ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 plays an essential role in KSHV lytic infection by promoting viral gene expression at the posttranscriptional level. Using bioinformatic and biochemical approaches, we determined that ORF57 contains two structurally and functionally distinct domains: a disordered nonstructural N-terminal domain (amino acids [aa] 1 to 152) and a structured α-helix-rich C-terminal domain (aa 153 to 455). The N-terminal domain mediates ORF57 interaction with several RNA-protein complexes essential for ORF57 to function. The N-terminal phosphorylation by cellular casein kinase II (CKII) at S21, T32, and S43, and other cellular kinases at S95 and S97 residues in proximity of the caspase-7 cleavage site, 30-DETD-33, inhibits caspase-7 digestion of ORF57. The structured C-terminal domain mediates homodimerization of ORF57, and the critical region for this function was mapped carefully to α-helices 7 to 9. Introduction of point mutations into α-helix 7 at ORF57 aa 280 to 299, a region highly conserved among ORF57 homologues from other herpesviruses, inhibited ORF57 homodimerization and led to proteasome-mediated degradation of ORF57 protein. Thus, homodimerization of ORF57 via its C terminus prevents ORF57 from degrading and allows two structure-free N termini of the dimerized ORF57 to work coordinately for host factor interactions, leading to productive KSHV lytic infection and pathogenesis.
IMPORTANCE KSHV is a human oncogenic virus linked to the development of several malignancies. KSHV-mediated oncogenesis requires both latent and lytic infection. The KSHV ORF57 protein is essential for KSHV lytic replication, as it regulates the expression of viral lytic genes at the posttranscriptional level. This report provides evidence that the structural conformation of the ORF57 protein plays a critical role in regulation of ORF57 stability. Phosphorylation by CKII on the identified serine/threonine residues at the N-terminal unstructured domain of ORF57 prevents its digestion by caspase-7. The C-terminal domain of ORF57, which is rich in α-helices, contributes to homodimerization of ORF57 to prevent proteasome-mediated protein degradation. Elucidation of the ORF57 structure not only enables us to better understand ORF57 stability and functions but also provides an important tool for us to modulate ORF57's activity with the aim to inhibit KSHV lytic replication.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 (also known as Mta) is expressed early in the KSHV lytic cycle and is required for the efficient expression of a subset of viral genes, including KSHV PAN, ORF59, K8, viral interleukin-6 (vIL-6), ORF47, and others (1–7). A KSHV genome lacking ORF57 expression is associated with a defective lytic cycle incapable of producing infectious virions (8, 9).
KSHV ORF57 functions as a posttranscriptional regulator of viral gene expression by affecting RNA stability (PAN, ORF59, and ORF47), splicing (ORF50 and K8), polyadenylation (ORF59), and translation (vIL-6) (1, 2, 4–7, 10) but appears not to promote RNA export (11, 12). Whether ORF57 directly promotes KSHV genome instability in infected cells (13) remains to be confirmed. Although all ORF57 functions involve ORF57 association with an RNA target, this association also requires cellular proteins to function as ORF57 cofactors (14, 15), and each of the ORF57-specific functions depends on a specific cofactor(s). This has been demonstrated by the observation that ORF57 stabilizes PAN RNA via interaction with PABPC1 (16), that ORF57 mediates K8 splicing by interaction with SRSF3 (7), that ORF57 enhances ORF59 expression by the suppression of SPEN-induced nuclear hyperpolyadenylation (4), and that ORF57 promotes vIL-6 translation by preventing Ago2, a major component of RISC complexes, from interacting with a microRNA binding site in vIL-6 RNA (6).
ORF57 interacts with Aly/REF (12, 14, 17, 18), a ubiquitously expressed nuclear protein which functions as a molecular chaperone to regulate dimerization, DNA binding, and transcriptional activity of basic region-leucine zipper (bZIP) proteins (19, 20). It was initially viewed as an RNA export cofactor (21, 22), but this interaction is not necessary for RNA export of viral intronless RNAs. Several lines of evidence support the latter conclusion. First, depletion of Aly/REF from HEK293 nuclear extract does not affect the ORF57 interaction with KSHV intronless ORF59 RNA, and small interfering RNA knockdown of Aly/REF from HeLa or HEK293 cells does not affect ORF57-mediated enhancement of ORF59 expression (14). Second, an ORF57 mutant with a deficiency in Aly/REF binding retains its ability to accumulate KSHV target mRNAs (12). Third, the Aly/REF-ORF57 interaction has been demonstrated to be nonessential for KSHV lytic replication but contributes to target RNA stability independently of effects in RNA export (23, 24). Fourth, a recent well-designed study concluded that ORF57 does not provide the specific RNA export function and is not a bona fide export factor for KSHV intronless RNAs (11). Finally, knockout of Aly/REF expression does not affect mRNA export in Drosophila melanogaster cells and Caenorhabditis elegans (25, 26) and in herpes simplex virus 1 (HSV-1) infection (27).
Posttranscriptional regulators with similar activities to KSHV ORF57 are also encoded by other members of the herpesvirus family. These include well-characterized HSV-1 ICP27 (28), human cytomegalovirus virus (HCMV) UL69 (29), Epstein-Barr virus (EBV) EB2 (or EB-SM) (30), and herpesvirus saimiri (HVS) ORF57 (31). While all homologues in the family share many common activities, they diverge with regard to specific functions and target specificities. Therefore, they do not complement each other's function to rescue virus infection by a homologue-deficient genome (9, 32). Although the functions of KSHV ORF57 and its homologues became more understood over recent decades, the regulation of their unique activities, in particular the contribution of their protein structures to their unique activities, remains to be elucidated. To date, there is no crystal structure for any member in this family. Because ORF57 has no homology to any known cellular proteins and bears only low amino acid sequence homology to its close homologues, it remains a great challenge to solve the ORF57 three-dimensional (3D) structure and correlate its structure with its function.
In this study, we conducted a comprehensive analysis of the ORF57 structure, both computationally and experimentally, and determined that the ORF57 protein contains two structurally and functionally distinct domains: an intrinsically disordered N-terminal domain and a structured C-terminal domain. We found that the distinct structures in the identified domains dictate ORF57 binding activities toward its self-association and interactions with cellular cofactors.
MATERIALS AND METHODS
In silico protein sequence analysis.
Protein secondary structures were predicted with PSIPRED version 3.0 (a PSI-BLAST-based secondary structure prediction tool publicly available at http://bioinf.cs.ucl.ac.uk/psipred/) (33). Links to all programs used for prediction of the intrinsically disordered region can be found at http://www.disprot.org/predictors.php. The kinase-specific eukaryotic protein phosphorylation sites were predicted by using the NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/) (34). The sequence homology analysis was performed by using ClustalW2, which is located at http://www.ebi.ac.uk/Tools/msa/clustalw2/.
Cells.
Human HEK293, HeLa, and monkey COS-1 cells were cultivated in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS; HyClone). KSHV-positive BCBL-1 (35) and JSC-1 (36) B cells were maintained in RPMI 1640 medium with 10% FBS. The KSHV lytic cycle was induced by treatment with 3 mM sodium butyrate (JSC-1 cells) or 0.6 mM sodium valproate (BCBL-1 cells) for 24 h. All transfections were carried out with Lipofectamine 2000 (Invitrogen) or LipoD293 (SignaGen Laboratories) as recommended by the manufacturers.
Plasmids and mutagenesis.
The eukaryotic expression vectors pFLAG-CMV-5.1 and p3×FLAG-CMV-14 (Sigma) and pEGFP-N1 (Clontech) were used. These previously described plasmids were used to express various forms of ORF57 protein and EBV EB2, as follows: pVM7 (ORF57-FLAG), pVM68 (ORF57-3×FLAG), pVM24 (ORF57-FLAG amino acids [aa] 1 to 251; ΔC wild type [wt]), pVM45-51 (ORF57-FLAG aa 1 to 251; ΔC with single, double, or triple nuclear localization signal [NLS] mutations), pVM35 (ORF57-GFP [green fluorescent protein]; mtNLS-1, -2, and -3 [mutated NLSs]), pVM20 (ORF57-GFP aa 167 to 455; ΔN), pcDNA-ORF57, and pGS113 (EB2-myc) (14, 37). The mutagenesis of conserved residues in the ORF57 helix 7 region was performed by overlapping PCR using the primers listed in Table 1. The amplicon with the designed mutation(s) in ORF57 was recloned into p3×FLAG-CMV-14 to generate the following plasmids: pVM83 (ORF57-3×FLAG, E287A, E288A), pVM84 (ORF57-3×FLAG, E287P, E288P), pVM85 (ORF57-3×FLAG, W292A), pVM86 (ORF57-3×FLAG, W292P), and pVM99 (ORF57-3×FLAG, K345A).
TABLE 1.
Oligonucleotides used in ORF57 mutagenesis
| Oligonucleotide | Sequencea | Mutation(s) | Plasmid |
|---|---|---|---|
| oVM193 | 5′-CTTTCGTGGCGGCACAAATGACGTGGGCC-3′ | E287A, E288A | pVM83 |
| oVM194 | 5′-CATTTGTGCCGCCACGAAAGCCCCAAGCG-3′ | E287A, E288A | pVM83 |
| oVM195 | 5′-CTTTCGTGCCGCCACAAATGACGTGGGCC-3′ | E287P, E288P | pVM84 |
| oVM196 | 5′-CATTTGTGGCGGCACGAAAGCCCCAAGCG-3′ | E287P, E288P | pVM84 |
| oVM197 | 5′-CAAATGACGGCGGCCCAGACGGTTGTGC-3′ | W292A | pVM85 |
| oVM198 | 5′-CTGGGCCGCCGTCATTTGTTCCTCCACGA-3′ | W292A | pVM85 |
| oVM199 | 5′-CAAATGACGCCGGCCCAGACGGTTGTGC-3′ | W292P | pVM86 |
| oVM200 | 5′-CTGGGCCGGCGTCATTTGTTCCTCCACGA-3′ | W292P | pVM86 |
| oVM273 | 5′-AGCACTGATCGCACAGGTGGCATATTTGGTA-3′ | K345A | pVM99 |
| oVM274 | 5′-ATGCCACCTGTGCGATCAGTGCTAGCTCGCT-3′ | K345A | pVM99 |
| oVM68b | 5′-TACTCAGAATTCACC/ATGGTACAAGCAATGATAGACATGG-3′ | ||
| oVM69b | 5′-ATCGTGGATCC/AGAAAGTGGATAAAAGAATAAACCCTTG-3′ |
Underlined portions of sequences indicate point mutations.
oVM68 is a forward primer with an EcoRI site, and oVM69 is a backward primer with a BamHI site (a slash divides the restriction cutting site from the viral primer sequence).
Antibodies and recombinant ORF57 protein.
Rabbit polyclonal antibody targeting the ORF57 N terminus (aa 119-PEKRPRRRPRDRLQ-132) was described before (8). To generate the antibody against the ORF57 C terminus, rabbits were immunized with a synthetic peptide corresponding to aa 394 to 412 (394-ARGQELFRTLLEYYRPGDV-412) of the ORF57 protein sequence. Other antibodies used in the study were anti-hnRNPU (Ab10297; Abcam), anti-RNase helicase A (Ab26271; Abcam), anti-nucleolin C23 (Ab13541; Abcam), anti-FLAG M2 (F1804; Sigma), anti-c-Myc (M4439; Sigma), anti-neomycin phosphotransferase II (06-747; Millipore), Lys48-specific anti-ubiquitin rabbit monoclonal antibody (clone Apu2; 05-1307; Millipore), and anti-β-tubulin (T5201; Sigma). Recombinant full-length ORF57 protein containing a C-terminal FLAG tag was expressed in Sf21 insect cells by using a baculovirus vector and purified by immunoaffinity column chromatography with an anti-FLAG antibody (38).
Limited proteolysis and Edman protein sequencing.
Lyophilized clostripain and proteinase K were obtained from Sigma and reconstituted as recommended by the manufacturer. The ORF57 limited proteolytic digestion was carried out in a 10-μl reaction mixture containing 4 μg of purified ORF57-FLAG protein or whole-cell extract resuspended in immunoprecipitation (IP) buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% NP-40) supplemented with increasing amounts of clostripain or proteinase K. The digestion reaction mixture was incubated for 3 min at room temperature and stopped by addition of 1 μl of 1 M protease inhibitor phenylmethylsulfonyl fluoride and 10 μl of 2× SDS protein sample buffer containing 10% (vol/vol) 2-mercaptoethanol (2-ME). After digestion, the samples were separated via SDS-PAGE and stained with GelCode Blue stain reagent (here referred to as Gel Blue; Thermo Scientific) or analyzed by Western blotting. To determine the N-terminal amino acid sequence of the ORF57 protease-resistant (PR) region, the digested ORF57 products with 0.1 U of clostripain were subjected to Edman degradation chemistry using an ABI Procise 494 sequencer (Alphalyse Inc.).
Immunoprecipitation and mass spectrometry.
The whole-cell extract used for IP was prepared by resuspending cell pellets in ice-cold RSB-150 buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40). After brief sonication, the extract was cleared by centrifugation for 10 min at 10,000 × g at 4°C, diluted in IP buffer, and incubated with antibody-coated agarose beads overnight at 4°C. After extensive washing, the immunoprecipitated complexes were eluted with IP buffer supplemented with 150 ng/μl of 3× FLAG peptide (Sigma) for 1 h at 4°C or with 2× SDS protein sample buffer supplemented with 5% (vol/vol) 2-ME. In some cases, the prepared cell extract was pretreated before IP with RNase A (Ambion) or DNase I (New England Biolabs) for 30 min at room temperature. Immunoprecipitated proteins were separated on SDS-PAGE and detected by Gel Blue staining or by Western blotting. The identity of immunoprecipitated proteins was determined by tandem mass spectrometry (MS/MS) after in-gel trypsin digestion (ProtTech, Norristown, PA).
In vitro phosphorylation and dephosphorylation.
ORF57 protein in vitro phosphorylation was carried out in a 10-μl reaction mixture containing 400 ng of recombinant ORF57-FLAG protein, 1× casein kinase II (CKII) buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2), 1 mM cold ATP or 25 μCi [γ-32P]ATP, and 125 U of recombinant CKII (New England BioLabs). The reaction mixture was incubated for 1 h at 30°C and stopped by addition of 10 μl of 2× SDS protein sample buffer with 10% (vol/vol) 2-ME. The 32P-labeled ORF57 was separated by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and detected by using a PhosphorImager (Molecular Dynamic). The band corresponding to phosphorylated ORF57 was then cut from the membrane for further analysis. In vitro dephosphorylation was performed in a 50-μl reaction mixture containing 10 μl of cell extract or 1 μg of recombinant ORF57-FLAG protein diluted in 1× protein phosphatase (PPase) buffer (50 mM HEPES [pH 7.5], 10 mM NaCl, 2 mM dithiothreitol [DTT], 0.01% Brij 35) supplemented with 1 mM MnCl2 and 1,000 U of Lambda bacteriophage λ-PPase (New England Biolabs). The reaction mixture was incubated at 30°C for 1 h. The λ-PPase activity was inhibited by addition of 10 mM sodium orthovanadate (Na3VO4) or 50 mM EDTA to the reaction mixture.
Mapping of ORF57 phosphorylation sites.
The phosphorylated residues in the ORF57 protein were mapped by using two complementary methods. First, recombinant ORF57-FLAG protein was subjected to tryptic digestion, and the resulting peptides were evaluated by matrix-assisted laser desorption ionization–time of flight analysis. A capillary liquid chromatography-electrospray ionization-MS (LC-ESI-MS) setup with an ion trap mass analyzer (ProtTech) was used to determine the exact position of phosphorylated residues. Second, the recombinant ORF57 protein was first in vitro phosphorylated with recombinant CKII in the presence of [γ-32P]ATP as described above. Radiolabeled ORF57 was separated by SDS-PAGE, transferred to a PVDF membrane, and detected with a PhosphorImager. The membrane containing the phosphorylated ORF57 protein was then subjected to proteolytic digestion with trypsin or endoproteinase GluC. The phosphopeptides carrying 32P labeling were isolated by reverse-phase high-performance liquid chromatography (HPLC) and subjected to phosphoamino amino acid analysis and phosphopeptide mapping by using Edman degradation as described previously (39, 40).
In vitro caspase cleavage assay.
ORF57 in vitro caspase cleavage was performed as described previously (38). Briefly, a 20-μl reaction mixture containing 200 ng of recombinant ORF57-FLAG protein diluted in 1× cleavage buffer {100 mM NaCl, 50 mM HEPES [pH 7.4], 10 mM DTT, 1 mM EDTA, 10% glycerol, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and 2.5 U of active recombinant caspases (EMD Millipore) was incubated for 4 h at 37°C. The reaction was stopped by addition of an equal amount of 2× SDS–10% 2-ME sample buffer, and the cleavage products were analyzed by Western blotting after separation by SDS-PAGE.
Chemical protein cross-linking.
All cross-linking experiments were performed with freshly made solution of disuccinimidyl suberate (DSS; Thermo Scientific) protein cross-linker dissolved in dimethyl sulfoxide (DMSO) as a 50 mM stock solution. For in vitro cross-linking, recombinant ORF57-FLAG protein was diluted in IP buffer. Alternatively, the cells expressing ORF57 protein for in vivo cross-linking were resuspended in phosphate-buffered saline (PBS). Both samples were then incubated with increasing amounts of DSS for 30 min at room temperature, along with DMSO (vehicle) used as a negative control. The cross-linking reaction was stopped by addition of 1 μl of 200 mM Tris-HCl (pH 7.4) and incubated for 15 min at room temperature to quench the cross-linker. The obtained protein complexes were separated by SDS-PAGE and analyzed by Gel Blue staining or by Western blotting.
Inhibition of ORF57 dimerization by synthetic peptides.
A dimerization interface of ORF57 homodimers was mapped by using a library of short synthetic peptides from the ORF57 C terminus (Table 2), which were synthesized by Peptide 2.0 Inc. (Chantilly, VA). The prepared peptides were dissolved in water to prepare a 5 mM stock solution. To allow the ORF57-peptide interaction at a ratio of 1:3, 200 ng (∼5 nM) of recombinant ORF57 protein was first preincubated with ∼15 nM (3 μl of 5 mM stock) of individual peptides in a 10-μl reaction mixture on ice for 30 min. Subsequently, for peptide dose-response evaluation, the same amount of recombinant ORF57 was incubated with increasing amounts of individual peptides. The formation of ORF57 homodimers was then tested by addition of DSS cross-linker at a final concentration of 500 μM. The cross-linking reaction was carried out as described above, and the cross-linked ORF57 complexes were analyzed by Western blotting.
TABLE 2.
ORF57-derived small synthetic peptides tested for inhibition of ORF57 homodimerization
| Peptide name | Sequence | Length (aa) | Positiona | Charge | pI |
|---|---|---|---|---|---|
| vm1 | IDGESPRFDDSIIPRHHGA | 19 | 183–201 | 0 (neutral) | 5.41 |
| vm2 | FTDRDITALIRAGGKDDE | 18 | 218–235 | −2 (acidic) | 4.19 |
| vm3 | NRLVEACNLLGEVKLNFR | 18 | 355–372 | 1 (basic) | 8.55 |
| vm4 | TSRHNQAYWVSCRRETAAAGG | 21 | 258–278 | 3 (basic) | 9.50 |
| vm5 | QTLGAFVEEQMTWAQTVVRH | 20 | 280–299 | 0 (neutral) | 5.48 |
| vm6 | TRFRLLHLSCVFDKQSE | 17 | 324–340 | 2 (basic) | 8.56 |
| vm7 | RRSISARGQELFRTLLEYYR | 20 | 389–408 | 3 (basic) | 11.09 |
| vm8 | EHHSLCRNSECAAATRAA | 18 | 422–439 | 2 (basic) | 7.42 |
| vm9 | TRAAMGSAKFNKGLFFYPLS | 20 | 436–455 | 3 (basic) | 10.90 |
The peptide position represents its amino acid residues in the KSHV ORF57 protein.
In vivo nuclear translocation assay.
The in vivo nuclear translocation assay was performed as previously described (41). Briefly, 1 × 105 COS-1 cells grown of glass coverslips in a 6-well plate were cotransfected with 200 ng of GFP-tagged “reporter” vector together with 400 ng of FLAG- or c-Myc-tagged “tester” plasmid. Twenty-four hours after transfection, the cells were fixed with cold ethanol at −20°C for 20 min, followed by immunofluorescent staining with anti-FLAG M2 or anti-c-Myc antibody (Sigma) in combination with a secondary anti-mouse Alexa Fluor 546-conjugated antibody. Cell nuclei were counterstained with Hoechst 33258 dye (Sigma) for 30 min at room temperature. The protein localization was determined by using a Zeiss LSM510 META laser-scanning confocal microscope (Zeiss), and the efficiency of translocation was determined as the percentage of double-positive cells with nuclear translocation ORF57-GFP protein.
RT-PCR.
The expression levels of ORF57 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNAs were determined by two-step reverse transcriptase PCR (RT-PCR) carried out on 1 μg of total RNA isolated with TriPure reagent (Roche) after treatment with Turbo DNase (Ambion). Reverse transcription was performed with murine leukemia virus (MULV) RT in the presence of random hexamers followed by PCR amplification using AmpliTaq (ABI) and the following gene-specific primers: oVM65 (5′-ATGCTCGCAGGGAGTCTGAG-3′) and oVM68 for ORF57 RNA and oZMZ269 (5′-GTCATCAATGGAAATCCCATCACC-3′) and oZMZ270 (5′-TGAGTCCTTCCACGATACCAAA-3′) for cellular GAPDH RNA for sample normalization.
Protein stability assay and proteasome inhibition.
To determine ORF57 protein stability, HeLa cells were transfected with vectors expressing the wild-type (pVM68) or dimerization-defective mutant (pVM85) form of ORF57 protein. Twenty-four hours after transfection, the cells were treated with 50 μg/ml of cycloheximide (CHX) for 0, 1, 2, or 4 h. At each time point, the cells were harvested directly in 2× SDS–2-ME protein sample buffer, and the level of the remaining ORF57 protein was determined Western blot analysis after normalization to neomycin phosphotransferase II or cellular β-tubulin. To inhibit ORF57 proteasome-mediated degradation, ORF57-expressing HeLa cells were treated 24 h after transfection with 20 μM proteasome inhibitor MG132 (Sigma) for 4 to 6 h or with DMSO (vehicle, negative control). The cells were then either lysed directly in SDS protein sample buffer for Western blotting or in RSB-150 buffer for IP analysis.
RESULTS
Secondary structure of KSHV ORF57 is highly conserved among its homologues.
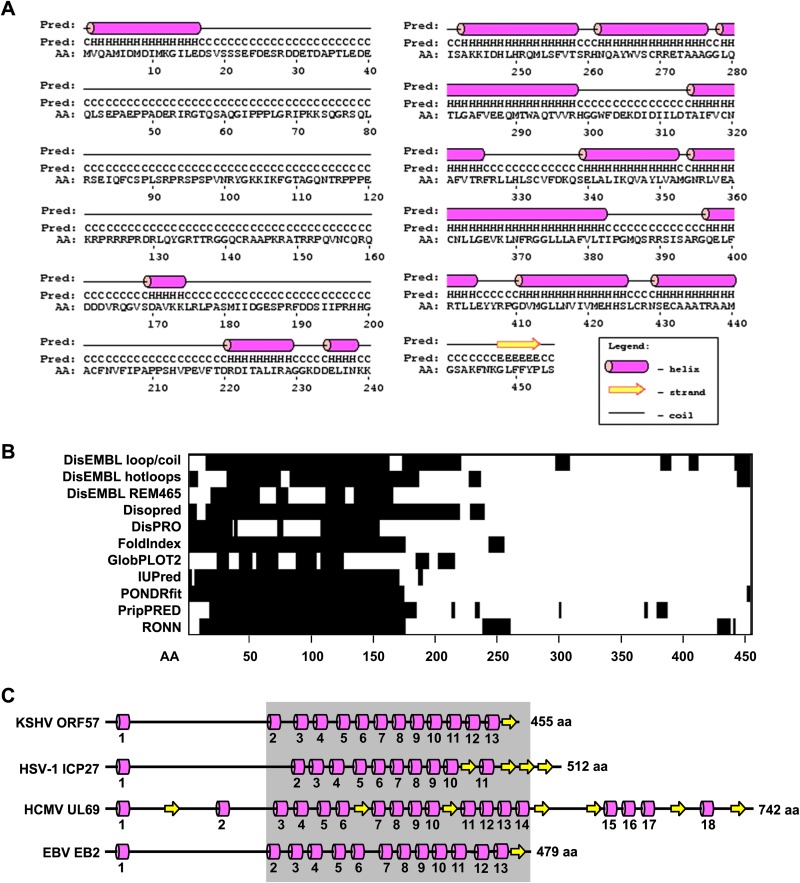
The secondary structure of the KSHV ORF57 protein (GenBank accession number YP_001129410.1) predicted by PSIPRED (version 3.0) displays 13 α-helices derived from 170 residues, representing ∼37.3% of ORF57 amino acid residues, and one β-sheet derived from only 6 aa residues, counting for ∼1.3% of ORF57 amino acid residues (Fig. 1A). The majority (279, or ∼66.3% of total ORF57 amino acid residues) of ORF57 are random coils lacking any secondary structure. The identified α-helices are unevenly distributed in the ORF57 polypeptide and are mostly clustered on the C-terminal half of ORF57, with only one α-helix at the very end (aa 2 to 16) of the N terminus of ORF57. This leaves the N-terminal half of the ORF57 protein mostly unstructured. We noticed that the predicted ORF57 structure is slightly different from that produced with other prediction algorithms (data not shown) and from the published one (15) because an updated version of PSIPRED (version 3) was used in this study.
FIG 1.
Predicted secondary structure of KSHV ORF57 protein and its conservation among ORF57 homologues. (A) Diagram of the predicted secondary structure of the ORF57 protein based on the PSIPRED algorithm. C, coil; H, α-helix; E, β-sheet/strand. (B) Positions and sizes of the intrinsically disordered regions (black boxes) in the ORF57 protein predicted by multiple algorithms (shown on the left). (C) Conservation of the secondary structure among ORF57 homologues HSV-1 ICP27, HCMV UL69, and EBV EB2, based on PSIPRED prediction (see also Fig. S1 in the supplemental material). Numbers under each α-helix represent the relative positions from the N to C terminus. Numbers at the end of each protein represent the protein length in amino acid residues. The gray box marks the region with increased sequence conservation. The drawings are not to scale.
A protein lacking the secondary structure generally indicates the presence of “intrinsically disordered” regions (IDR) with a characteristic sequence composition that contains high numbers of polar residues to cause low hydrophobicity and a high net charge. These features prevent the IDR from folding spontaneously into stable tertiary structures, thereby leading to “natively unfolded” domains with an extended conformation and high flexibility (for a review, see reference 42). Analyses of ORF57 by 11 available prediction algorithms (Fig. 1B) all consistently gave the same long IDR in the ORF57 N terminus covering approximately 200 aa residues. Also shown in Fig. 1B, a few shorter IDRs were predicted in the other parts of the ORF57 protein, but their predictions varied from one algorithm to another.
Despite some functional conservation, ORF57 homologues share very low amino acid sequence homology. To evaluate the possible structural similarities among ORF57 homologues, the secondary structures of HSV-1 ICP27 protein (GenBank accession number BAE44982.1) from the alphaherpesviruses, HCMV UL69 (GenBank accession number ACZ79980.1) from from the betaherpesviruses, and EBV EB2 (GenBank accession number YP_401659.1) from gammaherpesviruses were predicted by PSIPRED (see Fig. S1A to C in the supplemental material). The predicted secondary structures from these three ORF57 homologues (Fig. 1C) showed high similarity to the ORF57 structure and were characterized by a low density of structural motifs enriched in α-helices versus β-sheets at 11:4 in ICP27, 18:7 in UL69, and 13:1 in EB2 and an unevenly clustered distribution of the structural motifs in their C termini. As expected, the secondary structure of EBV EB2 closely resembles that of KSHV ORF57. ICP27 deviates slightly from ORF57, but UL69, the largest homologue, exhibits an additional 4 α-helices and 3 β-sheets from its unusually long C terminus. Together, we conclude that the KSHV ORF57 protein and its homologues all contain two structurally distinguishable domains, an “intrinsically disordered” N-terminal domain and a structured α-helix-rich C-terminal domain.
Limited proteolysis of the ORF57 protein.
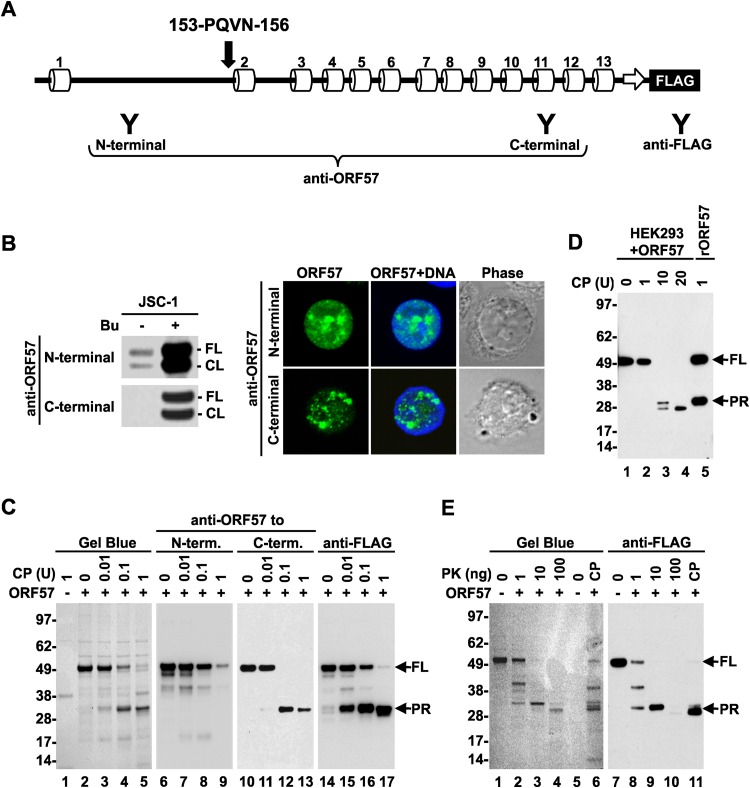
Limited proteolysis has been widely used to determine the conformation of both isolated proteins and protein complexes (43). The principle of this approach is to observe protease digestion of only flexible, unstructured protein regions, not the rigid globular domains, at low concentrations of proteases. This allows the identification of protein domains based on their distinct structures. The limited proteolysis of ORF57 was first carried out using purified recombinant ORF57-FLAG protein and clostripain, an arginine-specific protease (endoproteinase Arg-C). ORF57 contains 45 arginine residues, the most frequent residue, accounting for almost 10% of all amino acid residues in ORF57. These arginine residues are evenly distributed along the entire ORF57 polypeptide, with the longest interval between two arginines being ∼28 residues. After digestion with increasing amounts of clostripain, the cleavage products of ORF57 were resolved by SDS-PAGE (Fig. 2C) and stained with Gel Blue for total protein determination. This assay revealed a progressive digestion of the full-length (FL) ORF57 (∼50 kDa) by clostripain along with the appearance of a new protein band of approximately 30 kDa (Fig. 2C, lanes 3 to 5), which is smaller than clostripain, of size ∼38 kDa (Fig. 2C, lane 1). The data indicated that the 30-kDa band was from ORF57 and might represent the protease-resistant region (PR) of the ORF57 protein.
FIG 2.
Mapping of the ORF57 protein structure based on limited proteolysis. (A) Diagram of the ORF57 secondary structure based on PSIPRED prediction (see Fig. 1A), with a FLAG tag on the ORF57 C terminus and positions of the epitopes (Y) for separate anti-ORF57 and anti-FLAG antibody recognition. Positions and sequences of the residues as detected by Edman protein sequencing of the ORF57 PR region (PR) are shown above α-helix 2. (B) Western blot analysis of ORF57 in uninduced or 3 mM butyrate-induced JSC-1 cells with an anti-ORF57 N- or C-terminal antibody (left panel). FL, full-length; CL, cleaved. Indirect immunofluorescence staining of the butyrate-activated JSC-1 cells (right panel) was performed using an anti-N- or C-terminal ORF57 antibody in combination with an Alexa Fluor 488-conjugated secondary antibody (green). The cell nuclei were counterstained with Hoechst 33258 (blue). (C) Limited proteolysis of recombinant ORF57-FLAG protein with increasing amounts of clostripain (CP). ORF57 without digestion was used as a negative control. The cleavage products were separated by SDS-PAGE and visualized with Gel Blue staining (left panel) or by Western blotting using anti-ORF57 to the N terminus or C terminus and anti-FLAG antibodies (right panels). (D) Clostripain digestion of ectopic ORF57-FLAG protein in cell extract from HEK293 cells prepared 24 h after transfection with a pVM7 vector. The cleavage products were analyzed by Western blotting using an anti-FLAG antibody. rORF57, recombinant ORF57. (E) Cleavage of recombinant ORF57-FLAG protein with increasing amounts of proteinase K (PK). ORF57 cleaved with 1 U of clostripain served as a control. The cleaved products were analyzed with Gel Blue staining and by Western blotting using an anti-FLAG antibody as described for panel C. FL, full length. Numbers on the left are molecular markers (in kDa).
Subsequently, we analyzed the PR band derived from ORF57 by Western blotting using a set of anti-ORF57 antibodies specifically designed to target either the N- or C-terminal regions of ORF57 (Fig. 2A). Despite both antibodies recognizing full-length and caspase-7-cleaved ORF57 in butyrate-activated JSC-1 cells, based on a Western blotting assay (Fig. 2B, left panel) and immunofluorescent staining (Fig. 2B, right panel), only the anti-ORF57 C-terminal antibody, not the anti-ORF57 N-terminal antibody, reacted with the PR band (Fig. 2C, compare lanes 11 to 13 versus 7 to 9). We noticed that the full-length ORF57 level gradually decreased, along with the increasing doses of clostripain (Fig. 2C, lanes 6 to 13), and was accompanied by an increasing level of the PR product (Fig. 2C, lanes 10 to 13). These data demonstrate that the PR product from clostripain digestion represents the C terminus of the ORF57 protein. This conclusion was further verified with an anti-FLAG antibody against the FLAG tag fused to the ORF57 C terminus (Fig. 2C, lanes 14 to 17). To map the N terminus of the PR region, the cleaved PR products were sequenced by Edman protein sequencing; we identified PQVN as the last four N-terminal amino acid residues of the PR product, which mapped to aa 153 to 156 of ORF57, with two arginines at aa 151 to 152 immediately upstream, as the clostripain digestion site (Fig. 1A and 2A).
To confirm that ORF57 forms a similar structure under native conditions in cells, ORF57-FLAG was expressed in HEK293 cells and the whole-cell extract was used in a clostripain digestion at higher doses. Western blot analysis of the cleavage products with an anti-FLAG antibody revealed PR products with similar sizes as those derived from the recombinant ORF57 (Fig. 2D, compare lane 3 to lane 5), indicating that the ORF57 protein expressed in the cells in the presence of other cellular proteins displays a similar structure as recombinant ORF57 protein. However, we saw an additional band derived from further digestion of the PR products with increasing doses of clostripain (Fig. 2D, lanes 3 and 4). Finally, we confirmed the clostripain-specific digestion of ORF57 and PR production by the limited proteolysis of recombinant ORF57 protein by using a more promiscuous proteinase K, a broad-spectrum serine protease expected to cleave ORF57 at 201 sites, as predicted with the ExPASy PeptideCutter software (http://web.expasy.org/peptide_cutter). Similar to clostripain, the increased amounts of proteinase K led to progressive digestion of full-length ORF57 and increased production of the PR product similar in size to the one from clostripain digestion (Fig. 2E, compare lanes 4 and 6). Moreover, these PR products from proteinase K digestion were also recognizable with an anti-FLAG antibody (Fig. 2E, lanes 7 to 11). Collectively, our data from limited proteolysis using two proteases in combination with Edman protein sequencing indicate that the ORF57 conformation consists of an unstructured, protease-sensitive N terminus from aa 1 to 152 and a structured protease-resistant C terminus from aa 153 to 455.
The N terminus of ORF57 is essential for interaction with RNA binding proteins.
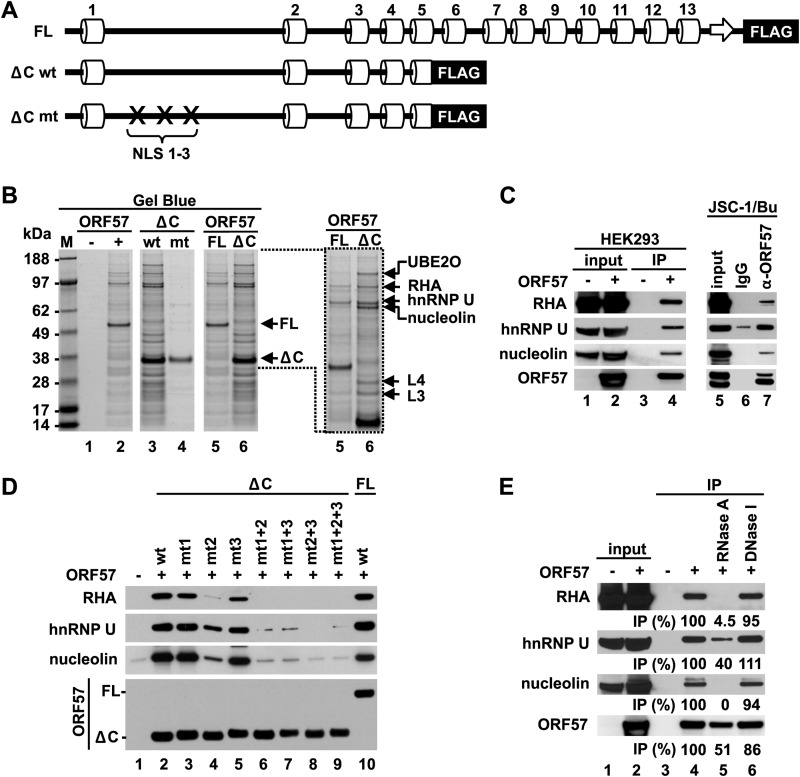
The flexibility and accessibility of the IDR would be characteristic for high binding potentials (44). To test the identified IDR in the ORF57 N terminus for its interaction with cellular proteins, the full-length (FL) ORF57 protein was compared with a C-terminal-truncated ORF57 (ΔC ORF57) containing only the N-terminal half (aa 1 to 251) (Fig. 3A) for the binding activities in the presence of HEK293 total cell extract by co-IP. We found that the protein binding profiles between FL ORF57 and ΔC ORF57 were similar (Fig. 3B), but a group of proteins subsequently identified by MS analysis, including UBE2O (ubiquitin-conjugating enzyme E2O), RHA (RNA helicase A), hnRNP U (heterogeneous nuclear ribonucleoprotein U), nucleolin, and ribosomal proteins L4 and L3, appeared to preferentially bind to ΔC ORF57 (Fig. 3B, lanes 5 and 6). The binding of such proteins may vanish by the introduction of point mutations into both NLS 1 and 2 of the ΔC ORF57 (Fig. 3B, compare lanes 4 and 3). We previously showed that the simultaneous mutations in any two of three ORF57's NLSs would be detrimental for ORF57 function, even though the mutant ORF57 (mtORF57) remains a nuclear protein (14). Together, these data indicate that the N-terminal IDR of ORF57 is a major domain that interacts with multiple cellular proteins, which might be affected minimally by the C-terminal structural domain of ORF57.
FIG 3.
The intrinsically disordered N terminus of the ORF57 protein mediates binding of ORF57 to cellular cofactors. (A) Diagrams of the secondary structure of full-length ORF57-FLAG (FL) and C-terminally truncated ORF57-FLAG (ΔC, aa 1 to 251) with wild-type (wt) or mutated (mt) NLS. (B) Binding activities of the ORF57 proteins described for panel A. Total cell extract from HEK293 cells transfected with individual ORF57-expressing vectors or an empty vector was used for IP. ORF57 complexes immunoprecipitated with an anti-FLAG antibody were resolved by SDS-PAGE and visualized by Gel Blue staining. M, molecular markers (in kDa). Enlarged portion from lanes 5 and 6 shows the most abundant proteins copurified with the ΔC ORF57 protein, identified by mass spectrometry. UBE2O, ubiquitin-conjugating enzyme E2O; RHA, RNA helicase A; hnRNPU, heterogeneous nuclear ribonucleoprotein U; L3 & L4, ribosomal proteins L3 and L4. (C) Interactions of ectopic ORF57 from HEK293 cells or endogenous ORF57 from JSC-1 cells with RHA, hnRNP U, and nucleolin, determined by IP with an anti-FLAG (left) or anti-ORF57 (right) antibody. The cells without ORF57 expression or with nonspecific immunoglobulins (IgG) for IP were used as negative controls. The immunoprecipitated proteins were analyzed by Western blotting using appropriate antibodies. Lytic KSHV infection in JSC-1 cells was activated by sodium butyrate (Bu). (D) Role of three nuclear localization signals (labeled as 1, 2, and 3) in the ORF57 interaction with RHA, hnRNP U, and nucleolin. Western blot analysis was conducted for RHA, hnRNP U, and nucleolin pulled down by an anti-FLAG antibody from HEK293 cells expressing three different forms of ORF57-FLAG fusion proteins (FL-wt, ΔC-wt, or ΔC-mt). The cells transfected with an empty vector were used as a negative control for IP specificity. (E) Role of RNA in ORF57 interaction with cellular cofactors. ORF57-associated proteins were immunoprecipitated from total cell extract of HEK293 cells expressing ORF57-FLAG by using an anti-FLAG antibody without or with prior treatment with RNase A or DNase I. Relative amount (as a percentage) of the immunoprecipitated proteins from nuclease-treated cell extract was determined by Western blotting compared to that from nuclease-untreated extract.
RHA, hnRNP U, and nucleolin were subsequently chosen for further verification. We confirmed their interactions with both ectopic ORF57 in HEK293 cells and native ORF57 in KSHV-infected JSC-1 cells by co-IP (Fig. 3C, lanes 4 and 7). Further analyses of ΔC ORF57 with point mutations in single, double, or triple NLS motifs for the binding of RHA, hnRNP U, and nucleolin demonstrated that, despite ΔC ORF57 exhibiting a comparable binding activity with FL ORF57 for these three proteins, the NLS2 in ΔC ORF57 appears to play a major role for binding of RHA and nucleolin, and introduction of point mutations into the NLS2 remarkably reduced the binding activities of ΔC ORF57 (Fig. 3D, lane 4). Introduction of point mutations into the NLS1 or NLS3 of ΔC ORF57 did not affect its interaction with any of the three proteins (Fig. 3D, compare lane 2 versus lanes 3 and 5). In contrast, ΔC ORF57 with combined mutations in both NLSs or all three NLSs lost the binding activity for all three proteins (Fig. 3D, lanes 6 to 9), which correlates with the reported activity of ORF57 in the enhancement of intronless KSHV ORF59 expression and of KSHV K8 splicing (5, 14).
Given that ORF57 in KSHV-infected cells is often associated with specific ribonucleoprotein complexes, including RNAs and proteins (15), the role of nucleic acids in the ORF57 interaction with RHA, hnRNP U, or nucleolin was tested in pulldown assays using HEK293 cell extract with or without RNase A or DNase I treatment. As expected, DNase I digestion of the cell extract produced no effect on the ORF57 association with RHA, hnRNP, or nucleolin (Fig. 3E, compare lane 6 to lane 4), but RNase A digestion of the cell extract almost completely disassociated ORF57 from RHA and nucleolin (Fig. 3E, compare lane 4 to lane 5), indicating that their interaction with ORF57 is RNA mediated. In contrast, such RNase A digestion of the cell extract led to ∼60% reduction of hnRNP U from association with ORF57 (Fig. 3E, lane 5), suggesting the presence of a partial protein-protein interaction between ORF57 and hnRNP U. Together, our data indicate that the disordered region in the ORF57 N terminus plays an essential role in ORF57 interactions with a wide range of cellular factors, either directly or indirectly.
The N-terminal region of ORF57 is posttranslationally modified by phosphorylation.
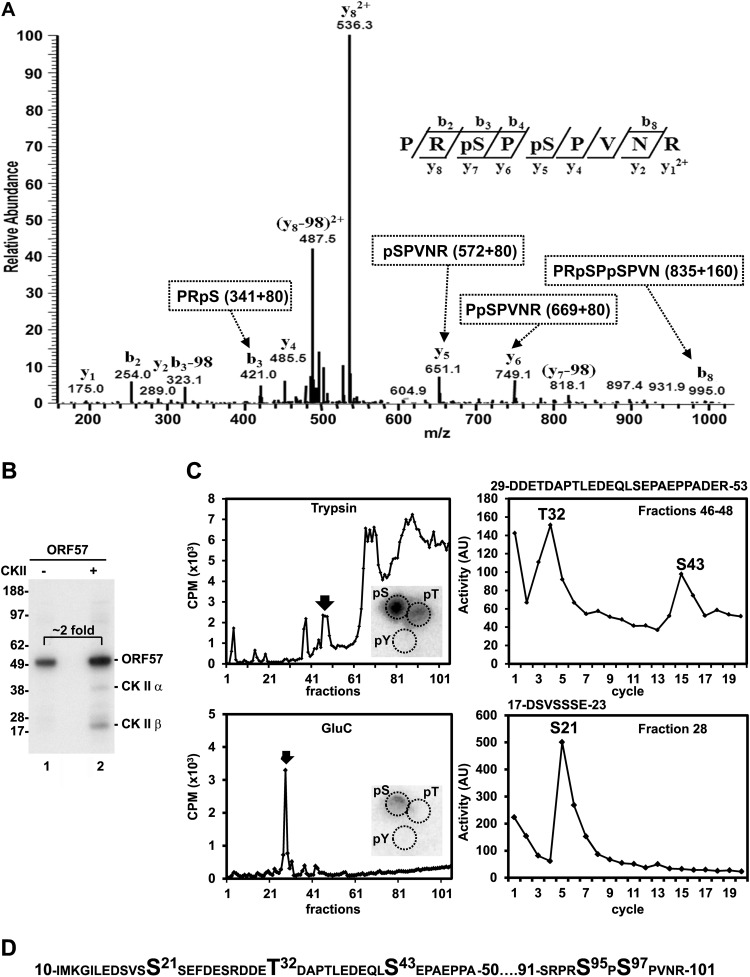
Posttranslational protein modification represents an important regulatory mechanism for many cellular processes. It has been demonstrated by proteome analysis that the IDR regions of a given protein are rich in posttranslational modifications, such as phosphorylation (45, 46). The KSHV ORF57 protein is phosphorylated by cellular CKII, but its phosphorylation sites remain to be identified (47). To experimentally map ORF57 phosphorylation sites, we took two complementary methods. First, a mass spectrometry analysis of phosphopeptides generated by trypsin digestion of recombinant ORF57 protein led to the identification of the phosphopeptide 93-PRpSPpSPVNR-101, which shows the phosphorylation of ORF57 at S95 and S97 residues (Fig. 4A). Web-based prediction of kinase phosphorylation sites in ORF57 by using the NetPhosK 1.0 server predicted glycogen synthase kinase 3 (GSK3) as a potential kinase responsible for the phosphorylation of both the S95 and S97 residues, p38MAPK kinase for S95 and ribosomal S6 kinase (RSK) for S97. Unexpectedly, neither of the two phosphorylation sites identified in ORF57 were predicted to be the sites phosphorylated by previously reported CKII (47).
FIG 4.
Phosphorylation sites identified in KSHV ORF57. (A) Identification of ORF57 phosphorylation sites by mass spectrometry. MS/MS analysis of abundance and mass of b and y ions obtained by fragmentation of ORF57 phosphopeptide aa 93 to 101 identified phosphorylated serine residues at S95 and S97. The protein sequences corresponding to individual ions and calculation of their masses are shown in the dashed boxes, with a mass of the phosphogroup equal to 80 Da. (B) SDS-PAGE autoradiograph of in vitro-phosphorylated recombinant ORF57-FLAG protein by recombinant CKII in the presence of radiolabeled [γ-32P]ATP. Numbers on the left represent the positions of protein markers (in kDa). (C) Mapping of CKII phosphorylation sites in ORF57. In vitro-phosphorylated recombinant ORF57 protein was digested with trypsin (top) or endoproteinase GluC (lower), and the resulting phosphopeptides were purified by reverse HPLC based on their activity (left). The phosphopeptides fractionated with high activity (marked by arrows in the left panels) were separately subjected to Edman protein sequencing (right panels) or phosphoamino acid analysis (insets). The activity (in arbitrary units [AU]) of each Edman sequencing cycle is shown on the right, together with the corresponding peptide sequence, which is shown above each panel. Insets in the left panels show phosphoamino acid analysis of the CKII-phosphorylated, trypsin-digested, or GluC-digested recombinant ORF57 fragments in the combined fractions from 46 to 48 or in the fraction 28 by 2D chromatography. pS, phosphoserine; pT, phosphothreonine; pY, phosphotyrosine. (D) A partial protein sequence of ORF57, with all five phosphorylated residues identified by two methods.
Therefore, in our next analysis, the recombinant ORF57 protein was first incubated in vitro with recombinant CKII in the presence of radiolabeled [γ-32P]ATP. This led to in vitro phosphorylation of ORF57 and autophosphorylation of CKII α and β subunits (Fig. 4B, lane 2). We also unexpectedly observed ORF57 phosphorylation in the absence of CKII, with ∼2-fold-lower intensity than that in the presence of CKII (Fig. 4B, compare lanes 1 and 2). This in vitro autophosphorylation of the ORF57 protein appeared to occur in the absence of any other phosphorylated protein (Fig. 4B, lane 1). To further map the positions of phosphorylation sites, the CKII-phosphorylated ORF57 protein was digested with trypsin or endoproteinase GluC, and the fractionated phosphopeptides with high activity were subjected to Edman peptide sequencing and phosphoamino acid analysis. This led to identification of three additional ORF57 phosphorylation sites at the S21, T32, and S43 residues. Phosphoamino acid analysis of the fractionated phosphopeptides confirmed the specific phosphorylation of serine (Ser or S) and threonine (Thr or T), but not tyrosine (Tyr or Y), which is in agreement with CKII being a serine/threonine-selective protein kinase (Fig. 4C, insets). Interestingly, the same analyses of autophosphorylated ORF57 protein in the absence of CKII also identified the phosphorylation of ORF57 at the S21, T32, and S43 residues (data not shown). The similarity of these mapped phosphorylation sites to the ones in CKII-phosphorylated ORF57 suggests ORF57 phosphorylation by a residual amount of insect CKII copurified with the recombinant ORF57 protein from Sf21 cells, as functional conservation of CKII has been reported among eukaryotes (48). Nevertheless, this study led us to experimentally identify five phosphorylation sites from the ORF57 N terminus (Fig. 4D) which are all localized in the IDR region, a preferential region for posttranslational modification of the ORF57 protein.
Phosphorylation and caspase-7 cleavage of KSHV ORF57.
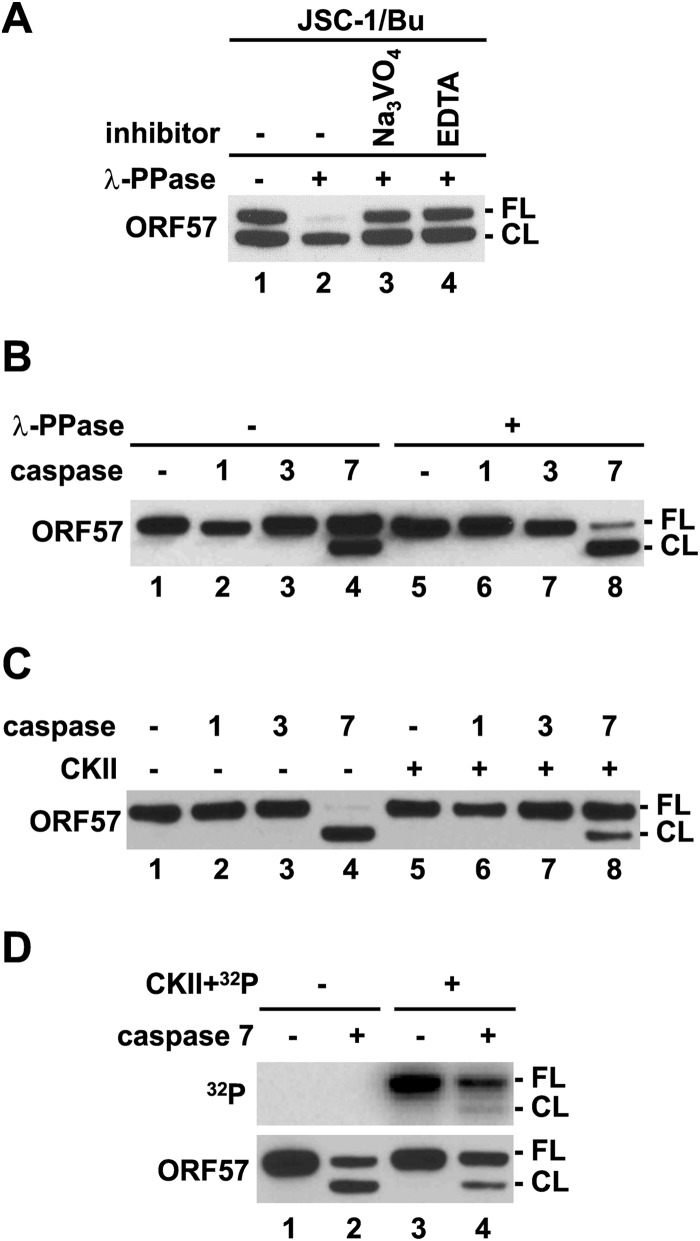
Protein phosphorylation is a reversible posttranslational modification which affects protein localization, folding, binding activities, stability, etc. It has been shown that phosphorylation modulates substrate recognition by cellular caspases (49). The KSHV ORF57 protein contains a caspase-7 cleavage site at aspartic residue D33, and its cleavage by caspase-7 produces a truncated, functionally attenuated form of ORF57 (38). Given that all identified CKII phosphorylation sites in ORF57 reside in close proximity to the caspase-7 cleavage site, with phosphorylation site T32 being positioned directly within the caspase-7 recognition site 30-DETD-33, we examined how phosphorylation and dephosphorylation of ORF57 regulate ORF57 cleavage by caspase-7. The whole-cell extract prepared from KSHV-infected JSC-1 cells expressing ORF57 after lytic reactivation by sodium butyrate was treated with nonspecific λ-PPase. Because KSHV reactivation in JSC-1 cells activates cellular caspases to cleave ORF57 protein (38), JSC-1 extract without λ-PPase treatment usually exhibits two forms of ORF57, an FL form and a faster-migrating cleaved form (CL), at a ratio of approximately 1:1 (Fig. 5A, lane 1). We found that λ-PPase treatment of the same extract strongly enhanced ORF57 cleavage (Fig. 5A, lane 2), and two λ-PPase inhibitors, sodium orthovanadate and EDTA, blocked the λ-PPase-induced ORF57 cleavage (Fig. 5A, compare lane 2 to lanes 3 and 4), indicating a direct effect of dephosphorylation on ORF57 cleavage. To further confirm this observation, recombinant ORF57 protein was treated with or without λ-PPase followed by in vitro cleavage assays with recombinant caspase-1, -3, or -7 (Fig. 5B). As expected, ORF57 in the absence of λ-PPase pretreatment was cleaved by caspase-7 by ∼50% (Fig. 5B, compare lane 1 to lane 4), whereas dephosphorylation of the ORF57 by λ-PPase significantly increased ORF57 cleavage (to ∼90%) by caspase-7 (Fig. 5B, compare lane 5 to lane 8). Under both conditions, ORF57 was not cleavable by caspase-1 or -3 (Fig. 5B, lanes 1 to 3 and 5 to 7). These data suggest that protein dephosphorylation promotes ORF57 cleavage specifically by caspase-7.
FIG 5.
Phosphorylation inhibits ORF57 cleavage by caspase-7. (A and B) Dephosphorylation of ORF57 promotes caspase cleavage of ORF57. (A) Cell extracts from JSC-1 cells induced with sodium butyrate (Bu) were treated with λ-PPase in the absence or presence of λ-PPase inhibitors orthovanadate (Na3VO4) or EDTA. Cell extract without any treatment served as a negative control. The cleaved ORF57 was detected by Western blotting using an anti-ORF57 N-terminal antibody. FL, full length ORF57; CL, cleaved ORF57. (B) Recombinant ORF57-FLAG protein with or without λ-PPase pretreatment was incubated with recombinant caspase-1, -3, or -7, and the cleavage products were detected by Western blotting as described for panel A. ORF57 protein without caspase digestion served as a negative control. (C and D) Phosphorylation of ORF57 inhibits caspase cleavage. Recombinant ORF57-FLAG protein without or with prior in vitro phosphorylation by recombinant CKII was cleaved with recombinant caspase-1, -3, or -7 and analyzed by Western blotting as described for panel A. Autoradiograph (upper panel) of caspase-7-digested recombinant ORF57 without or with phosphorylation by CKII plus [γ-32P]ATP (D, upper panel). The same membrane was then blotted with an anti-ORF57 antibody (D, lower panel). ORF57 without caspase digestion served as a control.
To further confirm the inhibitory effect of phosphorylation on ORF57 cleavage by capsase-7, recombinant ORF57 was first phosphorylated in vitro by CKII followed by caspase cleavage with recombinant caspase-1, -3, or -7 (Fig. 5C and D). Again, ORF57 without CKII treatment was cleaved by caspase-7, but not by caspase-1 or -3 (Fig. 5C, lanes 1 to 4, and D, lane 2). As expected, in vitro phosphorylation of ORF57 by CKII inhibited the cleavage of ORF57 by caspase-7 (Fig. 5C, lane 4 versus lane 8, and D, lane 2 versus lane 4). Moreover, we found that, in the presence of [γ-32P]ATP, CKII predominantly mediated the incorporation of γ-32P into the FL ORF57 to protect it from caspase-7 cleavage, and not much so into the cleaved ORF57 (Fig. 5D, compare top panel versus lower panel in lane 4), suggesting that other phosphorylation sites remaining in the cleaved ORF57 are less sensitive to CKII kinase. In summary, this study demonstrated that phosphorylation in the ORF57 N-terminal IDR inhibits ORF57 cleavage by cellular caspase-7 and plays an important role in regulation of ORF57 protein stability.
ORF57 homodimerization via its structural C terminus.
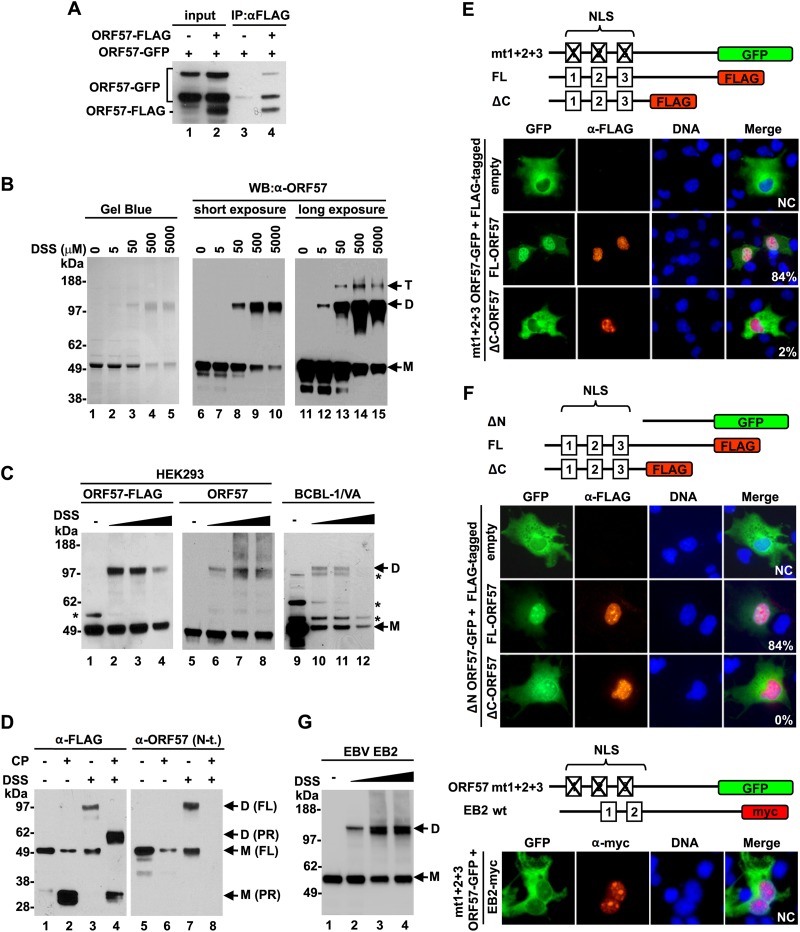
We next wished to determine the role of the structural C terminus in ORF57 self-interaction and function, because KSHV ORF57 and its homologues have been shown to form multimeric complexes by self-interaction via their C terminus (41, 50–52). However, these studies did not provide any hard evidence to show homodimerization of ORF57. To do this, we first repeated the association of two forms of ORF57 proteins tagged either with GFP or with FLAG by co-IP after their coexpression in HEK293 cells. Western blotting with an anti-ORF57 N terminus antibody that recognized both forms of ORF57 revealed efficient pulldown of ORF57-GFP by an anti-FLAG antibody from the cells cotransfected with ORF57-FLAG, but not from control cells cotransfected with an empty vector (Fig. 6A, compare lane 3 to lane 4), indicating the association of ORF57-GFP with ORF57-FLAG in the cells. To exclude the possibility that the observed association between two tagged ORF57 proteins was not mediated by other cellular proteins, we used recombinant ORF57 protein in an in vitro homodimerization assay. The recombinant ORF57 protein diluted in IP buffer was incubated at room temperature, and the expected ORF57 homodimers were stabilized by the addition of DSS, a noncleavable bivalent chemical cross-linker that is commonly used to detect direct protein-protein interactions. The resulting complexes were resolved by SDS-PAGE. Gel Blue protein staining showed ORF57 monomers (M) of ∼50 kDa without DSS (Fig. 6B, lane 1). With increasing doses of DSS, the intensity of ORF57 monomers reduced gradually along with the appearance of the predicted ORF57 homodimers, of ∼100 kDa (Fig. 6B, lanes 2 to 5). Both ORF57 monomers and dimers were specifically detectable with an anti-ORF57 N-terminal antibody (Fig. 6B, lanes 6 to 15). A longer exposure time for the Western blot assay revealed a small amount of ORF57 homotrimers of ∼150 kDa (Fig. 6B, lanes 13 to 15). Since DSS is membrane permeable, the formation of an ORF57 dimer was also examined in HEK293 cells expressing ectopic ORF57 with or without a C-terminal FLAG tag or in BCBL-1 cells with KSHV lytic infection and native ORF57 expression. As expected, Western blotting with an anti-ORF57 antibody showed that the samples without DSS contained only monomeric ORF57 (Fig. 6C, lanes 1, 5, and 9), whereas DSS treatment led to the formation of ORF57 homodimers in both types of cells (Fig. 6C, lanes 2 to 4, 6 to 8, and 10 and 11). We were unable to detect ORF57 homotrimers in the cells, presumably because of its much lower abundance. We noticed that DSS at 5 mM for in vivo cross-linking sometimes induced large protein aggregates that were hardly separable by SDS-PAGE.
FIG 6.
Homodimerization of ORF57 via the structured C-terminal domain. (A) Self-interaction of ORF57 in vivo. ORF57-GFP and ORF57-FLAG fusion proteins were coexpressed in HEK293 cells. ORF57-FLAG was immunoprecipitated with an anti-FLAG antibody, and proteins in the pulldown were analyzed by Western blotting with an anti-ORF57 N-terminal antibody. Cells cotransfected with ORF57-GFP and empty FLAG vectors (-) served as negative controls. (B) ORF57 homodimerization in vitro. Recombinant ORF57-FLAG protein was incubated with increasing amounts of the bivalent chemical cross-linker DSS. The cross-linked complexes were resolved by SDS-PAGE and analyzed with Gel Blue staining or by Western blotting using an anti-ORF57 N-terminal antibody. M, monomer; D, dimer; T, trimer. (C) ORF57 homodimerization in vivo. HEK293 cells expressing ORF57 with or without a C-terminal FLAG tag or KSHV-infected, valproic acid (VA)-activated BCBL-1 cells were incubated with increasing concentrations (50, 500, and 5,000 μM) of DSS, and the cross-linked proteins were analyzed by Western blotting with an anti-ORF57 N-terminal antibody. *, uncharacterized protein band. (D to F) Role of the structured C-terminal domain in ORF57 homodimerization. (D) Proteolytic digestion of recombinant ORF57-FLAG protein with clostripain (CP) with or without cross-linker DSS. The cleavage products were detected by Western blotting with an anti-FLAG or anti-ORF57 N-terminal (N-t.) antibody. ORF57 without CP digestion or DSS cross-linking served as negative controls. M, monomer; D, dimer; FL, full length. (E and F) Results of ORF57 translocation assays, with the upper panels showing the diagrams of ORF57 proteins fused with GFP or FLAG tags (color boxes). Numbered boxes in ORF57 are three NLSs, with a cross indicating its mutation. ORF57 localization in COS-1 cells cotransfected with a GFP-tagged ORF57 mutant (mt1+2+3 ORF57-GFP) or ΔN ORF57 (ΔN ORF57-GFP) as a GFP reporter together with a FLAG-tagged full-length or ΔC ORF57 vector. Cells cotransfected with the GFP reporters and an empty FLAG vector served as negative controls. Twenty-four hours after transfection, the cells were fixed and immunostained with an anti-FLAG antibody and then an Alexa Fluor 546-labeled secondary antibody (red). Localization of the ORF57-GFP fusion was detected with direct fluorescence (green) of the cells. The cell nuclei were counterstained with Hoechst 33258 dye (blue). Fifty cells positive for the coexpression of both ORF57-GFP and ORF57-FLAG proteins on each slide were examined for subcellular localization of the ORF57-GFP fusion. The percentage of cells positive for nuclear localization of ORF57-GFP due to dimerization of ORF57-GFP with ORF57-FLAG is shown in the right corner. NC, not counted. (G) EBV EB2 can be a homodimer, but it does not heterdimerize with KSHV ORF57. (Left) Formation of EB2 homodimer in HEK293 cells expressing c-myc-tagged EB2 protein. The cells at 24 h after transfection were harvested in PBS and incubated with increasing concentrations (50, 500, and 5,000 μM) of DSS for in vivo cross-linking. EB2 complexes were detected by Western blotting using anti-c-myc monoclonal antibody. (Right) Cytoplasmic ORF57-GFP translocation assays with myc-tagged EBV EB2. COS-1 cells were cotransfected with the cytoplasmic ORF57-GFP (mt1+2+3 ORF57-GFP) reporter along with c-myc-tagged EBV EB2 protein (EB2-myc). Twenty-four hours after transfection, nuclear localization of ORF57-GFP or EB2-myc was examined by immunofluorescent staining using an anti-FLAG or anti-c-Myc antibody in combination with an Alexa Fluor 546-labeled secondary antibody (red). Cell nuclei were counterstained with Hoechst dye (blue).
The contribution of the structural C terminus to ORF57 dimerization was further investigated in vitro by DSS cross-linking followed by clostripain digestion. We assumed that, if ORF57 homodimerization were mediated by its C terminus, proteolytic removal of the N terminus of ORF57 after DSS cross-linking would result in a smaller homodimer derived from the C terminus. Indeed, Western blot analysis of the cross-linked ORF57 after clostripain digestion with an anti-FLAG antibody to the ORF57 C-terminal FLAG tag identified a dimer ∼60 kDa derived from two cross-linked C termini (PR), each ∼30 kDa (Fig. 6D, compare lane 4 to lane 2). This dimer derived from the C termini after clostripain digestion was not detectable by Western blotting with an anti-ORF57 N terminus antibody (Fig. 6D, lane 8), further confirming its origin.
Using an in vivo nuclear translocation assay as our third approach to verify ORF57 dimerization in cells, we conducted the assay with a cytoplasmic “reporter” protein lacking a functional NLS due to a mutation (mt1+2+3 ORF57-GFP) or deletion of 166 aa from the ORF57 N terminus (ΔN ORF57-GFP) coexpressed with a “tester” protein that contained a functional NLS (ORF57-FLAG or ΔC ORF57-FLAG) in COS-1 cells (Fig. 6E and F protein diagrams) (14). Analysis of 50 cells positive for the coexpression of both ORF57-GFP and ORF57-FLAG proteins showed that ORF57-GFP reporters were efficiently (∼84%) translocated from the cytoplasm to the nucleus of COS-1 cells in the coexpression of the full-length ORF57-FLAG, but not much so in the coexpression of the ΔC ORF57-FLAG (Fig. 6E and F). The nuclear translocation was ORF57 specific, because EBV EB2, a close ORF57 homologue from human gammaherpesvirus, did not show such a function, although EBV EB2 homodimerizes in transfected cells (Fig. 6G). Together, these data provide convincing evidence that the ORF57 C terminus is essential for formation of ORF57 homodimers but incapable of forming a heterodimer with EBV EB2.
ORF57 homodimerization via the C-terminal α-helices 7 to 9.
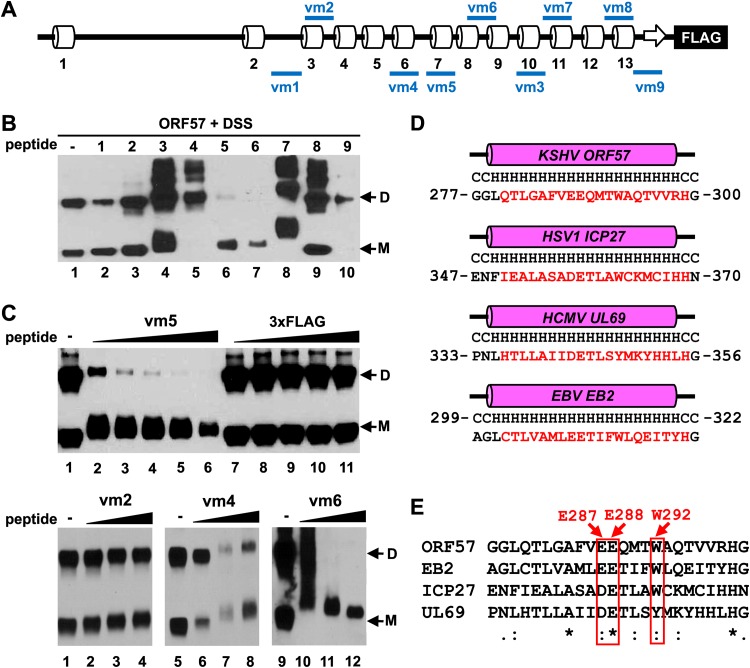
Considering that small synthetic peptides derived from interaction interface can inhibit protein-protein interactions (53, 54) and in the interest of identifying the ORF57 dimerization interface, we synthesized a set of 9 peptides from various regions of the ORF57 C terminus targeting both structural (α-helices) and unstructural (loops) regions (Fig. 7A; see also Fig. S2 in the supplemental material) using the following criteria: maximum length of 21 aa residues with a minimal 30% hydrophobic residues to enable high solubility in water (see Table 2 for details). The activity of individual peptides was tested in the in vitro dimerization assay described above, using recombinant ORF57 protein. As shown in Fig. 7B, peptides vm1 and vm2 exhibited no effect on ORF57 homodimerization (lanes 2 and 3), neither for peptides vm3, vm7, nor vm8, but the latter three peptides induced the formation of larger ORF57 complexes, presumably due to their excessive cross-linking to ORF57 (Fig. 7B, lanes 4, 8, and 9). Peptides vm4 and vm9 also displayed no effect on ORF57 homodimerization but appeared to induce the instability of monomeric ORF57 (lanes 5 and 10). Interestingly, peptide vm5, derived from ORF57 C-terminal aa 280 to 299 (α-helix 7), and peptide vm6, from aa 324 to 340 (an unstructural region between α-helices 8 and 9), were inhibitory to ORF57 homodimerization (lanes 6 and 7). By comparison with 3×FLAG peptide, serving as a negative control (Fig. 7C, lanes 7 to 11, upper panel), the peptide vm5 inhibited ORF57 homodimerization in a dose-dependent manner (Fig. 7C, lanes 2 to 6, upper panel) with a decreased mobility of ORF57 monomers by several kilodaltons, indicating cross-linking of the peptide vm5 (2.33 kDa) to ORF57. As expected, similar results were observed with the peptide vm6, but not with the peptide vm2 or vm4, which does not affect ORF57 homodimerization (Fig. 7C, three lower panels). The vm5-corresponding regions (α-helix 7) in ORF57 homologues, including HSV-1 ICP27, HCMV UL69, and EBV EB2, are a highly conserved region in structure and amino acid sequence composition (Fig. 7D and E). All homologues contain a 20-aa-long α-helix (helix 6 in ICP27 or EB2 and α-helix 9 in UL69) (Fig. 7D) and several fully conserved residues (Fig. 7E). The vm6 corresponding region among the homologues also has similar features (see Fig. S2 in the supplemental material). Taken together, our data indicate that ORF57 homodimerization requires the vm5 and vm6 corresponding regions.
FIG 7.
Inhibition of ORF57 homodimerization using small peptides. (A) Diagram of the predicted secondary structure of ORF57 protein along with positions of synthetic peptides vm1 to vm9 (blue lines). See Table 2 for peptide details. (B) Examination of individual peptides that affect ORF57 homodimerization. Recombinant ORF57-FLAG protein was first incubated with each peptide in a protein:peptide molar ratio of 1:3 (5 nM ORF57:15 nM peptide) for 30 min, followed by cross-linking with DSS (500 μM) and analysis by Western blotting with an anti-ORF57 N-terminal antibody. M, monomer; D, dimer. (C) Dose-dependent effect of peptide vm5 on ORF57 homodimerization. Western blotting of ORF57-FLAG protein after incubation with increasing concentrations (5, 10, 15, 20, and 25 nM) of peptide vm5 or with control 3×FLAG peptide was conducted with an anti-ORF57 N-terminal antibody. ORF57 without cross-linking with any peptide or in the presence of increasing concentrations (5, 10, and 15 nM) of peptide vm2, vm4, or vm6 was used as a control. (D and E) Conservation of the secondary structure (D) and amino acid sequence (E) of the peptide vm5-corresponding region in ORF57 homologues. PSIPRED prediction (D) and ClustalW2 alignment (E) were used to analyze KSHV ORF57, HSV-1 ICP27, HCMV UL69, and EBV EB2 proteins. Consensus symbols (asterisk, semicolon, and period) were used according to ClustalW2 methods. In the red boxes are conserved amino acids E287, E288, and W292, mutated for further analysis.
Disruption of ORF57 homodimerization via mutation of the peptide vm5 region.
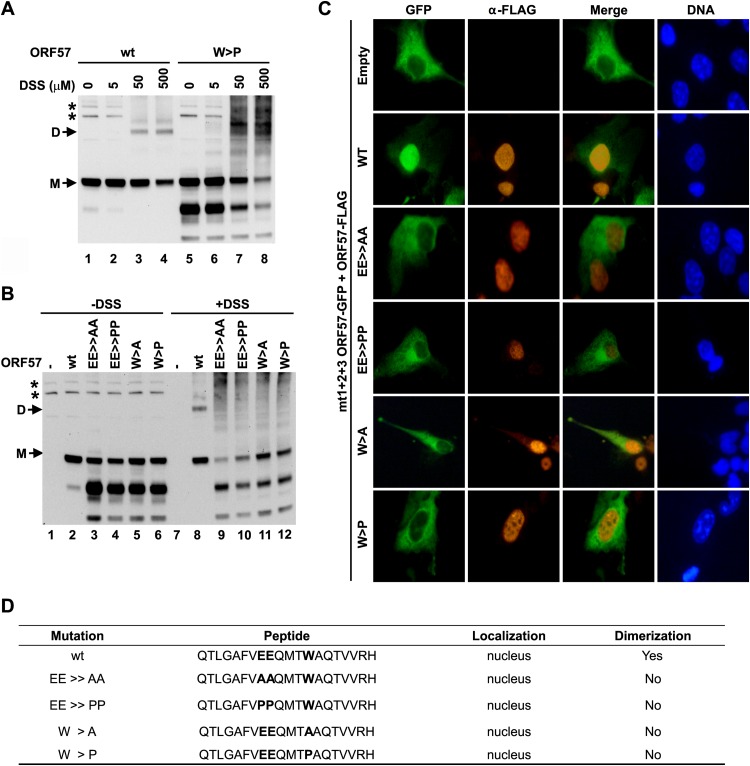
To further investigate the role of the peptide vm5 corresponding region in ORF57 dimerization, the conserved residues E287, E288, and W292 within α-helix 7 were mutated in the context of full-length ORF57. The selected amino acids were mutated to either alanine, to eliminate the amino acid side chain, or to helix-breaking proline (EE ≫ AA, EE ≫ PP, W > A, W > P). Resulting mutants were then tested for their ability to homodimerize after expression in HEK293 cells by DSS-mediated cross-linking. As shown in Fig. 8A, wt ORF57 homodimers were detectable in the samples with DSS (lanes 1 to 4), but the W > P mutant failed to efficiently homodimerize at all three tested doses of DSS (lanes 5 to 8). We noticed that, although the W > P mutant was unstable, lack of dimerization of the mutant was not a result of its instability and appeared even when the expression level of the mutant was justified to that of wt ORF57 (compare lanes 6 to 8 with lanes 2 to 4 for the monomeric ORF57 level). Similarly, none of the other mutants was able to form homodimers when tested in the same assay in the presence of DSS (Fig. 8B), with a justified protein level close to that of wt ORF57. It is worth noting that the mutant proteins lacking homodimerization might appear as a high-molecular-weight smear in HEK293 cells in the presence of high-dose DSS (Fig. 8A and B), indicating nonspecific cross-linking to other unknown cellular proteins. The formation of ORF57 homodimer was further verified by nuclear translocation assays using the cytoplasmic mt1+2+3 ORF57-GFP construct as a reporter. Compared with wt ORF57-FLAG, the mutants with all types of mutations in the peptide vm5 corresponding region displayed no ability to translocate the cytoplasmic mt1+2+3 ORF57-GFP into the nucleus, indicating dysfunctional dimerization of the mutants with the reporter (Fig. 8C). Their properties are summarized in Fig. 8D. In conclusion, the peptide vm5 corresponding region (aa 280 to 299) at α-helix 7 of the ORF57 protein is critical for ORF57 homodimerization mediated by the amino acid residues highly conserved among ORF57 homologues.
FIG 8.
Introduction of point mutations into the peptide vm5-corresponding region in ORF57 prevents ORF57 homodimerization. (A) Homodimerization capability of wt ORF57 and its W > P mutant in HEK293 cells at increasing concentrations of DSS. HEK293 cells at 5 × 105 cells in a 6-well plate were transfected with 0.2 μg (for wt) or 2 μg (for mutant) ORF57-FLAG vectors. The cells at 24 h after transfection were treated with 20 μM MG132 for an additional 6 h and harvested in PBS for in vivo cross-linking with DSS, and then cell lysates were detected by Western blotting with an anti-ORF57 N-terminal polyclonal antibody. *, a nonspecific protein band. (B) ORF57 protein carrying point mutations of E287A and E288A (EE > AA), E287P and E288P (EE > PP), W292A (W > A), and W292P (W > P) are all incapable of homodimerizing in HEK293 cells at 24 h after transfection in the presence of DSS (500 μM). Further deatils are provided in panel A. (C) In vivo translocation assays in COS-1 cells cotransfected with mt1+2+3 ORF57-GFP together with a wild-type (wt) or mutant ORF57-FLAG fusion, described for panel B. Localization of ORF57-GFP (green) and ORF57-FLAG (red) proteins was examined by immunofluorescence microscopy at 24 h after transfection. The cell nuclei were counterstained with Hoechst 33258 dye (blue). (D) Summary of various mutations in the peptide vm5-corresponding region that affect ORF57 homodimerization.
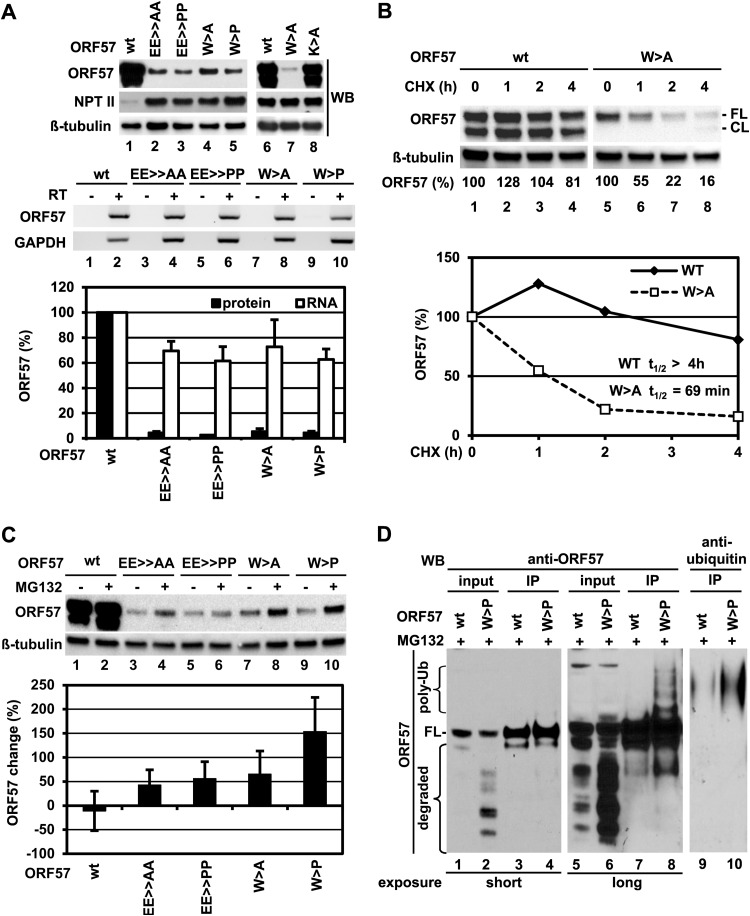
Inhibition of ORF57 homodimerization induces degradation of ORF57.
We noticed that introduction of point mutations into the peptide vm5 corresponding region led to a significant reduction of ORF57 protein expression in HeLa cells (Fig. 9A, compare lanes 2 to 5 to lane 1), but a dimerable K345A mutant which contained a point mutation outside the peptide vm6 corresponding region did not show such a reduction (Fig. 9A, compare lane 8 to lane 6). Compared to wt ORF57 RNA (Fig. 9A, middle panel) and its protein level (Fig. 9A, top panel), all ORF57 mutants showed a relative RNA level of ∼65% and a protein level of ∼5% of that for wt ORF57 (Fig. 9A, bar graphs). Obviously, the significantly reduced level of a mutant protein was not correlated to a minimally reduced RNA level and thus could not be caused at the transcriptional or posttranscriptional level.
FIG 9.
Protein dimerization is important for ORF57 stability. (A) Steady-state protein and RNA levels of wt ORF57 and its dimerization-defective mutants in HeLa cells. A dimerable K345A mutant (K > A) outside the vm6-corresponding region was used as a control. (Top) Western blot analysis of ORF57 proteins in HeLa cells transfected with an equal amount of individual ORF57 expression vectors, together with NPT II and β-tubulin, used as transfection efficiency and loading controls. The cell lysate sample from wt ORF57 was loaded only as a half (lanes 1 to 5) or an equal amount (lanes 6 to 8) of its mutant samples for Western blotting. (Middle) Relative levels of ORF57 RNA in the same sample set (lanes 1 to 5 for protein assays) determined by RT-PCR using ORF57-specific primers. (Bottom) Mean ± standard deviations of relative protein levels versus RNA levels of individual ORF57 proteins after normalization to β-tubulin (protein) or to GAPDH (RNA), based on three independent experiments. (B) Dimerization-defective mt ORF57 has a shorter half-life. HeLa cells were transfected with wt ORF57 or its W > A mutant for 24 h and then subjected to translation inhibition for the indicated time by using 50 μM CHX. The relative protein level of ORF57 at each time point (in hours) was determined by Western blotting using an anti-ORF57 N-terminal antibody. Cellular β-tubulin served as a loading control (top). The half-life of wt or mt ORF57 was determined from a line plot analysis, with the expression level at time zero set to 100%. (C) Degradation of ORF57 mutants can be blocked by MG132, a proteasome inhibitor. HeLa cells were first transfected with wt ORF57 or its dimerization-defective mutants for 24 h. Protein levels of ORF57 proteins in HeLa cells with or without MG132 (20 μM) for additional 4 h were examined by Western blotting using an anti-ORF57 N-terminal antibody (top). Bar graphs below the blot, with means ± standard deviations from three independent experiments, show the change (percentage) of each ORF57 protein in the presence of MG132 versus the absence of MG132, based on the signal intensity of each protein band after normalization to β-tubulin. (D) Accumulation of high-molecular-mass forms of mt ORF57 in the presence of MG132. FLAG-tagged ORF57 wt and the W > P mutant were immunoprecipitated with an anti-FLAG antibody from HeLa cells after 6 h of 20 μM MG132 treatment, followed by Western blotting with an anti-ORF57 N-terminal antibody (lanes 1 to 8) or K48-specific anti-polyubiquitin rabbit monoclonal antibody (lanes 9 and 10).
To examine whether an ORF57 mutant had a short half-life, we compared wt ORF57 with the W > A mutant for protein stability in HeLa cells treated with CHX in a time course manner. CHX is an inhibitor of protein biosynthesis in eukaryotes as it blocks protein translational elongation (55). Western blot analysis revealed that a large amount (81%) of wt ORF57 protein remained detectable even at 4 h after blocking translation by CHX (Fig. 9B, lanes 1 to 4). In contrast, the W > A mutant decreased over time, with only 16% of the protein left at 4 h after CHX blocking (Fig. 9B, lanes 5 to 8). From our calculation using a line plot analysis, we determined that the half-life (t1/2) of wt ORF57 is more than 4 h (the last time point for the sample collection in the study), and the mutant half-life is only about ∼1 h (Fig. 9B, line graph). Interestingly, we found that, although the W > A mutant was less stable, it resisted caspase cleavage (Fig. 9B, lanes 5 and 6), suggesting a correlation between ORF57 homodimerization and its caspase digestion.
As the majority of intracellular proteins in eukaryotic cells are degraded by cellular proteasomes (56), the effects of the proteasome inhibitor MG132 on the expression of wt ORF57 and its dimerization-defective mutants were tested in HeLa cells (Fig. 9C). Western blotting of protein samples showed an ∼50% increase in the protein levels of EE ≫ AA, EE ≫ PP, and W > A mutants and an ∼150% increase in the protein level of the W > P mutant in the presence of MG132 over that in the absence of MG132 (Fig. 9C, lanes 3 to 10 of top panel and the line graph in the lower panel), whereas wt ORF57 protein remained constant, exhibiting no visible changes under either condition (Fig. 9C, lanes 1 and 2, top panel), suggesting that the dimerization-defective mutants are prone to degradation via a cellular proteasome pathway. Given that proteasomic degradation is generally initiated by polyubiquitination of a target protein, the polyubiquitinated protein often exhibits an increase in protein molecular weight. However, the polyubiquitinated protein may not be detectable under physiological condition due to its rapid degradation. To examine this possible polyubiquitination in the dimerization-defective mutants, the W > P mutant was compared with wt ORF57 protein in HeLa cells treated with MG132 for 6 h. As expected, the mutant protein was heavily degraded with multiple degraded protein fragments easily detectable by Western blotting with an anti-ORF57 N terminus antibody (Fig. 9D, compare lanes 2 and 6 versus lanes 1 and 5 for input protein). Western blotting of wt ORF57 and the W > P mutant in the IP pulldown indicated that only the W > P mutant presented the higher-molecular-mass protein forms (polyubiquinated protein forms) recognizable by an anti-ORF57 N terminus antibody (Fig. 9D, compare lanes 8 versus 7) and by a K48-specific anti-polyubiquitin rabbit monoclonal antibody (Fig. 9D, lanes 9 and 10). The data demonstrate polyubiquitination of the dimerization-defective W > P mutant.
DISCUSSION
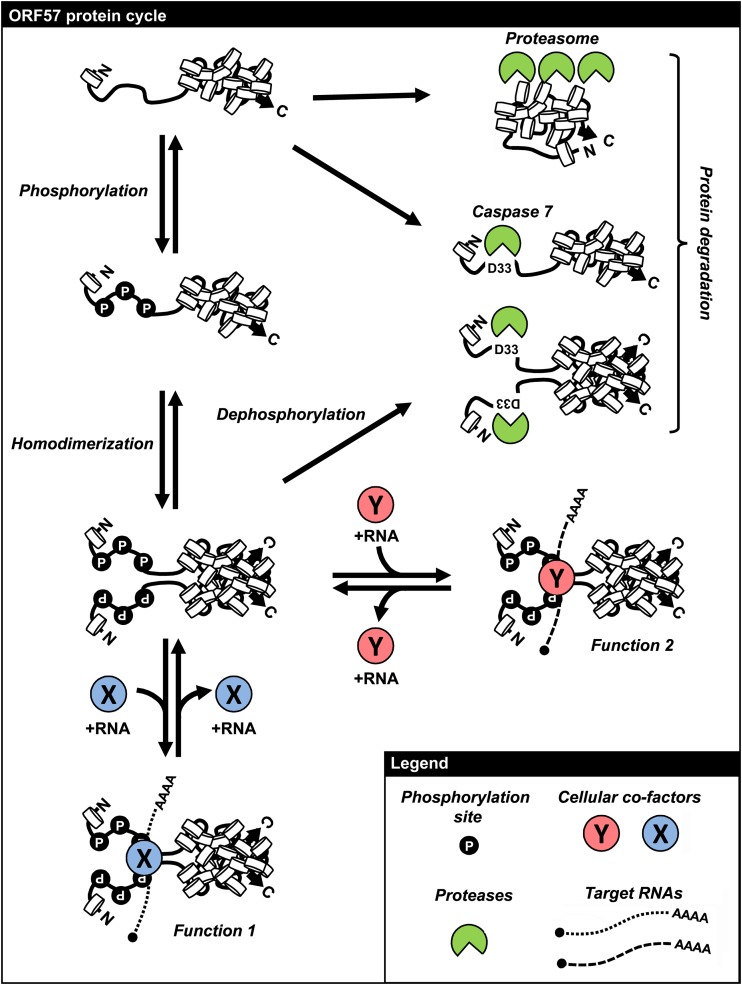
KSHV ORF57 belongs to a family of sequentially conserved poorly but functionally well-conserved homologues found in all herpesviruses (15). To date, for none of the family members has a 3D structure been solved by nuclear magnetic resonance (NMR) or X-ray crystallography, due to their poor expression in bacteria and precipitation during purification (unpublished data). In this study, we present the first comprehensive structural analysis of KSHV ORF57 protein using multiple computational prediction methods and experimental approaches. We demonstrated that the ORF57 protein displays two structurally distinct domains, an intrinsically disordered N-terminal domain and a structured α-helix-rich C-terminal domain. The N-terminal domain is essential for the ORF57 interaction with cellular cofactors and their associated RNAs, while the C-terminal domain mediates ORF57 homodimerization. This structure of ORF57 contributes to two divergent modes of protein stability. Phosphorylation of the unstructured N terminus by cellular CKII kinase prevents ORF57 cleavage by cellular caspase-7, while homodimerization of ORF57 monomer protects ORF57 from degradation by cellular proteasomes. With such structural features, the ORF57 protein has a long half-life to regulate viral gene expression at the posttranscriptional level for efficient KSHV replication and virus production.
The characteristic structures identified in KSHV ORF57 appear to be conserved among the analyzed ORF57 homologues in other members of the herpesvirus family (41, 57, 58) (Fig. 1C). A large-scale analysis of various proteomes conducted in the past decades discovered that the intrinsically disordered proteins (IDPs) or IDRs represent a group of highly dynamic structures for different conformations induced by binding to a specific ligand (42). With unique sequence composition enriched in charged and hydrophilic residues, IDPs and IDRs interact with numerous ligands to exert a high binding capacity and are common in multifunctional proteins, such as the ones in cell signaling and in the viral proteome (59). We confirmed in this study and based on other reports (3, 5, 7, 14, 16) that the identified N-terminal IDR within the first ∼250 aa residues of ORF57 is a major binding interface for ORF57 to interact with numerous cellular partners and RNAs and is highly conserved among other homologues as determined with the ANCHOR software (60, 61) (data not shown). While the structured C-terminal domain of ORF57 might play a role for a cofactor to efficiently bind to the ORF57 IDR, the finding that RBM15 and SRSF3 bind to ORF57 in a mutually exclusive manner due to both proteins binding to the same site in the N-terminal IDR (7) excludes such a possibility.
Phosphorylation is a reversible posttranslational modification in many fundamental cellular regulatory processes. Although CKII phosphorylation of KSHV ORF57 has been demonstrated via CKII β-subunit binding to the ORF57 IDR at aa 181 to 215 (47), we mapped for the first time in this study the ORF57 phosphorylation sites at S21, T32, S43, S95, and S97, all in the ORF57 N terminus. Consistently, the CKII-phosphorylated residues in ICP27 (S16, S18, and S114) (62) and in EB2 (S55, S56, and S57) (63) are also within the N termini. Given that IDRs bear predominant sites of protein posttranslational modification (45), we were able to correctly predict the majority of ORF57, ICP27, UL69, and EB2 phosphorylation sites (data not shown) by using DISPHOS, a disorder-enhanced phosphorylation sites predictor. Although CKII is a ubiquitous kinase responsible for phosphorylation of approximately a quarter of all phosphorylation sites identified in eukaryotic phosphoproteomes (64) and is a common kinase in phosphorylation of serine/threonine residues in ORF57 and its homologues, another kinase(s) may phosphorylate ORF57 at S95 and S97. In this study, recombinant ORF57 purified from Sf21 cells could be phosphorylated by itself in the presence of [γ-32P]ATP. Because the phosphorylation sites mapped in the self-phosphorylated protein were the same as the ones in CKII-phosphorylated ORF57 and CKII among eukaryotes is functionally conserved (48), we interpreted this self-phosphorylation to be a result of insect CKII copurified with recombinant ORF57 protein from Sf21 cells. This is in agreement with the observation that immunopurified ICP27 protein also fails to autophosphorylate (62).
Unexpectedly, we found that CKII phosphorylation prevents ORF57 from caspase-7 cleavage (38) and regulates ORF57 stability. Although all of the mapped CKII phosphorylation sites are within the N-terminal IDR of ORF57, in proximity to a caspase-7 cleavage site, 30-DETD-33 and, in particular, the CKII phosphorylation site T32 is positioned at the P2 site in the caspase-7 recognition site, T32 phosphorylation most likely contributes to the prevention of ORF57 caspase-7 cleavage. Consistently, other studies have also indicated that phosphorylation within or in close proximity to a caspase recognition motif can alter cellular protein sensitivity to caspase cleavage (49). Phosphorylation of threonine T58 at the P2 position in the caspase recognition motif by CKII or CKI prevents human Bid cleavage by caspase-8 (65). Phosphorylation of YY1 by CKIIα prevents cleavage by caspase-7 during apoptosis (66). CKII phosphorylation during virus infection plays an important role in regulation of ICP27 and EB2 binding activities to cellular cofactors and promotes virus replication of HSV-1 and EBV (63, 67, 68).
ORF57 and its homologues are self-interacting proteins via their C-terminal domains (41, 47, 50). However, little is known about the nature of the suggested protein dimer or even larger multimeric complexes (41). Using chemical cross-linking and limited clostripain proteolysis, we demonstrated that homodimers are the prevailing form of ORF57 both in vitro and in vivo. Although ORF57 also forms homotrimers, homodimers and monomers are the only two forms seen both in BCBL-1 cells with lytic KSHV infection and in other ORF57-transfected human cells. A FLAG tag attached to the ORF57 C terminus does not affect ORF57 dimerization. ORF57 does not form a heterodimer with the EBV EB2 protein, a closely related homologue that also forms a homodimer in mammalian cells. By comparing the N-terminal half to the C-terminal half of ORF57 in nuclear translocation of a cytoplasmic reporter protein, the C-terminal half of ORF57 was also identified for the homodimerization of ORF57 as reported previously (47, 52). By using small peptides derived from the ORF57 C terminus, we further identified the peptide vm5-corresponding α-helix 7, which is highly conserved among ORF57 homologues, as an interaction interface for the ORF57 homodimer. Introduction of point mutations into the conserved residues within the region blocked ORF57 homodimerization and triggered rapid degradation of ORF57. These data are in contrast to the report that the RGG2 motif (aa 372 to 374) in α-helix 10 is critical for ORF57 multimerization (52). Experimentally, we were unable to confirm the role of the RGG2 motif in α-helix 10 in ORF57 homodimerization because using a peptide vm3 corresponding to α-helix 10 did not affect dimerization of ORF57 (Fig. 7B). It might be that the RGG2 mutation changes the overall ORF57 structure, as those author suggested (52).
Homodimerization of KSHV ORF57 exerts two important functions: (i) preventing ORF57 from proteasomic degradation and (ii) making two flexible N-terminal IDRs in the dimer work coordinately to bind host factors (Fig. 10). One possible mechanism by which such homodimerization prevents ORF57 from degradation is to mask the ubiquitination sites located in the C terminus of ORF57. This presumption was partially supported by the observation that the dimerization-defective ORF57 mutants, but not wt ORF57, responded to treatment with MG132, a widely used proteasomal inhibitor. The similar regulation of protein stability by dimerization occurs for heterodimerization of PEBP2β with RUNX/AML1 and of ATF2 with c-Jun (69, 70). One caveat is that the dimerization-defective ORF57 mutants become resistance to cleavage by caspase-7 (38). This observation clearly indicates the existence of interplay between the N terminus and C terminus in monomeric ORF57. Although an intramolecular head-to-tail interaction has been observed in ICP27 (71), further study is needed to understand how this cross-talk between the two terminal domains of monomeric ORF57 affects its cleavage by caspase-7.
FIG 10.
ORF57 protein cycle. KSHV ORF57 features an intrinsically disordered N-terminal domain and a highly structured C-terminal domain. This protein is translated initially as a monomeric protein and undergoes N-terminal phosphorylation by host CKII or other kinases. The monomeric form of ORF57 is subjected to cleavage by caspase-7 or degradation via the proteasomal pathway. Two phosphorylated monomeric ORF57 proteins dimerize through C-terminal α-helix 7, resulting in the formation of an ORF57 homodimer. ORF57 homodimers interact with cellular protein-RNA complexes to exert various functions of ORF57 or otherwise are subject to caspase digestion.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the intramural research programs of the National Cancer Institute, National Institutes of Health. Calvin Chan and Nicholas Temkin took part in the NIH Summer Internship Program in Biomedical Research.
We thank Michal Kruhlak at the Experimental Immunology Branch of NCI for his technical support with confocal microscopy and Vladimir N. Uversky of the University of South Florida for his critical reading of the manuscript and suggestions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03721-14.
REFERENCES
- 1.Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int J Biol Sci 7:1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahin BB, Patel D, Conrad NK. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog 6:e1000799. doi: 10.1371/journal.ppat.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majerciak V, Deng M, Zheng ZM. 2010. Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology 407:206–212. doi: 10.1016/j.virol.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J, Pavlakis GN, Felber BK, Zheng ZM. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J Virol 85:1528–1540. doi: 10.1128/JVI.01709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. 2008. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol 82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol 85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majerciak V, Lu M, Li X, Zheng ZM. 2014. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 20:1747–1758. doi: 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8α, and K8.1 and the production of infectious virus. J Virol 81:1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Z, Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol 80:5251–5260. doi: 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J 29:1851–1864. doi: 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilkington GR, Majerciak V, Bear J, Uranishi H, Zheng ZM, Felber BK. 2012. Kaposi's sarcoma-associated herpesvirus ORF57 is not a bona fide export factor. J Virol 86:13089–13094. doi: 10.1128/JVI.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J Virol 81:9990–9998. doi: 10.1128/JVI.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson BR, Noerenberg M, Whitehouse A. 2014. A novel mechanism inducing genome instability in Kaposi's sarcoma-associated herpesvirus infected cells. PLoS Pathog 10:e1004098. doi: 10.1371/journal.ppat.1004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem 281:28365–28378. doi: 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- 15.Majerciak V, Zheng ZM. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front Biosci 14:1516–1528. doi: 10.2741/3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massimelli MJ, Majerciak V, Kruhlak M, Zheng ZM. 2013. Interplay between polyadenylate-binding protein 1 and Kaposi's sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J Virol 87:243–256. doi: 10.1128/JVI.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik P, Blackbourn DJ, Clements JB. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem 279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 18.Boyne JR, Colgan KJ, Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog 4:e1000194. doi: 10.1371/journal.ppat.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osinalde N, Olea M, Mitxelena J, Aloria K, Rodriguez JA, Fullaondo A, Arizmendi JM, Zubiaga AM. 2013. The nuclear protein ALY binds to and modulates the activity of transcription factor E2F2. Mol Cell Proteomics 12:1087–1098. doi: 10.1074/mcp.M112.024158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virbasius CM, Wagner S, Green MR. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol Cell 4:219–228. doi: 10.1016/S1097-2765(00)80369-X. [DOI] [PubMed] [Google Scholar]
- 21.Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt U, Im KB, Benzing C, Janjetovic S, Rippe K, Lichter P, Wachsmuth M. 2009. Assembly and mobility of exon-exon junction complexes in living cells. RNA 15:862–876. doi: 10.1261/rna.1387009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li DJ, Verma D, Swaminathan S. 2012. Binding of cellular export factor REF/Aly by Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication. J Virol 86:9866–9874. doi: 10.1128/JVI.01190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubbs SH, Hunter OV, Hoover A, Conrad NK. 2012. Viral factors reveal a role for REF/Aly in nuclear RNA stability. Mol Cell Biol 32:1260–1270. doi: 10.1128/MCB.06420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatfield D, Izaurralde E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longman D, Johnstone IL, Caceres JF. 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9:881–891. doi: 10.1261/rna.5420503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson LA, Li L, Sandri-Goldin RM. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J Virol 83:6335–6346. doi: 10.1128/JVI.00375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandri-Goldin RM. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci 13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- 29.Toth Z, Stamminger T. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front Biosci 13:2939–2949. doi: 10.2741/2899. [DOI] [PubMed] [Google Scholar]
- 30.Swaminathan S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J Cell Biochem 95:698–711. doi: 10.1002/jcb.20465. [DOI] [PubMed] [Google Scholar]
- 31.Boyne JR, Colgan KJ, Whitehouse A. 2008. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front Biosci 13:2928–2938. doi: 10.2741/2898. [DOI] [PubMed] [Google Scholar]
- 32.Moriuchi H, Moriuchi M, Smith HA, Cohen JI. 1994. Varicella-zoster virus open reading frame 4 protein is functionally distinct from and does not complement its herpes simplex virus type 1 homolog, ICP27. J Virol 68:1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 34.Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 35.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 36.Cannon JS, Ciufo D, Hawkins AL, Griffin CA, Borowitz MJ, Hayward GS, Ambinder RF. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J Virol 74:10187–10193. doi: 10.1128/JVI.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J Virol 80:11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majerciak V, Kruhlak M, Dagur PK, McCoy JP Jr, Zheng ZM. 2010. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J Biol Chem 285:11297–11307. doi: 10.1074/jbc.M109.068221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guszczynski T, Specht SI, Copeland TD. 2006. A microtiter plate fraction collector for the sequencing of radioactive phosphorylated peptides. Anal Biochem 356:151–153. doi: 10.1016/j.ab.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 40.Laronde-LeBlanc N, Guszczynski T, Copeland T, Wlodawer A. 2005. Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes. FEBS J 272:2800–2810. doi: 10.1111/j.1742-4658.2005.04702.x. [DOI] [PubMed] [Google Scholar]
- 41.Lischka P, Thomas M, Toth Z, Mueller R, Stamminger T. 2007. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J Gen Virol 88:405–410. doi: 10.1099/vir.0.82480-0. [DOI] [PubMed] [Google Scholar]
- 42.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. 2014. Classification of intrinsically disordered regions and proteins. Chem Rev 114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. 2004. Probing protein structure by limited proteolysis. Acta Biochim Pol 51:299–321. [PubMed] [Google Scholar]
- 44.Davey NE, Van RK, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. 2012. Attributes of short linear motifs. Mol Biosyst 8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 45.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. 2008. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol Cell Proteomics 7:1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Malik P, Clements JB. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res 32:5553–5569. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Dotan I, Ziv E, Dafni N, Beckman JS, McCann RO, Glover CV, Canaani D. 2001. Functional conservation between the human, nematode, and yeast CK2 cell cycle genes. Biochem Biophys Res Commun 288:603–609. doi: 10.1006/bbrc.2001.5804. [DOI] [PubMed] [Google Scholar]
- 49.Tozser J, Bagossi P, Zahuczky G, Specht SI, Majerova E, Copeland TD. 2003. Effect of caspase cleavage-site phosphorylation on proteolysis. Biochem J 372:137–143. doi: 10.1042/BJ20021901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhi Y, Sciabica KS, Sandri-Goldin RM. 1999. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 257:341–351. doi: 10.1006/viro.1999.9698. [DOI] [PubMed] [Google Scholar]
- 51.Bello LJ, Davison AJ, Glenn MA, Whitehouse A, Rethmeier N, Schulz TF, Barklie CJ. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J Gen Virol 80:3207–3215. [DOI] [PubMed] [Google Scholar]
- 52.Taylor A, Jackson BR, Noerenberg M, Hughes DJ, Boyne JR, Verow M, Harris M, Whitehouse A. 2011. Mutation of a C-terminal motif affects Kaposi's sarcoma-associated herpesvirus ORF57 RNA binding, nuclear trafficking, and multimerization. J Virol 85:7881–7891. doi: 10.1128/JVI.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lian W, Upadhyaya P, Rhodes CA, Liu Y, Pei D. 2013. Screening bicyclic peptide libraries for protein-protein interaction inhibitors: discovery of a tumor necrosis factor-alpha antagonist. J Am Chem Soc 135:11990–11995. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benyamini H, Friedler A. 2010. Using peptides to study protein-protein interactions. Future Med Chem 2:989–1003. doi: 10.4155/fmc.10.196. [DOI] [PubMed] [Google Scholar]
- 55.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 57.Sandri-Goldin RM. 2011. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol 6:1261–1277. doi: 10.2217/fmb.11.119. [DOI] [PubMed] [Google Scholar]
- 58.Tunnicliffe RB, Hautbergue GM, Kalra P, Jackson BR, Whitehouse A, Wilson SA, Golovanov AP. 2011. Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog 7:e1001244. doi: 10.1371/journal.ppat.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, Uversky VN, Longhi S. 2014. Structural disorder in viral proteins. Chem Rev 114:6880–6911. doi: 10.1021/cr4005692. [DOI] [PubMed] [Google Scholar]
- 60.Dosztanyi Z, Meszaros B, Simon I. 2009. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meszaros B, Simon I, Dosztanyi Z. 2009. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol 5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhi Y, Sandri-Goldin RM. 1999. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J Virol 73:3246–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Medina-Palazon C, Gruffat H, Mure F, Filhol O, Vingtdeux-Didier V, Drobecq H, Cochet C, Sergeant N, Sergeant A, Manet E. 2007. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J Virol 81:11850–11860. doi: 10.1128/JVI.01421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venerando A, Ruzzene M, Pinna LA. 2014. Casein kinase: the triple meaning of a misnomer. Biochem J 460:141–156. doi: 10.1042/BJ20140178. [DOI] [PubMed] [Google Scholar]
- 65.Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. 2001. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell 8:601–611. doi: 10.1016/S1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 66.Riman S, Rizkallah R, Kassardjian A, Alexander KE, Luscher B, Hurt MM. 2012. Phosphorylation of the transcription factor YY1 by CK2α prevents cleavage by caspase 7 during apoptosis. Mol Cell Biol 32:797–807. doi: 10.1128/MCB.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rojas S, Corbin-Lickfett KA, Escudero-Paunetto L, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants are defective in herpes simplex virus 1 replication and gene expression. J Virol 84:2200–2211. doi: 10.1128/JVI.00917-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corbin-Lickfett KA, Rojas S, Li L, Cocco MJ, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants display altered functional interactions with cellular export factors Aly/REF and TAP/NXF1 but are able to bind herpes simplex virus 1 RNA. J Virol 84:2212–2222. doi: 10.1128/JVI.01388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. 2001. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J 20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuchs SY, Ronai Z. 1999. Ubiquitination and degradation of ATF2 are dimerization dependent. Mol Cell Biol 19:3289–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez FP, Sandri-Goldin RM. 2010. Head-to-tail intramolecular interaction of herpes simplex virus type 1 regulatory protein ICP27 is important for its interaction with cellular mRNA export receptor TAP/NXF1. mBio 1(5):00268-10. doi: 10.1128/mBio.00268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.