ABSTRACT
The type III secretion system (T3SS) is a highly conserved protein delivery system found in multiple Gram-negative pathogens, including Yersinia pseudotuberculosis. Most studies of Yersinia species type III intoxication of host cells have focused on the bacterial determinants that promote assembly and function of the secretion system. In this study, we performed a pooled RNA interference (RNAi) screen to identify mammalian host proteins required for the cytotoxic effects associated with the Yersinia translocated substrate YopE, a GTPase-activating protein (GAP) that inactivates the small Rho GTPases. Cell populations were positively selected for short hairpin RNAs (shRNAs) that interfere with YopE activity using a combination of fluorescence resonance energy transfer (FRET) and flow cytometry, and the degree of enrichment was determined by deep sequencing. Analysis of the candidates identified by the enrichment process revealed that many were important for the initial step of Y. pseudotuberculosis T3SS function, YopB/D pore formation. These candidates included shRNA that depleted downstream effectors of RhoA signaling, coated pit formation, and receptors involved in cell signaling, including the chemokine receptor CCR5 (chemokine [C-C motif] receptor 5). Depletion of CCR5 in 293T cells yielded a defect in YopB/D pore formation and effector translocation, while both phenotypes could be complemented by overexpression of CCR5 protein. Yop effector translocation was also decreased in isolated primary phagocytic cells from a Ccr5−/− knockout mouse. We postulate that CCR5 acts to promote translocation by modulating cytoskeletal activities necessary for proper assembly of the YopB/D translocation pore. Overall, this study presents a new approach to investigating the contribution of the host cell to T3SS in Y. pseudotuberculosis.
IMPORTANCE
Many Gram-negative bacteria require type III secretion systems (T3SS) for host survival, making these highly specialized secretion systems good targets for antimicrobial agents. After the bacterium binds to host cells, T3SS deposit proteins into the cytosol of host cells through a needle-like appendage and a protein translocon channel. Translocation of proteins via this system is highly regulated, and the contribution of the host cell in promoting assembly and insertion of the channel into the plasma membrane, folding of the bacterial proteins, and trafficking of these substrates are all poorly characterized events. In this study, we identified host cell proteins important for activity of YopE, a Yersinia pseudotuberculosis T3SS-delivered protein. The results demonstrate that insertion and assembly of the translocon are complex processes, requiring a variety of membrane trafficking and cytoskeletal processes, as well as a surprising role for cell surface signaling molecules in supporting proper function.
INTRODUCTION
Type III secretion systems (T3SS) are critical determinants of virulence for a large number of Gram-negative pathogens (1, 2). Upon encountering a host cell, these highly conserved macromolecular complexes deliver unfolded substrate proteins from the bacterial cytosol through a needle-like apparatus into target eukaryotic host cells, allowing the pathogen to control a variety of host cell processes (3). The T3SS complex is comprised of three protein subgroups: the structural proteins that form the needle-like injectisome, substrate proteins that pass through the injectisome, and translocon proteins, which form a channel in the plasma membrane, allowing final passage into the host cell (2). Among the different Gram-negative pathogens possessing T3SS, there is high conservation in the structural proteins and translocator proteins. In contrast, although there is some sharing of individual translocated substrate proteins among pathogens, in general, these proteins have distinct catalytic activities to suit the respective pathogens encoding them (2). For example, Salmonella and Shigella species use T3S to inject proteins in order to promote their own uptake into nonphagocytic cells followed by establishing an intracellular replicative niche (4, 5). Conversely, Pseudomonas and Yersinia inject effectors by T3S in order to avoid phagocytosis by innate immune cells, thus impairing their function and promoting survival and persistence of bacteria in an extracellular locale (6).
All three Yersinia species that are pathogenic to humans, Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, carry genes on a 70-kb virulence plasmid that encode a T3SS (7). The Yersinia secretion apparatus is comprised of approximately 29 Ysc proteins that make up the export machinery as well as the needle-like injectisome (7). The needle is composed of YscF monomers with the scaffolding protein LcrV at the tip which forms a complex with the translocator Yops (Yersinia outer membrane proteins) YopB and YopD (8). YopB/D are then capable of forming pores in the host cell plasma membrane, leading to translocation of proteins into the host cytosol (9). Yersinia spp. translocate a group of either five or six Yops into the host cytosol to disrupt normal cell processes, including YopJ/YopP, YopM, YopO/YpkA, YopH, YopT, and YopE (10).
The role of the host cell in translocation, cellular trafficking, and subsequent localization of the Yops to the target sites is largely unknown, but evidence supports the hypothesis that host cell factors contribute to the translocation and activation of T3SS substrates. Previous studies of T3S in Shigella, Salmonella, Pseudomonas, and enteropathogenic Escherichia coli (EPEC) conclude that functional lipid rafts are critical for insertion of the T3SS translocon as well as subsequent translocation of proteins into host cells (11–13). Lipid rafts are domains within the plasma membrane, which are thought to coordinate signaling events since they contain a high concentration of protein receptors, signaling proteins, and cytoskeletal components (14). These highly organized signaling platforms have been shown to be important for G-protein-coupled receptor signaling, including chemokine receptor signaling, immune cell activation, membrane trafficking, and viral, bacterial, and bacterial toxin entry into cells (14).
A recent study of Pseudomonas T3SS concludes that an unidentified eukaryotic factor is responsible for triggering effector secretion, which is subsequently inactivated by the translocated substrate ExoS, a bifunctional protein that exhibits both RhoGAP activity and ADP ribosylation activity in cells (15). Furthermore, experiments reveal that in Y. pseudotuberculosis, insertion of the YopB/D translocon pore along with engagement of β1 integrins by Yersinia adhesins causes the activation of the Rho GTPases, stimulating accumulation of Yops within target cells. Consistent with ExoS, YopE and YopT activity downmodulates translocation by inactivating Rho family members (16). Last, experimental evidence investigating the EPEC T3SS demonstrated that there was a requirement for host cell factors in triggering secretion, translocation, and activation of the translocated Tir protein in cytoplasmic extracts (17).
In this study, an RNA interference (RNAi) knockdown screen was performed in order to investigate the contribution of the host cell during type III secretion. Combining fluorescence resonance energy transfer (FRET) of a Rho GTPase biosensor with flow cytometry, we were able to positively select short hairpin RNAs (shRNAs) that inhibit YopE inactivation to identify host proteins important for T3SS function. Validation of the shRNA candidates revealed that the chemokine receptor CCR5 (chemokine [C-C motif] receptor 5) is important for YopB/D translocon function. Overall, we have developed a unique screen to elucidate the role of host cell factors in T3S of Y. pseudotuberculosis. The results obtained from this screen provide critical new information on the molecular mechanism of T3SS protein translocation in Yersinia spp., which can also be extrapolated to T3SS in many other bacteria.
RESULTS
Flow cytometric assay for YopE GAP activity using a FRET readout.
We previously developed fluorescence resonance energy transfer (FRET) assays that allow image analyses of Y. pseudotuberculosis YopE GTPase-activating protein (GAP) action on the host Rho family members Rac1, Cdc42, and RhoG (18, 19). These previous assays used dual-plasmid systems with FRET donors and acceptors on separate plasmids to measure loss of GTP binding by the Rho family members. To perform a pooled shRNA screen, we adapted the microscopy assay to a population-based flow cytometry strategy, measuring YopE activity with a single-plasmid FRET system. To this end, a previously constructed plasmid encoding a single-chain four-protein fusion consisting of a Rho binding domain (RBD) fused upstream of cyan fluorescent protein (CFP), yellow fluorescent protein (YFP), and membrane-targeted RhoA (in order from the N to C terminus) (20). When activated, the GTP-loaded RhoA moiety binds the RBD, bringing CFP in close proximity to YFP. This results in FRET emission from YFP after excitation of CFP (Fig. 1A). Due to the fact that the RhoA/RBD FRET binding partners are physically linked in this biosensor, FRET can be accurately measured using flow cytometry in every transfected cell. When RhoA is in an inactive GDP-bound conformation, the fusion protein produces low YFP emission that fails to respond upon CFP excitation. In contrast, when RhoA is in the active GTP-bound conformation, the biosensor will produce a high FRET signal following excitation of CFP.
FIG 1 .
Cells can be sorted using a FRET readout for YopE activity. (A) Schematic representation of the Rho biosensor, adapted from reference 20 with permission. The N terminus (N Term) and C terminus (C Term) are indicated. (B) Measuring YopE activity within a population of cells by FRET using flow cytometry. (Top) 293T cells were transiently transfected with either the wild-type RhoA biosensor (RBS WT) or a mutant defective for GTP binding (RBS N19). (Bottom) Cells expressing RBS WT were incubated with Y. pseudotuberculosis YPIII yopE+ ΔyopHMOJ for 1 h at an MOI of 50. Cells were visualized using 405-nm excitation with 450-nm and 530-nm emission filters. Transiently transfected cells were gated away from untransfected cells (Materials and Methods). (C and D) Enrichment of FRET+ cells from a mixed population (Materials and Methods). (C) Cells expressing either RBS N19 (FRET−) or RBS WT cotransfected with pmCherry-N1 to mark the FRET+ cells were mixed together at a 20:1 ratio, respectively, prior to FACS enrichment. (D) RBS WT-expressing cells incubated with Y. pseudotuberculosis YPIII yopE+ ΔyopHMOJ for 1 h at an MOI of 50 (FRET−) and cells cotransfected with RBS WT with pmCherry-N1 to mark the FRET+ cells were mixed together at a 20:1 ratio, respectively, prior to FACS enrichment. Postsort analysis of populations (unsorted, FRET+, and FRET−) was performed using 405-nm excitation with 450-nm and 530-nm emission filters for CFP and YFP, respectively. mCherry expression was measured using 532-nm excitation with 610-nm emission filter (Materials and Methods). The numbers in the graphs are the percentages of cells at each respective gate.
Analyses using flow cytometry indicate that FRET emission in this system results from GTP loading of RhoA. The majority of cells that transiently expressed the wild-type RhoA biosensor (RBS WT) exhibited higher CFP→YFP FRET than cells transfected with a RhoA N19 mutant biosensor (RBS N19) that is defective in GTP binding (Fig. 1B). Similarly, cells that harbor RBS WT challenged with Y. pseudotuberculosis yopE+ resulted in low FRET emission levels, similar to that observed with the RBS N19 (Fig. 1B). These results predict that shRNA knockdown of host genes important for YopE function should prevent the inactivation of RhoA caused by active YopE and retain the FRET signal that is observed in uninfected cells. Furthermore, distinct populations can be positively selected, allowing cells with shRNA that interfere with YopE function to be isolated.
A proof-of-principle experiment was performed to demonstrate that a small population with detectable FRET emission could be enriched from the total population. Cells coexpressing the RBS WT and mCherry plasmids in order to mark the FRET-positive (FRET+) cells were mixed with cells expressing the GTP-loading defective RhoA N19 biosensor (FRET−) at a 1:20 ratio (mCherry-positive [mCherry+] RBS/RBS N19 ratio; Fig. 1C). Gates corresponding to FRET-positive and FRET-negative cells were established in order to collect cells by fluorescence-activated cell sorting (FACS), and the relative enrichment of the cells in the collected FRET+ population having mCherry was determined. The RBS WT (mCherry+) cells were enriched approximately 25-fold from the mixed population (Fig. 1C), which corresponded with a 15-fold enrichment of mCherry-expressing cells collected in the FRET-positive FACS gate. For further validation, cells coexpressing the RBS WT and mCherry were mixed at a 1:20 ratio with cells expressing the RBS WT incubated with Y. pseudotuberculosis yopE+ and sorted as described above (Fig. 1D). On the basis of the increase in the fraction of cells harboring mCherry, the FRET+ population was enriched approximately 7-fold. Overall, these data indicate that cells resistant to YopE activity should be found in the population having increased FRET emission and can be enriched from the total population in a pooled shRNA screen.
A genome-scale screen for host cell factors important for type 3-mediated YopE delivery.
A FACS-based pooled shRNA screen was carried out in 293T cells in order to identify host cell proteins that contribute to the translocation and activity of the Y. pseudotuberculosis type III secretion system substrate YopE. To perform the screen, plasmid libraries having 8 × 103 to 10 × 103 shRNAs were lentiviral packaged for delivery into 293T cells (Fig. 2A; also see Materials and Methods). The RBS WT was then transfected into shRNA-depleted cells 48 h after viral delivery, and approximately 24 h after transfection, the cells were incubated at a multiplicity of infection (MOI) of 50 with Y. pseudotuberculosis yopE+ for 1 h. To identify candidate shRNAs that interfere with YopE action, cells showing the highest levels of FRET emission were then positively selected by FACS. To identify the cell populations that should be collected, cells that harbor the RhoA biosensor without the shRNA bank were left uninfected or were incubated with Y. pseudotuberculosis yopE+ to identify the FRET+ (Fig. 2B, RBS) and FRET− (Fig. 2B, YPIII ΔHMOJ) sorting gates, respectively. Based on this information, FRET+ and FRET− sorting gates were set to allow collection of either population (Fig. 2B). shRNA depletion did not markedly change the distribution of FRET+ RhoA biosensor cells compared to cells expressing the RBS alone (Fig. 2B; compare RBS to RBS RNAi Library). Using these sorting gates, we found that the isolated FRET+ population was generally enriched approximately 10- to 20-fold for each individual experiment (Fig. 2B, FRET+).
FIG 2 .
A genome-scale screen for host cell proteins that support function of YopE protein. (A) Schematic representation of the screening procedure. 293T cells were transiently transfected with the wild-type Rho biosensor (RBS) 48 h after transduction with shRNA library. Cells were incubated with Y. pseudotuberculosis YPIII yopE+ ΔyopHMOJ for 1 h at an MOI of 50 at 24 h posttransfection. shRNAs were amplified and subjected to Illumina sequencing from an unsorted input control and the FRET+ and FRET− populations enriched by sorting (Materials and Methods). (B) FACS data from a representative experiment. FRET+ and FRET− gates were established using cells that were transfected with RBS WT in the absence of shRNA and were either not challenged with bacteria or challenged with YPIII yopE+ ΔyopHMOJ. Unsorted, FRET+, and FRET− populations were analyzed by FACS to observe enrichment after sorting. Postsort analysis was performed using 405-nm excitation with 450-nm and 530-nm emission filters for CFP and YFP. The numbers in the graphs are the percentages of cells at each respective gate. (C) Pie chart depicting the biological process categories of host factors significantly overrepresented among the shRNA candidates identified to inhibit YopE activity in the screen as calculated using the Panther database. The categories, the number of shRNAs identified in each category, and the P values are shown.
Genomic DNA (gDNA) was then extracted from three populations: (i) the collected FRET+ population; (ii) the population showing low levels of FRET emission (FRET−); and (iii) the unsorted input control (Fig. 2A). The shRNA sequences were PCR amplified from gDNA obtained from each of these three populations, subjected to deep sequencing, and aligned to the human genome hg_17 database to identify shRNA targets. The number of reads was normalized for each individual population before generating ratios of the FRET+/unsorted and FRET+/FRET− populations (Fig. 2A) for each shRNA (Materials and Methods). shRNAs that interfered with the action of YopE were defined either as shRNAs that were overrepresented in the FRET+ population compared to both the unsorted population and the FRET− population or shRNAs that were overrepresented in the FRET+ population compared to just the unsorted population. Using data from at least three independently performed enrichments, ZMAD values, indicating how many median absolute deviations (MAD) from the median, were calculated for both the FRET+/unsorted and FRET+/FRET− ratios for each shRNA (22). shRNAs were considered of interest for further analysis if they had a ZMAD of >2 in two independent experiments.
Overall, a total of 10,146 shRNAs that target 5,447 genes were screened at least twice. There were 389 identified shRNAs that were classified above the set threshold of ZMAD of >2 for both FRET+/unsorted and FRET+/FRET− (see Table S1 in the supplemental material). Increasing this threshold to an ZMAD of >5 or >10 reduced the number of shRNA candidates that passed this criterion to 121 or 69 for these ratios, respectively. Pathway analysis was performed using the Panther database to identify any statistically overrepresented biological processes among the genes identified in the screen, using 360 of the 389 shRNAs which mapped to the database (Fig. 2C; Table S1) (23). Overrepresented pathways identified included cellular communication, cellular adhesion, complement activation, and intracellular protein transport.
Candidate shRNAs caused defective YopB/D pore formation.
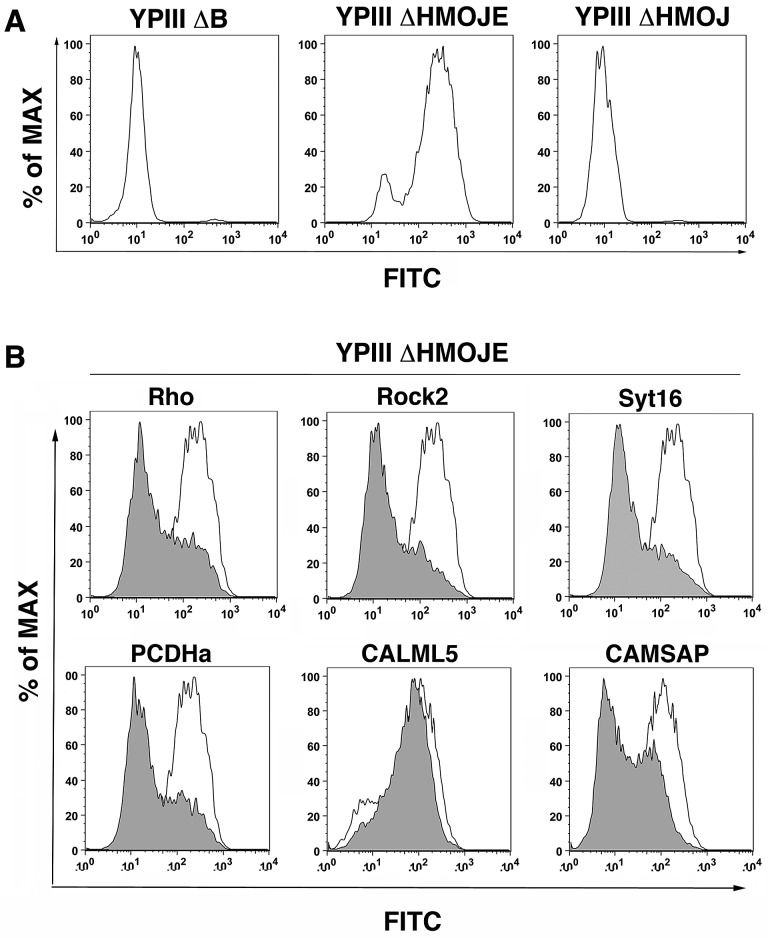
Since inhibition of YopE activity was the readout for the RNAi screen, candidate shRNAs identified may contribute to various aspects of T3SS function. Transfectants harboring shRNAs identified in the screen were analyzed to determine their effects on YopE function. We initially focused on insertion of the YopB/YopD translocon pore into the plasma membrane, which is the first step in movement of YopE into host cells. Pore formation by YopB/D requires activation of the GTPase RhoA. Subsequent delivery of YopE inactivates the Rho GTPase, resulting in closing of the translocon pore and cessation of translocation (16, 24). In the absence of YopE, YopB/D pores accumulate, leading to a compromised host cell membrane, as detected by release of lactate dehydrogenase (LDH) into the culture medium (24). As an alternative method to measure YopB/D pore formation, 293T cells were labeled with a membrane-impermeant amine-reactive fluorescent probe, allowing measurement of membrane integrity defects observed by increased fluorescence intensity relative to controls using FACS analysis. Cells probed after incubation with Y. pseudotuberculosis ΔyopHMOJE, which lacks all the effectors but can form YopB/D pores, caused a large shift in fluorescence intensity relative to the translocon-defective strain (YPIII ΔyopB) or the strain that has active YopE protein (YPIII ΔyopHMOJ) (Fig. 3A). This method, which is effective at detecting loss of membrane integrity at an MOI of 25 and allowed subpopulations to be analyzed, showed a higher sensitivity than that of the previously published LDH release assay, which used an MOI of 100 (24; data not shown).
FIG 3 .
Identification of shRNAs that result in defective YopB/D function. (A) YopB/D function correlates with membrane permeability. 293T cells were challenged with Y. pseudotuberculosis YPIII ΔyopB, YPIII ΔyopHMOJE, or YPIII yopE+ ΔyopHMOJ at an MOI of 25 for 2 h at 37°C. The cells were then stained using the FITC LIVE/DEAD cell viability kit (Invitrogen) for 30 min at room temperature (RT) and analyzed by flow cytometry. Representative histograms depicting relative FITC fluorescence after bacterial challenge are shown. (B) Depletion of host cell factors inhibit YopB/D function. 293T cells were transfected with individual shRNA plasmids coexpressing mCherry. After 72 h of knockdown, cells were incubated with YPIII ΔyopHMOJE for 2 h at an MOI of 25 followed by staining and flow cytometry analysis as described above for panel A. Representative histograms are overlays of mCherry+ cells from each labeled shRNA (gray) compared to vector control (white).
To analyze the effect on YopB/D pore formation of shRNAs identified in the screen, oligonucleotides were chosen based upon the shRNA sequence information obtained in the screen and introduced into an mCherry-expressing plasmid prior to transfection into 293T cells. After allowing 72-h knockdown, cells were challenged with Y. pseudotuberculosis ΔyopHMOJE to assess YopB/D pore formation. An shRNA designed against RhoA, which is required for pore formation, served as a control for reduced pore formation. As expected, cells expressing an shRNA for RhoA showed a decrease in fluorescence intensity by FACS, indicating that a large population of cells resist pore formation by YopB/D compared to cells expressing the empty shRNA vector control (Fig. 3B).
YopB/D pore formation was next assayed for selected shRNAs predicted to have a variety of phenotypes based on their ZMAD values obtained from the screen. The 35 shRNAs were chosen as follows: 23 shRNAs that were enriched in both the FRET+/FRET− (ZMAD > 2) and FRET+/unsorted populations (ZMAD > 2), 7 shRNAs that showed enrichment only in the FRET+/unsorted population, and 5 that were predicted to have no phenotype (ZMAD < 2 for both ratios) (Table 1). An shRNA directed against the RhoA downstream effector Rock2 interfered with YopB/D pore formation to a level similar to that of cells expressing the shRNA directed against RhoA (Fig. 3B; Table 1). Expression of candidate shRNAs directed toward either a member of the vesicle-tethering synaptotagmin family (SYT16) or the cell adhesion protein protocadherin 2 (PCDHA2) reduced YopB/D pore formation to levels indistinguishable from that of the RhoA control. Also, an shRNA directed against the calmodulin spectrin associated protein (CAMSAP) important for microtubule stablization, which was enriched only in the FRET+ sort compared to the unsorted pool (Table 1), also showed strong reduction. An shRNA directed toward a calmodulin-like signaling protein (CALML5), which was a clear candidate for lowering T3SS function (Table 1), had no observed defect in pore formation (Fig. 3B), indicating that it may be contributing to events downstream of pore formation. Overall, YopB/D pore formation was strongly reduced in cells expressing shRNAs for 22 of the 30 candidates predicted to reduce YopE activity, including 6 of the 7 shRNAs tested that showed enrichment only in the FRET+/unsorted ratio from the original screen (Table 1). As expected, expression of the majority of the shRNAs predicted to have no phenotype had no effect on YopB/D pore formation, with just 1 of the 5 shRNAs exhibiting a detectable defect (Table 1).
TABLE 1 .
Summary of pore formation and YopH-TEM translocation data for all shRNAs tested in this study
| Category and gene symbol | Pore formationa |
Stat. sign.b | YopH-TEM translocationa |
Stat. sign. | ||
|---|---|---|---|---|---|---|
| Avg (% vector control) | SEM | Avg (% vector control) | SEM | |||
| ZMAD > 2: FRET+/unsorted, FRET+/FRET− | ||||||
| CALML5 | 114.35 | 7.97 | NS | ND | ||
| CANX | 41.98 | 2.58 | **** | ND | ||
| CCR5 1 | 52.52 | 3.32 | **** | 3.73 | 9.77 | *** |
| CCR5 2 | 80.42 | 4.66 | NS | 41.37 | 13.04 | NS |
| CCR8 | 74.71 | 3.00 | ** | 81.04 | 15.64 | NS |
| CD59 | 56.52 | 3.16 | **** | 18.31 | 13.27 | ** |
| CLTC | 74.44 | 5.06 | * | ND | ||
| COPB1 | 47.38 | 3.66 | ** | ND | ||
| DNAJB6 | 56.17 | 2.42 | **** | 14.65 | 7.97 | ** |
| DNAJC13 | 76.73 | 14.12 | NS | ND | ||
| DNAJC27 | 44.73 | 1.92 | **** | 15.66 | 11.97 | ** |
| DYNC1LI1 | 204.50 | 13.01 | 119.67 | 13.31 | NS | |
| HSP90B1 | 49.85 | 1.93 | *** | ND | ||
| MYO6 | 43.65 | 2.15 | *** | 97.39 | 12.76 | NS |
| PCDHA2 | 31.39 | 2.05 | **** | 62.06 | 4.46 | ** |
| PRKCA | 55.03 | 4.00 | **** | 17.15 | 8.32 | ** |
| RAB27A | 106.91 | 3.84 | NS | 74.62 | 12.49 | NS |
| SEC24B | 81.88 | 4.95 | NS | 38.05 | 16.25 | * |
| SEC63 | 58.11 | 5.44 | ** | ND | ||
| SPTBN5 | 50.43 | 2.93 | **** | 24.89 | 16.79 | ** |
| SUMO3 | 96.86 | 7.82 | NS | 67.33 | 13.87 | NS |
| SYT16 | 38.24 | 4.03 | **** | 58.36 | 3.88 | ** |
| TRAPPC10 | 54.55 | 2.95 | **** | 81.68 | 16.63 | NS |
| ZMAD > 2: FRET/+unsorted | ||||||
| ARFIP2 | 41.19 | 6.69 | *** | 90.86 | 10.20 | NS |
| CAMSAP1 | 49.39 | 3.41 | *** | ND | ||
| DNAJC6 | 62.47 | 12.38 | * | 79.81 | 8.98 | NS |
| HIP1 | 41.87 | 4.35 | **** | ND | ||
| Rock2 | 22.85 | 2.24 | **** | 32.88 | 10.47 | * |
| TTPAL | 85.45 | 18.72 | NS | 80.35 | 4.96 | NS |
| UBQLN3 | 45.55 | 6.58 | *** | 67.91 | 14.74 | * |
| ZMAD < 2: FRET/+unsorted, FRET/+FRET− | ||||||
| APOD | 85.38 | 10.42 | NS | 98.57 | 9.04 | NS |
| COPS8 | 113.80 | 9.58 | NS | 124.32 | 7.88 | NS |
| CYP39A1 | 62.60 | 9.99 | * | 81.44 | 2.39 | NS |
| PRKCA | 127.24 | 18.13 | NS | 95.51 | 3.85 | NS |
| SPTAN1 | 69.24 | 11.74 | NS | 75.92 | 4.11 | NS |
| ARF1 | 38.63 | 5.49 | **** | ND | ||
| ARF6 | 35.14 | 2.77 | **** | ND | ||
| HSPA8 | 49.19 | 6.60 | *** | ND | ||
| RHOA | 25.55 | 2.85 | **** | ND | ||
Each shRNA was tested in each assay in triplicate, with experiments repeated at least twice. Data are given as means and standard errors of the means (SEM) of the vector control, pooling all the individual assays for each shRNA.
Statistical significance (Stat. sign.) was determined using an ANOVA (P < 0.0001) and a Dunnett’s posthoc test and are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant. Some means were not determined (ND).
Roles of membrane trafficking pathways in YopB/D pore formation.
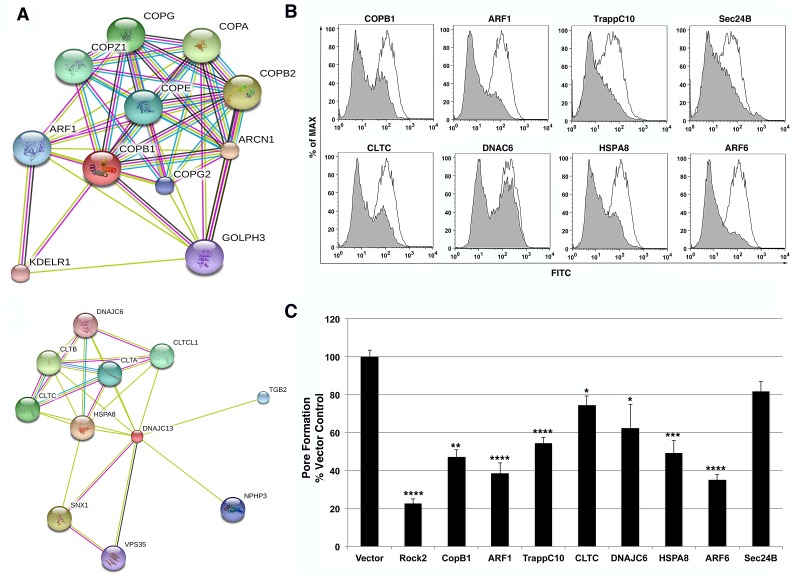
Intracellular membrane trafficking proteins, encompassing the intracellular protein transport, protein transport, and transport categories, were overrepresented among the candidate shRNAs identified in the screen, based on the Panther database (Fig. 2C) (23). Members of membrane trafficking pathways identified in the screen included proteins involved in formation of three major vesicle coat biogenesis pathways, COPI, COPII, and clathrin. COPI vesicles are responsible for retrograde transport through the Golgi apparatus to the endoplasmic reticulum (ER) and contain a distinct coatamer comprised of 7 protein subunits, including the βCop subunit (Copβ1) that was identified in the screen (25). Previous studies of depletion of the Copβ1 protein revealed that its expression is important for proper formation of the Golgi apparatus (26) with the consequence that its loss results in severely defective processing, sorting, and localization of proteins destined for membranous compartments. Cells expressing an shRNA that depletes Copβ1 were defective for pore formation when challenged with Y. pseudotuberculosis ΔHMOJE (Fig. 4B and C). The Copβ1 protein network in the String database included the 6 protein subunits that make up the COPI coat as well as the small GTPase ADP-ribosylation factor 1 (ARF1) (Fig. 4A) (27), which recruits COPI to vesicles budding from the Golgi apparatus (25). To gain further support that this pathway plays a role in YopB/D pore formation, an shRNA was designed toward ARF1, which was not present in the original shRNA library used in the screen. 293T cells knocked down with Arf1 shRNA also demonstrated reduced YopB/D pore formation compared to the shRNA vector control (Fig. 4B and C). Trafficking protein particle C10 (TrappC10), a component of the TRAPPII complex that tethers COPI-coated vesicles, was also a hit in the screen, and cells depleted of TrappC10 similarly exhibited reduced YopB/D-mediated pore formation (Fig. 4B and C). Therefore, depletion of a variety of proteins in the COPI pathway reduced YopB/D-mediated cytotoxicity, consistent with disruption of this pathway causing mislocalization of host factors important for T3SS function.
FIG 4 .
Intracellular protein trafficking pathways that support the function of the YopB/D translocon. (A) String analysis of COPB1 and CLTC allows identification of functional partners in each respective pathway. (B and C) Intracellular protein trafficking shRNAs were assessed in YopB/D functional assay. 293T cells were transfected with individual shRNA plasmids coexpressing mCherry. After 72 h of knockdown, cells were incubated with Y. pseudotuberculosis YPIII ΔyopHMOJE for 2 h at an MOI of 25. All cells were then stained using a FITC LIVE/DEAD cell viability kit (Invitrogen) for 30 min at RT and analyzed by flow cytometry. (B) Representative histograms depicting FITC fluorescence of mCherry+ cells are displayed as overlays for each labeled shRNA (gray) compared to vector control (white). (C) YopB/D pore formation was quantitated as the MFI of FITC for mCherry+ cells normalized to the vector control. Each shRNA was evaluated in this assay in triplicate in a minimum of three independent experiments. Data represented as means plus standard errors of the means (SEM) (error bars) for all experiments. Statistical significance was determined using an ANOVA (P < 0.0001) with Dunnett’s posthoc test. Values that are significantly different from the value for the vector control are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Clathrin-coated vesicles are the most predominant cargo vesicles within the cell and can be found budding from the plasma membrane during endocytosis or from the trans-Golgi network (TGN) during endocytic recycling. The protein coat of these vesicles is comprised of three clathrin heavy-chain and light-chain subunits (25). String analysis of the clathrin network identified the host cell chaperones auxilin (DNAJC6) and HSC70 (HSPA8) which are responsible for uncoating clathrin vesicles (Fig. 4A) (25, 27). In addition to its role in the COPI vesicle biogenesis, ARF1 also assists in clathrin vesicle formation at the TGN, while ARF6 promotes clathrin vesicle formation at the plasma membrane (28). Expression of shRNAs directed toward clathrin heavy chain (CLTC) and auxilin (DNAJC6), both identified in the screen, interfered with YopB/D-mediated pore formation (Fig. 4B and C). In addition, knockdown of the HSC70 (HSPA8) and ARF6 genes also produced defects in pore formation after bacterial challenge, further supporting the hypothesis that clathrin-dependent membrane trafficking pathways are involved in promoting YopB/D cytotoxicity (Fig. 4B and C). COPII vesicles, which include components Sec23 and Sec24 identified in the screen, promote anterograde transport of cargo from the ER to the Golgi apparatus (25). Although it is clear by FACS analysis that there is a decrease in fluorescein isothiocyanate (FITC) intensity by an shRNA targeting Sec24B (Fig. 4B), the defect in YopB/D-mediated pore formation in the presence of this shRNA did not show significance based on the statistical test we used (Fig. 4C; Materials and Methods). These data indicate that intracellular trafficking mediated by multiple vesicle coat complexes is required for the initial steps of YopB/D involved in T3SS-mediated cytotoxicity.
CCR5 expression stimulates YopB/D-mediated pore formation.
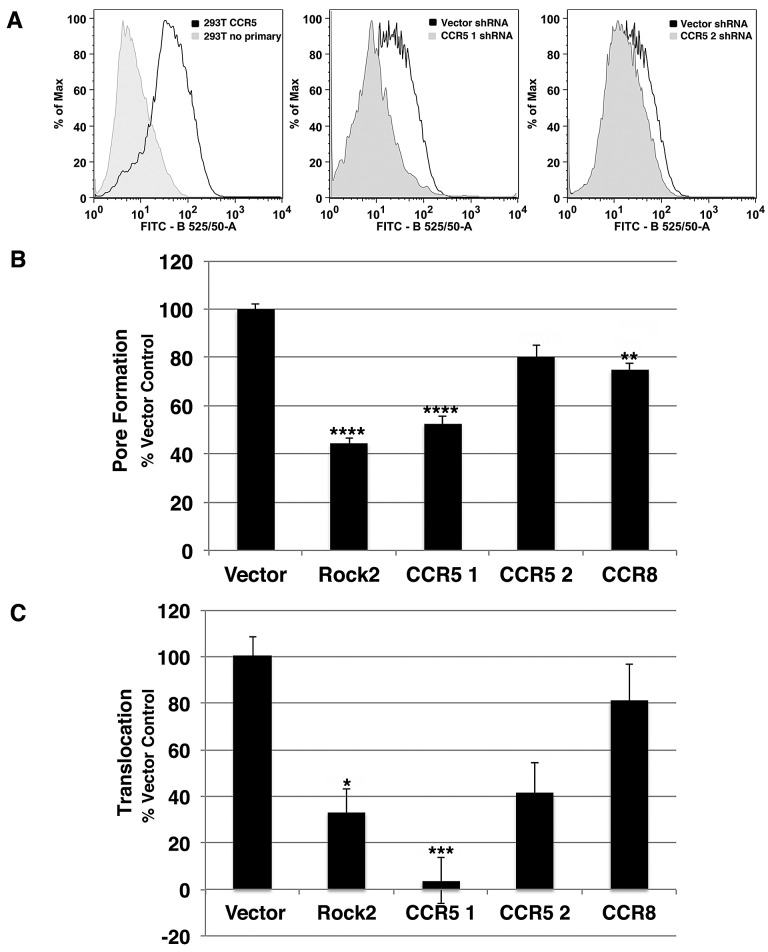
Due to the fact that depletion of proteins involved in intracellular membrane trafficking resulted in defective YopB/D pore formation, we hypothesized that improper membrane localization of an important host cell factor necessary for T3SS function could cause this defect. Furthermore, this protein has been hypothesized to play a role in Yersinia pestis pathogenesis, although there is little evidence to support this model (29, 30). The chemokine receptor CCR5 was investigated as a likely candidate for further analysis because two independent shRNAs were identified in the screen that target this protein (Fig. 5A, CCR5-1 and CCR5-2 shRNAs). 293T cells endogenously express low levels of CCR5 on their surface (Fig. 5A). Cells expressing either CCR5-1 or CCR5-2 shRNA, identified by mCherry coexpression, had reduced surface expression of CCR5 compared to cells expressing the shRNA vector control, confirming that the shRNA knockdown reduces CCR5 surface expression (Fig. 5A). The level of YopB/D pore formation in these cells was proportional to the magnitude of CCR5 expression when cells were challenged with Y. pseudotuberculosis ΔyopHMOJE (compare CCR5-1 to CCR5-2; Fig. 5A and B). Another chemokine receptor CCR8, which has 40% identity to CCR5, was also identified in the screen. Cells depleted of CCR8 with an shRNA directed against CCR8 showed reduced YopB/D activity compared to the control; however, the magnitude of defect was less severe than observed with CCR5 depletion (Fig. 5B).
FIG 5 .
CCR5 supports YopB/D function and YopH-TEM translocation into target cells. 293T cells were transfected with CCR5-1, CCR5-2, CCR8, and Rock2S shRNAs and the shRNA vector control coexpressing mCherry. All assays were performed 72 h after transfection. (A) Decreased surface expression of CCR5 in cells expressing CCR5-1 and CCR5-2 shRNAs. CCR5 surface expression of untransfected cells, cells transfected with CCR5-1 or CCR5-2 shRNA or shRNA vector control was measured by flow cytometry. (B) YopB/D function is suppressed upon CCR5 depletion. shRNA-transfected cells were incubated with Y. pseudotuberculosis YPIII ΔyopHMOJE for 2 h at an MOI of 25, staining using a FITC LIVE/DEAD cell viability kit (Invitrogen) for 30 min at RT, and analyzed by flow cytometry. YopB/D function was measured as the MFI of FITC for mCherry+ cells normalized to the vector control. (C) Depletion of CCR5 impairs YopH-TEM translocation. To measure translocation, cells were challenged with Y. pseudotuberculosis YPIII YopH-TEM for 1 h at an MOI of 1 and then incubated with CCF2-AM for 30 min at RT. Translocation was measured by flow cytometry as the ratio of the MFI at 450-nm emission/MFI at 530-nm emission after 405-nm excitation of mCherry+ cells, normalized to the vector control. Each experiment was performed in triplicate and repeated a minimum of three times. Data are means ± SEM for all experiments. Statistical significance was determined using an ANOVA (P < 0.0001) with Dunnett’s posthoc test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Depletion of CCR5 results in defective Yop translocation.
To ensure that defective YopB/D activity resulted in decreased Yop effector translocation, YopH translocation was assayed based on β-lactamase cleavage of the fluorescent dye CCF2 upon incubation of cells with Y. pseudotuberculosis YopH-TEM (31). After challenge of shRNA-depleted 293T cells at an MOI of 1 for 1 h, YopH-TEM translocation was quantitated by FACS analysis, determining the mean fluorescence intensity (MFI) of cleaved product (Materials and Methods). Consistent with the data on YopB/D activity (Fig. 4), cells expressing an shRNA against Rock2 exhibited an approximately 70% reduction in YopH-TEM translocation compared to cells expressing the shRNA vector control (Fig. 5C). Expression of either CCR5-1 or CCR5-2 shRNA also resulted in defective translocation, with the reduction of translocation proportional to their relative knockdown efficiencies (Fig. 5C). Cells transfected with an shRNA toward CCR8 exhibited a minor defect in YopH-TEM translocation, consistent with the small defect observed in YopB/D activity. These results are consistent with our analysis of the majority of the shRNAs tested in this assay, in which less YopB/D pore formation corresponded to a decrease in YopH-TEM translocation (Table 1).
Depletion of endogenous CCR5 can be reversed by reintroduction of the gene.
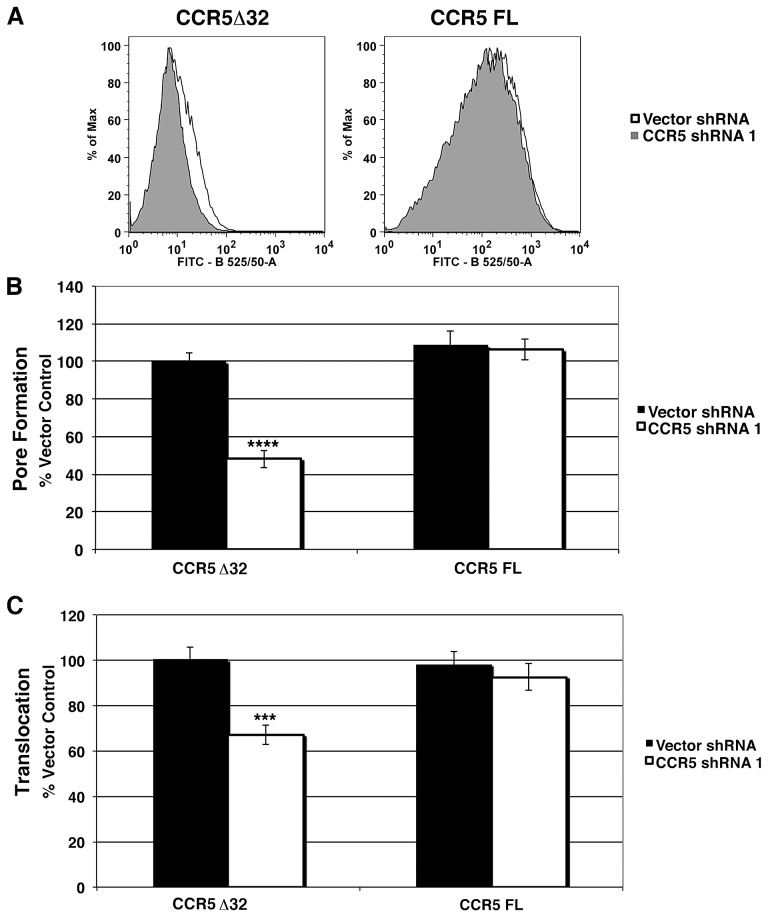
In order to ensure that the defects observed for CCR5 knockdown were not due to off-target effects, stable CCR5 transfectants, which overexpress the gene, were created that were resistant to shRNA knockdown (Fig. 6A) (Materials and Methods). 293T cells were found to be heterozygous for ccr5, having both the wild-type copy and a mutation previously characterized in the human population, a 32-bp frameshift deletion (CCR5Δ32) that prevents surface exposure (data not shown). Stable cell lines that express either full-length CCR5 or CCR5Δ32 were cotransfected with CCR5-1 shRNA or the shRNA vector control and challenged with Y. pseudotuberculosis ΔyopHMOJE in order to measure YopB/D activity. Cells expressing CCR5Δ32 were defective in YopB/D activity when transfected with the CCR5-1 shRNA compared to the shRNA vector control (Fig. 6A and B). In contrast, overexpression of full-length CCR5 was able to fully restore YopB/D activity in the presence of CCR5-1 shRNA (Fig. 6B). These results argue that expression of CCR5 rescues the defect in Yop B/D activity observed due to depletion of CCR5.
FIG 6 .
CCR5 overexpression complements the CCR5 knockdown defect in YopB/D function and YopE translocation. Stable cell lines overexpressing CCR5 or CCR5Δ32 were transfected with CCR5-1 shRNA or the shRNA vector control. All assays were performed 72 h after transfection. (A) CCR5 surface expression of cell lines expressing full-length (FL) CCR5 and CCR5Δ32 after transfection with CCR5-1 shRNA (gray) compared to the shRNA vector control (white) was measured by flow cytometry. (B) Defective YopB/D function is complemented by transfection of shRNA-insensitive CCR5. shRNA-transfected cells were incubated with Y. pseudotuberculosis YPIII ΔyopHMOJE for 2 h at an MOI of 25 followed by staining using a FITC LIVE/DEAD cell viability kit (Invitrogen) for 30 min at RT and flow cytometry. YopB/D function was measured as the MFI of FITC normalized to the CCR5Δ32 vector control. (C) Defective YopE translocation is complemented by transfection of shRNA-insensitive CCR5. To measure translocation, cells were challenged with YPIII YopE-TEM for 1 h at an MOI of 1 and incubated with CCF2-AM for 30 min at RT. Translocation was measured by flow cytometry as the ratio of the MFI at 450-nm emission/MFI at 530-nm emission after 405-nm excitation normalized to the CCR5 Δ32 vector control. Each experiment was performed in triplicate and repeated a minimum of three times. Data are means ± SEM for all experiments. Statistical significance was determined using an unpaired two-tailed t test assuming unequal variances (***, P < 0.001; ****, P < 0.0001).
CCR5 was also required for high-efficiency T3SS translocation. Stable cell lines expressing either full-length CCR5 or CCR5Δ32 were subjected to depletion with CCR5-1 shRNA or vector control followed by incubation with Y. pseudotuberculosis YopE-TEM, allowing translocation to be assayed by β-lactamase activity. CCR5-1 depletion of 293T cells that expressed CCR5Δ32 had reduced YopE-TEM translocation compared to the vector shRNA control (Fig. 6C). In contrast, overexpression of an intact ccr5 gene insensitive to the CCR5-1 shRNA fully restored YopE-TEM translocation to levels comparable to that of cells transfected with the shRNA vector control (Fig. 6C). Therefore, the observed defects due to CCR5 shRNA result from depletion of CCR5 and are not due to off-target effects.
Defective YopE-TEM translocation in cells from Ccr5−/− mice.
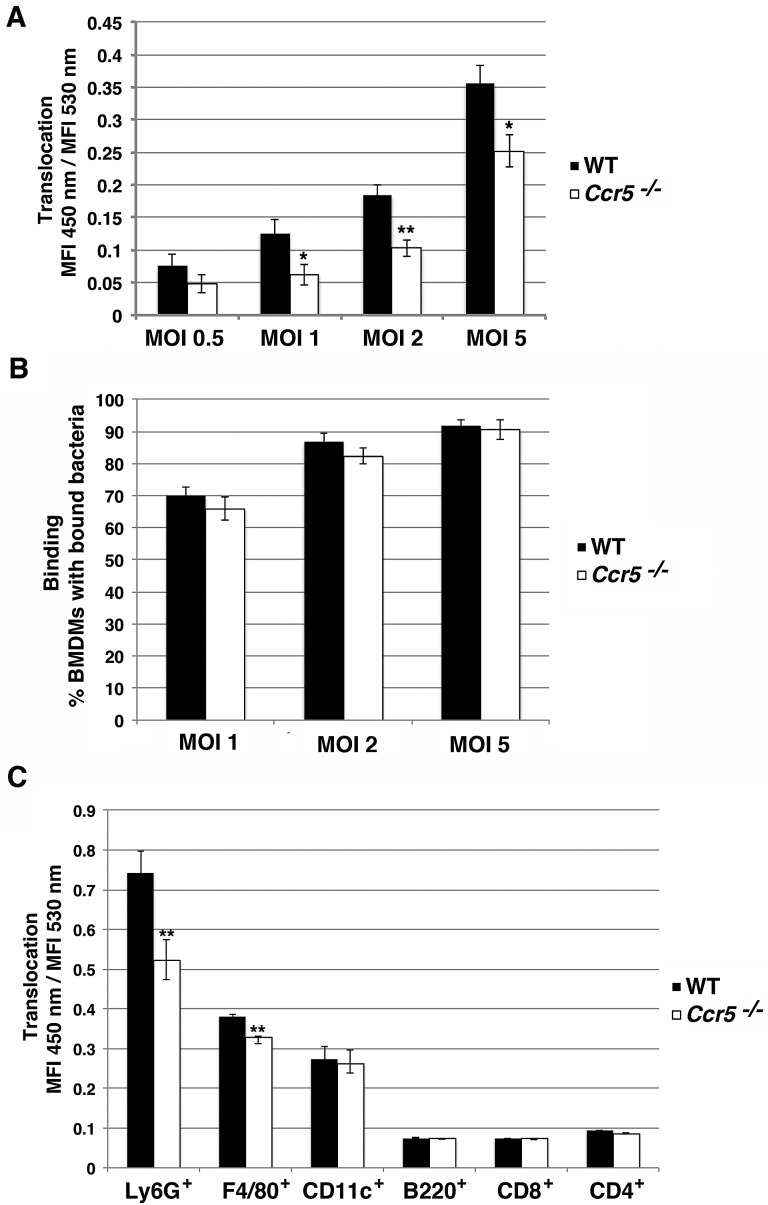
To further confirm that CCR5 contributes to T3SS function, bone marrow-derived macrophages (BMDMs) were isolated from both Ccr5−/− and parental C57BL/6 mice (32). Differentiated macrophages were seeded and challenged with Y. pseudotuberculosis YopE-TEM for 1 h at an MOI of 0.5, 1, 2, or 5, and translocation was assayed by FACS analysis of β-lactamase activity. Wild-type (WT) macrophages displayed a dose-dependent increase in YopE-TEM translocation corresponding to an increase in MOI (Fig. 7A). Ccr5−/− macrophages exhibited a significant defect in YopE-TEM translocation compared to wild-type macrophages (Fig. 7A). This observed defect was greater at lower MOIs of 1 and 2, but increasing the MOI could not completely rescue the defect (Fig. 7A). Additionally, analysis of binding of individual bacteria to BMDMs revealed no difference in Y. pseudotuberculosis gfp+ binding to WT and Ccr5−/− macrophages (Fig. 7B), indicating that the observed reduction in YopE-TEM translocation was not due to a decrease in bacterial binding to Ccr5−/− macrophages.
FIG 7 .
Primary immune cells from Ccr5−/− mice have defect in YopE-TEM translocation. (A) YopE-TEM translocation is attenuated in Ccr5−/− macrophages. Primary BMDMs from wild-type and Ccr5−/− C57BL/6 mice were challenged with Y. pseudotuberculosis YPIII YopE-TEM strain at the indicated MOI for 1 h at 37°C. The cells were then loaded with CCF2-AM for 30 min at RT, lifted, and analyzed on a BD LSRII flow cytometer. Translocation was measured at the MFI 450-nm emission/MFI 530-nm emission after 405-nm excitation. (B) Bacteria bind efficiently to both WT and Ccr5−/− macrophages. BMDMs were incubated with Y. pseudotuberculosis YPIII gfp+ at the indicated MOI for 15 min at 37°C, washed, fixed, and stained with Hoechst. Binding was determined by counting macrophages with GFP+ bound bacteria using fluorescence microscopy. (C) Translocation of YopE-TEM is attenuated in splenic neutrophils. Splenocytes harvested from wild-type and Ccr5−/− C57BL/6 mice were incubated with YPIII strain expressing YopE-TEM at an MOI of 0.5 for 45 min at 37°C. The cells were then treated with gentamicin, loaded with a CCF2-AM solution for 30 min at RT, followed by surface staining to the indicated surface markers and analysis by flow cytometry. Translocation was measured for individual cell types by measuring the MFI of 450-nm emission/MFI of 530-nm emission after 405-nm excitation within each gated population. Each experiment was performed in triplicate and repeated a minimum of three times. Data are means ± SEM for all experiments. Statistical significance was determined using an unpaired two tailed t test assuming unequal variances (*, P < 0.05; **, P < 0.01).
Previously, it has been shown that Yersinia preferentially targets and translocates effectors into professional phagocytes, including neutrophils, macrophages, and dendritic cells within splenic cell suspensions (33, 34). In order to investigate whether CCR5 expression was important for translocation in these cell types, spleens were harvested from uninfected wild-type and Ccr5−/− mice and homogenized to generate a single-cell suspension. Splenocytes were then challenged with Y. pseudotuberculosis YopE-TEM at an MOI of 0.5 for 45 min followed by incubation with CCF2-AM in the presence of antibiotics to halt translocation. The cells were then fluorescently labeled with antibodies in order to identify specific cell types: neutrophils (Ly6G+), macrophages (F4/80+), dendritic cells (CD11c+), B cells (B220+), T helper cells (CD4+), and cytotoxic T cells (CD8+). This procedure was followed by flow cytometry analysis to measure translocation into different cell types.
Splenocytes harvested from wild-type C57BL/6 mice exhibited the same preferential translocation into neutrophils, macrophages, and dendritic cells as previously reported (Fig. 7C) (34). In fact, neutrophils, which represent approximately 1% of the population in the spleen had the highest levels of YopE-TEM translocation (Fig. 7C). In Ccr5−/− splenocytes, there was a significant decrease in YopE-TEM translocation in the neutrophil population resulting in an approximately 30% reduction in translocation compared to wild-type splenocytes (Fig. 7C). A slight reduction in YopE-TEM translocation was also observed in macrophages in Ccr5−/− splenocytes, whereas there was no difference in translocation into dendritic cells, B cells, and T cells compared to wild-type splenocytes (Fig. 7C). Both wild-type and Ccr5−/− splenocytes also had similar percentages of each cell type measured within the total splenic population, suggesting that the observed defect in YopE-TEM translocation in neutrophils is specific for the loss of CCR5 and not because there are fewer neutrophils in the population.
DISCUSSION
In this study, a genetic screen was performed that identified host cell factors required for T3SS-mediated intoxication of target cells. This strategy used flow sorting to positively select shRNA-depleted cells that were insensitive to YopE inactivation of Rho biosensor based on retention of a positive FRET signal. Overall, 385 candidate genes were identified among 5,447 open reading frames (ORFs) after screening 10,146 shRNAs. Using inhibition of YopE activity as an endpoint in the screen allowed the identification of host factors that may contribute to several aspects of T3SS function, including insertion of the YopB/D translocon, as well as translocation, localization, and functional activation of YopE.
Reconstruction of several shRNAs revealed that the majority of the candidates caused defects in T3SS YopB/D pore formation. The relative abundance of such shRNAs may be a consequence of the strong defects predicted for YopB/D dysfunction, which should severely depress the concentration of YopE within host cells. The screen was not limited to identifying this step; it also identified shRNAs that showed no apparent defect in YopB/D function, as shRNAs targeting DYNC1LI1, Rab27A, Sumo3, and CALML5 revealed little loss of YopB/D activity. Consistent with the screen identifying defects in either step in the pathway, shRNAs directed toward Rab27A and Sumo3 were defective for YopH-TEM translocation, indicating these proteins work downstream of T3SS translocon insertion (Table 1).
Pathway analysis revealed that shRNAs depleting proteins involved in vesicle trafficking were significantly overrepresented among the candidates from the screen. In particular, depletion of proteins involved in controlling the clathrin, COPI, and COPII vesicle coats interfered with the function of the YopB/D translocon. Previous studies have shown that clathrin is required for actin pedestal formation induced by T3SS translocation of the EPEC effector Tir. Not determined, however, was whether depletion of clathrin resulted in a defect in the translocation of Tir (35, 36). Copβ1, on the other hand, was previously identified as important for T3SS-1 function during Salmonella enterica serovar Typhimurium uptake into cultured cells (37). The authors of that study showed that depletion of Copβ1 results in defective cholesterol and sphingolipid trafficking as well as an inability of Rac1 and Cdc42 GTPases to localize in the plasma membrane in response to bacterial contact (37). Our results indicate that inhibition of COPI vesicle trafficking interferes with pore formation and that this coat complex is a critical player in the earliest steps of T3SS-dependent translocon function. Therefore, the clathrin complex may also be a common host factor that supports the function of all T3SS translocons.
Two independent shRNAs identified in the screen targeted the gene for chemokine receptor CCR5. CCR5 depletion inhibited YopB/D activity as well as translocation of the YopH-TEM and YopE-TEM fusions (Fig. 5 and 6). Both translocation and pore formation defects were complemented by expression of CCR5 insensitive to knocking down by the shRNA. BMDMs derived from Ccr5−/− mice also exhibited reduced translocation (Fig. 7A). Overall, these data indicate that chemokine receptor function contributes to efficient T3SS-dependent translocon function.
The question of how CCR5 supports T3SS function in Y. pseudotuberculosis remains. One possibility is that CCR5 could be a receptor for the YopB/D translocon prior to insertion and pore formation into the plasma membrane. CCR5 was recently identified to be a receptor for the LukED pore-forming toxin from Staphylococcus aureus, whose primary target is neutrophils (38, 39). These cells are similarly targeted by Y. pseudotuberculosis during growth in deep tissue sites (34), and neutrophils appeared to be the cells in which loss of CCR5 resulted in the most profound defects in translocation (Fig. 7C). YopE-TEM translocation was only partially inhibited in Ccr5−/− primary cells, so it is possible that other chemokine receptors in addition to CCR5 may contribute to T3SS-mediated cytotoxicity. In fact, LukED can also target other chemokine receptors to intoxicate cells, CXCR1 (chemokine [C-X-C motif] receptor 1) and CXCR2 (40). Consistent with this model, the chemokine receptor CCR8 was also identified in the screen as depressing YopB/D pore formation. An alternative possibility is that CCR5 could play a role in activating the RhoA signaling pathway (16). Although it is known that engagement of integrin receptors results in activation of Rho family members, efficient assembly of the translocon channel could require supportive and simultaneous signaling from multiple cell surface molecules, including chemokine receptors.
CCR5 is best characterized as one of the coreceptors for HIV. Approximately 10% of northern Europeans are resistant to HIV due to a 32-base-pair deletion within CCR5 (CCR5Δ32), truncating the protein and preventing its localization at the cell surface. Although it is controversial, it has been widely postulated that the CCR5Δ32 mutation assumed prevalence in the northern European population due to a genetic selection event that allowed resistance to infectious diseases, such as bubonic plague or smallpox (41). Previous in vitro experiments revealed reduced uptake of Y. pestis and Y. pseudotuberculosis in Ccr5−/− macrophages, which suggests that the CCR5 may play a role in Yersinia pathogenesis (29). Conversely, in vivo experiments exhibited no observable difference in survival of wild-type and Ccr5−/− mice after challenge with Y. pestis, as well as similar colonization levels in Peyer’s patches and cecum after oral inoculation with Y. pseudotuberculosis (29, 30). However, Y. pseudotuberculosis mutants totally lacking a functional T3SS have little defect for colonization of these organs. Experiments have shown that Y. pseudotuberculosis T3SS mutants have the strongest colonization defect in the spleen (42). When we analyzed isolated splenocytes, we found that CCR5 was primarily involved in supporting translocation into neutrophils, the predominant cell type that Y. pseudotuberculosis targets within this organ site (Fig. 7C) (34). Therefore, the defective translocation exhibited by loss of this receptor could be organ or cell type specific.
Overall, this study describes a genetic screen that was able to identify a diverse set of host cell factors that are important for T3SS-mediated cytotoxicity. Included among the identified proteins are host factors previously known to contribute to T3SS function in other organisms, as well as new biochemical pathways not known to support the activity of this system. Future studies should elucidate the exact mechanism of these host factors in the Yersinia T3SS which can also be extrapolated to T3SS in a multitude of other bacterial pathogens. Additionally, many other bacterial pathogens commonly target the RhoA GTPase by a variety of different mechanisms during infection, thus broadening the potential applications of this screen to study other host-pathogen interactions in the future as well.
MATERIALS AND METHODS
Cell lines and bacterial strains.
293T cells obtained from ATCC were cultured at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen). The wild-type RhoA FLARE biosensor (RBS WT) and the GTP binding-defective mutant RhoA biosensor N19 (RBS N19) plasmids were obtained from Klaus Hahn (20). The RBS plasmid encodes a RhoA binding domain from rhotekin fused to cyan fluorescent protein (CFP), a linker region, followed by yellow fluorescent protein (YFP) and RhoA. Y. pseudotuberculosis YPIII strains obtained from Joan Mecsas at Tufts Medical School were previously described: YPIII gfp+ (43); YPIII ΔyopB, YPIII ΔyopHMOJ, and YPIII ΔyopHMOJE (44); YPIII HTEM (34); and YPIII ETEM (45). All Y. pseudotuberculosis strains were grown overnight at 26°C in 2×YT broth. To induce the Y. pseudotuberculosis T3SS, cultures were grown overnight, back-diluted to an A600 of approximately 0.2 into either Ca2+-depleted conditions (2×YT supplemented with 20 mM Na oxalate and 20 mM MgCl2) or Ca2+-replete conditions (2×YT supplemented with 5 mM CaCl2) grown at 26°C for 1.5 h and then shifted to 37°C for 1.5 h (46).
Rho biosensor flow cytometry analysis.
293T cells seeded in a 12-well tissue culture plate were transfected using Lipofectamine 2000 with either the RhoA biosensor WT or N19 plasmid at a 3:1 reagent/DNA ratio in Opti-MEM (Invitrogen). After 24 h of transfection, cells expressing the RhoA biosensor WT plasmid were challenged with a Y. pseudotuberculosis strain YPIII yopE+ ΔyopHMOJ at a multiplicity of infection (MOI) of 50 at 37°C in 5% CO2 for 1 h. Monolayers were then washed with phosphate-buffered saline (PBS), harvested, and resuspended in FACS buffer (PBS supplemented with 1 mM EDTA and 1% FBS). Samples were then analyzed by FACS on the LSRII flow cytometer (BD Biosciences) using a 405-nm excitation laser and 450-nm and 530-nm filters to visualize CFP and YFP emissions, respectively.
For reconstruction experiments to determine the efficiency of enriching for a population of cells that has higher levels of FRET (YFP) emission than the rest of the population, cells having high FRET levels that were marked with an mCherry plasmid were mixed with an excess of cells having low FRET levels but lacking the mCherry marker. The RBS WT plasmid was cotransfected with the pmCherry-N1 plasmid (Clontech) at a 9:1 (Rho biosensor/pmCherry) ratio in order to mark this FRET+ population. Cells either transfected exclusively with the RBS WT plasmid followed by incubation with Y. pseudotuberculosis strain YPIII yopE+ ΔyopHMOJ at an MOI of 50 at 37°C with 5% CO2 for 1 h or with the RBS N19 plasmid were used to establish the FRET− population. The FRET+ mCherry populations was then mixed with either cells expressing RBS N19 or the infected RBS WT cells at a 1:20 ratio (mCherry+ FRET− cells/FRET− cells). The cells were then sorted on the Cytopeia Influx flow cytometer using 405-nm excitation laser to separate the FRET+ and FRET− populations. Unsorted, FRET+, and FRET− populations were then analyzed on the BD LSRII flow cytometer as described above. Analysis of flow cytometry data was performed using FlowJo software (Tree Star).
Preparation of shRNA library.
The GeneNet human 50,000 (50K) short hairpin RNA (shRNA) library was obtained from System Biosciences (Mountain View, CA), which contains 200,000 shRNAs with coverage of 3 to 5 per annotated open reading frame (ORF) (21). The library was divided into 10 smaller pools by transforming into E. coli DH5α, plating out approximately 2 × 104 colonies on LB-carbenicillin plates, which were then scraped and collected in L broth. DNA was then extracted (Qiagen midiprep) from pelleted bacteria for each pool. The pools were packaged into virus by transfecting 3 × 106 293T cells with pooled DNA (4 µg) along with the packaging plasmids pFIV-34N (1.8 µg) and pVSV-G (VSV stands for vesicular stomatitis virus) (0.2 µg) (System Biosciences) using 18 µl Fugene HD (Promega) and incubating in DMEM culture medium in 10-cm dishes. After 48 h of transfection, supernatants were then collected, passed through a 0.45-μm filter, and stored at −80°C.
shRNA screen to enrich for transfectants insensitive to YopE GAP activity.
293T cells were transduced with the viral packaged pools of shRNA library (representing shRNA from 2 × 104 members of the library) at an MOI of 1. In each experiment, approximately 2 × 107 293T cells were challenged with virus in the presence of 10 µg/ml of Polybrene at approximately 80% confluence. After 24 h, the medium was changed to remove virus, and cells were replated at approximately 3 × 106 cells into 10-cm dishes with DMEM supplemented with 10% FBS. On day 3, 10 µg of the RBS WT plasmid was introduced into the cells by transfection using 30 µl of Lipofectamine 2000. After 24 h of further incubation, cells were then challenged with Y. pseudotuberculosis YPIII yopE+ ΔyopHMOJ at an MOI of 50 for 1 h at 37°C with 5% CO2. The cells were then lifted using 0.05% trypsin (Invitrogen), collected, and sorted using a Cytopeia Influx flow cytometer. Approximately 3 × 106 cells were set aside prior to sorting to be used as an unsorted shRNA input control. To determine the appropriate gates to collect cells, non-shRNA-depleted 293T cells expressing the RBS WT plasmid which were either left untreated or incubated with strain YPIII yopE+ ΔyopHMOJ at an MOI of 50 for 1 h at 37°C with 5% CO2 were used to establish the FRET+ and FRET− gates, respectively.
Genomic DNA (gDNA) was extracted from unsorted cells and cells collected from the gates corresponding to the FRET+ and FRET− gates. shRNA antisense sequences were then PCR amplified in two 100-µl reaction mixtures using 1 µg gDNA. The primers used were shRNA-IS F (F stands for forward) (5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGATCACACNNNNCTTCCTGTCAGA-3′ [the Illumina sequencing primer is shown in italic type, the bar code and random nucleotides are underlined, and the 12-bp shRNA loop is shown in bold italic type]) and shRNA-IS R (R stands for reverse) (5′-ATTTATTGTATCTGTGGGAGCCTC-3′) to amplify an approximately 200-bp fragment. PCR products were purified by gel extraction, concentrated, and subjected to sequencing on an Illumina HiSeq 2500 instrument.
Sequence analysis of pooled shRNA amplicons.
Illumina sequencing reads were sorted by bar codes, trimmed down in silico to the 27-bp antisense shRNA sequence (CLC sequence viewer; CLC Bio) and aligned to the human genome hg_17 database (47). The number of reads for each individual shRNA in a data set was normalized to the total number of reads in that respective data set (unsorted, FRET+, or FRET−). Any shRNAs that showed a total number of reads that represented less than 0.001% of both the unsorted and FRET+ data sets were discarded for each experiment. FRET+/unsorted and FRET+/FRET− ratios were calculated by dividing the normalized number of reads from the respective data sets for each shRNA. For each experiment, the median and the median absolute deviation (MAD) were calculated for both the FRET+/unsorted and FRET+/FRET− ratios. MAD was calculated as Mediani (|Xi − Medianj(Xj)|), in which Medianj is the median of all the deviations and Xj is the deviation of the sample. From these values, ZMAD was calculated for each shRNA in which ZMAD = (X − median)/MAD (22). Any shRNA with a ZMAD of >2 for both the FRET+/unsorted and FRET+/FRET− ratio was classified as a potential hit in the screen and was a candidate for further analysis.
Analysis of shRNA candidates.
To reconstruct shRNA clones, oligonucleotides were designed based on the shRNA sequence information obtained from the sequenced sorted and unsorted pools (see Table S2 in the supplemental material). The pSIF-H1 (puro) shRNA vector (System Biosciences) was modified by exchanging the puromycin resistance cassette with mCherry. Oligonucleotides covering both strands of the complete shRNA hairpin were phosphorylated, annealed, and ligated into either one or both the pSIF-H1 (puro) or pSIF-H1 (mCherry) shRNA vectors. Plasmid DNA was sequenced in order to verify sequences. Candidate shRNAs were then transfected into 293T cells using Fugene HD at a 3:1 reagent/DNA ratio, and transfected cells were analyzed 72 h later.
Assay for YopB/D function.
Approximately 2.5 × 105 293T cells that had been transfected with pSIF-H1 (mCherry) shRNA constructions for approximately 48 h were seeded overnight in a 24-well plate. The cells were challenged with Yersinia pseudotuberculosis strains YPIII ΔyopB, YPIII yopE+ ΔyopHMOJ, or YPIII ΔyopHMOJE at an MOI of 25 for 2 h at 37°C. The cells were then washed with PBS, stained using the fluorescein isothiocyanate (FITC) LIVE/DEAD cell viability kit (Invitrogen) for 30 min at room temperature in the dark, washed with PBS, and suspended in FACS buffer. The cells were then analyzed cytometrically (LSRII) using a 488-nm excitation laser and a 525-nm emission filter to measure FITC and a 532-nm excitation laser with a 610-nm emission filter for mCherry expression. A minimum of 2 × 104 events was analyzed using FlowJo software (Tree Star). YopB/D function for each shRNA was measured as mean fluorescence intensity (MFI) of FITC fluorescence as a percentage of the vector control. Statistical significance was measured using the appropriate test, either analysis of variance (ANOVA) followed by a Dunnett’s posthoc test (Prism) or unpaired two-tailed t test assuming unequal variances as denoted in the figure legends.
Assay for translocation of β-lactamase fusions.
Approximately 2.5 × 105 293T cells were seeded overnight in a 24-well plate. The cells were challenged with Yersinia YPIII YopH-TEM or YPIII YopE-TEM at an MOI of 1 for 1 h at 37°C with 5% CO2. Incubations were then continued for 30 min in the dark in the same medium containing 1 µg/ml of CCF2-AM and 1.5 mM probenecid (Invitrogen). The cells were then analyzed by FACS on the LSRII flow cytometer, collecting a minimum of 2 × 104 events per sample. FACS data analysis was performed using FlowJo (Tree Star), and translocation was measured as the ratio of MFI at 450-nm emission/MFI at 530-nm emission after 405-nm excitation after background subtraction from CCF2 uninfected control cells (31, 33, 34, 48, 49). Statistical significance was measured using the appropriate test, either ANOVA followed by a Dunnett’s posthoc test (Prism) or unpaired two-tailed t test assuming unequal variances as denoted in the figure legends.
Assay for bacterial binding.
Approximately 2.5 × 105 bone marrow-derived macrophages (BMDMs) were seeded overnight on coverslips in a 24-well plate. The cells were challenged with YPIII gfp+ strain at an MOI of 1, 2, or 5 for 15 min at 37°C with 5% CO2. The cells were then washed three times with PBS, fixed with 4% paraformaldehyde and stained with Hoechst (Sigma) at 1 µg/ml for 30 min at room temperature (RT), followed by placing ProLong Gold (Invitrogen) over the coverslips. Bacterial binding was then quantified using fluorescence microscopy by counting the macrophages with green fluorescent protein-positive (GFP+) bacteria bound. In each biological experiment, all experimental conditions were performed in triplicate counting a minimum of 100 cells per coverslip.
Construction of CCR5 and CCR5Δ32 stable cell lines.
The complete ORFs of CCR5 and the CCR5Δ32 mutant were amplified from the gDNA of 293T cells using the primers CCR5-BamHI F (5′-GAGGATCCGCCACCATGGATTATCAAGTGTCAAGTCC-3′) and CCR5-XbaI R (5′-GATCTAGATCACAAGCCCACAGATATTTCCTGC-3′). PCR products were digested with BamHI and XbaI and cloned into pcDNA3.1/Hygro+ (Hygro stands for hygromycin) vector (Invitrogen). Plasmid DNA was sequenced to identify full-length CCR5 and CCR5Δ32 clones. Plasmids linearized with SSP1 were transfected into 293T cells and selected with 100 µg/ml hygromycin (Invitrogen) to establish stable cell lines. Quantitation of CCR5 surface content was performed on cells fixed with 4% paraformaldehyde, probed with 1:100 dilution of human anti-CCR5 clone 2D7 (BD Biosciences), followed by detection with 1:500 dilution of FITC-labeled anti-mouse IgG. Fluorescence was quantitated by flow cytometry on a BD LSRII instrument.
Production and analysis of ex vivo-generated splenocytes and BMDMs.
C57BL/6 and C57BL/6 Ccr5−/− mice were obtained from Jackson Laboratories. Mice were euthanized using CO2 inhalation, and the spleens and femur and tibia bones were removed to isolate splenocytes or derive macrophages from the bone marrow. Bone marrow was isolated as previously described (50). Macrophages were differentiated in RPMI 1640 supplemented with FBS, L-cell-conditioned medium, and penicillin-streptomycin for 7 days prior to harvesting. Approximately 4 × 105 cells were seeded into 24-well non-tissue-culture-treated plates in RPMI 1640 with 10% FBS and L-cell-conditioned medium overnight. Macrophages were then challenged with YPIII YopE-TEM at the indicated MOI for 1 h at 37°C with 5% CO2, and translocation was measured as described above.
To isolate splenocyte populations, spleens were homogenized through a filter and treated with collagenase D (Roche) (1 mg/ml) for 30 min at 37°C, and red blood cells (RBCs) were lysed with 0.83% NH4Cl for 2 min at RT. Splenocytes were then resuspended in RPMI 1640 with 10% FBS at a density of 107 cells/ml and challenged for 45 min at an MOI of 0.5 with YPIII YopE-TEM at 37°C with 5% CO2. Splenocytes were then incubated in the dark for 30 min in medium containing 1 µg/ml of CCF2-AM, 1.5 mM probenecid (Invitrogen), and 100 µg/ml gentamicin (Sigma). Aliquots of cells (100-μl aliquots) were transferred to a 96-well plate and incubated with 50 µl of PBS supplemented with 1% FBS (PBS-1% FBS) containing a 1:200 dilution of mouse Fc Block (BD Biosciences) for 10 min at 4°C. The cells were then incubated with 50 µl of PBS-1% FBS containing fluorescent antibodies to Ly6G-PE-Cy7 (PE stands for phycoerythrin), F4/80-PE-Cy5, CD11C-APC (APC stands for allophycocyanin), B220-PE-Cy5, CD8-PE-Cy7, and CD4-PE-Cy7 (BD Biosciences) at dilutions of 1:75 (F4/80 dilution ratio of 2:75) for 30 min at 4°C. The probed cells were then washed once with PBS-1% FBS, pelleted at 1,000 rpm, resuspended in 200 µl PBS-1% FBS, and analyzed by flow cytometry on the BD LSRII instrument. A total of 2 × 105 cells were acquired per sample from three replicate biological experiments, and data were analyzed using FlowJo software (Tree Star).
SUPPLEMENTAL MATERIAL
Annotated gene list for the significant shRNA candidates identified in the screen. shRNAs having a ZMAD of >2 for both FRET+/unsorted and FRET+/FRET− in a minimum of two independent experiments are listed. The table contains gene identification (ID), reference and position of the shRNA on the chromosome, targeted sequence, calculated ZMAD values for both ratios from each experiment, UCSC Genome Browser reference IDs (47), and a description of each gene.
List of shRNA oligonucleotides used in this study
ACKNOWLEDGMENTS
We acknowledge Kip Bodi and the Tufts University Core Facility (TUCF) genomics core facility for technical advice and Illumina sequencing analysis, Stephen Kwok and Alan Parmelee from the Tufts FACS facility for assistance with sorting, Klaus Hahn for the RhoA biosensor plasmid, and Joan Mecsas for Yersinia strains and discussions. We thank K. Davis, A. Hempstead, S. Arasat, E. Geisinger, K. Kotewicz, and C. Murphy for critical review of the manuscript.
This work was supported by NIAID awards R21AI085250 and R56AI110684 to R.R.I. K.-L.S. was supported by an NRSA postdoctoral fellowship F32 AI 082914-01 and Natalie V. Zucker Fellowship. R.R.I. is an investigator of the Howard Hughes Institute.
Footnotes
Citation Sheahan K-L, Isberg RR. 2015. Identification of mammalian proteins that collaborate with type III secretion system function: involvement of a chemokine receptor in supporting translocon activity. mBio 6(1):e02023-14. doi:10.1128/mBio.02023-14.
REFERENCES
- 1.Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis GR, Van Gijsegem F. 2000. Assembly and function of type III secretory systems. Annu Rev Microbiol 54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 3.Büttner D, Bonas U. 2002. Port of entry—the type III secretion translocon. Trends Microbiol 10:186–192. doi: 10.1016/S0966-842X(02)02331-4. [DOI] [PubMed] [Google Scholar]
- 4.Gruenheid S, Finlay BB. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422:775–781. doi: 10.1038/nature01603. [DOI] [PubMed] [Google Scholar]
- 5.Schlumberger MC, Hardt WD. 2006. Salmonella type III secretion effectors: pulling the host cell’s strings. Curr Opin Microbiol 9:46–54. doi: 10.1016/j.mib.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Coburn B, Sekirov I, Finlay BB. 2007. Type III secretion systems and disease. Clin Microbiol Rev 20:535–549. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis GR. 2002. The Yersinia Ysc-Yop “type III” weaponry. Nat Rev Mol Cell Biol 3:742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 8.Mueller CA, Broz P, Müller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 9.Marenne MN, Journet L, Mota LJ, Cornelis GR. 2003. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog 35:243–258. doi: 10.1016/S0882-4010(03)00154-2. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis GR. 2002. Yersinia type III secretion: send in the effectors. J Cell Biol 158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. 2005. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol 56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- 12.Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. 2003. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. EMBO J 22:4957–4967. doi: 10.1093/emboj/cdg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Goot FG, Tran van Nhieu G, Allaoui A, Sansonetti P, Lafont F. 2004. Rafts can trigger contact-mediated secretion of bacterial effectors via a lipid-based mechanism. J Biol Chem 279:47792–47798. doi: 10.1074/jbc.M406824200. [DOI] [PubMed] [Google Scholar]
- 14.Simons K, Ehehalt R. 2002. Cholesterol, lipid rafts, and disease. J Clin Invest 110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cisz M, Lee PC, Rietsch A. 2008. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J Bacteriol 190:2726–2738. doi: 10.1128/JB.01553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mejía E, Bliska JB, Viboud GI. 2008. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog 4:e3. doi: 10.1371/journal.ppat.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swimm AI, Kalman D. 2008. Cytosolic extract induces Tir translocation and pedestals in EPEC-infected red blood cells. PLoS Pathog 4:e4. doi: 10.1371/journal.ppat.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong KW, Isberg RR. 2005. Yersinia pseudotuberculosis spatially controls activation and misregulation of host cell Rac1. PLoS Pathog 1:e16. doi: 10.1371/journal.ppat.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi S, Isberg RR. 2009. Yersinia pseudotuberculosis virulence determinants invasin, YopE, and YopT modulate RhoG activity and localization. Infect Immun 77:4771–4782. doi: 10.1128/IAI.00850-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertz O, Hodgson L, Klemke RL, Hahn KM. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 21.Guryanova OA, Makhanov M, Chenchik AA, Chumakov PM, Frolova EI. 2006. Optimization of a genome-wide disordered lentivector-based short hairpin RNA library. Mol Biol 40:396–405. doi: 10.1134/S002689330603006X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. 2008. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen 13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viboud GI, Bliska JB. 2001. A bacterial type III secretion system inhibits actin polymerization to prevent pore formation in host cell membranes. EMBO J 20:5373–5382. doi: 10.1093/emboj/20.19.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhausen T. 2000. Three ways to make a vesicle. Nat Rev Mol Cell Biol 1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Punj V, Sengupta D, Linstedt AD. 2008. Coat-tether interaction in Golgi organization. Mol Biol Cell 19:2830–2843. doi: 10.1091/mbc.E07-12-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. String v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Souza-Schorey C, Chavrier P. 2006. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 29.Elvin SJ, Williamson ED, Scott JC, Smith JN, Pérez De Lema G, Chilla S, Clapham P, Pfeffer K, Schlöndorff D, Luckow B. 2004. Evolutionary genetics: ambiguous role of CCR5 in Y. pestis infection. Nature 430:417. doi: 10.1038/nature02822. [DOI] [PubMed] [Google Scholar]
- 30.Mecsas J, Franklin G, Kuziel WA, Brubaker RR, Falkow S, Mosier DE. 2004. Evolutionary genetics: CCR5 mutation and plague protection. Nature 427:606. doi: 10.1038/427606a. [DOI] [PubMed] [Google Scholar]
- 31.Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol 186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuziel WA, Dawson TC, Quinones M, Garavito E, Chenaux G, Ahuja SS, Reddick RL, Maeda N. 2003. CCR5 deficiency is not protective in the early stages of atherogenesis in apoE knockout mice. Atherosclerosis 167:25–32. doi: 10.1016/S0021-9150(02)00382-9. [DOI] [PubMed] [Google Scholar]
- 33.Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. 2005. Plague bacteria target immune cells during infection. Science 309:1739–1741. doi: 10.1126/science.1114580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durand EA, Maldonado-Arocho FJ, Castillo C, Walsh RL, Mecsas J. 2010. The presence of professional phagocytes dictates the number of host cells targeted for Yop translocation during infection. Cell Microbiol 12:1064–1082. doi: 10.1111/j.1462-5822.2010.01451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veiga E, Guttman JA, Bonazzi M, Boucrot E, Toledo-Arana A, Lin AE, Enninga J, Pizarro-Cerdá J, Finlay BB, Kirchhausen T, Cossart P. 2007. Invasive and adherent bacterial pathogens co-opt host clathrin for infection. Cell Host Microbe 2:340–351. doi: 10.1016/j.chom.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonazzi M, Vasudevan L, Mallet A, Sachse M, Sartori A, Prevost MC, Roberts A, Taner SB, Wilbur JD, Brodsky FM, Cossart P. 2011. Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. J Cell Biol 195:525–536. doi: 10.1083/jcb.201105152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, Rout S, Stark M, von Mering C, Pelkmans L, Hardt WD. 2011. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol 7:474. doi: 10.1038/msb.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alonzo F III, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, Torres VJ. 2013. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonzo F III, Benson MA, Chen J, Novick RP, Shopsin B, Torres VJ. 2012. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol Microbiol 83:423–435. doi: 10.1111/j.1365-2958.2011.07942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes-Robles T, Alonzo F III, Kozhaya L, Lacy DB, Unutmaz D, Torres VJ. 2013. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe 14:453–459. doi: 10.1016/j.chom.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan SR, Scott S, Duncan CJ. 2005. Reappraisal of the historical selective pressures for the CCR5-Delta32 mutation. J Med Genet 42:205–208. doi: 10.1136/jmg.2004.025346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balada-Llasat JM, Mecsas J. 2006. Yersinia has a tropism for B and T cell zones of lymph nodes that is independent of the type III secretion system. PLoS Pathog 2:e86. doi: 10.1371/journal.ppat.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crimmins GT, Mohammadi S, Green ER, Bergman MA, Isberg RR, Mecsas J. 2012. Identification of MrtAB, an ABC transporter specifically required for Yersinia pseudotuberculosis to colonize the mesenteric lymph nodes. PLoS Pathog 8:e1002828. doi: 10.1371/journal.ppat.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logsdon LK, Mecsas J. 2003. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect Immun 71:4595–4607. doi: 10.1128/IAI.71.8.4595-4607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maldonado-Arocho FJ, Green C, Fisher ML, Paczosa MK, Mecsas J. 2013. Adhesins and host serum factors drive Yop translocation by Yersinia into professional phagocytes during animal infection. PLoS Pathog 9:e1003415. doi: 10.1371/journal.ppat.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mecsas J, Raupach B, Falkow S. 1998. The Yersinia Yops inhibit invasion of Listeria, Shigella and Edwardsiella but not Salmonella into epithelial cells. Mol Microbiol 28:1269–1281. doi: 10.1046/j.1365-2958.1998.00891.x. [DOI] [PubMed] [Google Scholar]
- 47.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien RY. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 49.Harmon DE, Davis AJ, Castillo C, Mecsas J. 2010. Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob Agents Chemother 54:3241–3254. doi: 10.1128/AAC.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med 200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotated gene list for the significant shRNA candidates identified in the screen. shRNAs having a ZMAD of >2 for both FRET+/unsorted and FRET+/FRET− in a minimum of two independent experiments are listed. The table contains gene identification (ID), reference and position of the shRNA on the chromosome, targeted sequence, calculated ZMAD values for both ratios from each experiment, UCSC Genome Browser reference IDs (47), and a description of each gene.
List of shRNA oligonucleotides used in this study