ABSTRACT
The human microbiome influences and reflects the health or disease state of the host. Periodontitis, a disease affecting about half of American adults, is associated with alterations in the subgingival microbiome of individual tooth sites. Although it can be treated, the disease can reoccur and may progress without symptoms. Without prognostic markers, follow-up examinations are required to assess reoccurrence and disease progression and to determine the need for additional treatments. To better identify and predict the disease progression, we aim to determine whether the subgingival microbiome can serve as a diagnosis and prognosis indicator. Using metagenomic shotgun sequencing, we characterized the dynamic changes in the subgingival microbiome in periodontitis patients before and after treatment at the same tooth sites. At the taxonomic composition level, the periodontitis-associated microorganisms were significantly shifted from highly correlated in the diseased state to poorly correlated after treatment, suggesting that coordinated interactions among the pathogenic microorganisms are essential to disease pathogenesis. At the functional level, we identified disease-associated pathways that were significantly altered in relative abundance in the two states. Furthermore, using the subgingival microbiome profile, we were able to classify the samples to their clinical states with an accuracy of 81.1%. Follow-up clinical examination of the sampled sites supported the predictive power of the microbiome profile on disease progression. Our study revealed the dynamic changes in the subgingival microbiome contributing to periodontitis and suggested potential clinical applications of monitoring the subgingival microbiome as an indicator in disease diagnosis and prognosis.
IMPORTANCE
Periodontitis is a common oral disease. Although it can be treated, the disease may reoccur without obvious symptoms. Current clinical examination parameters are useful in disease diagnosis but cannot adequately predict the outcome of individual tooth sites after treatment. A link between the subgingival microbiota and periodontitis was identified previously; however, it remains to be investigated whether the microbiome can serve as a diagnostic and prognostic indicator. In this study, for the first time, we characterized the subgingival microbiome of individual tooth sites before and after treatment using a large-scale metagenomic analysis. Our longitudinal study revealed changes in the microbiota in taxonomic composition, cooccurrence of subgingival microorganisms, and functional composition. Using the microbiome profiles, we were able to classify the clinical states of subgingival plaque samples with a high accuracy. Follow-up clinical examination of sampled sites indicates that the subgingival microbiome profile shows promise for the development of diagnostic and prognostic tools.
INTRODUCTION
Microbes live symbiotically with humans and play important roles in human health and disease. Changes in the microbiome contribute to the pathogenesis of many diseases (1, 2) and reflect the health or disease state of the host. Therefore, monitoring the changes in the microbiome is a promising potential new application in disease diagnosis and prognosis (3–5).
Periodontitis is a disease associated with alterations in the subgingival microbiome. This significant oral disease affects approximately 47% of American adults and more than 740 million people in the world with an increasing prevalence as people age (6, 7). Although not fatal, it can result in tooth loss and significantly reduce the quality of life. During disease onset and progression, pathogenic bacteria colonize the periodontal sulcus/pocket to form subgingival oral biofilms, which adhere primarily to the tooth root surface causing inflammation in the periodontal tissues (8). In the severe forms of the disease, destruction of periodontal tissues leads to progressive attachment loss, bone loss, and ultimately, tooth loss. Chronic periodontal disease infection elicits the host immune system to mount an inflammatory response, and the inflammatory response is associated with a number of systemic and chronic diseases (9). Although the disease can be treated, periodontal health needs to be continuously monitored after initial treatment and resolution of disease, because the disease may reoccur and progress without obvious symptoms in some individuals (10). Risk factors and clinical indicators for tooth loss, a surrogate marker of periodontal disease, have been suggested for prognostication (11, 12). The authors found that commonly used clinical parameters for predicting tooth loss were useful but did not adequately predict the relationship between initial findings and disease progression. With no reliable prognostic markers available to predict disease progression, identification of patients with treatment needs occurs only after tissue destruction is evident. Hence, development of effective prognostic tools is essential to identify those patients in need of close follow-up and treatment.
Studies of the subgingival microbial community identified a link between taxonomic composition and disease pathogenesis. The classic study by Socransky et al. revealed disease associations of specific bacterial organisms, including Porphyromonas, Treponema, and Tannerella, which were classified as the red complex organisms (13). Recent studies using 16S rRNA gene sequencing analysis provided a more comprehensive view of the subgingival community in health and disease. By analyzing samples pooled from different tooth sites and/or individuals, several case-control studies revealed distinct differences in the taxonomic composition for healthy and diseased states (14–17). Additionally, two longitudinal studies compared the taxonomic composition of the subgingival microbiome before and after treatment (18, 19). These two studies, albeit limited to two and four subjects, respectively, suggested an altered microbiome following treatment. Since periodontitis is a disease of individual subgingival pockets, the use of pooled samples in these previous studies provided an incomplete overall assessment that did not distinguish between the individual microbiomes represented by each affected site. Detailed knowledge of the dynamic changes in the microbiome within each pocket for different clinical states will be extremely useful in diagnosis and prognosis of individual tooth sites in clinical practice.
Although the taxonomic composition of the subgingival community has been characterized, the functional potentials encoded in the microbiome largely remain to be characterized. By comparing pooled samples from periodontitis patients to those from healthy individuals, Li et al. (17) and Liu et al. (15) revealed associations between disease and several metabolic pathways and virulence factors. However, it remains to be investigated whether the functional profiles of the subgingival microbiome are different in the healthy and disease states. Furthermore, it is unclear whether differences in the functional profile would be reflective of disease progression and could thus serve as markers in disease diagnosis and prognosis.
To better understand the polymicrobial mechanism of periodontitis, in this study, we investigated the dynamic changes in the subgingival microbiome within individual pockets before treatment (diseased state) and after treatment (resolved state). Both the taxonomic composition and functional composition of the microbiome were determined using metagenomic shotgun sequencing analysis. Additionally, we evaluated whether the subgingival microbiome profile can be used to classify and predict the clinical states of the periodontium for disease diagnosis and prognosis.
RESULTS
Sample collection and sequencing.
We performed a longitudinal study investigating the dynamic changes in the subgingival microbiome. We recruited 12 systemically healthy adults with chronic periodontitis and sampled multiple tooth sites per subject before and after nonsurgical initial therapy. The therapy consisted of scaling and root planing (SRP). Oral hygiene instructions were also given to the subjects. There is substantial evidence indicating that successful nonsurgical therapy is compatible with sustained disease resolution (20). Only tooth sites that were clinically resolved after the initial therapy were selected for the longitudinal study. On average, we analyzed samples from two sites per subject. Forty-eight subgingival samples were sequenced using a metagenomic shotgun approach with a total of 7.43 billion reads. An average of 155 million reads was analyzed for each sample.
Taxonomic composition of the subgingival microbiome.
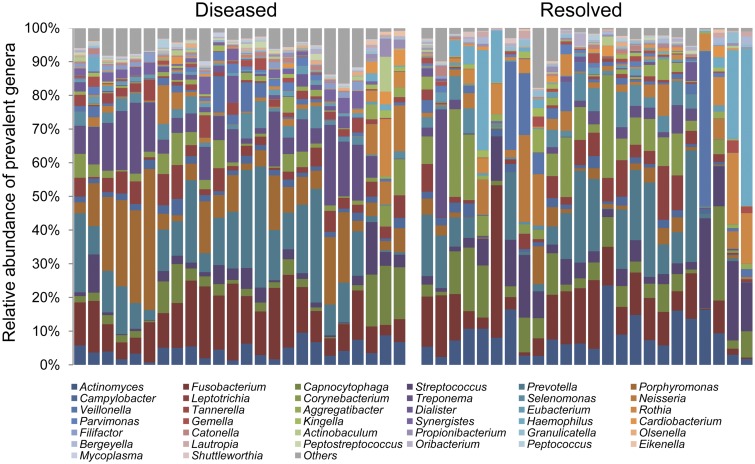
To determine the taxonomic composition of the subgingival microbiome, we extracted 16S rRNA sequences from the metagenomic shotgun sequencing data. We mapped the sequences against the SILVA rRNA database (21) and two human oral microbiome databases, the Human Oral Microbiome Database (HOMD) (22) and the Ohio State University (OSU) CORE database (23). On average, 48,140 16S rRNA sequence reads were obtained for each sample. These reads provided sufficient sequencing depth for detection of microorganisms in all samples as shown by the rarefaction curves (see Fig. S1 in the supplemental material). In total, 38 bacterial genera that had an abundance of greater than 1% in samples from at least two subjects were identified (Fig. 1). Prevotella and Fusobacterium were among the most abundant genera. All 38 genera have previously been associated with the human oral cavity (22).
FIG 1 .
Taxonomic composition of the subgingival microbiome. Thirty-eight genera were identified with a relative abundance of greater than 1% in samples from at least two subjects. The paired samples, collected before and after treatment at the same tooth sites, are ordered in the same way in the diseased group (before treatment) (left) and the resolved group (after treatment) (right).
To verify the above results, we used two other methods to determine the taxonomic composition of the subgingival microbiome. We established a set of oral microbial reference genomes, which consisted of 298 bacterial species from 103 genera (see Text S1 in the supplemental material). Using a method similar to that of Schloissnig et al. (24), instead of analyzing only the 16S rRNA reads, we mapped all metagenomic shotgun sequence reads against our reference genome set. The taxonomic composition of the microbiome was determined based on the mapped reference genomes with a breadth of greater than 0.4. Additionally, as confirmation, we constructed full-length 16S rRNA clone libraries for 8 of the 48 samples and sequenced by the Sanger method (Text S1 in the supplemental material). The taxonomic compositions determined by the three methods were highly consistent with each other (see Fig. S2 in the supplemental material).
Dynamic changes in the subgingival microbiome.
Our longitudinal comparison of the subgingival microbiome revealed significant shifts in the taxonomic composition from the diseased state to the resolved state within each periodontal site. We found that the microbial community evenness and richness (alpha diversity) significantly decreased after treatment (P < 0.029 [see Fig. S3 in the supplemental material]). This is in agreement with the hypothesis that periodontal disease is associated with alterations of the complex microbial community rather than dominance of a single pathogen. This observation of decreased diversity in the resolved state is consistent with previous case-control studies where the microbiome in the naive healthy state had lower diversity than in the diseased state (14–16).
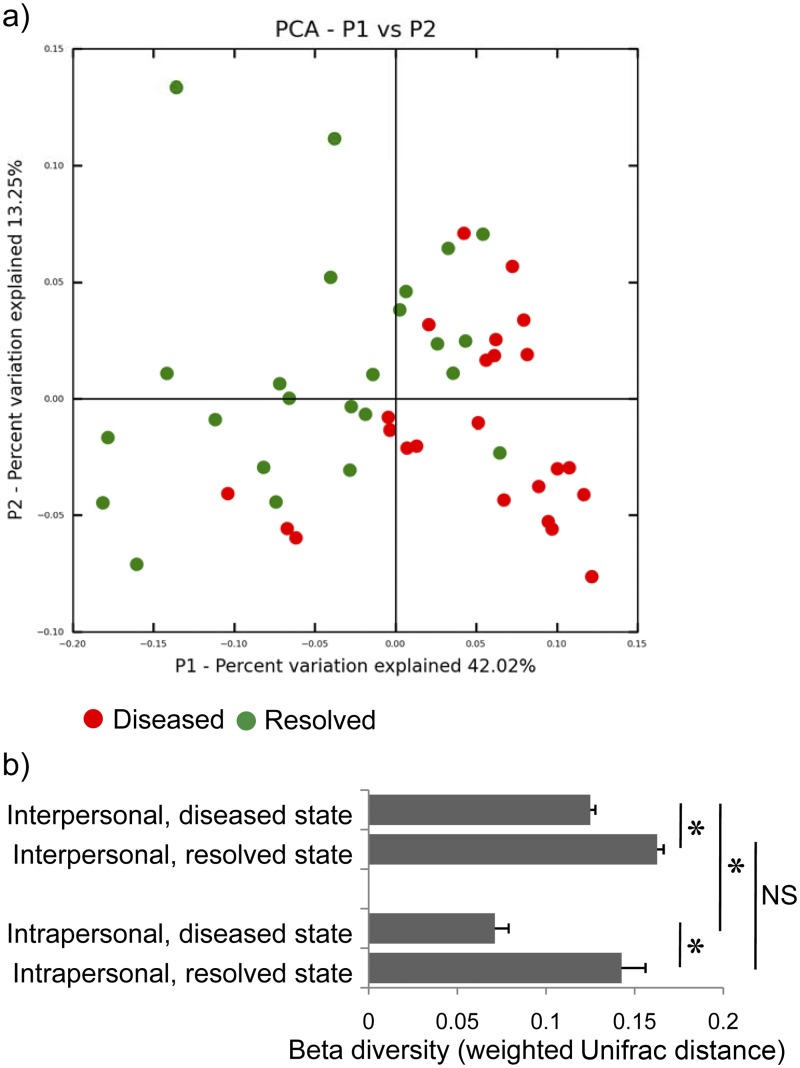
The microbial community structure was significantly different in diseased and resolved states as indicated by their differences in beta diversity (P < 0.001 by analysis of similarity [ANOSIM]; Fig. 2a). In the diseased state, intrapersonal samples (different tooth sites from the same individual) were more similar to each other than interpersonal samples, and this was statistically significant (P < 10−5). However, this trend is not observed for the resolved state (P = 0.17; Fig. 2b), suggesting that health-associated subgingival microbiomes can be highly variable in composition even within the same individual. Diseased samples were more similar to each other than resolved samples in both intrapersonal comparison (P < 0.0002) and interpersonal comparison (P < 10−18; Fig. 2b). These results suggest that a group of pathogenic microorganisms is common to diseased sites in individuals (13). Resolved sites, in contrast, can have site-specific groups of health-associated organisms.
FIG 2 .
Interpersonal and intrapersonal beta diversity of the subgingival microbiome. (a) Principal coordinate analysis (PCoA) shows that the microbiomes of diseased samples and resolved samples were significantly different. (b) The diseased samples were more similar to each other than the resolved samples in both interpersonal and intrapersonal comparisons. Sites from the same individual were significantly more similar to each other in the diseased state (before treatment), but not in the resolved state (after treatment). Data are represented as means plus SEMs (error bars). Values that are significantly different (P < 0.01) are indicated by a bar and asterisk; values that are not significantly different (P > 0.05) are indicated by a bar labeled NS.
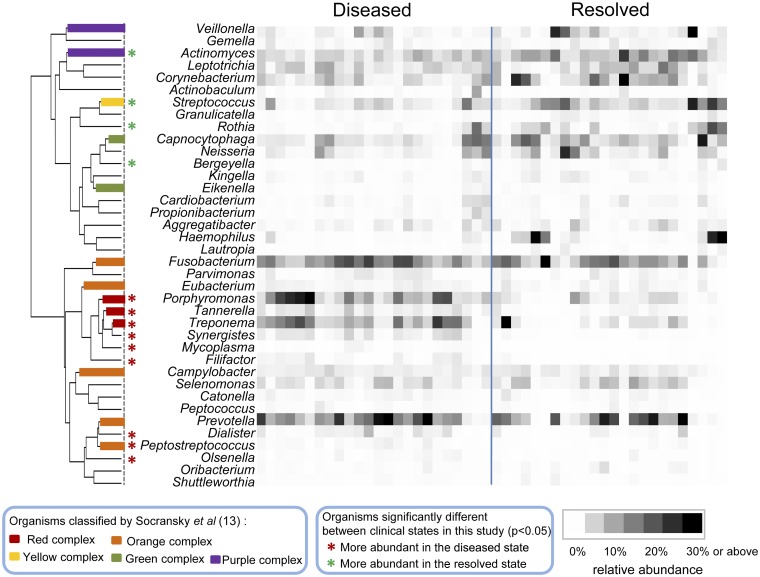
On the basis of the relative abundance profiles of the bacterial genera, using hierarchical clustering, we found two distinct groups of bacterial organisms—disease-associated and health-associated organisms (Fig. 3). The subgingival microbiome was dominated by anaerobes in the diseased state and by facultatively anaerobic or aerobic organisms in the resolved state. Among the 38 bacterial genera, 8 were significantly more abundant in the diseased state (see Table S1 in the supplemental material), including Porphyromonas, Treponema, and Tannerella, which are the major Gram-negative periodontal pathogens designated as the red complex by Socransky et al. (13). Additionally, Peptostreptococcus, Synergistes, Filifactor, Mycoplasma, and Olsenella were significantly more abundant in the diseased state. Peptostreptococcus is a member of the orange complex and is known as a potential pathogen associated with periodontitis (13). While Synergistes, Filifactor, Mycoplasma, and Olsenella were not part of the complexes classified by Socransky and coworkers (13), they were found to be associated with periodontitis in previous studies (14–16). On the other hand, four genera were significantly more abundant in the resolved state, including representatives of the purple complex, Actinomyces, and the yellow complex, Streptococcus (13), as well as Rothia and Bergeyella. These four genera were previously found to be more abundant in healthy subgingival pockets than in diseased pockets in case-control studies (14–16). Our longitudinal comparison between diseased and resolved states confirmed the previous findings in microbiome differences in healthy and diseased individuals, suggesting that the microbiome in the resolved state resembles the healthy state to a large extent. Consistent with previous studies, we showed that disease is associated with shifts of the complex microbial community rather than by a single pathogen.
FIG 3 .
Disease-associated and health-associated subgingival microorganisms. On the basis of their relative abundance, the subgingival microorganisms were clustered into disease-associated and health-associated groups, as shown by the dendrogram on the left. The branches in the dendrogram are colored according to the bacterial complex designations of Socransky et al. (13). Genera are indicated with asterisks if they were significantly more abundant in the diseased state (red) or in the resolved state (green). The paired samples, collected before and after treatment at the same tooth sites, are ordered in the same way in the diseased group and the resolved group. Relative abundance is indicated by a gradient of grey shades from white to black.
Cooccurrence of periodontitis-associated organisms in the subgingival microbiome.
Since periodontitis is associated with shifts of the microbial community in the subgingival pocket, it is important to investigate the coordinated interactions among the subgingival microorganisms. This is essential to understand the polymicrobial pathogenesis mechanism of the disease and to better predict disease progression based on the microbiome profile. Socransky et al. previously studied the cooccurrence of subgingival microorganisms in the diseased state based on the presence/absence patterns of a selected group of organisms in the plaque samples collected from periodontitis patients (13). Bik et al. reported the cooccurrence of subgingival microorganisms in healthy periodontium based on pooled samples from multiple tooth sites of 10 individuals (25).
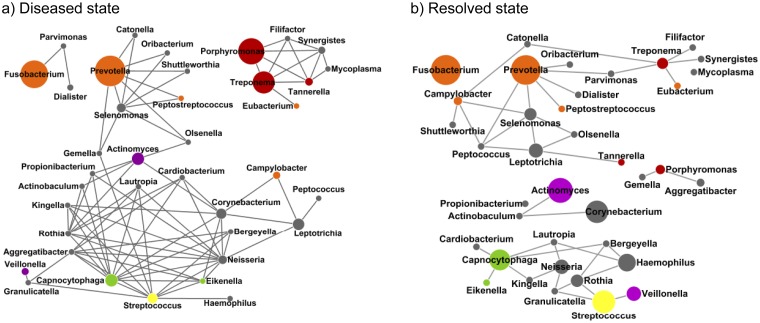
In this study, we investigated whether disruptions of the coordinated interactions among organisms occur during the transition from the diseased state to the resolved state. Using a method similar to Qin et al. (26), we constructed cooccurrence networks of the microorganisms in the diseased or resolved state based on the relative abundances of the major genera found in the samples. We found that the strength of the microbial cooccurrence was significantly greater for the diseased state than for the resolved state (P < 0.029 [see Fig. S4 in the supplemental material]). In the diseased state, both disease- and health-associated organisms cooccur more often within each subnetwork than in the resolved state (Fig. 4). This suggests that coordinated microbial interactions in early and late colonization are essential to disease pathogenesis (8). Six disease-associated genera, including the red complex organisms Porphyromonas, Treponema, and Tannerella (13), as well as Synergistes, Filifactor, and Mycoplasma, had higher relative abundances and were all highly correlated with each other in the diseased state, forming a subnetwork with fully connected topology. This result suggests that Synergistes, Filifactor, and Mycoplasma should be considered expanded members of the red complex. In the resolved state, the topology of the cooccurrence network was altered, and the number of connections among the organisms was significantly reduced. Not only were the relative abundances of red complex members reduced, they no longer correlated with each other. This suggests that the connections between these pathogenic organisms are disrupted after treatment. On the other hand, the relative abundances of health-associated organisms were increased than those in the diseased state. In both diseased and resolved states, the relative abundances of disease-associated organisms (red and orange complexes) did not correlate with health-associated organisms (yellow, green, and purple complexes), confirming the previous findings that an antagonistic relationship exists between the two groups of organisms regardless of the clinical state (27, 28).
FIG 4 .
Microbial cooccurrence in the subgingival microbiome. Cooccurrence networks were constructed on the basis of the relative abundance profiles of subgingival microorganisms in the diseased state (a) and in the resolved state (b). Each node represents a genus and is colored according to the bacterial complex designations of Socransky et al. (13). The gray nodes represent the organisms that were not classified by Socransky et al. The sizes of the nodes are proportional to the average relative abundances of the genera (if >1%) in each state. Each edge represents a positive correlation between the two genera with a Pearson’s correlation coefficient of >0.4 (26).
Periodontitis-associated functional pathways.
The dynamic shifts in the functional potentials encoded in genes in the subgingival microbiome in healthy and diseased states are poorly understood. Although disease-associated microorganisms have been identified, examinations of potential virulence factors shared by red complex members have not yielded clear associations with the disease (29). The large amount of metagenomic shotgun sequencing data obtained in this longitudinal study allowed us to determine the shifts in the functional pathways encoded in the metagenome in diseased and resolved states.
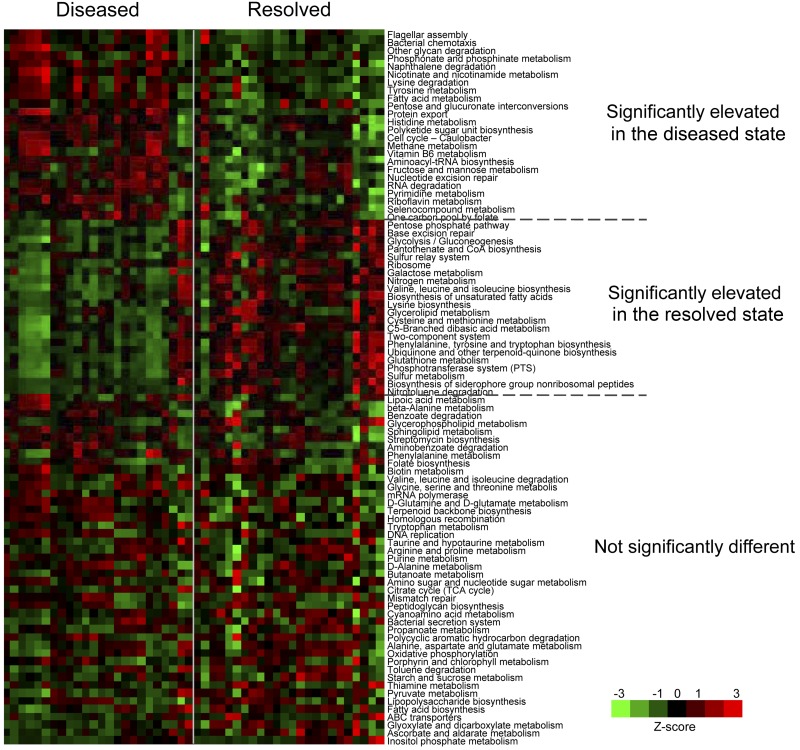
We aimed to investigate whether the functional profiles of the microbiome are distinct in the diseased state and resolved state and thus can be used for disease diagnosis and prognosis. Using the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous group annotation, we identified significant differences in the diseased and resolved states at the functional pathway level. Among the 90 bacterial pathways found in the microbiome, 24 were significantly overrepresented and 22 were significantly underrepresented in the diseased state (Fig. 5). We observed that genes encoding components involved in flagellar assembly and bacterial chemotaxis, which are virulence factors in periodontitis (30, 31), were significantly more abundant in the diseased state. This is consistent with previous observations that flagellated motile species in the oral cavity, including Treponema, Selenomonas, and Campylobacter (32), were found associated with periodontitis. Chemotaxis-guided motility enhances the nutrient intake of pathogens favoring their growth and plays an important role in the colonization of pathogens by facilitating them to penetrate oral epithelial cells (30–33). Additionally, we found that the disease-associated subgingival microbiome encodes more metabolic degradation processes than biosynthesis processes. For example, lysine degradation was significantly overrepresented in the diseased state, while lysine biosynthesis was significantly underrepresented. In the diseased state, disruptions of host tissues provide a different pool of nutrients than that of the healthy state, and thus, a microbial community with enhanced abilities to employ metabolic degradation processes may be selected. Taken together, our analysis suggests that the subgingival microorganisms change their relative abundances with altered functional potentials to adapt to the changes in the host environment.
FIG 5 .
Functional profiles of the subgingival microbiome. The relative abundances of 46 KEGG functional pathways were significantly different in the diseased state and in the resolved state. The paired samples are ordered in the same way in the diseased and resolved groups, respectively.
Classification of clinical states based on the subgingival microbiome profile.
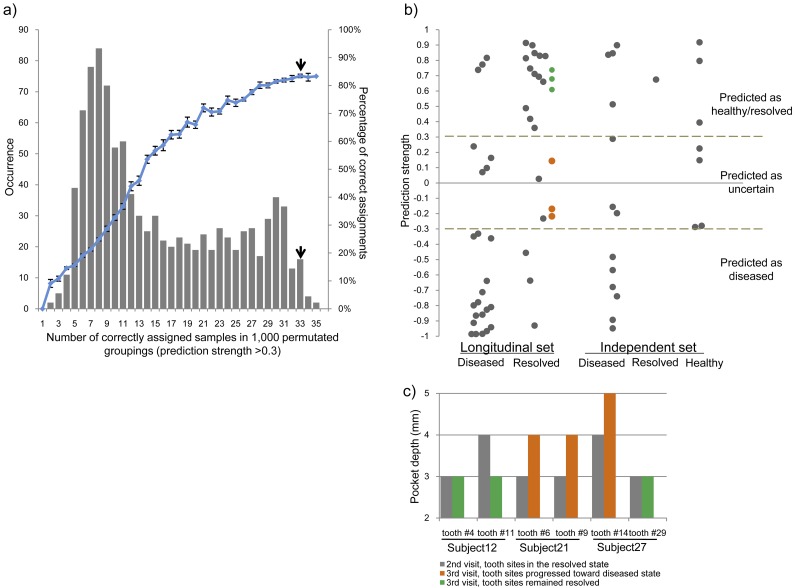
Our taxonomic and functional analyses demonstrate distinct differences in the subgingival microbiome in diseased and resolved states. To examine whether the microbiome profile can be used to classify the clinical state of the periodontium in disease diagnosis, we determined the robustness of the microbiome functional profiles in correctly assigning samples to their clinical state. We applied a supervised classification method on our 48 longitudinal samples using a weighted gene-voting algorithm and leave-one-out cross-validation (34, 35). With the use of the clinically defined grouping, the classifier based on the microbiome profile correctly classified 33 of 39 assigned samples with a prediction strength of >0.3 (Fig. 6b). Nine samples could not be assigned due to lower prediction strength. The number of correctly assigned samples based on the clinical grouping was significantly higher than those in 1,000 randomly permutated groupings (P = 0.022 [Fig. 6a]). The clinical grouping had a significantly higher prediction accuracy (84.6%) among all permutated groupings (P = 0.04). We obtained similar results (a prediction accuracy of 83.8%) when we used the taxonomic profiles of the subgingival microbiome to classify the clinical state of the samples.
FIG 6 .
Classification of clinical states based on the subgingival microbiome profile. (a) Prediction accuracy using a weighted gene-voting algorithm and leave-one-out cross-validation. The number of correctly assigned samples based on the clinical grouping (indicated by the top short black arrow) was significantly higher than those in permutated groupings. The clinical grouping also had a significantly higher prediction accuracy (84.6%). (b) Classification of 48 longitudinal samples and an independent set of 21 samples based on the subgingival microbiome profile. The independent set consisted of 13 samples collected in the diseased state and one sample collected in the resolved state. Additionally, we included seven samples obtained from healthy subjects in the Human Microbiome Project (36). (c) Follow-up examination demonstrates our ability to predict disease progression. Six tooth sites in three patients were examined again during the third clinic visit. The directions of the clinical progressions of all six sites were correctly predicted based on the preceding microbiome profile at the second clinic visit. The tooth sites that remained healthy at the third visit were predicted as resolved (shown in green [Fig. 6b]), and the tooth sites that progressed toward or became diseased were predicted to be between the resolved and diseased states (shown in orange [Fig. 6b]).
We further confirmed the discriminatory power of the classification approach using an independent set of subgingival samples. This set consisted of 13 diseased samples and 1 resolved sample that we collected. They were sequenced in the same way as the longitudinal samples. Additionally, we included seven subgingival samples of healthy individuals from the Human Microbiome Project (36). This yielded a total of 21 independent samples. Among these, seven could not be classified due to their weak prediction strength. Ten of the remaining 14 samples were assigned correctly using our classifier (Fig. 6b). Overall, our classifier assigned all samples to their clinical state with an accuracy of 81.1%.
Among the 26 samples that were classified to the diseased state, 23 were correctly assigned (precision of 88.5%). The other three samples were considered resolved on the basis of the clinical observation. It is possible that although the diseased sites were resolved clinically, the subgingival microbiome had not completely shifted to a healthy state. Alternatively, it is also possible that the resolved sites had started to progress to disease and that the dysbiosis of the microbiome had already shifted to a diseased state before clinical symptoms manifested.
Follow-up clinical examination of sampled subgingival sites can provide further evaluation on the predictive power of the microbiome profile on disease progression. Three of the 12 patients recruited in our longitudinal study had a third visit to the clinic for routine periodontal maintenance. The previously sampled tooth sites in these three subjects (2 sites per patient) were examined again. This allowed us to determine whether our predictions of the clinical progression based on the microbiome profile at the resolved state (the second visit) were consistent with the clinical observations at the third visit. Compared to the clinical parameters obtained at the second and third visits, three of the six tooth sites remained healthy with no increases in pocket depth, two tooth sites progressed toward the diseased state with increased pocket depths (from 3 mm to 4 mm) but still were defined as resolved, and one tooth site became diseased with a pocket depth of 5 mm (Fig. 6c). On the basis of the preceding microbiome profile at the second clinic visit, we were able to correctly predict the direction of the clinical progressions of all six sites. The three tooth sites that remained resolved at the third visit were predicted to be resolved (shown in green [Fig. 6b]), and the tooth sites that progressed toward or became diseased were predicted to be between resolved and diseased states (shown in orange [Fig. 6b]). This suggests that these three tooth sites had already shifted away from the resolved state at the second visit and moved toward the diseased state. This also suggests that the microbiome had already reflected the changes before clinical manifestation. Furthermore, we observed that individual sites from the same subject (subject 27) progressed in different directions (Fig. 6c), further supporting the idea that periodontitis is a disease of individual sites and thus has strong associations with alterations of the microbiome in each distinct subgingival pocket. Although more samples need to be tested to confirm the predictive power, our ability to assess the risk of future disease progression of individual tooth sites based on the microbiome profile in these three cases (six sites) suggests that monitoring the microbiome of individual periodontal pockets could be useful for disease diagnosis, prognosis, and early intervention.
DISCUSSION
Periodontitis is the most common disease associated with the subgingival microbiota. Although the disease can be accurately diagnosed through clinical examinations, reliable prognosis is still difficult to achieve (11, 12). A link between the taxonomic composition of the subgingival microbiota and periodontitis pathogenesis has been established by several previous studies using traditional methods or 16S rRNA gene profiling (14–17). In this study, by analyzing the dynamics of the subgingival microbiome from individual tooth sites before and after periodontal treatment, we aimed to determine whether the taxonomic and/or functional composition could be used as a marker for improved disease diagnosis and prognosis.
The complexity of the subgingival microbial community has been recognized since early microscopic examinations of dental plaques in the 17th century (37). The recent large-scale study of the healthy subgingival microbiome by the Human Microbiome Project reinforced the early observations that the microbiota is composed of hundreds of microorganisms and revealed apparent differences among different tooth sites even within the same individuals (36). However, the dynamic changes in the subgingival microbiome of individual tooth sites during disease development have not been well characterized. Most of the previous subgingival microbiota studies were performed using pooled plaque samples from multiple individuals or from multiple tooth sites of the same individuals. Intrapersonal variations of the subgingival microbiome were unknown and thus were not considered in evaluating the disease associations of the subgingival microbial community. Because individual tooth sites are likely to have independent clinical states and unique microbial communities in subgingival pockets, longitudinal examination of the microbiome at each individual tooth site during disease development is essential to understand the role of the oral microorganisms in disease pathogenesis.
Using a large-scale metagenomic shotgun sequencing analysis, our longitudinal study presented here determined dynamic changes in the subgingival microbiome of individual tooth sites within the same individuals in the diseased and resolved states (Fig. 1). This study presents an important advancement from the Human Microbiome Project (36). We revealed that the microbial community shifted significantly in the taxonomic composition in the two different clinical states (Fig. 2). In the diseased state, the microbiome is similar among different tooth sites and among different individuals; however, in the resolved state, it is highly variable, even among tooth sites within the same individual. This suggests that while a common group of pathogenic microorganisms is responsible for the disease, a healthy periodontium can accommodate a variety of health-associated microorganisms.
We also found that the subgingival microbial community in the two clinical states differed not only in the taxonomic composition but also in the cooccurrence patterns of periodontal microorganisms (Fig. 4). This suggests that correlated interactions among pathogens are critical to disease pathogenesis and is consistent with the theory that the oral microorganisms form an intricate complex with layers of organisms colonizing the periodontium sequentially during biofilm formation (8). Our cooccurrence networks indicate that in the diseased state, pathogenic organisms need to be present in correlation with each other in order to promote disease, while in the healthy and resolved states, the oral microorganisms are less correlated in relative abundance and can vary significantly among different tooth sites and different individuals. Investigations on how to disrupt the synergistic interactions of periodontopathogens may have potential applications on effective prevention of periodontal diseases.
Periodontitis can reoccur and may progress without symptoms even after treatment. Lacking a good prognosis indicator, current clinical practice demands that individual patients and tooth sites must be examined regularly with no distinction between individuals, regardless of whether sites are successfully resolved or relapsing. Therefore, prognostic tools are essential in practice to identify those patients in need. Such tools would allow early identification and early intervention, minimizing not only disease destruction but also the extent of treatment required while increasing predictability as well. In this study, we demonstrated that the functional composition of the subgingival microbiome shifted significantly from the diseased state to the resolved state (Fig. 5). On the basis of the functional profile of the microbiome, we built a classifier, with which we were able to assign the clinical states of the sampled tooth sites with high accuracy (Fig. 6). Furthermore, using the classifier, we were able to predict the disease progression of individual tooth sites. For the first time, we demonstrated potential applications of the subgingival microbiome analysis in periodontitis diagnosis and prognosis. This can give rise to potential future development of useful diagnostic and prognostic tools that enable more specific, individualized therapies, with better follow-up strategies, more effective treatment, and reduced cost for periodontal care.
MATERIALS AND METHODS
Subjects and sample collection.
Twelve adult volunteers with chronic periodontitis were recruited for the study. The patients gave consent, and the study protocol was approved by the UCLA Institutional Review Board. The age of the patients ranged from 37 to 65 years with an average age of 53 years. Subjects with a history of antibiotic treatment in the past 6 months, history of smoking, or diabetes were excluded from the study. For each patient, clinical parameters of the gingival index, recession of gums, attachment level, pocket depth, and bleeding on probing were measured for all sampled sites to assess the state of periodontal disease. Following a 24-h period of no oral hygiene, subgingival plaques were collected from two affected tooth sites per subject on average. The area to be sampled was isolated and dried, and the supragingival plaque was removed. The subgingival plaque samples were then collected using sterile curettes (Hu-Friedy Mfg. Co., Inc., Chicago, IL). The sampled plaques were suspended directly in ATL buffer (Qiagen, Inc., Valencia, CA) containing 0.1-mm glass beads (BioSpec Products, Inc., Bartlesville, OK) and immediately transported to the laboratory for further processing. Conventional initial periodontal therapy was subsequently conducted, including scaling and root planing (SRP), and oral hygiene instructions were given to the patients. The patients had a second clinic visit 4 to 19 weeks (on average 60 days) after completion of initial therapy. The same tooth sites were examined and resampled if the sites had resolved (presenting with a probing depth of ≤4 mm and without gingival inflammation). On average, the probing depth of sampled sites decreased from 6 mm prior to treatment to 3 mm at the second visit. Three of the 12 subjects were followed up with a third clinic visit 7 to 14 months after the second visit. The same tooth sites were examined again, and clinical parameters were recorded.
Genomic DNA extraction.
Genomic DNA was extracted from the plaque samples using the QIAamp DNA microkit (Qiagen, Inc.) with a modified “Isolation of Genomic DNA from Tissues” protocol. Bead beating for maximal bacterial cell lysis was added. All procedures were completed in a laminar flow hood (NuAire Inc., Plymouth, MN) with RNase-free materials. The extracted genomic DNA was eluted with EB buffer (Qiagen, Inc.) and stored at −20°C for short-term storage and at −80°C for long-term storage.
Sequencing data collection.
The genomic DNA samples were sequenced using the metagenomic shotgun method on the Illumina sequencing platform (Illumina, Inc., San Diego, CA). Paired-end reads of 100 bp were obtained. A data cleaning process was applied to all samples, following the procedure used in the Human Microbiome Project (36). Briefly, human DNA sequences were filtered out first. They accounted for 59.7% ± 3.4% of the raw sequence data. Duplicate reads, low-quality reads, and low-compositional-complexity reads were then removed. This resulted in an average of 42.3 million reads of cleaned data per sample, which were used in the downstream analyses.
Taxonomic composition analysis.
We used a mapping-based method to extract 16S rRNA sequences from the shotgun sequencing data. We aligned cleaned sequence reads against the SILVA rRNA database (nonredundant SSU Ref data set, release 108) (21), HOMD (16S rDNA RefSeq version 11.0) (22), and OSU CORE database (updated 9 February 2012) (23). The alignments were performed using Bowtie (38) to search sequences, with the best hits having ≥97% nucleotide identity with reference sequences. We required that the paired-end reads align to the same reference sequence in different strand directions and within 800 bp. The extracted 16S rRNA sequences were annotated at the genus level by cross-referencing the alignment results from all three databases. The integration of the alignment results from three databases improved the accuracy of taxonomic assignment; 96.8% of the 16S rRNA sequences were annotated on the basis of consistent alignment results from at least two databases (see Fig. S5 in the supplemental material), and 92.2% of the 16S rRNA sequences can be uniquely assigned to a classified genus. The relative abundances of genera in each sample were calculated on the basis of the 16S rRNA sequences with unique assignment of the classified genera. The sequencing depths in all samples were assessed by the rarefaction analysis using the function rarefy in the R package Vegan (39).
Analysis of microbial community structure.
Alpha diversity (Shannon index), beta diversity (weighted UniFrac), and principal coordinate analysis (PCoA) were calculated using QIIME (40). Nonparametric multivariate analysis of changes in community structure (analysis of similarity [ANOSIM]) (41) was conducted using Mothur (42) with default parameters to test whether the microbiome similarities within groups is statistically significantly higher than the similarities between groups.
Correlation and cooccurrence analyses of the subgingival microorganisms.
Hierarchical clustering was performed on the relative abundance profiles of the genera found in samples with Cluster 3.0 (43). The distance measurement was based on the Spearman rank correlation between samples. The heatmap was generated using Java Treeview (44). To construct the cooccurrence networks, we calculated pairwise intergenus correlations based on the relative abundance profiles of the genera in the diseased state or in the resolved state. To exclude coabsence, cooccurrences were not counted between genus pairs if both had an average relative abundance of <1% in the same state. The networks were then visualized using Cytoscape v2.8 (45).
Functional profile analysis.
Metagenomic shotgun sequencing reads were annotated with KEGG orthologous groups (KOs) by mapping them to the KEGG database (version 54; E value of <10−5, bit score of >50, and identity of >50%) (46, 47). UBLAST in the USEARCH package was used (48). On average, 14.9% of the metagenomic shotgun sequencing reads can be assigned to KOs, and 99.8% of the assigned reads had the best hit on a unique KO. Read counts were normalized within each sample to represent the relative abundance of functional genes. The relative abundance of each functional pathway was calculated by summing all the functional genes involved in the functional pathway. Pathways with a relative abundance of >0.1% in samples from at least two subjects were chosen. To avoid overrepresentation, pathways with two or fewer KO genes identified were not counted.
Classification of clinical states based on the microbiome profile.
We performed a supervised classification using the weighted gene-voting algorithm with minor modification (the Student's t score used as the weighting factor) and leave-one-out cross-validation as described by Golub et al. (34) and Bleharski et al. (35). The sample classification based on the clinical grouping was compared to those based on 1,000 permutation groupings. In each permutation, 48 longitudinal samples were randomly assigned to two groups (diseased and resolved) with equal number of samples in each group.
Statistical analysis.
Data are represented as means ± standard errors of the means (SEMs) unless otherwise indicated. Paired Student’s t test with two-tailed distribution was used in statistical testing of all comparisons of differences between the longitudinally paired samples unless otherwise indicated. The P values were adjusted for multiple testing with p.adjust in R using the false discovery rate (49).
Nucleotide sequence accession numbers.
The sequence data from this study have been submitted to NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject) under accession no. 255922.
SUPPLEMENTAL MATERIAL
Supplementary material. Download
Rarefaction curve of the shotgun 16S rRNA sequences. Data are represented as means plus SEMs. Download
Comparison of the taxonomic compositions determined based on the three approaches. The taxonomic compositions were determined based on the shotgun 16S rRNA-based and shotgun reference genome-based (Ref Genome-based) approaches. As a validation, we constructed traditional 16S rRNA clone libraries on 8 samples combined with Sanger sequencing. Data are represented as means plus SEMs. Download
The alpha diversity of the subgingival microbiome in the diseased and resolved states. Data are represented as means plus SEMs. Download
Strength distribution of the microbial cooccurrence. The strength was determined based on the relative abundances of the major genera found in samples in each clinical state using a method similar to Qin et al. (26). The cooccurrence of subgingival microorganisms was identified if the positive correlation between the two genera with a Pearson’s correlation coefficient was above 0.4. The strength of the microbial cooccurrence was significantly higher in the diseased state than in the resolved state. Download
The shotgun 16S rRNA sequences were annotated at the genus level by cross referencing the alignment results from multiple databases. 96.8% of the shotgun 16S rRNA sequences could be annotated based on consistent alignment results from at least two databases. Data are represented as means plus SEMs. Download
Taxonomic composition of the subgingival microbiome in healthy individuals from the Human Microbiome Project. Download
Principal coordinate analysis of the subgingival microbiome in the diseased state (red), resolved state (green), and healthy state from the Human Microbiome Project (HMP) (blue). Weighted UniFrac distances were used in the analysis. Download
Taxonomic composition of the subgingival microbiome.
ACKNOWLEDGMENTS
We thank Shuta Tomida and Karthik Kota for their help and suggestions on data analysis. We also thank Carlos Sermeno for his help in sample collection.
This study was supported by NIH/NIDCR grants RC1 DE020298 and R01 DE021574.
Footnotes
Citation Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, Sodergren E, Weinstock GM, Haake SK, Li H. 2015. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio 6(1):e01926-14. doi:10.1128/mBio.01926-14
REFERENCES
- 1.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pflughoeft KJ, Versalovic J. 2012. Human microbiome in health and disease. Annu Rev Pathol 7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 3.Statnikov A, Alekseyenko AV, Li Z, Henaff M, Perez-Perez GI, Blaser MJ, Aliferis CF. 2013. Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci Rep 3:2620. doi: 10.1038/srep02620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 5.Huang S, Li R, Zeng X, He T, Zhao H, Chang A, Bo C, Chen J, Yang F, Knight R, Liu J, Davis C, Xu J. 2014. Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME J 8:1768–1780. doi: 10.1038/ismej.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 7.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, Abraham J, Ackerman I, Aggarwal R, Ahn SY, Ali MK, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Bahalim AN, Barker-Collo S, Barrero LH, Bartels DH, Basanez MG, Baxter A, Bell ML, Benjamin EJ, Bennett D, Bernabe E, Bhalla K, Bhandari B, Bikbov B, Bin Abdulhak A, Birbeck G, Black JA, Blencowe H, Blore JD, Blyth F, Bolliger I, Bonaventure A, Boufous S, Bourne R, Boussinesq M, Braithwaite T, Brayne C, Bridgett L, Brooker S, Brooks P, Brugha TS, et al.. 2012. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 9.Linden GJ, Lyons A, Scannapieco FA. 2013. Periodontal systemic associations: review of the evidence. J Clin Periodontol 40(Suppl 14):S8–S19. doi: 10.1111/jcpe.12064. [DOI] [PubMed] [Google Scholar]
- 10.Machtei EE, Dunford R, Hausmann E, Grossi SG, Powell J, Cummins D, Zambon JJ, Genco RJ. 1997. Longitudinal study of prognostic factors in established periodontitis patients. J Clin Periodontol 24:102–109. doi: 10.1111/j.1600-051X.1997.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 11.McGuire MK. 1991. Prognosis versus actual outcome: a long-term survey of 100 treated periodontal patients under maintenance care. J Periodontol 62:51–58. doi: 10.1902/jop.1991.62.1.51. [DOI] [PubMed] [Google Scholar]
- 12.McGuire MK, Nunn ME. 1996. Prognosis versus actual outcome. II. The effectiveness of clinical parameters in developing an accurate prognosis. J Periodontol 67:658–665. doi: 10.1902/jop.1996.67.7.658. [DOI] [PubMed] [Google Scholar]
- 13.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr.. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 14.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang YC, Li S, Stine OC, Hasturk H, Kasif S, Segrè D, Pop M, Amar S. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, He J, He Z, Zhou Y, Yuan M, Xu X, Sun F, Liu C, Li J, Xie W, Deng Y, Qin Y, Vannostrand JD, Xiao L, Wu L, Zhou J, Shi W, Zhou X. 2014. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J 8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jünemann S, Prior K, Szczepanowski R, Harks I, Ehmke B, Goesmann A, Stoye J, Harmsen D. 2012. Bacterial community shift in treated periodontitis patients revealed by ion torrent 16S rRNA gene amplicon sequencing. PLoS One 7:e41606. doi: 10.1371/journal.pone.0041606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laksmana T, Kittichotirat W, Huang Y, Chen W, Jorgensen M, Bumgarner R, Chen C. 2012. Metagenomic analysis of subgingival microbiota following non-surgical periodontal therapy: a pilot study. Open Dent J 6:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews D. 2005. Conclusive support for mechanical nonsurgical pocket therapy in the treatment of periodontal disease. How effective is mechanical nonsurgical pocket therapy? Evid Based Dent 6:68–69. doi: 10.1038/sj.ebd.6400338. [DOI] [PubMed] [Google Scholar]
- 21.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, Vriesendorp B, Faust RA, Janies DA, Leys EJ. 2011. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One 6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, Kota K, Sunyaev SR, Weinstock GM, Bork P. 2013. Genomic variation landscape of the human gut microbiome. Nature 493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA. 2010. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grenier D. 1996. Antagonistic effect of oral bacteria towards Treponema denticola. J Clin Microbiol 34:1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Hoogmoed CG, Geertsema-Doornbusch GI, Teughels W, Quirynen M, Busscher HJ, Van der Mei HC. 2008. Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol Immunol 23:43–48. doi: 10.1111/j.1399-302X.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 29.Curtis MA, Zenobia C, Darveau RP. 2011. The relationship of the oral microbiota to periodontal health and disease. Cell Host Microbe 10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charon NW, Goldstein SF. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genet 36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 31.Dashper SG, Seers CA, Tan KH, Reynolds EC. 2011. Virulence factors of the oral spirochete Treponema denticola. J Dent Res 90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lux R, Shi W. 2004. Chemotaxis-guided movements in bacteria. Crit Rev Oral Biol Med 15:207–220. doi: 10.1177/154411130401500404. [DOI] [PubMed] [Google Scholar]
- 33.Lux R, Miller JN, Park NH, Shi W. 2001. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect Immun 69:6276–6283. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 35.Bleharski JR, Li H, Meinken C, Graeber TG, Ochoa MT, Yamamura M, Burdick A, Sarno EN, Wagner M, Röllinghoff M, Rea TH, Colonna M, Stenger S, Bloom BR, Eisenberg D, Modlin RL. 2003. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 301:1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 36.The Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter JR. 1976. Antony van Leeuwenhoek: tercentenary of his discovery of bacteria. Bacteriol Rev 40:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2012. vegan: community ecology package. R package version 2.0-4. http://cran.r-project.org/web//packages/vegan/index.html.
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 44.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 45.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Stat Methodol 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. Download
Rarefaction curve of the shotgun 16S rRNA sequences. Data are represented as means plus SEMs. Download
Comparison of the taxonomic compositions determined based on the three approaches. The taxonomic compositions were determined based on the shotgun 16S rRNA-based and shotgun reference genome-based (Ref Genome-based) approaches. As a validation, we constructed traditional 16S rRNA clone libraries on 8 samples combined with Sanger sequencing. Data are represented as means plus SEMs. Download
The alpha diversity of the subgingival microbiome in the diseased and resolved states. Data are represented as means plus SEMs. Download
Strength distribution of the microbial cooccurrence. The strength was determined based on the relative abundances of the major genera found in samples in each clinical state using a method similar to Qin et al. (26). The cooccurrence of subgingival microorganisms was identified if the positive correlation between the two genera with a Pearson’s correlation coefficient was above 0.4. The strength of the microbial cooccurrence was significantly higher in the diseased state than in the resolved state. Download
The shotgun 16S rRNA sequences were annotated at the genus level by cross referencing the alignment results from multiple databases. 96.8% of the shotgun 16S rRNA sequences could be annotated based on consistent alignment results from at least two databases. Data are represented as means plus SEMs. Download
Taxonomic composition of the subgingival microbiome in healthy individuals from the Human Microbiome Project. Download
Principal coordinate analysis of the subgingival microbiome in the diseased state (red), resolved state (green), and healthy state from the Human Microbiome Project (HMP) (blue). Weighted UniFrac distances were used in the analysis. Download
Taxonomic composition of the subgingival microbiome.