ABSTRACT
Subcellular organization of the bacterial nucleoid and spatiotemporal dynamics of DNA replication and segregation have been studied intensively, but the functional link between these processes remains poorly understood. Here we use quantitative time-lapse fluorescence microscopy for single-cell analysis of chromosome organization and DNA replisome dynamics in Mycobacterium smegmatis. We report that DNA replication takes place near midcell, where, following assembly of the replisome on the replication origin, the left and right replication forks colocalize throughout the replication cycle. From its initial position near the cell pole, a fluorescently tagged chromosomal locus (attB, 245° from the origin) moves rapidly to the replisome complex just before it is replicated. The newly duplicated attB loci then segregate to mirror-symmetric positions relative to midcell. Genetic ablation of ParB, a component of the ParABS chromosome segregation system, causes marked defects in chromosome organization, condensation, and segregation. ParB deficiency also results in mislocalization of the DNA replication machinery and SMC (structural maintenance of chromosome) protein. These observations suggest that ParB and SMC play important and overlapping roles in chromosome organization and replisome dynamics in mycobacteria.
IMPORTANCE
We studied the spatiotemporal organization of the chromosome and DNA replication machinery in Mycobacterium smegmatis, a fast-growing relative of the human pathogen Mycobacterium tuberculosis. We show that genetic ablation of the DNA-binding proteins ParB and SMC disturbs the organization of the chromosome and causes a severe defect in subcellular localization and movement of the DNA replication complexes. These observations suggest that ParB and SMC provide a functional link between chromosome organization and DNA replication dynamics. This work also reveals important differences in the biological roles of the ParABS and SMC systems in mycobacteria versus better-characterized model organisms, such as Escherichia coli and Bacillus subtilis.
INTRODUCTION
During the bacterial cell cycle, chromosome replication and segregation must be coordinated with cell division. Bacteria typically contain a single circular chromosome. DNA replication initiates at a unique origin (ori) (1), proceeds bidirectionally around the chromosome, and terminates in the terminus region (ter) opposite to ori (2). Different models of DNA replication have been described in bacteria based on the subcellular localization and dynamics of the sister replisomes that replicate the left and right chromosome arms. In Bacillus subtilis and Pseudomonas aeruginosa, the sister replisomes colocalize around midcell, forming a “replication factory,” and the chromosome is spooled through the replisome complex during the process of replication (3, 4). In sharp contrast, in Escherichia coli, the replisomes first assemble at ori in the midcell region and then separate and move to opposite cell halves before returning to midcell for termination (5). In Caulobacter crescentus and Helicobacter pylori, the replisomes assemble at ori near the old cell pole before moving jointly toward midcell (6, 7); Myxococcus xanthus displays similar replisome dynamics, although the two replication forks often split and merge during the DNA replication cycle (8). In these species, the nonrandom movement of sister replisomes during the process of replication, which has been conserved in distantly related species (9–12), has led to a model in which the DNA polymerases that replicate the left and right replicores function independently of each other and use the chromosome as both a template and a track.
Visualization of fluorescently tagged chromosomal loci in live cells reveals that bacterial chromosomes are spatially organized with individual chromosomal loci occupying specific subcellular addresses along the cell length (13, 14). This spatial chromosome arrangement is restored in the daughter cells after completion of chromosome replication and segregation. Proteins involved in the organization of the chromosome arms along the cell length include the chromosome partitioning system (ParABS) and the structural maintenance of chromosome (SMC) protein (14, 15).
The ParABS system, which is present in diverse bacteria (16), comprises a DNA binding protein (ParB), a Walker-type ATPase (ParA), and centromere-like parS sequences located in the ori-proximal region of the chromosome. Upon binding to parS sites, ParB oligomerizes to form large nucleoprotein complexes called segrosomes. Segrosomes organize the ori-proximal regions of newly replicated chromosomes and facilitate their proper positioning in the future daughter cells, either at the cell center (B. subtilis) or close to the cell poles (C. crescentus) (14). This process is followed by segregation and condensation of other regions of the chromosome (14). The formation and positioning of ParB complexes depend on ParA. Filaments of ParA undergo ParB-dependent polymerization and depolymerization and may provide motive force for segrosomes within bacterial cells (14, 17), although more recently this interpretation has been contested (18). With the exception of C. crescentus (19) and M. xanthus (8), deletion of parA or parB is not lethal. In some species, ParA/ParB deficiency causes only minor defects in chromosome organization and segregation, which might imply redundancy in the systems that control positioning of the chromosome. In other species, such as Pseudomonas putida (20) and Mycobacterium smegmatis (21, 22), lack of ParA or ParB results in a severe defect in chromosome inheritance, as indicated by the large fraction of anucleate cells (up to 10%). In M. smegmatis, ParB binds two ori-proximal parS sites (20) and ParA and ParB together may localize ori around the quarter-cell positions (21).
SMC protein complexes, as well as SMC-like MukBEF proteins in E. coli and related bacteria, have been shown to play an important role in chromosome organization, condensation, and segregation (14, 15). SMC deficiency usually leads to global chromosome decondensation and increased formation of anucleate cells (15). In all of the bacteria studied so far, SMC forms variable numbers of discrete subcellular foci (15). In B. subtilis (23, 24) and Streptococcus pneumoniae (25), SMC is enriched in chromosomal regions surrounding ori. ParB may assist the loading of SMC at ori, possibly through direct protein-protein interactions (23–25). In P. aeruginosa (26) and M. smegmatis (27), SMC deficiency results in a weak or no discernible phenotype, respectively.
The spatial organization of DNA replication in bacteria mirrors the organization of the chromosome within the nucleoid, and the replication process itself could play a role in establishing the organization of the chromosome (5, 28). In E. coli, the subcellular localization of chromosome replication initiation is dependent on MukB, which mediates the positioning of ori and replication forks through an unknown mechanism (6, 29, 30). In B. subtilis, Soj (ParA homolog) and Spo0J (ParB homolog) are also involved in the initiation of DNA replication (31, 32). To date, however, the relationship between subcellular chromosome organization and spatiotemporal localization of the replication forks has not being extensively investigated. In a previous study, we measured the single-cell dynamics of chromosome replication, cell growth, and cell division in M. smegmatis with a dual-reporter strain expressing fluorescent markers of the cell division septum (Wag31-GFP) and the DNA replisome (mCherry-DnaN) (33). Here, we use fluorescent reporter strains, microfluidics, and quantitative time-lapse microscopy to investigate the spatiotemporal localization of DNA replication forks relative to specific chromosomal loci in single cells of M. smegmatis. We find that DNA replication takes place at midcell, where the two replication forks colocalize. A fluorescently tagged chromosomal locus (attB, situated 245° from ori) is actively pulled to the replisome just prior to its duplication, and the newly duplicated loci segregate to mirror-symmetric positions relative to midcell. We also find that genetic ablation of proteins involved in the maintenance of chromosome organization (ParB and SMC) results in a severe defect in subcellular localization and movement of DNA replisomes. These observations reveal a functional link between subcellular chromosome organization and spatiotemporal localization of DNA replication in mycobacteria.
RESULTS
Subcellular localization of DNA replisome complexes.
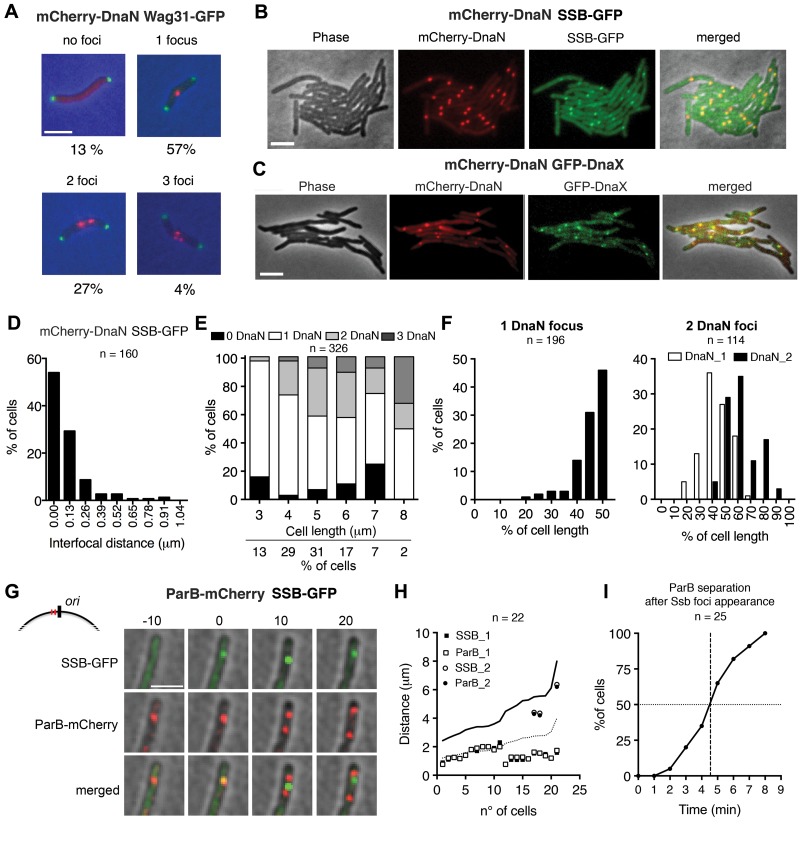
We tracked the spatiotemporal dynamics of DNA replication and cytokinesis in M. smegmatis by using a dual-reporter strain expressing fluorescent markers of the DNA replisome (mCherry-DnaN) and cell division septum (Wag31-green fluorescent protein [GFP]) (Fig. 1A) (33). The mCherry-dnaN gene replaces the dnaN gene at the native chromosomal locus and encodes a functional protein, as evidenced by normal (wild-type) growth kinetics and cell morphology (Fig. 1A; see Fig. S1A in the supplemental material). The mCherry-DnaN fusion protein forms bright diffraction-limited foci that dynamically assemble and disassemble inside the cells, signaling the onset and completion of DNA replication, respectively (Fig. 1A) (33). We compared the localization of mCherry-DnaN with other replisome components by GFP-tagging SSB (single-strand DNA-binding protein) (Fig. 1B; see Fig. S1B in the supplemental material) or DnaX (DNA replisome tau subunit) (Fig. 1C; see Fig. S1B). In both cases, we found that mCherry-DnaN foci localize close to the SSB-GFP foci (Fig. 1B and D) or DnaX-GFP foci (Fig. 1C; see Fig. S1C). Microscopic analysis of wild-type cultures reveals cells with zero (13%), one (57%), two (27%), or three (4%) mCherry-DnaN foci (Fig. 1A and E). The cell fraction with zero foci is enriched for short cells (≤3 µm) and long cells (6 to 7 µm), presumably corresponding to the prereplication B period and postreplication D period of the cell cycle, respectively. Cells containing one or more foci, corresponding to the C period of the cell cycle, are predominantly of intermediate length (between 3 and 6 µm). In the small number of cells with three foci, which are most abundant in the fraction of very large cells (>7 µm), one is bright and the other two are relatively dim. mCherry-DnaN foci are located near midcell in cells with one focus (43% ± 6% of the cell length) or two foci (39% ± 12% and 56% ± 11% of the cell length), and in the latter case, the spatial distribution is broad (Fig. 1F).
FIG 1 .
Subcellular localization of DNA replisome complexes. (A) Representative snapshots of wild-type cells with zero, one, two, or three mCherry-DnaN foci (red) and their frequencies (n = 645). Green foci, Wag31-GFP. Scale bar, 3 µm. (B) Colocalization of mCherry-DnaN foci with SSB-GFP foci. Scale bar, 3 µm. (C) Colocalization of mCherry-DnaN foci with DnaX-GFP foci. Scale bar, 3 µm. (D) Distribution of interfocal distances between mCherry-DnaN and SSB-GFP foci (n = 160). (E) Number of mCherry-DnaN foci relative to cell length (n = 326). (F) Position of mCherry-DnaN foci in cells with one (n = 196) or two (n = 114) foci. (G) Time-lapse analysis of cells expressing ParB-mCherry and SSB-GFP. Images were recorded at 10-min intervals. As shown in the schematic, ParB binds two parS sequences (red lines) located in the origin-proximal region (black line). Newly formed SSB-GFP foci colocalize with ParB-mCherry (0 min). Duplication and segregation of ParB-mCherry foci occur shortly (10 min) after replisome assembly, and segregation continues thereafter (20 min). Scale bar, 3 µm. (H) Positional correlation between ParB-mCherry and SSB-GFP foci at the first appearance of SSB-GFP foci (n = 22). DNA replication initiates prior to division in some cells. Solid line, cell length; dotted line, midcell. (I) Time-lapse analysis of cells expressing ParB-mCherry and SSB-GFP. Images were recorded at 1-min intervals. Time intervals between the first appearance of SSB-GFP foci and the separation of ParB-mCherry foci are plotted (n = 25).
The M. smegmatis ParB protein binds specifically to two parS sequences located near ori, multimerizes, and spreads out over the adjacent DNA (21). We tracked the subcellular localization of ori in space and time by using a fluorescent reporter strain expressing a ParB-mCherry fusion protein, which forms diffraction-limited foci in the vicinity of ori. The parB-mCherry gene replaces the parB gene at the native chromosomal locus and encodes a functional protein, as evidenced by normal (wild-type) growth kinetics and cell morphology (data not shown). In newborn cells with a single ParB-mCherry focus located around midcell, duplication of the focus is followed by rapid segregation of the two foci toward opposite cell poles until they reach the quarter-cell positions, where they remain for the duration of the cell cycle (see Fig. S1D in the supplemental material). In dual-reporter cells expressing SSB-GFP (replisome marker) and ParB-mCherry (ori marker), the first appearance of the SSB-GFP focus, indicating assembly of the DNA replisome complex, is strongly correlated with the position of the ParB-mCherry focus (Fig. 1G and H). Two physically separated ParB-mCherry foci become visible, on average, 5 min after the first appearance of the SSB-GFP focus (Fig. 1I). Immediately thereafter, the newly separated ParB-mCherry foci move rapidly away from the SSB-GFP focus (Fig. 1G). Similar results were obtained with a dual-fluorescent-reporter strain expressing ParB-mCherry (ori marker) and GFP-DnaN (replisome marker) fusion proteins (see Fig. S1E). These observations suggest that ori is replicated within 5 min after the first appearance of the replisome focus; therefore, replication initiation must occur within 0 to 5 min after replisome assembly. These data are also consistent with the notion that the newly duplicated ori sequences remain cohered for no more than a few minutes after ori is replicated.
Spatiotemporal tracking of DNA replisomes in single cells.
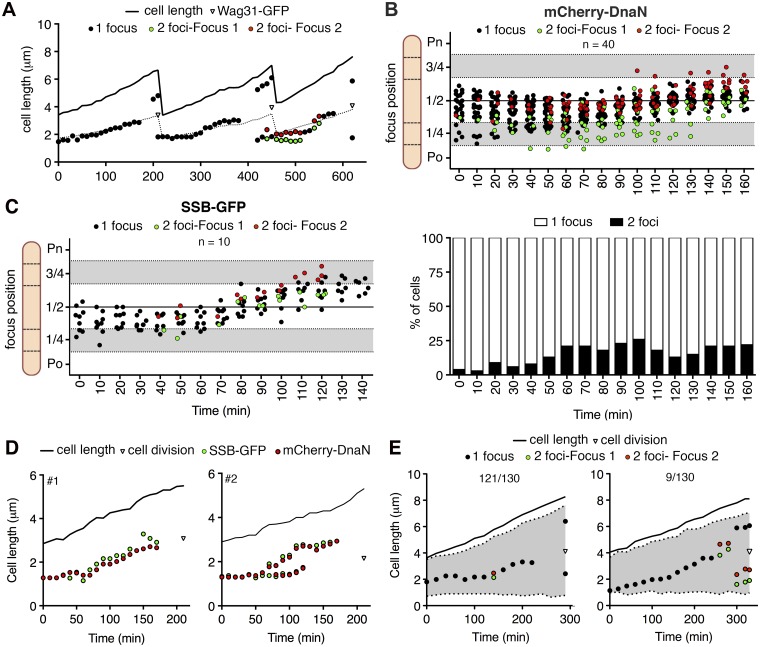
The spatiotemporal distribution of mCherry-DnaN foci suggests that the replication machinery preferentially localizes around midcell and the replication forks colocalize during a large fraction of the cell cycle. We tracked the dynamic behavior of mCherry-DnaN foci in 40 randomly selected cells at 10-min intervals from their appearance to their disappearance, corresponding to replication initiation and termination, respectively (Fig. 2A; see Fig. S2A in the supplemental material) (33). On average, mCherry-DnaN foci are present for 162 ± 26 min, compared with an average interdivision time of 198 ± 32 min, indicating that the replication forks progress with an average speed of about 400 bp/s. At the initiation of DNA replication, a single mCherry-DnaN focus appears around midcell (44% ± 11% of the cell length) and the replication forks remain colocalized as a single focus for most of the C period, although dynamic splitting and rejoining of replisomes sometimes occur (Fig. 2A and B). When split, the two mCherry-DnaN foci usually remain close to each other around midcell. However, in a small number of cells (3/40), the two foci occupy different cell halves for much of the DNA replication cycle (see Fig. S2A). On average, the net distance traveled by the replisome from assembly to disassembly is about 1/10 of the cell length. For replisomes that exhibit splitting, movement toward the quarter-cell positions is more pronounced (Fig. 2A and B; see Fig. S2A and B). The replisome shows a similarly dynamic localization pattern when SSB-GFP foci are tracked separately in wild-type cells (Fig. 2C) or in parallel with mCherry-DnaN in a dual-reporter strain (Fig. 1B and 2D; see Fig. S2C).
FIG 2 .
Spatiotemporal tracking of DNA replisomes in single cells. (A) Representative time traces of subcellular positions of overlapping (black circles) or split (green and red circles) mCherry-DnaN foci and Wag31-GFP foci (triangles) relative to the old cell pole. Images were recorded at 10-min intervals. Also see Fig. S2 in the supplemental material. (B) mCherry-DnaN foci during a single DNA replication cycle in 40 time-lapse series. Time zero corresponds to the first appearance of mCherry-DnaN foci. Upper panel, positions of overlapping (black circles) or split (green and red circles) mCherry-DnaN foci; lower panel, fraction of cells with one or two mCherry-DnaN foci at each time point. Po, old cell pole; Pn, new cell pole. (C) Positions of overlapping (black circles) or split (green and red circles) SSB-GFP foci during one DNA replication cycle in 10 time-lapse series. Time zero corresponds to the first appearance of SSB-GFP foci. (D) Two representative time traces of positions of mCherry-DnaN (red circles) and SSB-GFP (green circles) foci relative to the old cell pole. Images were recorded at 10-min intervals. Also see Fig. S2. (E) two representative time traces of the positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci and nucleoid stained with SYTO green (grey-shaded area) relative to the old cell pole. Images were recorded at 20-min intervals. Data are representative of 130 time-lapse series in which 121/130 cells show pattern 1 (left panel) and 9/130 cells show pattern 2 (right panel).
Movement of the replisome foci coincides with replication of the chromosome. We therefore used time-lapse microscopy to track the spatiotemporal dynamics of mCherry-DnaN foci relative to the nucleoid, which can be visualized by staining with SYTO green. In ~92% of the cells, mCherry-DnaN foci track along the middle of the cells and the nucleoid is equally distributed around them (Fig. 2E, cell 1). In the remaining ~8% of the cells, the mCherry-DnaN foci first appear at the edge of the nucleoid closest to the old cell pole, and as the C period progresses, they move toward the middle of the cell (Fig. 2E, cell 2). The latter pattern is similar to the replisome dynamics described for C. crescentus (6) and M. xanthus (8).
Subcellular localization of ori requires ParB.
The observation that ParB marks the position of the chromosomal origin and the location of replisome assembly suggests that ParB deficiency might affect the subcellular localization of the replication machinery. As reported previously (21), we found that a parB deletion mutant (ΔparB) has a discernible growth defect (see Fig. S3A in the supplemental material) and 15% of the cells are anucleate (see Fig. S3B), suggesting a defect in chromosome segregation. The distributions of interdivision times are similar for wild-type and ΔparB strains (see Fig. S3C), suggesting that the mutant’s growth defect is not due to delayed cell division but rather to the formation of nonviable anucleate cells.
In wild-type cells, the cell division septum is positioned with a narrow spatial distribution that is skewed toward the new cell pole (52 to 58% of the cell length) (see Fig. S3D) (33). In contrast, the division septum can be located anywhere along the cell length in ΔparB cells, except for 30% of the cell length closest to the old cell pole and 20% of the cell length closest to the new cell pole (see Fig. S3D), which accounts for the broader distribution of cell birth lengths for ΔparB cells than for wild-type cells (see Fig. S3E). Thus, ParB is important for subcellular septum positioning but has little impact on the timing of septum formation.
ParB deficiency also has an impact on nucleoid structure (see Fig. S3B) (21). The compact and condensed nucleoid bodies observed in wild-type cells appear to be more decondensed, stretched, or elongated in ΔparB cells. Moreover, in ΔparB cells, the nucleoid structure is highly irregular and cells with increased DNA content or guillotined chromosomes also occur (see Fig. S3B). Consistent with these observations, some ΔparB cells contain nucleoids that are longer than wild-type nucleoids (see Fig. S3F).
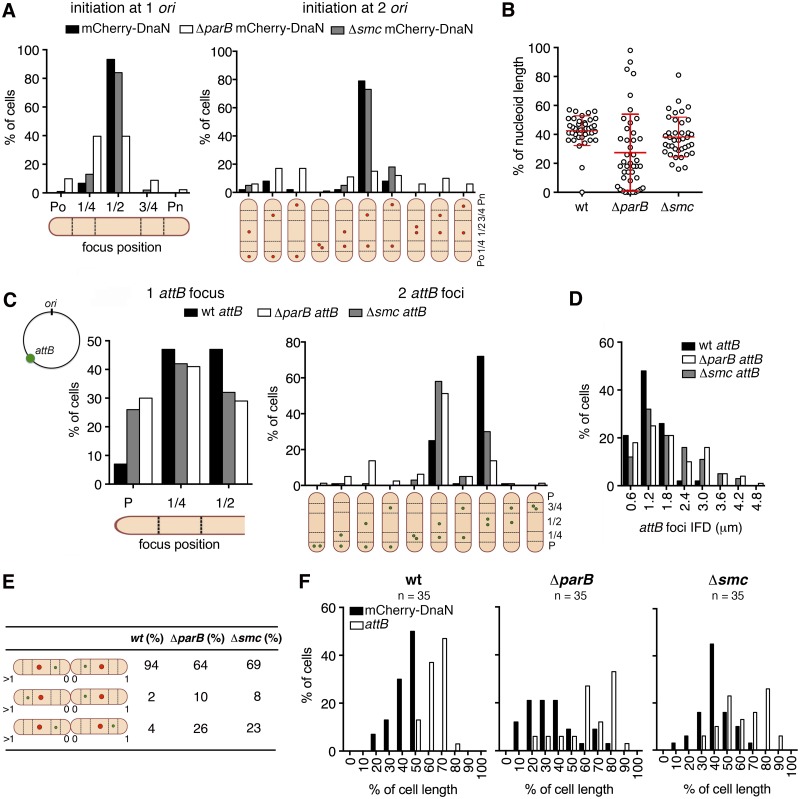
We investigated the impact of ParB deficiency on the localization of the chromosomal origin at the time of replication initiation. The subcellular positions of newly assembled mCherry-DnaN foci can be used to infer ori localization in wild-type and ΔparB cells because the replisome colocalizes with ori at the initiation of chromosome replication (Fig. 1G and H; see Fig. S1E in the supplemental material). The Wag31-GFP marker can be used to distinguish between initiation of DNA replication after (one ori) or before (two ori) cytokinesis (33). We found that newly assembled mCherry-DnaN foci are frequently mislocalized in ParB-deficient cells. In newborn cells with one ori, mCherry-DnaN foci appear at midcell in ~90% of wild-type cells compared to just ~40% of ΔparB cells, in which mCherry-DnaN foci more frequently appear at the quarter-cell (~50%) and polar (~10%) positions (Fig. 3A; see Fig. S4A). In newborn cells with two ori, mCherry-DnaN foci appear at the quarter-cell positions in ~80% of wild-type cells compared to just ~16% of ΔparB cells, in which mCherry-DnaN foci more frequently appear at the polar positions (~50%) (Fig. 3A; see Fig. S4B). The aberrant localization of replisomes observed in ΔparB cells reflects a mispositioning within the nucleoid bodies. In ParB-deficient cells, mCherry-DnaN foci assemble close to the edge of the nucleoid rather than the center of the nucleoid, as observed in wild-type cells (Fig. 3B). Although elongated nucleoids are evident in ~22% of ΔparB cells, average nucleoid length at initiation of chromosome replication is not significantly different for wild-type and ΔparB cells (see Fig. S4C). These observations indicate that ParB plays an important role in subcellular positioning of ori and replisome assembly, as well as chromosome organization and segregation.
FIG 3 .
Subcellular localization of ori and replisome assembly require ParB but not SMC. (A) Distribution of positions of mCherry-DnaN foci relative to the old cell pole (Po) at initiation of DNA replication in wild-type (black bars), ΔparB (white bars), and Δsmc (grey bars) cells (n = 100 per strain) that initiate replication after cytokinesis (one ori, left panel) or before cytokinesis (two ori, right panel). Timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell. Cells that initiate DNA replication before cytokinesis are classified on the basis of the positions of the two mCherry-DnaN foci (schematic). (B) Positions of mCherry-DnaN foci at initiation of DNA replication relative to the SYTO green-stained nucleoid length in wild-type (wt), ΔparB, and Δsmc cells (n = 40 per strain). (C) Distribution of positions of FROS-attB foci in wild-type (wt; black bars, n = 135), ΔparB (white bars, n = 112), and Δsmc (grey bars, n = 196) cells containing one FROS-attB focus (left panel) or two FROS-attB foci (right panel). Cells with two FROS-attB foci are classified on the basis of the positions of the two foci (schematic). (D) Distribution of IFDs of FROS-attB foci in wild-type (wt; black bars, n = 305), ΔparB (white bars, n = 351), and Δsmc (grey bars, n = 261) cells with two FROS-attB foci. (E) Positions of mCherry-DnaN foci (red circle) and FROS-attB foci (green circle) at the time of replisome assembly in sibling cells. Three scenarios and their relative frequencies are shown for wild-type (wt), ΔparB, and Δsmc cells. (F) Distribution of positions of mCherry-DnaN foci (black bars) and FROS-attB foci (white bars) at the time of replisome assembly relative to the old cell pole (pole age, ≥1) in wild-type (wt), ΔparB, and Δsmc cells (n = 35 per strain).
Subcellular localization of ori does not require SMC.
The SMC protein plays an important role in chromosome organization, condensation, and segregation in B. subtilis (23, 24, 34, 35) and C. crescentus (36). The M. smegmatis genome encodes a single SMC homolog, MSMEG_2423 (referred to as MSMEG_2422 in reference 27). A previous study demonstrated that SMC deficiency did not result in any discernible growth defects, but the effect of SMC deficiency on chromosome organization and segregation was not reported (27). We confirmed that deletion of the smc gene (Δsmc) does not significantly affect bacterial growth at the population level (data not shown), although single-cell imaging reveals the presence of small numbers of anucleate cells (~0.3%) and cells containing elongated nucleoids (see Fig. S3F in the supplemental material). We also found that Δsmc cells are slightly defective in septum localization, showing a broader distribution of septum positions than wild-type cells (see Fig. S3D). Subcellular localization of replisome assembly (first appearance of mCherry-DnaN foci) relative to cell length (Fig. 3A; see Fig. S4A and S4B) or nucleoid length (Fig. 3B; see Fig. S4C) is similar in wild-type and Δsmc cells, although the distributions are slightly broader in mutant cells. Similarly, we found that subcellular localization of ParB-mCherry foci (ori marker) is indistinguishable in wild-type and Δsmc cells (see Fig. S4D). These observations indicate that SMC plays only a minor role in subcellular positioning of ori and replisome assembly, as well as chromosome organization and segregation.
Subcellular localization of the chromosomal attB locus requires ParB and SMC.
Fluorescently tagged chromosomal loci can be imaged in live cells by using fluorescent repressor-operator systems (FROS) comprising an integrated array of tetO operator sequences and a TetR-GFP fusion protein that binds specifically to the tetO array. We used a mycobacteriophage L5 integrase-dependent plasmid (see Fig. S4E in the supplemental material) to integrate the tetO array at the chromosomal attB locus, which is located on the left arm of the chromosome at 245° relative to ori (37) (see Fig. S4F). The FROS-attB reporter strain has no discernible growth defect compared to wild-type cells (data not shown).
Wild-type cells containing one (~57%) or two (~43%) FROS-attB foci (see Fig. S4G) average 4.4 ± 1.0 µm and 5.7 ± 0.8 µm in length, respectively (see Fig. S4H). A large fraction (~15%) of ΔparB cells exhibit no visible FROS-attB foci, consistent with the formation of anucleate cells, and most of these cells are shorter than average (see Fig. S4H). Excluding anucleate cells, the fractions of ΔparB cells with one FROS-attB focus (~55%) or two FROS-attB foci (~43%) are similar to those of wild-type cells, but the mutant cells are longer, on average, at 4.7 ± 1.1 and 6.4 ± 1.2 µm, respectively (see Fig. S4H). Very long cells (average, 8.7 ± 2.0 µm) with three or even four FROS-attB foci comprise a small fraction (~2%) of ΔparB cells but are never observed among wild-type cells (see Fig. S4H).
The subcellular localization of FROS-attB foci is also impaired in ParB-deficient cells. In wild-type cells containing a single FROS-attB focus, the focus is usually localized at the midcell or quarter-cell position or less frequently (~6%) at a polar position (Fig. 3C). In ΔparB cells containing a single FROS-attB focus, the focus is more frequently (~30%) localized at a polar position (Fig. 3C). In wild-type cells containing two FROS-attB foci, the foci are usually distributed symmetrically relative to midcell, with average positions at 37% ± 5% and 54% ± 6% of the cell length (Fig. 3C; see Fig. S4I). In ΔparB cells containing two FROS-attB foci, the foci are usually distributed asymmetrically relative to midcell, with average positions at 23% ± 9% and 49% ± 13% of the cell length (Fig. 3C; see Fig. S4I). The distribution of interfocal distances (IFDs) between the two FROS-attB foci is also broader in ΔparB cells than in wild-type cells (Fig. 3D). Similar localization patterns are reflected in the position of FROS-attB foci relative to the nucleoid length in cells with one focus (see Fig. S4J) or two foci (see Fig. S4K). In ΔparB cells, the FROS-attB foci are located close to the edge of the nucleoid (average position, 21% ± 13% relative to the closest cell pole) rather than farther inside the nucleoid, as observed in wild-type cells (average position, 38% ± 9% relative to the closest pole) (see Fig. S4K).
Although SMC deficiency has only minor effects on subcellular positioning of ori and replisome assembly (see above), we found that subcellular localization of FROS-attB foci is markedly altered in Δsmc cells. Indeed, the localization of FROS-attB foci in Δsmc cells with one focus (average position, 27% ± 12% of cell length) or two foci (average positions, 28% ± 9% and 53% ± 9% of cell length) is similar to that in ΔparB cells (Fig. 3C). However, the distribution of subcellular positioning of FROS-attB foci is less heterogeneous in Δsmc cells than in ΔparB cells and no cells with more than two foci occur among Δsmc cells (Fig. 3C and D; see Fig. S4I).
We examined the subcellular localization of the FROS-attB locus relative to the replisome at the time of replisome assembly (first appearance of mCherry-DnaN foci) in sibling cell pairs. Three distinct scenarios were observed (Fig. 3E). In wild-type cells at the time of replisome assembly, ~94% of sibling cells show a mirror-symmetric distribution of FROS-attB foci relative to the division septum DnaN-attB–septum–attB-DnaN. The fraction of cells exhibiting an asymmetric distribution of FROS-attB foci relative to the division septum DnaN-attB–septum–DnaN-attB is greater in ΔparB cells (~36%) and Δsmc cells (~31%) than that in wild-type cells (~6%). In all three strains, asymmetric distribution of FROS-attB foci is more common among new-pole siblings (pole age, 1-0) compared to old-pole siblings (pole age, >1-0). On average, the IFD between mCherry-DnaN and FROS-attB foci is greater in ΔparB cells than in wild-type cells, whereas Δsmc cells show an intermediate phenotype due to mislocalization of FROS-attB foci (Fig. 3F).
ParB/SMC deficiency affects the number of replisomes per cell.
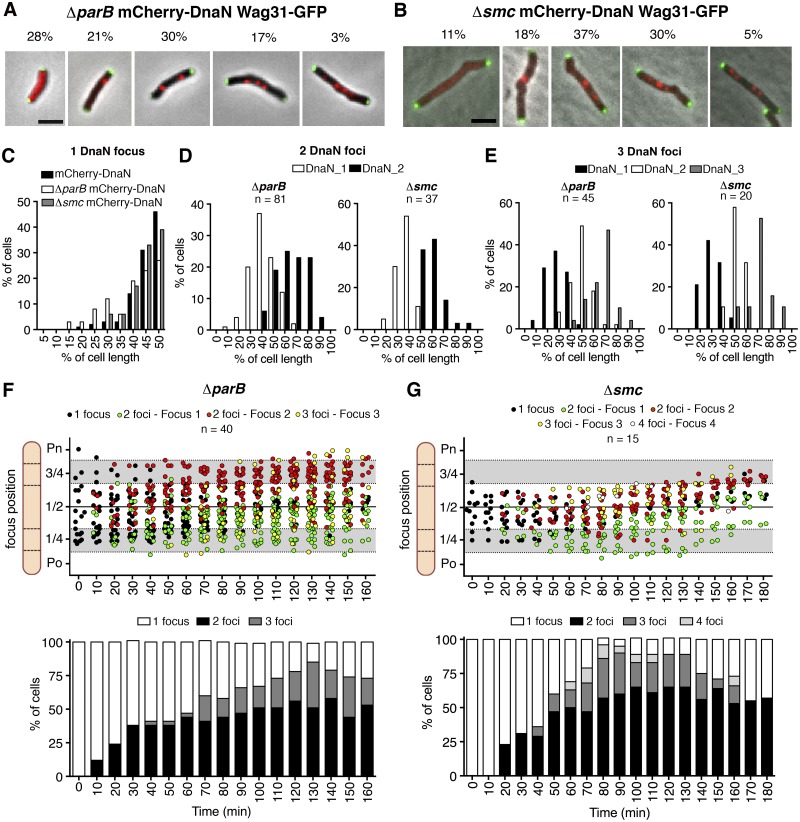
Because ParB and SMC seem to play important roles in chromosome organization, we investigated whether these factors might also affect the number and subcellular localization of replisomes. Microscopic analysis of ΔparB cultures revealed cells with zero, one, two, three, or four mCherry-DnaN foci (Fig. 4A; cf. Fig. 1A for wild-type cells). The fraction of cells with zero mCherry-DnaN foci is greater among ΔparB cells (28%) than wild-type cells (13%) and comprises mainly short cells (2 to 3 µm) and long cells (≥7 µm), presumably corresponding to the prereplication B period and postreplication D period of the cell cycle, respectively (see Fig. S5A in the supplemental material). The fraction of cells with one mCherry-DnaN focus is smaller among ΔparB cells (21%) than among wild-type cells (57%) and comprises mainly cells of intermediate length (between 3 and 6 µm) (see Fig. S5A). The fraction of cells containing two mCherry-DnaN foci is similar in ΔparB cells (30%) and wild-type cells (27%), whereas the fraction of cells containing three mCherry-DnaN foci is higher among ΔparB cells (17%) than among wild-type cells (4%). Also, ΔparB cells include a small fraction of long cells containing four usually rather dim mCherry-DnaN foci (3%), which are never observed among wild-type cells. The spatial distribution of mCherry-DnaN foci is broader in ΔparB cells containing one, two, or three foci (Fig. 4C to E) than in wild-type cells (Fig. 1F), suggesting that ParB plays an important role in replisome positioning. Similar perturbations of the distribution of replisome numbers (Fig. 4B) and their subcellular localization (Fig. 4C to E) occur among Δsmc cells, except that the fraction of cells with zero mCherry-DnaN foci is smaller (11%) and the fraction of cells with three mCherry-DnaN foci is greater (30%) than in ΔparB cells.
FIG 4 .
ParB and SMC deficiency affects replisome dynamics. (A and B) Representative snapshots of ΔparB (A) and Δsmc (B) cells with zero, one, two, three, or four mCherry-DnaN foci and their frequencies (n = 494 ΔparB cells, n = 150 Δsmc cells). Green foci, Wag31-GFP. Scale bar, 3 µm. (C) Distribution of positions of mCherry-DnaN foci in wild-type (n = 196), ΔparB (n = 130), and Δsmc (n = 140) cells with one focus. (D) Distribution of positions of mCherry-DnaN foci in ΔparB (n = 81) and Δsmc (n = 37) cells with two foci. (E) Distribution of positions of mCherry-DnaN foci in ΔparB (n = 45) and Δsmc (n = 20) cells with three foci. (F and G) Subcellular positions of mCherry-DnaN foci in 20 time-lapse series of ΔparB cells (F) and 15 time-lapse series of Δsmc cells (G). Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Upper panels, overlapping replication forks (black circles) and split replication forks (green and red circles); lower panels, fractions of cells with one, two, three, or four mCherry-DnaN foci at each time point. Po, old cell pole; Pn, new cell pole.
ParB/SMC deficiency affects replisome dynamics.
Previously we reported that wild-type M. smegmatis cells do not undergo multifork replication, in contrast to cells of better-characterized species such as B. subtilis and E. coli (33). At the population level, the ori-ter ratios are similar in wild-type and ΔparB strains (D. Jakimovic, personal communication), suggesting that ParB deficiency probably does not result in replication reinitiation and multifork replication. We therefore used time-lapse microscopy to identify the reason for the increased number of mCherry-DnaN foci observed in ΔparB cells.
Lineage analysis of time-lapse series revealed that ~21% of newborn ΔparB cells contain two chromosomes as a consequence of missegregation prior to division. In many of these cells, only one replisome assembles and three FROS-attB foci are visible at the time of replisome disassembly, indicating that only one of the two chromosomes undergoes replication. Subsequently, these cells divide into daughter cells that inherit one or two chromosomes. Eventually, cells containing two chromosomes at birth divide without undergoing another round of replication, thereby generating two daughter cells with normal ploidy. ParB-deficient cells include a small fraction (~2%) of elongated cells with three or even four chromosomes (see Fig. S5B in the supplemental material). These polyploid cells are characterized by simultaneous rounds of DNA replication on different chromosomes, resulting in the appearance of several mCherry-DnaN foci that often split and converge.
Although chromosome missegregation events may contribute to the increased number of replisomes observed in ΔparB cells, they cannot account for the large fraction of ΔparB cells (~80%) with normal ploidy but increased replisome numbers (see Fig. S5C). Nor can missegregation events account for the increased number of mCherry-DnaN foci in Δsmc cells, in which anucleate and polyploidy cells are uncommon (~0.3%). Time-lapse microscopy of ΔparB cells containing one chromosome at the time of replisome assembly reveals that the fraction of cells with two or three mCherry-DnaN foci increases during the elongation phase of DNA replication (Fig. 4F; see Fig. S6A and Movie S1). In ~30% of ΔparB cells undergoing DNA replication, the mCherry-DnaN focus splits into two separate foci around 20 to 30 min after initiation. The separated foci are mobile and frequently localize to different cell halves, and many of them split and converge repeatedly, generating cells with three or four foci (Fig. 4F). Similar replisome dynamics occur in Δsmc cells, although the distribution of subcellular positions of mCherry-DnaN foci tends to be narrower (Fig. 4G; see Fig. S6B and Movie S2). On average, the IFD of mCherry-DnaN foci during one DNA replication cycle is greater in ΔparB cells (1.4 ± 0.8 µm; P < 0.0001 by Fisher’s test) and Δsmc cells (1.1 ± 0.8 µm; P < 0.01 by Fisher’s test) than in wild-type cells (0.8 ± 0.4 µm) (see Fig. S5D). In a small fraction of mutant cells (ΔparB and Δsmc), the replisomes do not split, or split and converge only rarely, during the DNA replication cycle (see Fig. S5E). These observations suggest that ParB and SMC might be involved in the maintenance of cohesion between twin replication forks (and possibly leading- and lagging-strand replisomes) and restriction of the replication machinery to a specific subcellular address, thus generating the “factory-like” replisome organization observed in wild-type cells.
We conclude that the increased numbers of mCherry-DnaN foci observed in ΔparB and Δsmc cells are due not to multifork replication but rather to polyploidy (ΔparB cells) and splitting of sister replisomes (ΔparB and Δsmc cells).
Evidence for a “replication factory” near midcell.
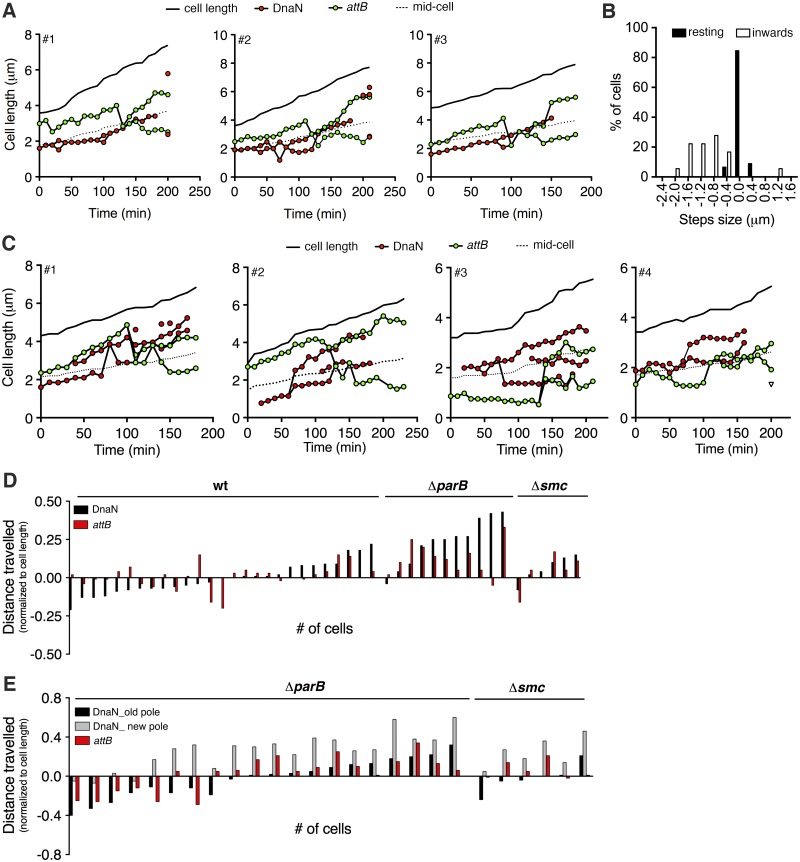
Two models of bacterial DNA replication have been proposed (38). In the first model, the replisomes move by tracking along the DNA, which is stationary; in the second model, the replisomes form a localized “replication factory” through which the DNA is spooled as it is replicated. We attempted to distinguish between these models by tracking the movement of replisome (mCherry-DnaN) foci relative to FROS-attB foci in wild-type cells (Fig. 5A and B). At the beginning of the C period, the FROS-attB focus and mCherry-DnaN focus localize close to the new cell pole and around midcell, respectively. Duplication of the FROS-attB focus later in the C period is preceded by a sudden inward movement of the FROS-attB focus toward the mCherry-DnaN focus until the two foci colocalize (Fig. 5A). The two newly duplicated FROS-attB foci then separate and move from midcell to ~40% and ~60% of the cell length, similar to the positions observed in snapshots of wild-type cells with two FROS-attB foci (Fig. 3C). Colocalization of FROS-attB and mCherry-DnaN foci occurs, on average, 109 ± 12 min after replication initiation at ori. Assuming a constant replication velocity, this value is in good agreement with the expected time of FROS-attB replication (~104 min postinitiation), based on the average C-period duration of 164 ± 16 min measured by time-lapse microscopy, the total length of the chromosome (6,988,192 bp), and the distance between attB and ori (2,236,678 bp). Separation of newly duplicated FROS-attB foci occurs, on average, 12 ± 8 min after colocalization of FROS-attB and mCherry-DnaN foci, and termination (replisome disassembly) occurs, on average, 52 ± 10 min after colocalization of FROS-attB and mCherry-DnaN foci, which is also in good agreement with the expected time to replicate the stretch of DNA between attB and ter (~57 min). During the early C period, FROS-attB foci move randomly with a speed of ~0.1 µm/min, which increases up to ~0.8 µm/min during the inward movement of FROS-attB toward the replisome (Fig. 5B). These observations are consistent with a “replication factory” model, in which the replisome remains positioned near midcell while DNA is pulled toward the replisome, spooled through it, duplicated, and extruded (3).
FIG 5 .
Altered movement of mCherry-DnaN and FROS-attB foci in ParB- and SMC-deficient cells. (A) Three representative time traces of positions of mCherry-DnaN (red circles) and FROS-attB (green circles) foci relative to the old cell pole. Images were recorded at 10-min intervals. Solid line, cell length; dotted line, midcell. See also Fig. S3B in the supplemental material. (B) Distribution of step sizes of FROS-attB foci. Images were recorded at 2-min intervals. Black bars, distribution of steps during the resting period prior to movement toward the replisome (n = 168). White bars, distribution of steps during movement toward replisome (n = 18). (C) Four representative time traces of positions of mCherry-DnaN (red circles) and FROS-attB (green circles) foci relative to the old cell pole in ΔparB cells. Images were recorded at 10-min intervals. Solid line, cell length; dotted line, midcell. See also Fig. S3B in the supplemental material. (D) Net movement of mCherry-DnaN foci (black bar) relative to FROS-attB foci (red bars) during the 90-min period after the first appearance of the replisome in wild-type, ΔparB, and Δsmc cells with unsplit mCherry-DnaN foci. (E) Net movement of mCherry-DnaN foci (grey and black bars) relative to FROS-attB foci (red bars) during the 90-min period after the first appearance of the replisome in wild-type (wt), ΔparB, and Δsmc cells with split mCherry-DnaN foci that move independently.
Increased replisome movement in ParB- and SMC-deficient cells.
Increased replisome mobility and splitting in ParB- and SMC-deficient cells suggests that, in the absence of constraints that restrict the replisome to a specific subcellular position, the split replisomes might move independently along the DNA. We addressed this possibility by tracking the movement of replisome (mCherry-DnaN) foci relative to FROS-attB foci in ΔparB cells. We found that in some cells the FROS-attB locus localizes near the old cell pole prior to duplication, and newly duplicated FROS-attB loci sometimes segregate asymmetrically with respect to midcell (for example, cells 3 and 4 in Fig. 5C). In these cases, one locus moves rapidly toward the new cell pole while the other locus shows little change in its subcellular position over time. In contrast to wild-type cells (Fig. 5B), in ~50% of ΔparB cells, the FROS-attB focus does not show any sudden movement toward the replisome before its duplication (Fig. 5C). Instead, the FROS-attB focus remains localized around the quarter-cell position while one of the two replisome foci (presumably the one replicating the left arm of the chromosome) moves toward the FROS-attB focus and colocalizes with it just prior to its duplication (for example, cell 2 in Fig. 5C).
These observations suggest that in ΔparB cells the normal positioning of the replisome at midcell is lost and the replisome tracks along the DNA (6). Consistent with this interpretation, the net average movement of the mCherry-DnaN and FROS-attB foci during the 90-min period after replication initiation is 0.08% ± 0.06% and 0.05% ± 0.05% of the cell length in wild-type cells (Fig. 5D), compared to 0.24% ± 0.14% and 0.13% ± 0.09% of the cell length in ΔparB cells where the replisome does not undergo splitting (Fig. 5E). Movement of the mCherry-DnaN and FROS-attB foci is similarly increased in the fraction of ΔparB cells where the replisome does undergo splitting and the split foci move independently (Fig. 5E). In these cells, net movement of the split mCherry-DnaN foci is 0.47% ± 0.05% of the cell length, whereas movement of the FROS-attB foci is 0.09% ± 0.13% of the cell length, similar to that in wild-type cells (Fig. 5E).
The Δsmc strain behaves somewhat differently. In the fraction of Δsmc cells with split replisome foci, movement of the split foci relative to each other is 0.47% ± 0.07% of the cell length, whereas movement of the FROS-attB foci is 0.08% ± 0.06% of the cell length, similar to that in ΔparB cells (Fig. 5E). However, in the fraction of Δsmc cells with a single (unsplit) replisome focus, the movement of mCherry-DnaN and FROS-attB foci is 0.08% ± 0.06% and 0.05% ± 0.05% of the cell length, respectively, similar to that in wild-type cells (Fig. 5D).
Despite the striking heterogeneity of single-cell behavior, these observations suggest that in a large fraction of ParB- and SMC-deficient cells the replisome undergoes splitting and the split replisomes move through the cytoplasm as if tracking along the DNA. We have never observed this dynamic behavior in wild-type cells, where the replisome remains unsplit and positioned around midcell, appearing to spool the DNA through it rather than tracking along the DNA.
ParB is required for the normal subcellular localization of SMC.
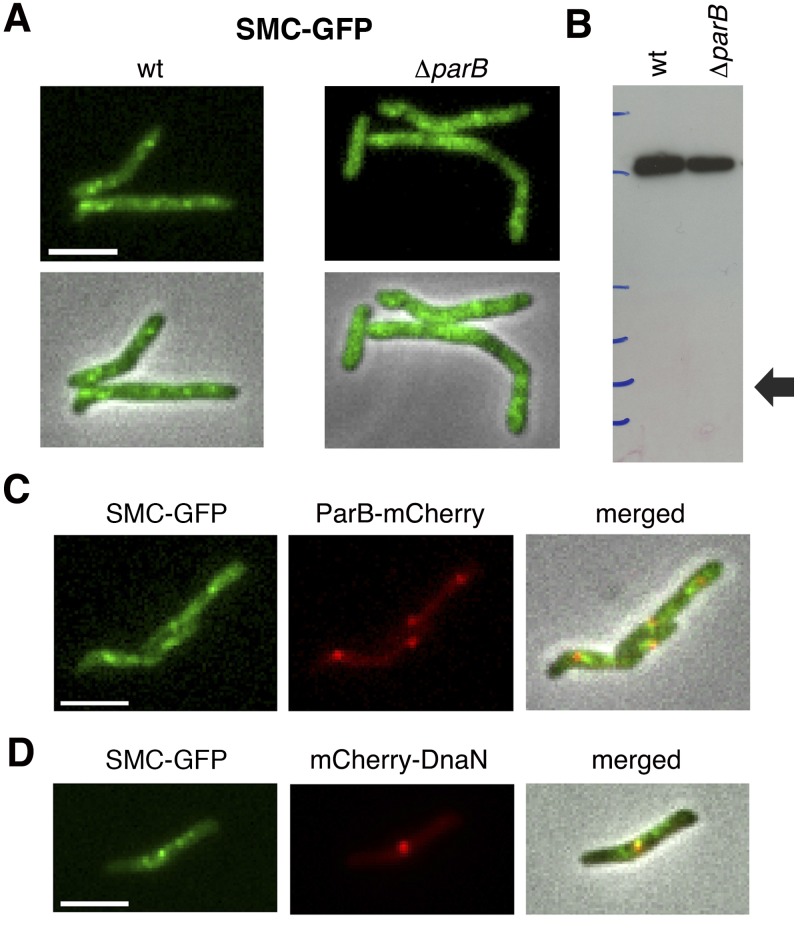
Phenotypic similarities between the M. smegmatis ΔparB and Δsmc mutants suggested that ParB and SMC might have overlapping roles in chromosome organization and replisome positioning. This interpretation is consistent with the observation that SMC foci colocalize with ParB foci in B. subtilis (24). We tracked the spatiotemporal dynamics of SMC localization in M. smegmatis by using reporter strains expressing an smc-gfp fusion gene, which replaces the wild-type smc gene at the native chromosomal locus. The fusion protein appears to be functional, as evidenced by the normal (wild-type) growth kinetics of the SMC-GFP reporter strain (see Fig. S3G in the supplemental material).
In wild-type cells, a variable number of discrete SMC-GFP foci are distributed throughout the cell, except at the polar positions (Fig. 6A). In ParB-deficient cells, the SMC-GFP fusion protein does not form bright fluorescent foci but rather appears to be dispersed in patches throughout the cytoplasm (Fig. 6A). The more diffuse pattern of SMC-GFP fluorescence in ΔparB cells is not due to cleavage of the SMC-GFP fusion protein and release of free GFP, as confirmed by immunoblot analysis (Fig. 6B). Although we found that SMC-GFP and ParB-mCherry foci seldom colocalize, they are frequently observed in close proximity to each other (Fig. 6C). Similarly, although SMC-GFP foci are frequently observed in close proximity to mCherry-DnaN foci, colocalization is uncommon (Fig. 6D).
FIG 6 .
ParB deficiency affects subcellular localization of SMC-GFP. (A) Representative images of SMC-GFP foci in wild-type (wt) and ΔparB cells. Signal intensity is normalized to facilitate comparison. (B) Immunoblot assay of SMC-GFP in wild-type (wt) and ΔparB strains with anti-GFP antibody. The arrow shows the predicted position of GFP. (C) Representative images of SMC-GFP (green) and ParB-mCherry (red) foci in exponentially growing wild-type cells. Scale bar, 3 µm. (D) Representative images of SMC-GFP (green) and mCherry-DnaN (red) foci in exponentially growing wild-type cells. Scale bar, 3 µm.
DISCUSSION
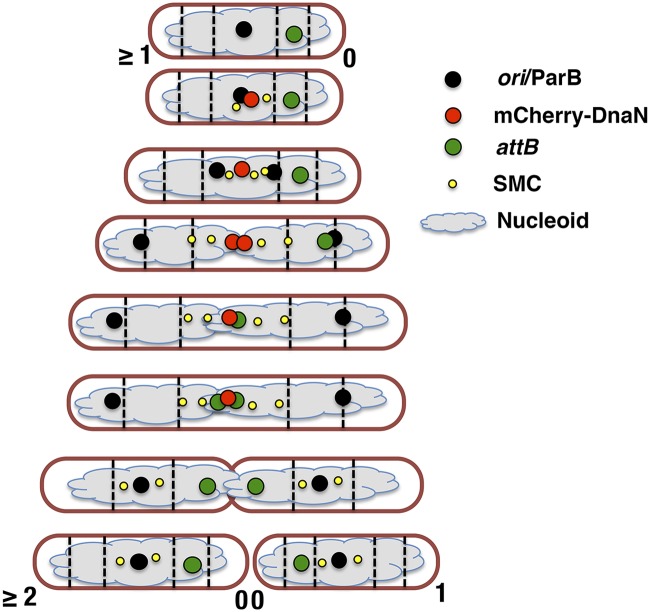
In this study, we report that ParB and SMC coordinate chromosome organization and spatial localization of DNA replication in M. smegmatis. In wild-type cells, the replisome is positioned near midcell, the two replication forks colocalize throughout the DNA replication cycle, and the chromosomal attB locus moves rapidly from cell pole to midcell immediately before its duplication. These observations are consistent with the “replication factory” model proposed for B. subtilis and P. aeruginosa (3, 4). However, movement of the attB locus from cell pole to midcell is faster in M. smegmatis (~2 to 4 min) than the movement of a fluorescently tagged chromosomal locus in B. subtilis over a similar distance (~30 min) (3). We hypothesize that this movement could be due to the replisome functioning as a DNA-translocating molecular motor that “reels in” the attB locus once the slack in the intervening stretch of DNA has been taken up by the process of replication.
Prior to replication, the M. smegmatis chromosome is arranged in space such that ori is positioned near midcell while the attB locus on the left arm of the chromosome is in close proximity to the new cell pole, suggesting a speculative R-ori-L configuration (Fig. 7) (39). Confirmation of this configuration will require additional experiments with multiple fluorescently tagged loci on both the left and right arms of the chromosome. Maintenance of the wild-type chromosome configuration, which is recapitulated with remarkable fidelity after each cell division, is somehow dependent on the concerted action of the ParB and SMC proteins. Although ParB-deficient cells exhibit enhanced cell-to-cell phenotypic variation, including a minority of cells with nearly wild-type behavior, in the majority of ΔparB cells, ori localization (and, consequently, replisome assembly) is biased toward cell pole or quarter-cell positions (Fig. 3A, initiation at two ori). Although parB is not essential for viability, the large fraction of anucleate cells formed by the ΔparB strain indicates that ParB is required for the high fidelity of ori positioning and segregation exhibited by wild-type cells.
FIG 7 .
Schematic of chromosome organization and replisome dynamics in M. smegmatis.
Although the mechanisms responsible for ori localization and movement in M. smegmatis are unknown, ParB might play a role in the observed segregation of the ori-proximal region immediately after its duplication and subsequent maintenance of the duplicated ori-proximal regions around the quarter-cell positions, which will become the midcell positions in the daughter cells following cell division. Increased IFD between the ori and attB loci, as observed in ParB-deficient cells, suggests that in the absence of ParB-mediated constraints on ori positioning, the chromosome might adopt a longitudinal organization similar to that in C. crescentus (40, 41) or MukB-deficient E. coli (42). ParA has also been implicated in these processes, as indicated by the mislocalization of ParB in a ParA-deficient strain of M. smegmatis (22). However, the mechanism of action of the ParABS system in M. smegmatis is unclear. In C. crescentus, it has been proposed that ParA filaments extend across the cell, attach to the chromosome via interactions with the ParB-parS complex, and pull the ori-proximal region of the chromosome toward the new cell pole by depolymerization-mediated contraction, analogous to the role of the mitotic spindle in segregating eukaryotic chromosomes (43). However, an alternative “DNA relay” model has recently been proposed, in which an asymmetric spatial gradient of ParA molecules (with the highest concentration at the new cell pole) rectifies the otherwise symmetric elastic dynamics of the chromosome through transient interactions with the ParB-parS complex (18).
In contrast to the SMC protein of B. subtilis (34, 35), that of M. smegmatis seems to be involved in neither localization nor segregation of ori, although the aberrant subcellular positioning of the attB locus observed in SMC-deficient cells suggests that SMC does play a role in chromosome organization. In the absence of SMC, DNA synthesis continues and replicated foci are resolved and segregated, but in many cells, the subcellular chromosome organization differs from that in the wild type. We hypothesize that the SMC foci observed in cells expressing an SMC-GFP fusion protein serve as centers of SMC activity, as proposed in B. subtilis (24). ParB seems to play a role in the focal recruitment of SMC to the chromosome, as SMC foci are reduced or eliminated in ParB-deficient cells. In wild-type cells, SMC foci are often found in close proximity to but usually do not colocalize with ParB, suggesting that ParB might recruit SMC to the ori-proximal region and from there, SMC might diffuse along the chromosome, as proposed in B. subtilis and S. pneumoniae (23, 25). Moreover, it has been suggested that SMC might dictate the overall organization of the bacterial chromosome by condensing lengthwise along the two arms of the chromosome, thereby generating a stiffer structure into which newly synthesized DNA is folded (15, 44). Consistent with this idea, we found that the mirror-symmetrical localization of attB loci observed in wild-type sibling cells is partially lost in SMC-deficient cells, in which an asymmetric distribution of attB loci relative to midcell is more frequently observed. We hypothesize that in wild-type sibling cells, the chromosomes adopt an R-L–L-R configuration, which may switch to an R-L–R-L configuration in the absence of SMC.
In a small fraction of wild-type cells the replisome undergoes splitting and the split replisomes localize to opposite cell halves for the remainder of the replication cycle. The frequency of cells containing split replisomes is strongly increased in ParB- and SMC-deficient strains, for two reasons: replisome splitting in a large fraction of ΔparB and Δsmc cells and transient polyploidy in a small fraction of ΔparB cells because of chromosome missegregation events during cell division. Both phenotypes might be attributable to impaired subcellular chromosome organization in the absence of ParB or SMC. Consistent with this interpretation, we found that the replisome can assemble at almost any subcellular position in ParB-deficient cells, whereas in wild-type and SMC-deficient cells, replisome assembly is confined to the midcell region. In ParB- and SMC-deficient cells, the split replisomes usually move toward the edge of the nucleoid, similar to those in E. coli (5), although in a small fraction of cells, the replisomes move longitudinally through the cell, similar to those in C. crescentus (6). In these species the pattern of replisome migration matches the orientation of the chromosome: L-ori-R for E. coli (39) and ori(out)-ter(in) for C. crescentus (40, 41). It is possible that the altered replisome dynamics observed in ΔparB and Δsmc cells might be linked to impaired subcellular chromosome organization in these mutants. Thus, chromosomal regions that are normally proximal to each other in wild-type cells might be spatially separated in ParB- and SMC-deficient cells, resulting in separation of replisomes engaged in replicating the left and right chromosome arms. Consistent with this interpretation, in ParB- and SMC-deficient cells, the split replisomes seem to track along the DNA following the subcellular disposition of the nucleoid.
Time-lapse analysis of ParB- and SMC-deficient strains sometimes revealed cells containing three or even four DnaN foci, which arise by transient splitting of the two main replisomes into smaller and fainter foci. We hypothesize that these additional foci might be engaged in the synthesis of Okazaki fragments on the lagging strand, consistent with the observation that these small and dim DnaN foci often colocalize with SSB foci. It is possible that the separation of leading- and lagging-strand replisomes might become transiently visible because of loss of local chromosome compaction in ParB- and SMC-deficient cells. An alternative explanation is suggested by the recent demonstration that the DnaN clamp zone provides a platform for spatiotemporal coupling of DNA replication and mismatch detection in B. subtilis (45). However, the latter interpretation is unlikely to be correct because SMC deficiency does not result in a mutator phenotype and SMC does not seem to play an important role in DNA repair (27).
Although localization of replisomes around midcell is apparently not essential for DNA replication and cell cycle progression, we hypothesize that replisome localization might play a role in positioning of the cell division septum. This idea arises from the observation that the division septum is often mislocalized in ParB- and SMC-deficient cells; this defect is particularly pronounced in ParB-deficient cells, which generate small anucleate progeny because of mispositioning of the division septum. Time-lapse microscopy of dual-reporter strains expressing fluorescent markers of the replisome (mCherry-DnaN) and the division septum (Wag31-GFP) also supports the idea that the replisome might play an important role in division site selection (our unpublished data). Linking division site selection to the spatiotemporal dynamics of chromosome replication and segregation might serve as an alternative positioning mechanism in mycobacteria, which do not encode homologs of the minicell or nucleoid occlusion proteins responsible for division site selection in other bacteria (46).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. smegmatis mc2155 (wild type) and derivative strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 0.5% albumin, 0.2% glucose, 0.085% NaCl, 0.5% glycerol, and 0.05% Tween 80. Cultures were grown at 37°C with aeration to mid-log phase (optical density at 600 nm [OD600] of ~0.5). Aliquots were stored in 15% glycerol at −80°C and thawed at room temperature before use; individual aliquots were used once and discarded.
Oligonucleotides, plasmids, and bacterial strains.
The oligonucleotides, plasmids, and bacterial strains used in this study are listed in Table S1.1-S1.3 in the supplemental material. Details of plasmid and strain construction are provided in Text S1.
Single-cell microscopy.
For snapshot microscopy, bacteria were grown to mid-log phase (OD600 of ~0.5) in 7H9 medium, collected by centrifugation (2,400 × g, 5 min), concentrated 10-fold in fresh 7H9 medium (37°C), and passed through a 5-µm-pore-size polyvinylidene difluoride syringe filter (Millipore) to remove clumps. The declumped bacteria were transferred to a glass slide with a 1% agarose pad soaked in LB medium and covered with a coverslip. Nucleoid staining was done by incubating cells for 5 min with SYTO 17 red or SYTO 13 green (Invitrogen) diluted 1:2,000 prior to microscopy at room temperature.
For time-lapse microscopy, bacteria were prepared as described above, except that the declumped cell suspension was spread on a glass coverslip, covered with a semipermeable membrane and cultured in a custom-made microfluidic device with a continuous flow of 7H9 medium at 37°C (flow rate, 25 µl/min), as previously described (47). Nucleoid staining was done by adding SYTO green (diluted 1:2,000) to the flow medium.
Bacteria were imaged with a DeltaVision personalDV microscope (Applied Precision) equipped with a 100× oil immersion objective and an environmental chamber maintained at 37°C. Images were recorded on phase-contrast and fluorescence channels (490/20-nm excitation filter and 528/38-nm emission filter for GFP, 575/25-nm excitation filter and 632/60-nm emission filter for mCherry) with a CoolSnap HQ2 camera. Images were processed with Softworx software (Applied Precision). Cell length was measured as the sum of short linear segments tracking along the center line of an individual cells in order to accommodate irregularities in cell shape. Interdivision time (It) was defined as the time interval between the first appearance of septal Wag31-GFP (birth) and the next appearance of septal Wag31-GFP (division), as previously described (33).
Immunoblotting.
Bacteria were grown to mid-log phase (OD600 of ~0.5), collected by centrifugation (2,400 × g, 5 min), and resuspended in lysis buffer (50 mM Tris-Cl, pH 7.5) containing protease inhibitors (Roche). Proteins were extracted in a FastPrep-24 instrument (MP Biomedicals) with 0.1-mm zirconia beads, separated on NuPAGE Novex 3 to 5% Tris-acetate gels (Invitrogen), and electrotransferred to a nitrocellulose membrane. A mouse anti-GFP monoclonal antibody was used as the primary antibody (1:1,000). A rabbit anti-mouse antibody conjugated to horseradish peroxidase (Dako) was used as the secondary antibody (1:1,000).
SUPPLEMENTAL MATERIAL
Materials and Methods. Download
Oligonucleotides, plasmids, and bacterial strains used in this study.
Replisome localization in single cells. (A) Growth curves of wild-type M. smegmatis (●) and reporter strains expressing mCherry-DnaN (red circle) or mCherry-DnaN and Wag31-GFP (green circle). Data are representative of three independent experiments. (B) Growth curves of wild-type M. smegmatis (●) and reporter strains expressing SSB-GFP (green circle), mCherry-DnaN (red circle), mCherry-DnaN and SSB-GFP (blue circle), or mCherry-DnaN and GFP-DnaX (■). Data are representative of three independent experiments. (C) Distribution of IFDs between mCherry-DnaN and GFP-DnaX foci (n = 70). (D) Two representative time traces of positions of ParB-mCherry foci in a strain expressing ParB-mCherry and Wag31-GFP. ParB-mCherry foci are orange (old-pole side) or blue (new-pole side). Images were recorded at 10-min intervals. Po, old pole; Pn, new pole. (E) Time-lapse image series of cells expressing ParB-mCherry and GFP-DnaN. Images were recorded at 10-min intervals. Initially, there are no GFP-DnaN foci visible (0 min). Newly formed GFP-DnaN foci colocalize with ParB-mCherry foci (10 min). Duplication and segregation of ParB-mCherry foci occur shortly after replisome assembly (20 min), and segregation continues thereafter (30 min). Scale bar, 3 µm. Download
Spatiotemporal tracking of DNA replisomes in single cells. (A) Nine representative time traces of subcellular positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci and Wag31-GFP foci (triangles) relative to the old cell pole. Images were recorded at 10-min intervals. Solid line, cell length; dotted line, midcell position. (B) Average positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci during one DNA replication cycle in 40 cells (mean values ± standard errors of the means). Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Po, old pole; Pn, new pole. (C) Positions of mCherry-DnaN (red circles) and SSB-GFP (green circles) foci during one DNA replication cycle in 10 cells. Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Po, old pole; Pn, new pole. Download
ParB deficiency impairs chromosome segregation and cell division. (A) Upper panel, growth curves of wild-type and ΔparB strains; lower panel, growth curves of mCherry-DnaN, ΔparB mCherry-DnaN, ΔparB mCherry-DnaN Wag31-GFP, and ΔparB mCherry-DnaN FROS-attB strains. Bacteria were grown in 7H9 liquid medium at 37°C. Data are representative of three experiments with similar results. (B) Nucleoids of wild-type (left) and ΔparB (right) cells stained with SYTO green (pseudocolored red). Anucleate ΔparB cells (white asterisks) and ΔparB cells with guillotined chromosomes (white arrows) are indicated. (C) Distribution of interdivision times in wild-type cells (black bars, n = 452) and ΔparB cells (white bars, n = 356). (D) Distribution of septum positions in wild-type (black bars, n = 87), ΔparB (white bars, n = 153), and Δsmc (grey bars, n = 93) cells. (E) Distribution of birth lengths (Lb) in wild-type (black bars), ΔparB (grey bars), and Δsmc (white bars) cells (n = 100 per strain). (F) Distribution of nucleoid lengths in wild-type (black bars), ΔparB (grey bars), and Δsmc (white bars) cells (n = 50 per strain). (G) Growth curves of wild-type and SMC-GFP strains. Download
ParB/SMC deficiency affects nucleoid organization. (A) Positions of mCherry-DnaN foci relative to the old cell pole in wild-type, ΔparB, and Δsmc cells (n = 100 per strain) that initiate DNA replication after cytokinesis (one ori). The timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell (33). Fisher’s exact test: NS, not significant; *, P = 0.01; **, P = 0.008. (B) Distribution of positions of mCherry-DnaN foci in cells that initiate DNA replication before cytokinesis (two ori’s) in wild-type (black bars, n = 100), ΔparB (white bars, n = 100), and Δsmc (grey bars, n = 22) cells. The timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell (33). (C) Nucleoid lengths at the time of replisome assembly (first appearance of mCherry-DnaN foci) in wild-type, ΔparB, and Δsmc cells (n = 40 per strain). The nucleoid was stained with SYTO green. (D) Spatial positions of duplicated ParB-mCherry foci in wild-type (n = 74) and Δsmc (n = 42) cells. Fisher’s exact test: NS, not significant. (E) Schematic of pIS280, an attB-integrating FROS plasmid that contains 256 tetO repeats (blue) and expresses TetR-GFP (green) from the hsp60 promoter. Gn, gentamicin resistance; HygR, hygromycin resistance; L5, bacteriophage L5 integrase gene; attP, bacteriophage L5 attachment site. (F) Schematic of M. smegmatis genome showing the location of the attB locus (green circle) at 245°. (G) Representative phase-contrast and fluorescence images (merged) of M. smegmatis with one or two FROS-attB foci (green). Cells were stained with the fluorescent membrane dye FM4-64. Scale bar, 3 µm. (H) Number of FROS-attB foci relative to cell length in wild-type (n = 326, left panel) and ΔparB (n = 313, right panel) cells. (I) Distribution of positions of FROS-attB foci in cells with two foci in wild-type (n = 70), ΔparB (n = 78), and Δsmc (n = 94) cells. (J and K) Distribution of FROS-attB foci relative to nucleoid length in wild-type and ΔparB cells with one (J) or two (K) foci. Nucleoid stained with SYTO green. Download
ParB/SMC deficiency alters replisome localization and dynamics. (A) Numbers of mCherry-DnaN foci relative to cell length in ΔparB (n = 300) and Δsmc (n = 300) cells. (B) Two representative pedigree trees derived from time-lapse microscopy of ΔparB mCherry-DnaN FROS-attB dual-reporter cells. Images were recorded at 10-min intervals. Pedigree trees represent the behavior of all of the descendants of a single progenitor cell through five generations. Black vertical lines indicate cell divisions. Red horizontal lines indicate the C period, defined by appearance and disappearance of mCherry-DnaN foci. At each division, the number of FROS-attB foci indicates the number of chromosomes inherited by each sibling cell: siblings with a single chromosome each (1/1) or one anucleate sibling and one diploid sibling (0/2). The majority (77%) of anucleate cells arise from the new-pole half of the mother cells. (C) Representative time-lapse image series of ΔparB mCherry-DnaN FROS-attB dual-reporter cells recorded on the phase-contrast and fluorescence channels (merged) at the time points indicated (minutes). At division (black arrow at 0 min), each daughter cell inherits one chromosome, as indicated by a single FROS-attB focus in each cell. After the initial appearance of a single mCherry-DnaN focus (asterisk at 30 min), the replisome splits into two or three mCherry-DnaN foci (asterisks). Duplication of the FROS-attB focus is visible at 210 min (white arrows). At division, each cell inherits a single FROS-attB focus (division site marked by black arrow at 260 min). (D) Distribution of IFDs between mCherry-DnaN foci in wild-type (black bars, n = 160), ΔparB (white bars, n = 150), and Δsmc (grey bars, n = 160) cells. (E) Number of times mCherry-DnaN foci split during a single DNA replication cycle, expressed as a fraction of the length of the C period, in wild-type (blue line), ΔparB (orange line), and Δsmc (black line) cells (n = 100 cells per strain). Download
Temporal tracking of subcellular positions of mCherry-DnaN foci in ΔparB and Δsmc cells. (A and B) Eight representative time traces showing positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci relative to the old cell pole in ΔparB cells (A) and Δsmc cells (B). When present, third and fourth mCherry-DnaN foci are indicated (yellow and white circles). Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Download
Time-lapse microscopy of ΔparB mCherry-DnaN Wag31-GFP dual-reporter cells. Representative time-lapse microscopy of ΔparB cells expressing mCherry-DnaN (marks the DNA replisome) and Wag31-GFP (marks the cell division septum and cell poles). Bacteria were cultivated at 37°C in a custom-made microfluidic device (6) and fed with a continuous flow of Middlebrook 7H9 medium. Phase-contrast and fluorescence images were acquired at 10-min intervals with a 100× oil immersion objective. Subcellular localization and movement of mCherry-DnaN foci were tracked at the single-cell level. The interdivision time is defined by two successive events of Wag31-GFP localization to the septum position near midcell (1). The C period is defined by the appearance (replication initiation) and disappearance (replication termination) of mCherry-DnaN foci. Download
Time-lapse microscopy of Δsmc mCherry-DnaN Wag31-GFP dual-reporter cells. Representative time-lapse microscopy of Δsmc cells expressing mCherry-DnaN (marks the DNA replisome) and Wag31-GFP (marks the cell division septum and cell poles). Bacteria were cultivated at 37°C in a custom-made microfluidic device (6) and fed by a continuous flow of Middlebrook 7H9 medium. Phase-contrast and fluorescence images were acquired at 10-min intervals with a 100× oil immersion objective. Subcellular localization and movement of mCherry-DnaN foci were tracked at the single-cell level. The interdivision time is defined by two successive events of Wag31-GFP localization to the septum position near midcell (1). The C period is defined by the appearance (replication initiation) and disappearance (replication termination) of mCherry-DnaN foci. Download
ACKNOWLEDGMENTS
This work was funded in part by grants to J.D.M. from the Swiss National Science Foundation (310030_156945) and the Innovative Medicines Initiative (115337), a joint undertaking of the European Union Seventh Framework Programme and EFPIA (http://www.imi.europa.eu/).
We thank David Sherratt (Oxford University) for providing the pLAU44 and pLAU53 plasmids and Djenet Bousbaine (EPFL) for assistance with data analysis. We are grateful to Neeraj Dhar, Dagmara Jakimowicz, Paul Murima, Shogo Ozaki, and Antonio Porro for valuable discussions and critical comments on the manuscript. We have no conflicts of interest to declare.
Footnotes
Citation Santi I, McKinney JD. 2015. Chromosome organization and replisome dynamics in Mycobacterium smegmatis. mBio 6(1):e01999-14. doi:10.1128/mBio.01999-14.
Contributor Information
William Margolin, University of Texas Medical School at Houston.
Steven J. Norris, University of Texas-Houston Medical School.
REFERENCES
- 1.Kaguni JM. 2011. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol 15:606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendrickson H, Lawrence JG. 2007. Mutational bias suggests that replication termination occurs near the dif site, not at ter sites. Mol Microbiol 64:42–56. doi: 10.1111/j.1365-2958.2007.05596.x. [DOI] [PubMed] [Google Scholar]
- 3.Lemon KP, Grossman AD. 2000. Movement of replicating DNA through a stationary replisome. Mol Cell 6:1321–1330. doi: 10.1016/S1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 4.Vallet-Gely I, Boccard F. 2013. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet 9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen RB, Wang SC, Shapiro L. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J 20:4952–4963. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma A, Kamran M, Verma V, Dasgupta S, Dhar SK. 2014. Intracellular locations of replication proteins and the origin of replication during chromosome duplication in the slowly growing human pathogen Helicobacter pylori. J Bacteriol 196:999–1011. doi: 10.1128/JB.01198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms A, Treuner-Lange A, Schumacher D, Søgaard-Andersen L. 2013. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet 9:e1003802. doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kongsuwan K, Dalrymple BP, Wijffels G, Jennings PA. 2002. Cellular localisation of the clamp protein during DNA replication. FEMS Microbiol Lett 5:255–262. doi: 10.1016/S0378-1097(02)01023-6. [DOI] [PubMed] [Google Scholar]
- 10.Migocki MD, Lewis PJ, Wake RG, Harry EJ. 2004. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol Microbiol 54:452–463. doi: 10.1111/j.1365-2958.2004.04267.x. [DOI] [PubMed] [Google Scholar]
- 11.Bates D, Kleckner N. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 17:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkmen MB, Grossman AD. 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol 62:57–71. doi: 10.1111/j.1365-2958.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- 13.Toro E, Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol 2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Montero Llopis P, Rudner DZ. 2013. Organization and segregation of bacterial chromosomes. Nat Rev Genet 14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolivos S, Sherratt D. 2014. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev 38:380–392. doi: 10.1111/1574-6976.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Possoz C, Junier I, Espeli O. 2012. Bacterial chromosome segregation. Front Biosci 17:1020–1034. doi: 10.2741/3971. [DOI] [PubMed] [Google Scholar]
- 18.Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. 2014. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife 3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohl DA, Easter J, Gober JW. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol 42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 20.Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537–548. [DOI] [PubMed] [Google Scholar]
- 21.Jakimowicz D, Brzostek A, Rumijowska-Galewicz A, Zydek P, Dołzbłasz A, Smulczyk-Krawczyszyn A, Zimniak T, Wojtasz L, Zawilak-Pawlik A, Kois A, Dziadek J, Zakrzewska-Czerwińska J. 2007. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology 153:4050–4060. doi: 10.1099/mic.0.2007/011619-0. [DOI] [PubMed] [Google Scholar]
- 22.Ginda K, Bezulska M, Ziółkiewicz M, Dziadek J, Zakrzewska-Czerwińska J, Jakimowicz D. 2013. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol 87:998–1012. doi: 10.1111/mmi.12146. [DOI] [PubMed] [Google Scholar]
- 23.Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/Spo0J promotes chromosome segregation in B. subtilis. Cell 15:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 15:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. 2011. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol 81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- 26.Petrushenko ZM, She W, Rybenkov VV. 2011. A new family of bacterial condensins. Mol Microbiol 81:881–896. doi: 10.1111/j.1365-2958.2011.07763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Güthlein C, Wanner RM, Sander P, Böttger EC, Springer B. 2008. A mycobacterial smc null mutant is proficient in DNA repair and long-term survival. J Bacteriol 190:452–456. doi: 10.1128/JB.01315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson D, Wang X, Rudner DZ. 2012. Spatio-temporal organization of replication in bacteria and eukaryotes (nucleoids and nuclei). Cold Spring Harb Perspect Biol 4:a010389. doi: 10.1101/cshperspect.a010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitao T, Dasgupta S, Nordström K. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol Microbiol 38:392–400. doi: 10.1046/j.1365-2958.2000.02138.x. [DOI] [PubMed] [Google Scholar]
- 30.Adachi S, Kohiyama M, Onogi T, Hiraga S. 2005. Localization of replication forks in wild-type and mukB mutant cells of Escherichia coli. Mol Genet Genomics 274:264–271. doi: 10.1007/s00438-005-0023-6. [DOI] [PubMed] [Google Scholar]
- 31.Lee PS, Grossman AD. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol 60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 32.Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 33.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun 4:2470. doi: 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 34.Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. 2014. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol 24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Tang OW, Riley EP, Rudner DZ. 2014. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol 24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz MA, Shapiro L. 2011. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol Microbiol 82:1359–1374. doi: 10.1111/j.1365-2958.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peña CE, Stoner JE, Hatfull GF. 1996. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J Bacteriol 178:5533–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bates D. 2008. The bacterial replisome: back on track? Mol Microbiol 69:1341–1348. doi: 10.1111/j.1365-2958.2008.06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Liu X, Possoz C, Sherratt DJ. 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev 20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A 101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umbarger MA, Toro E, Wright MA, Porreca GJ, Baù D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, Shapiro L, Dekker J, Church GM. 2011. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. 2007. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol 65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. 2010. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marko JF. 2009. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E 79:051905. doi: 10.1103/PhysRevE.79.051905. [DOI] [PubMed] [Google Scholar]
- 45.Lenhart JS, Sharma A, Hingorani MM, Simmons LA. 2013. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol Microbiol 87:553–568. doi: 10.1111/mmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monahan LG, Liew AT, Bottomley AL, Harry EJ. 2014. Division site positioning in bacteria: one size does not fit all. Front Microbiol 5:19. doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods. Download
Oligonucleotides, plasmids, and bacterial strains used in this study.
Replisome localization in single cells. (A) Growth curves of wild-type M. smegmatis (●) and reporter strains expressing mCherry-DnaN (red circle) or mCherry-DnaN and Wag31-GFP (green circle). Data are representative of three independent experiments. (B) Growth curves of wild-type M. smegmatis (●) and reporter strains expressing SSB-GFP (green circle), mCherry-DnaN (red circle), mCherry-DnaN and SSB-GFP (blue circle), or mCherry-DnaN and GFP-DnaX (■). Data are representative of three independent experiments. (C) Distribution of IFDs between mCherry-DnaN and GFP-DnaX foci (n = 70). (D) Two representative time traces of positions of ParB-mCherry foci in a strain expressing ParB-mCherry and Wag31-GFP. ParB-mCherry foci are orange (old-pole side) or blue (new-pole side). Images were recorded at 10-min intervals. Po, old pole; Pn, new pole. (E) Time-lapse image series of cells expressing ParB-mCherry and GFP-DnaN. Images were recorded at 10-min intervals. Initially, there are no GFP-DnaN foci visible (0 min). Newly formed GFP-DnaN foci colocalize with ParB-mCherry foci (10 min). Duplication and segregation of ParB-mCherry foci occur shortly after replisome assembly (20 min), and segregation continues thereafter (30 min). Scale bar, 3 µm. Download
Spatiotemporal tracking of DNA replisomes in single cells. (A) Nine representative time traces of subcellular positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci and Wag31-GFP foci (triangles) relative to the old cell pole. Images were recorded at 10-min intervals. Solid line, cell length; dotted line, midcell position. (B) Average positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci during one DNA replication cycle in 40 cells (mean values ± standard errors of the means). Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Po, old pole; Pn, new pole. (C) Positions of mCherry-DnaN (red circles) and SSB-GFP (green circles) foci during one DNA replication cycle in 10 cells. Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Po, old pole; Pn, new pole. Download
ParB deficiency impairs chromosome segregation and cell division. (A) Upper panel, growth curves of wild-type and ΔparB strains; lower panel, growth curves of mCherry-DnaN, ΔparB mCherry-DnaN, ΔparB mCherry-DnaN Wag31-GFP, and ΔparB mCherry-DnaN FROS-attB strains. Bacteria were grown in 7H9 liquid medium at 37°C. Data are representative of three experiments with similar results. (B) Nucleoids of wild-type (left) and ΔparB (right) cells stained with SYTO green (pseudocolored red). Anucleate ΔparB cells (white asterisks) and ΔparB cells with guillotined chromosomes (white arrows) are indicated. (C) Distribution of interdivision times in wild-type cells (black bars, n = 452) and ΔparB cells (white bars, n = 356). (D) Distribution of septum positions in wild-type (black bars, n = 87), ΔparB (white bars, n = 153), and Δsmc (grey bars, n = 93) cells. (E) Distribution of birth lengths (Lb) in wild-type (black bars), ΔparB (grey bars), and Δsmc (white bars) cells (n = 100 per strain). (F) Distribution of nucleoid lengths in wild-type (black bars), ΔparB (grey bars), and Δsmc (white bars) cells (n = 50 per strain). (G) Growth curves of wild-type and SMC-GFP strains. Download
ParB/SMC deficiency affects nucleoid organization. (A) Positions of mCherry-DnaN foci relative to the old cell pole in wild-type, ΔparB, and Δsmc cells (n = 100 per strain) that initiate DNA replication after cytokinesis (one ori). The timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell (33). Fisher’s exact test: NS, not significant; *, P = 0.01; **, P = 0.008. (B) Distribution of positions of mCherry-DnaN foci in cells that initiate DNA replication before cytokinesis (two ori’s) in wild-type (black bars, n = 100), ΔparB (white bars, n = 100), and Δsmc (grey bars, n = 22) cells. The timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell (33). (C) Nucleoid lengths at the time of replisome assembly (first appearance of mCherry-DnaN foci) in wild-type, ΔparB, and Δsmc cells (n = 40 per strain). The nucleoid was stained with SYTO green. (D) Spatial positions of duplicated ParB-mCherry foci in wild-type (n = 74) and Δsmc (n = 42) cells. Fisher’s exact test: NS, not significant. (E) Schematic of pIS280, an attB-integrating FROS plasmid that contains 256 tetO repeats (blue) and expresses TetR-GFP (green) from the hsp60 promoter. Gn, gentamicin resistance; HygR, hygromycin resistance; L5, bacteriophage L5 integrase gene; attP, bacteriophage L5 attachment site. (F) Schematic of M. smegmatis genome showing the location of the attB locus (green circle) at 245°. (G) Representative phase-contrast and fluorescence images (merged) of M. smegmatis with one or two FROS-attB foci (green). Cells were stained with the fluorescent membrane dye FM4-64. Scale bar, 3 µm. (H) Number of FROS-attB foci relative to cell length in wild-type (n = 326, left panel) and ΔparB (n = 313, right panel) cells. (I) Distribution of positions of FROS-attB foci in cells with two foci in wild-type (n = 70), ΔparB (n = 78), and Δsmc (n = 94) cells. (J and K) Distribution of FROS-attB foci relative to nucleoid length in wild-type and ΔparB cells with one (J) or two (K) foci. Nucleoid stained with SYTO green. Download
ParB/SMC deficiency alters replisome localization and dynamics. (A) Numbers of mCherry-DnaN foci relative to cell length in ΔparB (n = 300) and Δsmc (n = 300) cells. (B) Two representative pedigree trees derived from time-lapse microscopy of ΔparB mCherry-DnaN FROS-attB dual-reporter cells. Images were recorded at 10-min intervals. Pedigree trees represent the behavior of all of the descendants of a single progenitor cell through five generations. Black vertical lines indicate cell divisions. Red horizontal lines indicate the C period, defined by appearance and disappearance of mCherry-DnaN foci. At each division, the number of FROS-attB foci indicates the number of chromosomes inherited by each sibling cell: siblings with a single chromosome each (1/1) or one anucleate sibling and one diploid sibling (0/2). The majority (77%) of anucleate cells arise from the new-pole half of the mother cells. (C) Representative time-lapse image series of ΔparB mCherry-DnaN FROS-attB dual-reporter cells recorded on the phase-contrast and fluorescence channels (merged) at the time points indicated (minutes). At division (black arrow at 0 min), each daughter cell inherits one chromosome, as indicated by a single FROS-attB focus in each cell. After the initial appearance of a single mCherry-DnaN focus (asterisk at 30 min), the replisome splits into two or three mCherry-DnaN foci (asterisks). Duplication of the FROS-attB focus is visible at 210 min (white arrows). At division, each cell inherits a single FROS-attB focus (division site marked by black arrow at 260 min). (D) Distribution of IFDs between mCherry-DnaN foci in wild-type (black bars, n = 160), ΔparB (white bars, n = 150), and Δsmc (grey bars, n = 160) cells. (E) Number of times mCherry-DnaN foci split during a single DNA replication cycle, expressed as a fraction of the length of the C period, in wild-type (blue line), ΔparB (orange line), and Δsmc (black line) cells (n = 100 cells per strain). Download
Temporal tracking of subcellular positions of mCherry-DnaN foci in ΔparB and Δsmc cells. (A and B) Eight representative time traces showing positions of overlapping (black circles) and split (green and red circles) mCherry-DnaN foci relative to the old cell pole in ΔparB cells (A) and Δsmc cells (B). When present, third and fourth mCherry-DnaN foci are indicated (yellow and white circles). Images were recorded at 10-min intervals. Time zero corresponds to the first appearance of mCherry-DnaN foci. Download
Time-lapse microscopy of ΔparB mCherry-DnaN Wag31-GFP dual-reporter cells. Representative time-lapse microscopy of ΔparB cells expressing mCherry-DnaN (marks the DNA replisome) and Wag31-GFP (marks the cell division septum and cell poles). Bacteria were cultivated at 37°C in a custom-made microfluidic device (6) and fed with a continuous flow of Middlebrook 7H9 medium. Phase-contrast and fluorescence images were acquired at 10-min intervals with a 100× oil immersion objective. Subcellular localization and movement of mCherry-DnaN foci were tracked at the single-cell level. The interdivision time is defined by two successive events of Wag31-GFP localization to the septum position near midcell (1). The C period is defined by the appearance (replication initiation) and disappearance (replication termination) of mCherry-DnaN foci. Download
Time-lapse microscopy of Δsmc mCherry-DnaN Wag31-GFP dual-reporter cells. Representative time-lapse microscopy of Δsmc cells expressing mCherry-DnaN (marks the DNA replisome) and Wag31-GFP (marks the cell division septum and cell poles). Bacteria were cultivated at 37°C in a custom-made microfluidic device (6) and fed by a continuous flow of Middlebrook 7H9 medium. Phase-contrast and fluorescence images were acquired at 10-min intervals with a 100× oil immersion objective. Subcellular localization and movement of mCherry-DnaN foci were tracked at the single-cell level. The interdivision time is defined by two successive events of Wag31-GFP localization to the septum position near midcell (1). The C period is defined by the appearance (replication initiation) and disappearance (replication termination) of mCherry-DnaN foci. Download