP2BR2 segment, and there are 10 basic residues in AAP2mt22 (Fig. 1). Therefore, the extra positive electric charges added to GFP likely explain why the GFP-AAP2144-156, GFP-AAP2144–184mt22, and GFP-AAP2144–184mt26 fusion proteins were still capable of nucleolar retention while AAP2mt26 was not. Taken together, although there is no inconsistency in the NLS roles of AAP2BR1 and AAP2BR2 between the AAP2 mutants and the GFP fusion proteins, their NoLS roles are context dependent.
Capsid assembly-promoting functions of AAP2BR mutants.
A series of studies have demonstrated that nucleolar localization of viral components is important for the AAV life cycle (24–27). Indispensable roles of the nucleolus in the virus life cycle have also been demonstrated in other viruses (28–30). Having created a panel of AAP2 mutants whose intracellular localizations are characterized, we addressed how aberrant localization of AAP2 might affect its assembly-promoting function. To this end, we performed a transcomplementation assay in which AAV2 VP3-only particles were produced in HEK 293 cells in the presence of the wild-type AAP2 or a series of AAP2 mutants and quantified AAV2 particle yields using an AAV2 intact-particle-specific ELISA. There was a strong correlation between aberrant intracellular localization of AAP2 and attenuated AAV2 capsid production (Fig. 6A). In particular, AAP2 mutants with nucleolar exclusion consistently showed a 3-log-unit or more reduction of capsid production compared to the wild type. Interestingly, AAP2mt8, -mt9, -mt10, and -mt13, which showed exclusive nuclear localization with enhanced nucleolar association similar to that of the wild type, exhibited a more than 20- to 100-fold decrease in capsid production (Fig. 6A). Thus, normal intracellular localization of AAP2 is a prerequisite but is not sufficient for effective capsid production. The impaired function observed in AAP2 mutants showing wild-type-like intracellular localization implies that AAP2BRs are not only the redundant organelle-targeting signals, but also play a direct or indirect role in the capsid assembly process, which we address below.
FIG 6.
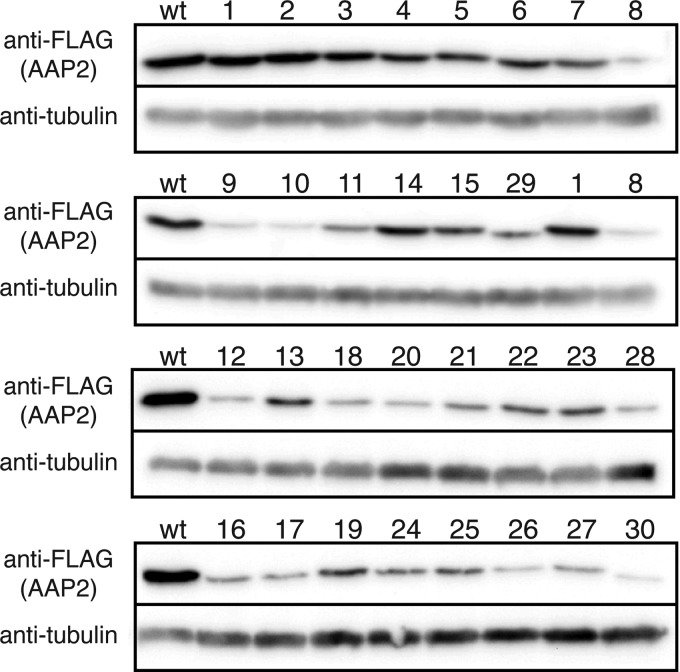
AAV2 VP3 capsid production titers and wild-type and mutant AAP2 protein expression levels in HEK 293 cells. HEK 293 cells seeded on 6-cm dishes were transfected with pAAV2-RepVP3, pCMV-FLAG-cmAAP2 expressing either the full-length wild-type AAP2 or a full-length AAP2 mutant with an AAP2BR mutation(s), and pHelper, using PEI (12). The cells were harvested at 48 h posttransfection and made into a 100-μl crude cell lysate in HEPES-buffered saline after three cycles of freezing and thawing. The AAV2 particle concentration in each crude lysate was quantified by an A20 antibody-based ELISA. (A) Viral titers relative to the titer obtained with the wild-type AAP2 (black bars). A relative titer of 1.0 corresponds to 7.2 × 1012 particles/ml. The thick horizontal black line between the graph and the x axis labels indicates the wild type and AAP2 mutants that showed exclusive nuclear localization with enhanced nucleolar association. The thick horizontal gray line indicates the mutants showing nucleolar exclusion. The data were collected from biologically triplicate experiments. *, P < 0.05 (two-tailed Welch's t test) compared to the wild-type values. In a separate experiment, HEK 293 cells seeded on 6-cm dishes were transfected with pCMV-FLAG-cmAAP2 (the wild type or mutant) using PEI. The cells were harvested 48 h posttransfection, and mutant AAP2 protein expression levels relative to the level of the wild type were determined by a quantitative Western blot analysis using biologically duplicate samples (gray bars). The error bars represent standard errors of the mean (SEM) (black bars) or the difference between each value and the mean value (gray bars). (B) Correlations between AAP2 protein levels of the wild type and mutants indicated by the thick horizontal black line in panel A and AAV2 VP3 capsid production titers are shown in a scatter plot. All values are relative to the levels of the wild type.
AAP2BRs maintain AAP2 expression levels for effective capsid assembly.
During the microscopic analysis, we had noticed that the numbers of transfected HeLa cells and the signal intensities in many AAP2 mutants were lower than those from the wild type to various degrees. Since AAP2BR mutations may have affected not only intracellular localization, but also protein expression levels, we performed a quantitative Western blot assay on lysates from HEK 293 cells transfected with either the wild-type or mutant pCMV-FLAG-cmAAP2 plasmid. The molecular mass of the FLAG-tagged AAP2 protein was determined to be 27 kDa by SDS-PAGE (26 kDa for AAP2), which is slightly higher than a calculated molecular mass of 24 kDa. Mutation or deletion of the AAP2BR regions decreased AAP2 protein expression levels to various degrees between 0.02 and 0.64 compared to 1.0 for the wild type (Fig. 6A and 7). The expression levels of all the mutants showing aberrant localization were reduced substantially, with the relative levels ranging from 0.02 to 0.14. The reduction of protein levels was particularly pronounced in the nucleolar-excluded mutants. The AAP2BR1 or AAP2BR2 mutants showing localization similar to that of the wild type had greater quantities of protein on average than the mutants showing aberrant localization; however, they also showed attenuated AAP2 expression ranging from 0.03 to 0.64. Importantly, the results showed a very nice correlation between intracellular AAP2 levels, nucleolar accumulation of AAP2, and AAV capsid production. When AAP2 mutants retained the ability to localize normally (these mutants are indicated by the horizontal black line in Fig. 6A), AAV capsid yields were positively correlated with the AAP2 mutant protein levels, with a Pearson correlation coefficient of 0.94 (Fig. 6B). When AAP2 mutants were excluded from the nucleolus, AAV capsid yields were disproportionately lower than the AAP2 mutant protein levels (these mutants are indicated by the horizontal gray line in Fig. 6A). Although the underlying mechanism has yet to be elucidated, these results indicate that AAP2BRs play a pivotal role in maintaining the AAP2 levels for effective capsid assembly. The AAP2BRs' role in this respect well explains why some of the AAP2BR1 or AAP2BR2 mutants showing wild-type localization failed to produce capsids effectively.
FIG 7.
Western blot analysis of the wild-type and mutant AAP2 proteins expressed in HEK 293 cells. Cells seeded on 6-cm dishes were transfected with pCMV-FLAG-cmAAP2 (wild type or mutant) using PEI. The same quantity of crude lysates prepared at 48 h posttransfection were separated on an 8% SDS-PAGE gel and blotted onto a membrane, which was then probed with both anti-FLAG and anti-α-tubulin antibodies. The AAP2 mutant numbers are shown above the lanes.
The AAP2 NoLS is important for nucleolar accumulation of AAV2 VP3.
The data presented above provided insights into the AAP2BRs' functional roles in intracellular localization of AAP2 and capsid assembly; however the data did not address the AAP2 role as a VP3 transporter. Since the nucleolus has been thought to be important for AAV2 capsid assembly and nucleolar-excluded AAP2BR mutants showed a substantially reduced ability to produce viral capsids, it is possible that AAP2BR mutants would not associate with VP3, leading to impaired nuclear transport and/or failure to accumulate VP3 in the nucleolus even if VP3 is actively localized to the nucleus. To address this, HeLa cells were transfected with pCMV-AAV2VP3 and pCMV-FLAG-cmAAP2 (either the wild type or AAP2mt26), and intracellular localization of AAP2 and VP3 was analyzed by immunofluorescence microcopy at 48 h posttransfection. As Sonntag et al. have previously shown, VP3 diffusely localizes to the cytoplasm and nucleoplasm but remains outside the nucleus unless AAP2 is present (3) (Fig. 8A and B). In the presence of AAP2mt26, which had the ability to exclusively translocate to the nucleus but was not able to accumulate in the nucleolus, VP3 was seen predominantly in the nucleoplasm, colocalizing with AAP2mt26 and leaving only a small quantity of VP3 proteins in the cytoplasm. Importantly, VP3 failed to accumulate in the nucleolus (Fig. 8C). We have also found, using a plasmid carrying an AAV2 cap gene that does not express AAP2, that expression of the AAV2 VP1, VP2, and VP3 proteins does not result in nucleolar localization of any of the VP proteins in HeLa cells when AAP2 is not coexpressed (Earley and Nakai, unpublished). Taken together, these observations indicate that the NoLS in AAP2 plays a primary role in targeting capsid VP proteins to the nucleolus.
FIG 8.
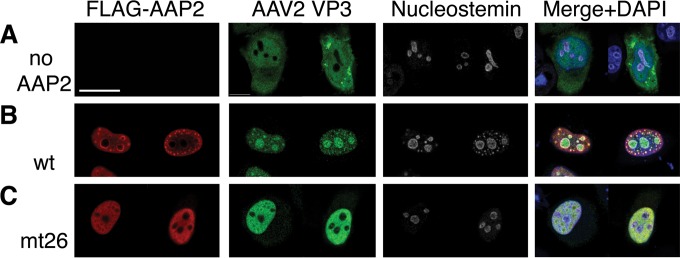
Intracellular localization of AAV2 VP3 in the presence or absence of wild-type AAP2 or in the presence of the nuclear-targeted, nucleolar-excluded AAP2 mutant, AAP2mt26. HeLa cells were transfected with the following plasmids: pCMV-AAV2VP3 only (A), pCMV-AAV2VP3 and pCMV-FLAG-cmAAP2 (wild type) (B), and pCMV-AAV2VP3 and pCMV-FLAG-cmAAP2mt26 (C). The cells were fixed at 48 h posttransfection and immunostained with anti-FLAG, anti-AAV VP1/VP2/VP3, and anti-nucleostemin antibodies. Scale bar, 20 μm.
DISCUSSION
The NLS in AAP2 had remained unidentified in a series of studies reported by Kleinschmidt's group (1–3), which prompted us to seek to determine the amino acid residues responsible for nuclear translocation of AAP2. The consensus sequences of the classical, importin-α-binding NLSs have been well defined. The monopartite NLSs have the consensus sequence K(K/R)X(K/R), while the bipartite NLSs show a consensus sequence composed of two short basic amino acid clusters separated by a linker sequence, such as (K/R)(K/R)X10–12(K/R)3/5, where the C-terminal side cluster contains at least three basic amino acids within 5 consecutive residues (14, 18, 19). Many noncanonical, importin-α-binding NLSs have also been identified, including bipartite NLSs with a long linker of up to 29 residues (14, 18, 19). In light of this knowledge about various types of NLSs, one could assume that AAP2 has one classical monopartite NLS in AAP2BR1 (KSKRSRR) and/or one noncanonical bipartite NLS with a long linker in AAP2BR1-X30-AAP2BR5 (RRIK). Our experimental data, however, do not support this assumption. None of the AAP2BRs were capable of serving as a monopartite NLS. Substantial nuclear enrichment required the presence of AAP2BR1-AAP2BR2 (KSKRSRR-X3-RRR) or at least four AAP2BRs when either AAP2BR1 or AAP2BR2 is absent (i.e., AAP2BP1-AAP2BP3-AAP2BP4-AAP2BP5 and AAP2BP2-AAP2BP3-AAP2BP4-AAP2BP5). Thus, we identified at least three different motifs that can function as an NLS and overlap within the AAP2144–184 region. This redundant nature of the AAP2 NLS explains why a pervious investigation concluded that AAP2BR1 was not involved in nuclear localization even though the region contained a classical NLS (1).
In nucleolar-localizing proteins, an NLS and an NoLS can reside in separate locations, but they often coreside in a region highly rich in lysine and arginine residues. The NLS and NoLS of AAP2 that we identified is a joint NLS-NoLS, because all the AAP2BR regions play either, or both, of the NLS and NoLS roles. Many such joint NLS-NoLSs have been reported as NLSs in the literature, mainly due to incomplete investigation for nucleolar accumulation, and therefore, joint NLS-NoLSs have been overlooked in many instances (17). A recent attempt to extract commonalities of 48 human NoLSs, including joint NLS-NoLSs, has revealed that NoLSs are highly basic; are predominantly located near the N or C terminus of proteins, in α-helices or coils and rarely in β-strands; and are solvent accessible (17). In this regard, the AAP2144–184 region containing the joint NLS-NoLSs is consistent with these general trends. That is, in addition to the high proportion of basic amino acids, the NLS-NoLS resides near the C terminus; the AAP2144–184 residues are exclusively in α-helices (49%) or coils (51%) according to the Jpred3 prediction (31); and the proportions of buried residues in the AAP2144–184 region predicted by Jpred3 are 37%, 10%, and 2% at prediction thresholds of 25%, 5%, and 0%, respectively, which are lower than in the full-length protein control in the study reported by Scott et al. (17) The AAP2144–184 region also contains the previously identified (K/R)(K/R)X(K/R) NoLS motif (15). Through a literature search, we could identify a joint NLS-NoLS that resembles the AAP2 NLS-NoLS. Liu et al. reported that a stretch of 18 amino acid residues, 2744KKKMKKHKNKSEAKKRKI2761, is part of a joint NLS-NoLS found in 1A6/downregulated in metastasis (1A6/DRIM), a large nucleolar-targeting protein (2,785 amino acids) involved in rRNA processing (32). This amino acid sequence is reminiscent of 154RRRLPITLPARFRCLLTRSTSSRTSSARRIK184, one of the complete joint NLS-NoLSs that we identified in AAP2. Although the observations obtained from the arginine/lysine-to-alanine mutagenesis experiments were not exactly the same in the Liu et al. study and ours, both identified basic amino acid clusters that play a dual NLS-NoLS role. In addition, both studies showed that a decrease in electric charge in the signals resulted in nucleolar exclusion. As demonstrated with AAP2 and many other nucleolar-localizing proteins (17), NLSs and NoLSs are inherently difficult to clearly define, particularly when they reside within the same region with a portion of the signal playing a dual role.
How NoLSs perform their role in nucleolar targeting has not been fully understood. Unlike NLSs, NoLSs do not have well-defined consensus sequences, although they share common characteristics to some degree, as described above. The nucleolus does not have any compartmentalizing membranous structures, and therefore, nucleolar accumulation presumably does not require sophisticated machinery that actively targets the organelle, like the nuclear transport machinery. Rather, nucleolar accumulation of proteins has been thought to be mediated by a retention mechanism through interactions with nucleolar components, such as RNA and other nucleolar proteins (33). RNA-binding motifs have been identified in many nucleolar proteins, including nucleolin, fibrillarin, and viral RNA-binding proteins (34–36). Nucleophosmin, a putative shuttle protein that transports cargos between the cytoplasm and the nucleolus (37–40), has been shown to mediate nucleolar retention of other proteins through the interaction between two highly acidic regions (120EDAESEDEEEED132 and 161DEDDDDDDEEDDDEDDDDDDFDDEEAEE188) in nucleophosmin and positively charged amino acid regions in the other proteins (40). Thus, nucleolar accumulation is most likely mediated by electrostatic interaction rather than by a defined set of particular amino acid sequences (9). Concordant with this notion, we could only vaguely define the AAP2 NoLS, as opposed to its NLSs, and found that the abundance of positively charged AAP2BR regions, rather than a specific amino acid sequence or a specific combination of AAP2BRs, confers on AAP2 the ability to translocate from the nucleoplasm to the nucleolus. This is in line with the notion that the nucleolar accumulation of AAP2 results from nonspecific interaction with negatively charged nucleolar components and also raises the possibility that the degree of accumulation is determined by the net electric charge of the AAP2144–184 region. The electric charge of AAP2144–184 is 15, and at least 12 has been found to be required for efficient nucleolar accumulation of the AAP2BR mutants (Fig. 1). The net electric charges of the AAP2144–184-corresponding regions of various serotype AAPs (AAP1 to -13) (1) vary from 8 to 15, with AAP5 being the lowest and AAP2 and AAP3B being the highest. Assuming that nucleolar accumulation is determined by the abundance of the net positive charges within this region, AAP5 would likely be devoid of an NoLS and excluded from the nucleolus. If this is true, AAP5 would not be able to transport AAV5 capsid VP proteins to the nucleolus, the organelle that is believed to be important for AAV capsid assembly, at least for AAV2 (24–27). How would AAV5 viral capsid proteins assemble if they are not transported to the nucleolus? Although our study demonstrates that the nucleolar localization of AAP2 strongly correlates with the ability of AAV2 to form capsids, it is intriguing to speculate that the nucleolar accumulation of AAP2 might merely be a coincidental, nonspecific consequence associated with a function(s) other than nucleolar targeting and that this targeting might not be functionally important. Such nonspecific nucleolar accumulation with no functional role has been demonstrated in the human histone H2B protein (9). Future studies on the subcellular localizations of AAPs derived from various serotypes will address this potential paradox and elucidate the biological significance of the presence of an NoLS in AAPs. Nonetheless, our observations corroborate the driving role of the electric charge in nucleolar targeting and support the idea that NoLSs pay a lower cost for the acquisition of their role by intermingling their signals with NLSs that already have high electric charges. This is particularly advantageous to small viruses that have a limited capacity to store genetic information.
Surprisingly, we could identify at least three complete NLSs that overlap within the same AAP2144–184 region. They are AAP2BR1 plus -BR2, AAP2BR1 plus -BR3 plus -BR4 plus -BR5, and AAP2BR2 plus -BR3 plus -BR4 plus -BR5. Redundant NLSs, as either separate NLSs or overlapping NLSs, have been identified in many proteins, including those derived from viruses. For example, the polyomavirus large T antigen has two separate NLSs, VSRKRPR and PKKARED, each of which mediates nuclear localization in the absence of the other (41, 42); the nonstructural protein 1 of influenza A virus (NS1A) has a functionally competent monopartite NLS, DRLRRDQKSLR, and a classical bipartite NLS-like sequence, KRKMARTARSKVRRDKMAD (43), which also conveys nucleolar localization; and the bovine adenovirus 3 33,000-molecular-weight (33K) protein has a 40-residue-long region where two karyopherin-binding motifs, an importin-α5-binding motif and a transportin 3-binding motif, overlap (44). Utilization of different nuclear transport pathways through the interactions between an NLS and multiple karyopherins have also been demonstrated in human immunodeficiency virus (HIV) Rev protein (45). The straightforward inference about the NLS redundancy is that it ensures and promotes efficient nuclear entry by increasing the avidity for the nuclear transport system and/or by using more than one nuclear entry pathway. However, there are also many instances in which the amino acid residues composing an NLS perform functions distinct from those in nuclear translocation. For example, the unconventional NLS in the nucleoprotein (NP) of the influenza A virus plays a crucial role in nuclear import of the viral genomic RNA (46), and the second NLS in the transcription factor Fli-1 also functions as a DNA-binding domain (47). In fact, DNA-binding domains frequently overlap NLSs (47), and this has led to speculation about the possible coevolutionary origins of the two motifs (48, 49). In the case of AAP2, our present and previous studies provide clues to understanding the necessity of redundancy. In our previous study, we used a directed-evolution approach to experimentally and computationally evolve functionally competent mutant AAP2s from a randomly mutagenized library (12). We demonstrated that the functionally optimal AAP2 mutants harbor a strongly basic and hydrophilic heptapeptide, (R/S)7, in the AAP2BR1 region that remarkably resembles the native sequence, KSKRSRR (12). Since all the AAP2 mutants used in the previous study had a completely functional NLS-NoLS composed of AAP2BR2, -3, -4, and -5, AAP2BR1 is most likely not simply a redundant sequence composing an NLS-NoLS but has an important non-NLS-NoLS role. In keeping with this notion, mutations in this region could substantially reduce AAP2 protein levels without affecting its intracellular localization and could result in significantly impaired capsid assembly (AAP2mt8, -mt9, and -mt10). The reduction of capsid production by an AAP2BR1 mutation has also been reported by Naumer et al. (1). Although the mechanism for the decreased protein expression has yet to be determined, it could be either due to intrinsic instability caused by protein misfolding or due to decreased protein-protein or protein–nucleic-acid interactions that stabilize AAP2. Taken together, the redundancy of the NLS-NoLS in AAP2 is likely a necessity for optimal capsid production and presumably results from their alternative functional roles.
In summary, this study identifies a joint NLS-NoLS near the C terminus of AAP2. The identified basic amino acid region is important, not only for nuclear and nucleolar localization, but also for maintaining AAP2 levels, indicating a non-NLS-NoLS role in the region. The system described here can readily be applied to AAPs derived from other serotypes. Further studies on interactions between AAP, capsid, and cellular proteins will help us understand the mechanism of AAV capsid assembly and may lead to improved AAV vector production for gene therapy.
ACKNOWLEDGMENTS
We thank Stefanie Kaech Petrie and Aurelie Snyder at the OHSU Advanced Light Microscope Core for their technical expertise, Hiroyuki Ido for technical assistance in molecular cloning of plasmids, Michael R. Lasarev for his advice on statistical analyses, and Michael S. Chapman for critical reading of the manuscript.
This work was supported by Public Health Service grants (R01 DK078388, R01 NS088399, T32 GM071338, and T32 AI007472) and a Sponsored Research Fund from Takara Bio Inc.
REFERENCES
- 1.Naumer M, Sonntag F, Schmidt K, Nieto K, Panke C, Davey NE, Popa-Wagner R, Kleinschmidt JA. 2012. Properties of the adeno-associated virus assembly-activating protein. J Virol 86:13038–13048. doi: 10.1128/JVI.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonntag F, Schmidt K, Kleinschmidt JA. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chook YM, Suel KE. 2011. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta 1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalderon D, Roberts BL, Richardson WD, Smith AE. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 6.Robbins J, Dilworth SM, Laskey RA, Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623. doi: 10.1016/0092-8674(91)90245-T. [DOI] [PubMed] [Google Scholar]
- 7.Dingwall C, Laskey RA. 1991. Nuclear targeting sequences—a consensus? Trends Biochem Sci 16:478–481. doi: 10.1016/0968-0004(91)90184-W. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Efraim I, Gerace L. 2001. Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J Cell Biol 152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musinova YR, Lisitsyna OM, Golyshev SA, Tuzhikov AI, Polyakov VY, Sheval EV. 2011. Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim Biophys Acta 1813:27–38. doi: 10.1016/j.bbamcr.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Chen B, Li Y, Zhou D, Chen S. 2011. PNRC accumulates in the nucleolus by interaction with B23/nucleophosmin via its nucleolar localization sequence. Biochim Biophys Acta 1813:109–119. doi: 10.1016/j.bbamcr.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szebeni A, Herrera JE, Olson MO. 1995. Interaction of nucleolar protein B23 with peptides related to nuclear localization signals. Biochemistry 34:8037–8042. doi: 10.1021/bi00025a009. [DOI] [PubMed] [Google Scholar]
- 12.Kawano Y, Neeley S, Adachi K, Nakai H. 2013. An experimental and computational evolution-based method to study a mode of co-evolution of overlapping open reading frames in the AAV2 viral genome. PLoS One 8:e66211. doi: 10.1371/journal.pone.0066211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. 2005. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol 79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem 282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber JD, Kuo ML, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, Sherr CJ. 2000. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol 20:2517–2528. doi: 10.1128/MCB.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viranaicken W, Gasmi L, Chaumet A, Durieux C, Georget V, Denoulet P, Larcher JC. 2011. L-Ilf3 and L-NF90 traffic to the nucleolus granular component: alternatively-spliced exon 3 encodes a nucleolar localization motif. PLoS One 6:e22296. doi: 10.1371/journal.pone.0022296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. 2010. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res 38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. 2010. Expanding the definition of the classical bipartite nuclear localization signal. Traffic 11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin α. J Biol Chem 284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 20.Svistunova DM, Musinova YR, Polyakov VY, Sheval EV. 2012. A simple method for the immunocytochemical detection of proteins inside nuclear structures that are inaccessible to specific antibodies. J Histochem Cytochem 60:152–158. doi: 10.1369/0022155411429704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosugi S, Yanagawa H, Terauchi R, Tabata S. 2014. NESmapper: accurate prediction of leucine-rich nuclear export signals using activity-based profiles. PLoS Comput Biol 10:e1003841. doi: 10.1371/journal.pcbi.1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu SC, Imai K, Horton P. 2011. Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res 39:e111. doi: 10.1093/nar/gkr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto G, Fullaondo A, Rodriguez JA. 2014. Prediction of nuclear export signals using weighted regular expressions (Wregex). Bioinformatics 30:1220–1227. doi: 10.1093/bioinformatics/btu016. [DOI] [PubMed] [Google Scholar]
- 24.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol 71:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bevington JM, Needham PG, Verrill KC, Collaco RF, Basrur V, Trempe JP. 2007. Adeno-associated virus interactions with B23/Nucleophosmin: identification of sub-nucleolar virion regions. Virology 357:102–113. doi: 10.1016/j.virol.2006.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu J, Brown KE. 1999. A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology 257:373–382. doi: 10.1006/viro.1999.9664. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JS, Samulski RJ. 2009. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol 83:2632–2644. doi: 10.1128/JVI.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiscox JA. 2002. The nucleolus—a gateway to viral infection? Arch Virol 147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews D, Emmott E, Hiscox J. 2011. Viruses and the nucleolus, p 321–345 InOlson M. (ed), The nucleolus. Springer, New York, NY. [Google Scholar]
- 31.Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Du X, Ke Y. 2006. Mapping nucleolar localization sequences of 1A6/DRIM. FEBS Lett 580:1405–1410. doi: 10.1016/j.febslet.2006.01.064. [DOI] [PubMed] [Google Scholar]
- 33.Olson MO, Dundr M. 2005. The moving parts of the nucleolus. Histochem Cell Biol 123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]
- 34.Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt-Zachmann MS, Nigg EA. 1993. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci 105:799–806. [DOI] [PubMed] [Google Scholar]
- 36.Aris JP, Blobel G. 1991. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc Natl Acad Sci U S A 88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli UK. 1991. Specific complex of human immunodeficiency virus type 1 Rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol Cell Biol 11:2567–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 39.Valdez BC, Perlaky L, Henning D, Saijo Y, Chan PK, Busch H. 1994. Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J Biol Chem 269:23776–23783. [PubMed] [Google Scholar]
- 40.Adachi Y, Copeland TD, Hatanaka M, Oroszlan S. 1993. Nucleolar targeting signal of Rex protein of human T-cell leukemia virus type I specifically binds to nucleolar shuttle protein B-23. J Biol Chem 268:13930–13934. [PubMed] [Google Scholar]
- 41.Richardson WD, Roberts BL, Smith AE. 1986. Nuclear location signals in polyoma virus large-T. Cell 44:77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- 42.Howes SH, Bockus BJ, Schaffhausen BS. 1996. Genetic analysis of polyomavirus large T nuclear localization: nuclear localization is required for productive association with pRb family members. J Virol 70:3581–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melen K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol 81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulshreshtha V, Ayalew LE, Islam A, Tikoo SK. 2014. Conserved arginines of bovine adenovirus-3 33K protein are important for transportin-3 mediated transport and virus replication. PLoS One 9:e101216. doi: 10.1371/journal.pone.0101216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold M, Nath A, Hauber J, Kehlenbach RH. 2006. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J Biol Chem 281:20883–20890. doi: 10.1074/jbc.M602189200. [DOI] [PubMed] [Google Scholar]
- 46.Cros JF, Garcia-Sastre A, Palese P. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 47.Cokol M, Nair R, Rost B. 2000. Finding nuclear localization signals. EMBO Rep 1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu W, Philips AS, Kwok JC, Eisbacher M, Chong BH. 2005. Identification of nuclear import and export signals within Fli-1: roles of the nuclear import signals in Fli-1-dependent activation of megakaryocyte-specific promoters. Mol Cell Biol 25:3087–3108. doi: 10.1128/MCB.25.8.3087-3108.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaCasse EC, Lefebvre YA. 1995. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res 23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]