ABSTRACT
The genomes of four novel marine Actinobacteria have been assembled from large metagenomic data sets derived from the Mediterranean deep chlorophyll maximum (DCM). These are the first marine representatives belonging to the order Acidimicrobiales and only the second group of planktonic marine Actinobacteria to be described. Their streamlined genomes and photoheterotrophic lifestyle suggest that they are planktonic, free-living microbes. A novel rhodopsin clade, acidirhodopsins, related to freshwater actinorhodopsins, was found in these organisms. Their genomes suggest a capacity to assimilate C2 compounds, some using the glyoxylate bypass and others with the ethylmalonyl-coenzyme A (CoA) pathway. They are also able to derive energy from dimethylsulfopropionate (DMSP), sulfonate, and carbon monoxide oxidation, all commonly available in the marine habitat. These organisms appear to be prevalent in the deep photic zone at or around the DCM. The presence of sister clades to the marine Acidimicrobiales in freshwater aquatic habitats provides a new example of marine-freshwater transitions with potential evolutionary insights.
IMPORTANCE
Despite several studies showing the importance and abundance of planktonic Actinobacteria in the marine habitat, a representative genome was only recently described. In order to expand the genomic repertoire of marine Actinobacteria, we describe here the first Acidimicrobidae genomes of marine origin and provide insights about their ecology. They display metabolic versatility in the acquisition of carbon and appear capable of utilizing diverse sources of energy. One of the genomes harbors a new kind of rhodopsin related to the actinorhodopsin clade of freshwater origin that is widespread in the oceans. Our data also support their preference to inhabit the deep chlorophyll maximum and the deep photic zone. This work contributes to the perception of marine actinobacterial groups as important players in the marine environment with distinct and important contributions to nutrient cycling in the oceans.
INTRODUCTION
The bottleneck of pure culture has revealed only a partial glimpse of the actinobacterial world (1, 2). In recent years, a vast uncultured world of genuinely planktonic Actinobacteria has been discovered. These newly discovered microbes are in complete contrast to their better-studied soil counterparts in their size and genomic features. With the advent of 16S rRNA cloning, it became possible to interrogate microbial community composition without the bias of culture, and one of the first such surveys in the marine habitat (3) identified nearly identical sequences from high-GC gram-positive organisms from the Pacific and Atlantic oceans. Phylogenetic analyses of these sequences also suggested that they represented a deep-branching clade, quite different from known organisms. It was not until 1997 that full-length 16S rRNA sequences became available for these microbes (OM1 from Cape Hatteras, North Atlantic coast, and SAR432 from the Sargasso Sea) and enabled a robust taxonomic placement, reinforcing their novelty (4). A subsequent follow-up study, with additional sequences (including Actinobacteria of terrestrial origin), established that microbes harboring these sequences were sufficiently different to warrant a new subclass within the phylum Actinobacteria (5). This group was referred to as the marine actinobacterial clade (MAC). Interestingly, in that work, the authors stated that the GC content of the 16S rRNA gene for these marine sequences was slightly lower (51.9 to 53.1%) than the typical values of other Actinobacteria (55 to 64%), providing the first indication of the lower GC content that characterizes planktonic Actinobacteria (2, 6). More information about the MAC was later revealed by studies on samples from the Bermuda Atlantic Time-series (BATS) location in the Sargasso Sea. In 2005, Morris and collaborators used terminal restriction fragment length polymorphism (T-RFLP), among other techniques, to examine spatial and temporal patterns across a decade of sampling (7). This study showed for the first time that, like SAR11, marine Actinobacteria (represented at this time by the MAC) increased in abundance following convective overturn at a depth of 200 m, suggesting postmixing blooming. Another recent study has also provided substantial evidence of the association of the MAC with the deep chlorophyll maximum (DCM) (8), the zone of maximum phytoplankton concentration in marine stratified water columns.
Recently, by sequencing and assembly of metagenomic fosmids, the nearly complete genome of members of the MAC group has been obtained (6). The reconstructed genome was remarkably small (estimated to be <1 Mb) and had very low genomic GC content, only 32%. Both values are the lowest for any Actinobacteria described until then. The estimated cell volume was only 0.006 to 0.024 µm3, making them smaller even than “Candidatus Pelagibacter ubique” and the smallest free-living cells described until then. A new taxon (subclass “Candidatus Actinomarinidae,” order “Candidatus Actinomarinales”) was proposed for the organisms belonging to this clade, and the organism itself was referred to as “Candidatus Actinomarina minuta.” All previously recovered sequences, including BDA1-5, OM1, SAR432, and D92-32, described as belonging to the MAC are comprised within “Ca. Actinomarinidae.” Two incomplete single-cell amplified genomes (SAGs) that are closely related to “Ca. Actinomarina minuta” (SAG D07 and SAG M09) have also been published recently (9), adding further representatives of this new subclass.
The existence of another different marine actinobacterial group was also detected by 16S rRNA analysis of clone libraries and metagenomic data sets (10) that indicated the presence of members of the Acidimicrobiales, an order with very few representative isolates (11). Subsequently, in an extensive study analyzing seasonal and vertical changes in the BATS (12), sequences related to the acidimicrobial microbe “Ca. Microthrix parvicella” were found. These “marine Microthrix” were restricted to the DCM in particular, blooming occasionally during summer stratification. Their population dynamics appeared similar to that of the SAR11 1b ecotype. Therefore, at this point, two taxonomically distinct planktonic actinobacterial groups are known to be present in the marine habitat. Both appear to be associated with the zone of maximal photosynthetic production, but genomic information is only available for one of them, the “Ca. Actinomarinales.”
The deep chlorophyll maximum is the site of maximal phytoplanktonic density in the stratified marine water column. It is located permanently at the depth of ~100 m in tropical oceans and at a variable depth (~50 to 100 m) during summer in temperate latitudes. As such, it is one of the largest habitats on earth. In the present work, using assembly from deep metagenomic sequencing from the DCM of the Mediterranean, we describe four nearly complete genomes of marine Actinobacteria belonging to the order Acidimicrobiales. These reconstructed genomes are essentially composites, each likely originating from several coexisting clonal lineages, and provide insights into the lifestyle of this diverse and important group of microbes, complementing the genomic information obtained before about the “Ca. Actinomarinales.”
RESULTS AND DISCUSSION
Identification of Acidimicrobiales contigs and genome reconstruction.
A broad overview of the phylogenetic composition of the unassembled data set was obtained by retrieving 16S rRNA fragments (ca. 41,000) from the raw reads. A comparison of the community structure determined this way with those of two other DCM data sets (one of them a 454 run [13]) from the same site is shown in Fig. S1 in the supplemental material. Specifically, the contributions of Actinobacteria in these samples appear to be rather similar (1.6 to 3%).
Since our target for this work was the marine Acidimicrobiales from the DCM, we specifically selected long contigs (>10 kb) in which the majority of genes gave best hits to organisms of the subclass Acidimicrobidae. This allows a simple but stringent selection method toward identifying the target group. A total of 160 contigs with a combined length of 6.6 Mb (the longest contig was 431 kb) were thus identified as putative Acidimicrobiales genomic fragments. Using GC content, coverage, and principal component analysis of tetranucleotide frequencies (see Materials and Methods), 151 contigs could be further classified into four groups. Each of these groups likely comes from cells that belong to a single species, although, as is always the case with metagenomic assemblies, the clonal purity of the fragments cannot be guaranteed. For the sake of simplicity, we will use the term “genome” to refer to these composites and we will refer to them as the MedAcidi group. Some summary statistics about these genomes are provided in Table 1. Three genomes had GC contents of between 40 and 45% and were predicted to have maximum sizes in the range of 1.7 to 2 Mb, while the fourth had slightly higher values (50% GC content and a 2.33-Mb genome). With one exception (MedAcidi-G2A), the genomes were predicted to be nearly 90% complete.
TABLE 1.
Summary statistics of the reconstructed genomes
| Genome | No. of contigs | GC% | Total length (Mb) | Completeness (%) | Estimated size (Mb) |
|---|---|---|---|---|---|
| MedAcidi-G1 | 43 | 37–44 | 1.68 | 84–91 | 1.85–2 |
| MedAcidi-G2A | 48 | 41–46 | 1.37 | 71–79 | 1.73–1.93 |
| MedAcidi-G2B | 28 | 44–46 | 1.44 | 88–95 | 1.52–1.64 |
| MedAcidi-G3 | 32 | 50–52 | 2.1 | 90–96 | 2.19–2.33 |
Three of the genomes had contigs that contained 16S rRNA. Figure 1a shows the reconstructed phylogeny of the 16S rRNA of our genomes together with several reference genomes. They were more related to each other and to other uncultured microbes than to any of the cultured and sequenced representatives of this subclass (e.g., members of “Ca. Microthrix” and Ilumatobacter). Related 16S rRNA sequences came from diverse marine environments around the world, including the Pacific and the Atlantic oceans, suggesting a widespread distribution. We also used a concatenate of 106 proteins that are conserved between these and 150 actinobacterial genomes for a more robust phylogenetic affiliation and, also, to be able to place MedAcidi-G2A, which did not have a 16S rRNA in the contigs. The resulting tree confirms the Acidimicrobiales affiliation of these genomes (Fig. 1b). In both trees, the nearest neighbors were Ilumatobacter coccineum (a beach isolate) and the recently described genome acAcidi, from a freshwater reservoir (14). Their similarity with the MedAcidi genomes was rather low (average nucleotide identity, ~60%), suggesting that these belong to different genera. Therefore, our reconstructed genomes provide the first genomic information about genuine marine and planktonic Acidimicrobiales.
FIG 1 .
Phylogeny of the MedAcidi group of marine Actinobacteria. (a) Maximum-likelihood phylogeny of 16S rRNA of the assembled MedAcidi genomes and related sequences from clone libraries. Sequences from the order Actinomycetales are used to root the tree. The 16S rRNAs from the MedAcidi groups are highlighted by colored boxes. (b) Maximum-likelihood phylogeny using a concatenation of 106 conserved proteins: the assembled genomes of the MedAcidi group are highlighted by colored boxes. Bootstrap values (%) are indicated at each node. Reference organisms shown in both trees are connected by dashed lines.
Lifestyle insights.
The sample from which these microbes were assembled (75-m depth in offshore Mediterranean waters) already indicates that they come from an oligotrophic habitat (15). However, even in oligotrophic open ocean waters, some microbes have opportunistic copiotrophic lifestyles, taking advantage of microheterogeneity (particulate organic matter, for example) or sporadic nutrient inputs (such as upwelling or deep mixing). In the case of the MedAcidi genomic fragments described here, several lines of evidence support a real planktonic and oligotrophic lifestyle. Perhaps the most prominent feature of oligotrophic marine planktonic free-living microbes is that they possess very streamlined genomes (9, 16). In addition to the reduction in the number of genes, they also display a characteristic reduction in the length of intergenic spacers. Along these lines, planktonic Actinobacteria, including the MedAcidi genomes, display strikingly short intergenic spacers (see Fig. S2 in the supplemental material), in the range of 3 bp (“Ca. Actinomarina minuta”) to 32 bp for the freshwater actinobacterium acMicro-1 (14). The smallest median intergenic spacer sizes in these new genomes were found to be in the range of 14 to 23 bp (MedAcidi-G2B and MedAcidi-G3, respectively). Another indication of streamlined genomes is a reduction in regulatory sigma factors. For example, the streamlined genome of “Ca. Pelagibacter ubique” encodes only two sigma factors (17). Only two sigma factors were found in the genomes of MedAcidi-G1 and MedAcidi-G2B, four in MedAcidi-G2A, and a maximum of five in MedAcidi-G3. Only one sigma factor (sigma-70) was found in the “Ca. Actinomarina minuta” genome. However, this is not yet definite, as the genome is estimated to be ~70% complete. No sigma factors were found in the SAG genomes of “Ca. Actinomarinales” either. In contrast, the model pathogenic actinobacterium Mycobacterium tuberculosis H37Rv encodes 13, implying a much more complex response to the environment. Another feature of truly planktonic microbes is a lack of any apparatus for motility, and indeed, no flagellar genes were found in any of these new Acidimicrobiales genomes, suggesting that they are nonmotile.
Acidirhodopsins: a new rhodopsin clade shared between marine and freshwater environments.
Photoheterotrophy is a common feature of both planktonic marine and freshwater Actinobacteria (6, 14, 18, 19). Of the four genomes described here, one (MedAcidi-G1) was found to harbor a rhodopsin-coding gene, which showed the highest sequence similarity (82%) to a recently described rhodopsin from an Acidimicrobiales bacterium (uncultured actinobacterium acAcidi) from freshwater (14). Both sequences appear to be related to actinorhodopsins (Fig. 2). Searches against the global ocean sampling (GOS) database using the rhodopsin of MedAcidi-G1 revealed several hits to sequences from diverse samples originating from the Sargasso Sea, Indian Ocean, and Galapagos Islands. Moreover, these sequences reciprocally gave the best BLAST hits to the freshwater sequence, as did the rhodopsin of MedAcidi-G1. In cases where adjacent genes were found on the GOS reads, they also gave the best hits to the same freshwater Acidimicrobiales bacterium. Although actinorhodopsins are known to be widespread in freshwater Actinobacteria, they have not been identified in the marine environment (19, 20). Moreover, the recently described “Ca. Actinomarina minuta” harbors rhodopsin sequences that form a distinct clade (MACrhodopsins) in the rhodopsin phylogeny (6). Though sequence similarity suggested a close relationship to freshwater actinorhodopsins, phylogenetic analysis indicates that these new sequences form a sister clade to known actinorhodopsins, xanthorhodopsins, and eukaryal rhodopsins. The new cluster is sufficiently different from other known branches that we propose a new name, “acidirhodopsins,” for this branch of the rhodopsin tree (Fig. 2). Analysis of the critical amino acids of these sequences suggests they absorb in the green region of the spectrum (20). This is only the second example of a rhodopsin that has close representatives in marine and freshwater habitats, the other being the similar rhodopsins of marine alphaproteobacterial SAR11 and the freshwater LD12 clades. The other three MedAcidi genomes did not contain rhodopsins or any gene that suggested the presence of one, although this might be due to the incompleteness of the genomes. All four genomes encoded photolyases and UvrABC systems for correcting UV light-induced DNA damage, which are typical for microbes inhabiting the photic zone. This, together with the rhodopsin found in MedAcidi-G1 and the sample from which they were obtained, suggests that they inhabit the photic zone (see below).
FIG 2 .
Rhodopsin phylogeny. A maximum-likelihood tree of all known types of rhodopsins is shown. Sequences belonging to the new clade of acidirhodopsins are highlighted. Sequences originating from marine and freshwater habitats are indicated by colored squares. Bootstrap values (%) are shown on the nodes.
Carbon and energy metabolism.
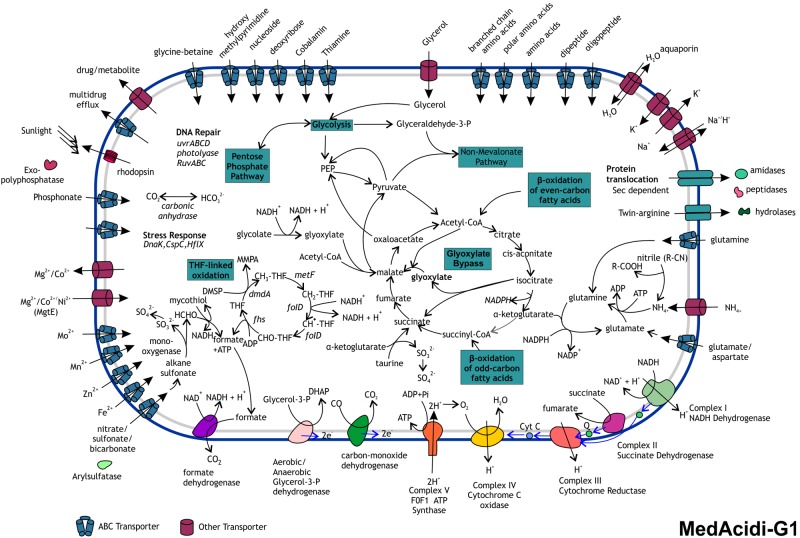
The genomes provided solid evidence for the MedAcidi being heterotrophs (or in the case of MedAcidi-G1, a photoheterotroph). All of the genomes encoded transporters for organic matter, e.g., glycerol, sugars, amino acids, and peptides (Fig. 3 and Fig. 4; see Fig. S3 and S4 in the supplemental material). Most of the steps for both glycolysis and the TCA cycle were found in all of the genomes, suggesting that these pathways are essentially complete (see Data Set S1 in the supplemental material). Apart from using these typical carbon and energy sources, all genomes appeared to be able to oxidize even- and odd-carbon number fatty acids as well (to acetyl-CoA and propionyl-CoA, respectively). In the absence of complex carbon sources like glucose, the glyoxylate bypass allows the utilization of C2 carbon units from acetate (21) and the acetyl-CoA generated by the oxidation of fatty acids. Both of the critical enzymes of the glyoxylate pathway (isocitrate lyase and malate synthase) were found in MedAcidi-G1, MedAcidi-G2A, and MedAcidi-G2B, showing that they can use C2 compounds. However, the MedAcidi-G3 genome did not encode the genes for the glyoxylate cycle. The recently described ethylmalonyl pathway has an equivalent role in C2 assimilation (22). Nearly all of the enzymes of this pathway were found in MedAcidi-G3 (including the marker enzyme crotonyl-CoA carboxylase/reductase) (Fig. S5). A common C2 compound in the marine environment is glycolate, a highly labile compound that is a byproduct of photorespiration (23). All of the MedAcidi genomes contained glycolate oxidase genes, suggesting the ability to use this compound.
FIG 3.
Metabolic overview of the marine actinobacterium MedAcidi-G1. Major pathways are indicated in boxes. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate; DHAP, dihydroxyacetone phosphate.
FIG 4.
Metabolic overview of the marine actinobacterium MedAcidi-G3. Major pathways are indicated in boxes. Some reactions that have not been found in this genome but are found in the other MedAcidi genomes are shown in grey. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate; ggt, gamma-glutamyl transpeptidase; DHAP, dihydroxyacetone phosphate.
The complete tetrahydrofolate (THF)-linked pathway was detected in the MedAcidi-G1 genome, and parts of it were found in the others, suggesting that these microbes might be capable of producing energy using dimethylsulfopropionate (DMSP). The first key enzyme, dimethylsulfoniopropionate-dependent demethylase (DmdA), which removes a methyl group from DMSP and yields methylmercaptopropionate (MMPA), was found in all MedAcidi genomes (24, 25). DMSP is a commonly occurring osmolyte in the marine habitat, particularly in marine algae (26, 27). Microbial degradation of DMSP can lead to the release of dimethylsulfide (DMS), a highly volatile compound that is released to the atmosphere in large amounts (28). It has been suggested that nearly a third of the surface ocean microbes are capable of performing this transformation (24), and several can incorporate the DMSP-derived sulfur into amino acids (methionine) (29). It has been shown that “Ca. Pelagibacter ubique” is able to oxidize MMPA-derived methyl groups via this pathway to produce energy (30).
Another common energy source for marine microbes is carbon monoxide. Carbon monoxide dehydrogenase (CODH) genes were found in all of the MedAcidi genomes. Carbon monoxide produced by photolysis of dissolved organic matter (31, 32) is readily available in the marine habitat. Moreover, evidence from genomic and metagenomic analyses is also increasingly revealing the widespread distribution of aerobic carbon monoxide dehydrogenases in the seas (33–36). Two types of these heterotrimeric enzymes are recognized, with the largest subunit (CoxL) possessing characteristic catalytic sites. Form I CODH enzymes, originally described in Oligotropha carboxydovorans, are fast CO oxidizers, while form II enzymes (e.g., those found in Bradyrhizobium, Mesorhizobium, and Sinorhizobium species) are several times slower (10 to 1,000 times). A total of 19 candidate CoxL genes were found in the MedAcidi genomes, although only 8 of them had the catalytic-site pattern of the form II CODH. However, all the genomes encoded at least one potentially active form II enzyme. No form I CODH catalytic site pattern was identified, making them slow CO oxidizers. A CODH has also been found in acAcidi. However, whether or not all form II CODH genes actually are responsible for CO oxidation has not been conclusively demonstrated. For example, in the case of Roseobacter species, even though form II CODH genes were readily identifiable, CO oxidation was not observed (37).
The MedAcidi-G1 genome encoded a transporter for alkanesulfonates and, also, an alkanesulfonate monooxygenase (also found in MedAcidi-G2B). The alkanesulfonate monooxygenase is used to split the C-S bond in the sulfonates and release formaldehyde and sulfite. Sulfonates are organosulfur compounds that are widely distributed in nature (38–41), and several bacteria are known to use sulfonates as a source of sulfur (42). One widespread form of organosulfonates is methanesulfonate, produced from the chemical oxidation of biogenic DMS in the atmosphere (39). While the sulfite may undergo spontaneous or enzymatic oxidation to sulfate, some organisms, e.g., methylotrophs, can utilize the formaldehyde as a carbon and energy source via the serine pathway, but this pathway was not found in the MedAcidi genomes. In any case, intracellular formaldehyde must be detoxified. In Actinobacteria, formaldehyde can spontaneously combine with the Actinobacteria-specific thiol mycothiol to form S-hydroxymethylmycothiol, which is the substrate for the mycothiol-dependent formaldehyde dehydrogenase. The gene coding for this enzyme was found in MedAcidi-G1 and MedAcidi-G2B. The use of mycothiol (a pseudodisaccharide) as a cofactor in place of the universal tripeptide thiol glutathione is characteristic of this particular enzyme (43). This enzyme converts S-hydroxymethylmycothiol to S-formylmycothiol, and in a second step, a thiol esterase yields formate and the original mycothiol (44). Additional oxidation of formate by formate dehydrogenases yields CO2 and reducing equivalents in the form of NADH. Formate dehydrogenase can also be linked to the THF-linked oxidation pathway (see above), and genes coding for this enzyme were found in all four genomes.
Nitrogen metabolism.
We found ammonium transporters in all of the genomes except MedAcidi-G2B (its absence in this genome could again reflect incompleteness). Additionally, transporters for glutamine and glutamate/aspartate were also found in MedAcidi-G1 and MedAcidi-G3. All genomes encoded more general amino acid, dipeptide, and oligopeptide transporters (Fig. 3 and 4; see Fig. S3 and S4 in the supplemental material). We also found genes coding for the hydrolysis of nitriles, which can be a potential source of both carbon and nitrogen, in three of these genomes. Nitriles are naturally occurring compounds produced by plants and are also released into the environment by human industrial activity, e.g., petrochemical refineries, mining, etc. (45). Nitrile-hydrolyzing bacteria have been sought after not only because of their ability to degrade these environmentally hazardous cyanide-containing compounds but also because of the successful commercial application of nitrile-hydrolyzing enzymes as biocatalysts for the commercial production of a variety of organic compounds, e.g., acrylamides, carboxylic acids, etc. (46). Actually, Actinobacteria with such metabolic capacities have been reported from terrestrial and deep sea sediments (47). Though there are several described pathways for nitrile degradation (45), we found two in these actinobacterial genomes, both of which yield carboxylate and ammonium as end products, which can be used as carbon and nitrogen sources, respectively. In the first pathway, found in MedAcidi-G1 and MedAcidi-G2B, a single-step hydrolysis using nitrilases yields these final compounds directly. In the second, found in MedAcidi-G3, a two-step hydrolysis is predicted, where a nitrile hydratase first transforms nitriles to amides, which are then processed by amidases to yield the same end products.
Distribution of marine actinobacterial groups.
Fragment recruitment analysis of genomes provides the most accurate estimate of the presence and abundance of the corresponding microbes in different habitats. However, to perform these analyses, large sequence data sets are required and, as yet, very few are available, particularly of depth profiles in the ocean. We have three metagenomes (samples taken in 2007 at a depth of 50 m, in 2012 at 75 m, and in 2013 at 55 m), all from the depth of the DCM at the same site from which the genomes were assembled. The recruitment results are shown in Fig. 5 and indicate that all four microbes are more abundant in the sample of origin (2012), as should be expected since long contigs are easier to assemble in a sample in which they are very prevalent. However, their presence in the other high-coverage data set (2013, 55 m) attests to their permanence in this habitat. They were barely detectable in the smaller 454 data set from 2007. Moreover, these genomes also recruited very little from the BATS (48) and the Hawaii Ocean Time-series (HOTS) (49) data sets (Fig. 5). In comparison, “Ca. Actinomarina minuta,” represented by 43 assembled fosmids (6) and two SAGs (9), recruited consistently across these data sets. When similar analyses was performed using the Global Ocean Sampling (GOS) data set (50), similar results were found, where the genomes of “Ca. Actinomarinales” recruited much more than the marine Acidimicrobiales (see Fig. S6 in the supplemental material). However, recruitments across the depth profile of HOTS (Fig. S7) indicate that the marine Acidimicrobiales are represented much more at the deep chlorophyll maximum (DCM) than at the surface.
FIG 5.
Metagenomic fragment recruitment. (a) Relative abundances of genomes of marine Acidimicrobiales and “Ca. Actinomarinales” across two data sets from the Mediterranean deep chlorophyll maximum, Hawaii Ocean Time-series (HOTS), Bermuda Atlantic Time-series (BATS), and the Red Sea. Data are expressed as RPKG (reads recruited per Kb of genome per Gb of metagenome). (b) Recruitment plots of two representative Acidimicrobiales genomes (MedAcidi-G1 and MedAcidi-G3) and one “Ca. Actinomarinales” genome (“Ca. Actinomarina minuta”) compared to two metagenomes (MedDCM-2012 and MedDCM-2013) are shown. The dashed horizontal line indicates 95% nucleotide sequence identity. (c) Relative abundances of 16S rRNA sequences of marine Acidimicrobiales and “Ca. Actinomarinales” across the same data sets. All data sets that are from a deep chlorophyll maximum are marked with a green circle. Data are expressed as hits/Gb of metagenome.
Given the scarcity of fragments recruited by the genomes, we have also used several 16S rRNA sequences from each group to assess their relative levels of abundance across the marine habitat (see Materials and Methods) (Fig. 5c; see Fig. S8 in the supplemental material). This method allows a much better assessment of their presence in low-coverage data sets, although it is much less reliable. The results again show a wide variation among different data sets but indicate that, not only are the marine Acidimicrobiales detectable at DCM depths but they may also be found at greater depths (e.g., HOTS 500 m and 4,000 m). Low but nearly equal abundances were found for both groups in additional metagenomic data sets from the deep Sea of Marmara at 1,000 m (51) and the Puerto Rico Trench at 4,000 m (52). Results from the GOS data set (Fig. S8) and from the Red Sea (53) also indicate, similar to the results from genome recruitments, that “Ca. Actinomarinales” representatives are relatively more abundant than Acidimicrobiales. On the other hand, marine Acidimicrobiales 16S rRNA sequences were detectable in the polar latitudes, both in the Antarctic (54) and the Arctic (55), while “Ca. Actinomarinales” appeared to be restricted to temperate and tropical latitudes (Fig. S8), suggesting that at least some members of this new group might be psychrotolerant.
The partial MedAcidi genomes described here provide the first glimpses into the physiology and lifestyle of the marine acidimicrobia, hitherto known only from cloned 16S rRNA sequences and denominated the “marine Microthrix” group (12). They belong to representatives of the second major phylogenetic group of planktonic marine Actinobacteria to be described at this level, after a similar description became available for the “Ca. Actinomarinales” (6), formerly known as the “marine actinobacterial clade” (MAC). Both groups have streamlined genomes and are heterotrophs, sometimes supplemented by phototrophic energy derived from rhodopsins. They seem to have free-living bona fide planktonic cells (as opposed to particle associated) and are well adapted to the oligotrophic marine waters, using organic matter that is predicted to be common in this habitat. Genome recruitment indicated that these microbes appear to be more abundant in the deeper photic zone than at the surface and are probably more prevalent around DCM depths (50 to 120 m, also favored by the “Ca. Actinomarinales”). These depths are very productive in the stratified ocean and are free from the damaging UV light of shallower depths. Recent developments in the field of freshwater Actinobacteria have revealed the presence of related acidimicrobial groups (14). This situation is reminiscent of those of the marine “Ca. Pelagibacterales” and the freshwater LD12 groups. In both cases, the microbes are distantly related and also share phylogenetically related rhodopsins, suggesting that the acquisition of photoheterotrophy predates the marine-to-fresh water transition. The “Ca. Actinomarinales” do not seem to have sister clades in freshwater.
MATERIALS AND METHODS
Sampling, sequencing, assembly, and annotation.
Sampling was done on 20 July 2012 from the DCM depth (75 m) at a distance of ~20 nautical miles off the coast of Alicante, Spain (38°4′6.64″N, 0°13′55.18″E). The deep chlorophyll maximum was determined from a water column fluorescence profile by using a Seabird SBE 19 multiprobe profiler. Two hundred liters of seawater was filtered through a series of 20-µm, 5-µm, and 0.22-µm polycarbonate filters (Millipore). DNA from the 0.22-µm fraction was extracted as described previously (56). Sequencing was performed using the Illumina HiSeq 2000 PE (Macrogen, South Korea). Two libraries of different insert sizes, 300 bp and 3 kb, were created. In total, 11.9 Gb (135 million reads) and 5.3 Gb (62 million reads) of data were obtained for the two libraries, respectively. This data set is referred to as the MedDCM-2012 data set in this work. Both data sets (197 million reads) were assembled together using the IDBA assembler (57). The second metagenome used here, referred to as MedDCM-2013, was sampled from the same site on 6 September 2013 and processed in identical fashion. Only the unassembled reads from this data set are used in this work. Protein-coding genes in the assembled contigs were predicted using Prodigal in metagenomic mode (58), and tRNAs were predicted using tRNAscan-SE (59). Ribosomal rRNA genes were identified using ssu-align (60) and meta_rna (61). Additional annotation of genes was done by comparing against the NCBI NR, COG (62), and TIGRfam (63) databases. In addition, genomes were also annotated using the RAST server (64). Additional local BLAST searches against the latest NCBI-NR database were performed whenever necessary.
16S rRNA classification.
A nonredundant version of the RDP database (65) was prepared by clustering the ca. 2.3 million 16S rRNA sequences into ~800,000 sequences at the 90% identity level using UCLUST (66). Comparisons against this representative 16S rRNA database were used to identify candidate 16S rRNA sequences in the complete Illumina data sets. A sequence matching this database at an e value of <1e−5 was considered a potential 16S sequence. Candidate 16S rRNA sequences were further examined using ssu-align, which uses hidden Markov models (HMMs) to separate these into archaeal, bacterial, and eukaryal 16S/18S rRNA or non-16S rRNA sequences (60). These bona fide sequences were finally compared to the complete RDP database and classified into a high-level taxon if the sequence identity was ≥80% and the alignment length was ≥90 bp. For 454 sequences, the alignment length was relaxed to 100 bp. Sequences failing these thresholds were discarded.
Identification of Acidimicrobiales contigs and genome reconstruction.
We only used contigs of >10 kb for all analyses. A contig was considered to originate from the order Acidimicrobiales if the majority of genes gave best blast hits to this order. Further clusters were made using taxonomy, principal component analysis of tetranucleotide frequencies, GC content, and coverage in both the metagenomes as described previously (67–69). Tetranucleotide frequencies were computed using the wordfreq program in the EMBOSS package (70). Principal component analysis was performed using the FactoMineR package in R (71).
Genomic phylogenetic trees and genome size estimation.
A set of 97 conserved proteins found in a set of 155 complete actinobacterial genomes were used to estimate genome completeness for these genomes. For whole-genome phylogeny, a smaller set of representative actinobacterial genomes was chosen in order to maximize the number of common proteins, though all available genomes from the order Acidimicrobiales were included. Using the COG database (62), 106 conserved proteins were found in these five genomes and the other reference genomes. COG assignments were made using an e value of <1e−5, >80% query coverage, and >30% identity. These proteins were concatenated and used to create the whole-genome phylogeny of these new microbes. The alignment was performed using Kalign (72) and trimmed using trimAL (73). A maximum-likelihood tree was constructed with FastTree2 (74), using a JTT+CAT model, a gamma approximation, and 100 bootstrap replicates.
Single gene trees.
All 16S sequences were trimmed using ssu-align (60), and multiple sequence alignments were created using MUSCLE (75). 16S rRNA phylogenetic trees were constructed using FastTree2 (74) with a GTR+CAT model, a gamma approximation, and 100 bootstrap replicates. The sequences of rhodopsins were also aligned using MUSCLE, and a maximum-likelihood tree was constructed with FastTree2 (74), using a JTT model, a gamma approximation, and 100 bootstrap replicates.
Metagenomic recruitment.
Recruitments were performed using BLASTN (76), and a hit was considered only when it was at least 50 nucleotides long, with an identity of >95% and with an e value of ≤1e−5. These hits were used to compute the RPKG (reads recruited per kilobase of genome per gigabase of metagenome) values, which provide a normalized value that is comparable across various metagenomes. For 16S rRNA abundance comparison of the two groups, 13 sequences from the marine Acidimicrobiales group and 7 from “Ca. Actinomarinales” were compared to the 16S rRNA sequences in the metagenomic data sets mentioned here. Hits were assigned to each group if they had more than 95% nucleotide sequence identity (species-level cutoffs) and alignment lengths of >100 bp. The number of unique hits for each group was normalized by dividing by the data set size.
Data accession numbers.
The metagenomic data have been submitted to NCBI SRA and are accessible under the BioProject identifier PRJNA257723. The assembled genome sequences have been deposited to DDBJ/EMBL/GenBank and can be accessed using the accession numbers JUEM00000000, JUEN00000000, JUEO00000000, and JUEP00000000.
SUPPLEMENTAL MATERIAL
Excel file containing important pathways and transporters found in the MedAcidi genomes. Download
16S rRNA classification of the Mediterranean deep chlorophyll maximum data sets. y axis indicates percentage of reads attributed to each taxon. Color key is at top right. Download
(a) Genome size versus GC content. (b) Median intergenic spacer size versus GC content. 1386 prokaryotic genomes are shown here. The marine actinobacterial genomes assembled in this work and another marine actinobacterial genome (“Ca. Actinomarina minuta”) are labeled in white boxes. Genomes of freshwater Actinobacteria are labeled. All other genomes are shown in grey. Download
Metabolic overview of the marine Actinobacteria member MedAcidi-G2A. Reactions that have not been found but are likely to be present are shown in grey. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate. Download
Metabolic overview of the marine Actinobacteria member MedAcidi-G2B. Reactions/enzymes that have not been found but are likely to be present are shown in grey. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate. Download
The ethylmalonyl pathway in the genome of MedAcidi-G3. Enzymes that were found in the genome are shown in blue boxes, and those not found are colored in grey. Download
Metagenomic fragment recruitment across the Global Ocean Sampling (GOS) data set and a transect across the Southern Ocean. The two groups of marine Actinobacteria, the “Ca. Actinomarinales” (3 representatives), and the marine Acidimicrobiales (4 representatives) are shown. y axis indicates RPKG (reads recruited per kb per Gb of metagenome). Color key is at top right. Download
Metagenomic fragment recruitment along the depth profile of the Hawaii Ocean Time-series data set. The two groups of marine Actinobacteria, the “Ca. Actinomarinales” (3 representatives), and the marine Acidimicrobiales (4 representatives) are shown. y axis indicates depth (in meters), and x axis indicates RPKG (reads recruited per kb per Gb of metagenome). M. parvicella and Ilumatobacter sp. strain Y16-304 are non-marine representatives of Acidimicrobidae and are shown for reference. Prochlorococcus marinus NATL1A is a genuine deep chlorophyll maximum inhabitant and is shown as a reference for “Ca. Actinomarinales” genomes. Color key is at top right. Download
Relative abundances of 16S rRNA sequences belonging to marine Acidimicrobiales and “Ca. Actinomarinales” in metagenomic data sets. (a) Global Ocean Sampling data set. Open ocean data sets and coastal data sets are shown separately. (b) Southern Ocean (Antarctic) transect, an Arctic metagenome, and two metagenomes from the deep ocean: Marmara (1,000 m) and Puerto Rico Trench (4,000 m). Color keys are at top right. Download
ACKNOWLEDGMENTS
This work was supported by projects MICROGEN (Programa CONSOLIDER-INGENIO 2010 CSD2009-00006) from the Spanish Ministerio de Ciencia e Innovación, MEDIMAX BFPU2013-48007-P from the Spanish Ministerio de Economía y Competitividad, MaCuMBA 311975 of the European Commission FP7, ACOMP/2014/024, AORG 2014/032, and PROMETEO II/2014/012 from the Generalitat Valenciana.
R.G. and F.R.-V. conceived the study. All authors performed the sample collection and filtration. C.M.M. and R.G. analyzed the data. R.G. and F.R.-V. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Citation Mizuno CM, Rodriguez-Valera F, Ghai R. 2015. Genomes of planktonic Acidimicrobiales: widening horizons for marine Actinobacteria by metagenomics. mBio 6(1):e02083-14. doi:10.1128/mBio.02083-14.
REFERENCES
- 1.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghai R, McMahon KD, Rodriguez-Valera F. 2012. Breaking a paradigm: cosmopolitan and abundant freshwater Actinobacteria are low GC. Environ Microbiol Rep 4:29–35. doi: 10.1111/j.1758-2229.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- 3.Fuhrman JA, McCallum K, Davis AA. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol 59:1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappé MS, Kemp PF, Giovannoni SJ. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceangr 42:811–826. doi: 10.4319/lo.1997.42.5.0811. [DOI] [Google Scholar]
- 5.Rappé MS, Gordon DA, Vergin KL, Giovannoni SJ. 1999. Phylogeny of Actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol 22:106–112. doi: 10.1016/S0723-2020(99)80033-2. [DOI] [Google Scholar]
- 6.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. 2013. Metagenomics uncovers a new group of low GC and ultra-small marine actinobacteria. Sci Rep 3:2471. doi: 10.1038/srep02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris RM, Vergin KL, Cho J, Rappé MS, Carlson CA, Giovannoni SJ. 2005. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceangr 50:1687–1696. doi: 10.4319/lo.2005.50.5.1687. [DOI] [Google Scholar]
- 8.Morris RM, Frazar CD, Carlson CA. 2012. Basin-scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the south Atlantic. Environ Microbiol 14:1133–1144. doi: 10.1111/j.1462-2920.2011.02694.x. [DOI] [PubMed] [Google Scholar]
- 9.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A 110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen PR, Lauro FM. 2008. An assessment of actinobacterial diversity in the marine environment. Antonie Van Leeuwenhoek 94:51–62. doi: 10.1007/s10482-008-9239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stackebrandt E, Rainey FA, Ward-Rainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 12.Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, Giovannoni SJ. 2009. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J 3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- 13.Ghai R, Martin-Cuadrado A-B, Molto AG, Heredia IG, Cabrera R, Martin J, Verdú M, Deschamps P, Moreira D, López-García P, Mira A, Rodriguez-Valera F. 2010. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- 14.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. 2014. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol Ecol 23:6073–6090. doi: 10.1111/mec.12985. [DOI] [PubMed] [Google Scholar]
- 15.Estrada M, Marrasé C, Latasa M, Berdalet E, Delgado M, Riera T. 1993. Variability of deep chlorophyll maximum characteristics in the northwestern Mediterranean. Mar Ecol Prog Ser 92:289–300. doi: 10.3354/meps092289. [DOI] [Google Scholar]
- 16.Giovannoni SJ, Cameron Thrash J, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J 8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 18.Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R, Grossart H-P, Woyke T, Warnecke F. 2013. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J 7:137–147. doi: 10.1038/ismej.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. 2008. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol 10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 20.Sharma AK, Sommerfeld K, Bullerjahn GS, Matteson AR, Wilhelm SW, Jezbera J, Brandt U, Doolittle WF, Hahn MW. 2009. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J 3:726–737. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg HL. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci U S A 104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leboulanger C, Oriol L, Jupin H, Desolas-gros C. 1997. Diel variability of glycolate in the eastern tropical Atlantic Ocean. Deep Sea Res Part I Oceanogr Res Pap 44:2131–2139. [Google Scholar]
- 24.Howard EC, Henriksen JR, Buchan A, Reisch CR, Bürgmann H, Welsh R, Ye W, González JM, Mace K, Joye SB, Kiene RP, Whitman WB, Moran MA. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 25.Reisch CR, Moran MA, Whitman WB. 2008. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol 190:8018–8024. doi: 10.1128/JB.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otte ML, Wilson G, Morris JT, Moran BM. 2004. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J Exp Bot 55:1919–1925. doi: 10.1093/jxb/erh178. [DOI] [PubMed] [Google Scholar]
- 27.Stefels J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res 43:183–197. doi: 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- 28.Kettle AJ, Andreae MO. 2000. Flux of dimethylsulfide from the oceans: a comparison of updated data sets and flux models. J Geophys Res Atmosphere 105:26793–26808. doi: 10.1029/2000JD900252. [DOI] [Google Scholar]
- 29.Kiene RP, Linn LJ, González J, Moran MA, Bruton JA. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl Environ Microbiol 65:4549–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE, Landry ZC, Giovannoni SJ. 2011. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS One 6:e23973. doi: 10.1371/journal.pone.0023973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zafiriou OC, Andrews SS, Wang W. 2003. Concordant estimates of oceanic carbon monoxide source and sink processes in the Pacific yield a balanced global “blue-water” CO budget. Global Biogeochem Cycles 17. doi: 10.1029/2001GB001638. [DOI] [Google Scholar]
- 32.King GM, Weber CF. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5:107–118. doi: 10.1038/nrmicro1595. [DOI] [PubMed] [Google Scholar]
- 33.Martin-Cuadrado A-B, Ghai R, Gonzaga A, Rodriguez-Valera F. 2009. CO dehydrogenase genes found in metagenomic fosmid clones from the deep Mediterranean Sea. Appl Environ Microbiol 75:7436–7444. doi: 10.1128/AEM.01283-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP, Henriksen JR, King GM, Belas R, Fuqua C, Brinkac L, Lewis M, Johri S, Weaver B, Pai G, Eisen JA, Rahe E, Sheldon WM, Ye W, Miller TR. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 35.Tolli JD, Taylor CD. 2005. Biological CO oxidation in the Sargasso Sea and in Vineyard Sound, Massachusetts. Limnol Oceangr 50:1205–1212. doi: 10.4319/lo.2005.50.4.1205. [DOI] [Google Scholar]
- 36.Tolli JD, Sievert SM, Taylor CD. 2006. Unexpected diversity of bacteria capable of carbon monoxide oxidation in a coastal marine environment, and contribution of the Roseobacter-associated clade to total CO oxidation. Appl Environ Microbiol 72:1966–1973. doi: 10.1128/AEM.72.3.1966-1973.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunliffe M. 2011. Correlating carbon monoxide oxidation with cox genes in the abundant marine Roseobacter clade. ISME J 5:685–691. doi: 10.1038/ismej.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdlenbruch BN, Kelly DP, Murrell JC. 2001. Alkanesulfonate degradation by novel strains of Achromobacter xylosoxidans, Tsukamurella wratislaviensis and Rhodococcus sp., and evidence for an ethanesulfonate monooxygenase in A. xylosoxidans strain AE4. Arch Microbiol 176:406–414. doi: 10.1007/s002030100340. [DOI] [PubMed] [Google Scholar]
- 39.Kelly DP, Murrell JC. 1999. Microbial metabolism of methanesulfonic acid. Arch Microbiol 172:341–348. doi: 10.1007/s002030050770. [DOI] [PubMed] [Google Scholar]
- 40.Vairavamurthy A, Zhou W, Eglinton T, Manowitz B. 1994. Sulfonates: a novel class of organic sulfur compounds in marine sediments. Geochim Cosmochim Acta 58:4681–4687. doi: 10.1016/0016-7037(94)90200-3. [DOI] [Google Scholar]
- 41.Autry AR, Fitzgerald JW. 1990. Sulfonate-S—a major form of forest soil organic sulfur. Biol Fertil Soils 10:50–56. [Google Scholar]
- 42.Cook AM, Laue H, Junker F. 1998. Microbial desulfonation. FEMS Microbiol Rev 22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 43.Misset-Smits M, van Ophem PW, Sakuda S, Duine JA. 1997. Mycothiol, 1-O-(2′-[N-acetyl-l-cysteinyl]amido-2′-deoxy-alpha-d-glucopyranosyl)-d-myo-inositol, is the factor of NAD/factor-dependent formaldehyde dehydrogenase. FEBS Lett 409:221–222. doi: 10.1016/S0014-5793(97)00510-3. [DOI] [PubMed] [Google Scholar]
- 44.Rawat M, Av Gay Y. 2007. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol Rev 31:278–292. doi: 10.1111/j.1574-6976.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 45.Ebbs S. 2004. Biological degradation of cyanide compounds. Curr Opin Biotechnol 15:231–236. doi: 10.1016/j.copbio.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Yamada H, Kobayashi M. 1996. Nitrile hydratase and its application to industrial production of acrylamide. Biosci Biotechnol Biochem 60:1391–1400. doi: 10.1271/bbb.60.1391. [DOI] [PubMed] [Google Scholar]
- 47.Brandão PF, Bull AT. 2003. Nitrile hydrolysing activities of deep-sea and terrestrial mycolate actinomycetes. Antonie Van Leeuwenhoek 84:89–98. doi: 10.1023/A:1025409818275. [DOI] [PubMed] [Google Scholar]
- 48.Coleman ML, Chisholm SW. 2010. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci U. S. A. 107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 50.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quaiser A, Zivanovic Y, Moreira D, López-García P. 2011. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5:285–304. doi: 10.1038/ismej.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eloe EA, Fadrosh DW, Novotny M, Zeigler Allen L, Kim M, Lombardo MJ, Yee-Greenbaum J, Yooseph S, Allen EE, Lasken R, Williamson SJ, Bartlett DH. 2011. Going deeper: metagenome of a hadopelagic microbial community. PLoS One 6:e20388. doi: 10.1371/journal.pone.0020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson LR, Field C, Romanuk T, Ngugi D, Thompson LR, Field C, Romanuk T, Ngugi D, Siam R, El Dorry H, Stingl U. 2013. Patterns of ecological specialization among microbial populations in the Red Sea and diverse oligotrophic marine environments. Ecol Evol 3:1780–1797. doi: 10.1002/ece3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, Lauro FM, Cavicchioli R. 2013. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev 37:303–335. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- 55.Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay J-É, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaes. Proc Natl Acad Sci U S A 109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martín-Cuadrado AB, López-García P, Alba JC, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodríguez-Valera F. 2007. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng Y, Leung HC, Yiu S-M, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 58.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nawrocki EP. 2009. Structural RNA homology search and alignment using covariance models. Ph.D. thesis Washington University, Saint Louis, MO. [Google Scholar]
- 61.Huang Y, Gilna P, Li W. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 67.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 68.Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F. 2011. New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135. doi: 10.1038/srep00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 70.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology Open software suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 71.Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. [Google Scholar]
- 72.Lassmann T, Sonnhammer EL. 2005. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file containing important pathways and transporters found in the MedAcidi genomes. Download
16S rRNA classification of the Mediterranean deep chlorophyll maximum data sets. y axis indicates percentage of reads attributed to each taxon. Color key is at top right. Download
(a) Genome size versus GC content. (b) Median intergenic spacer size versus GC content. 1386 prokaryotic genomes are shown here. The marine actinobacterial genomes assembled in this work and another marine actinobacterial genome (“Ca. Actinomarina minuta”) are labeled in white boxes. Genomes of freshwater Actinobacteria are labeled. All other genomes are shown in grey. Download
Metabolic overview of the marine Actinobacteria member MedAcidi-G2A. Reactions that have not been found but are likely to be present are shown in grey. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate. Download
Metabolic overview of the marine Actinobacteria member MedAcidi-G2B. Reactions/enzymes that have not been found but are likely to be present are shown in grey. DMSP, dimethylsulfopropionate; MMPA, methylmercaptopropionate; THF, tetrahydrofolate. Download
The ethylmalonyl pathway in the genome of MedAcidi-G3. Enzymes that were found in the genome are shown in blue boxes, and those not found are colored in grey. Download
Metagenomic fragment recruitment across the Global Ocean Sampling (GOS) data set and a transect across the Southern Ocean. The two groups of marine Actinobacteria, the “Ca. Actinomarinales” (3 representatives), and the marine Acidimicrobiales (4 representatives) are shown. y axis indicates RPKG (reads recruited per kb per Gb of metagenome). Color key is at top right. Download
Metagenomic fragment recruitment along the depth profile of the Hawaii Ocean Time-series data set. The two groups of marine Actinobacteria, the “Ca. Actinomarinales” (3 representatives), and the marine Acidimicrobiales (4 representatives) are shown. y axis indicates depth (in meters), and x axis indicates RPKG (reads recruited per kb per Gb of metagenome). M. parvicella and Ilumatobacter sp. strain Y16-304 are non-marine representatives of Acidimicrobidae and are shown for reference. Prochlorococcus marinus NATL1A is a genuine deep chlorophyll maximum inhabitant and is shown as a reference for “Ca. Actinomarinales” genomes. Color key is at top right. Download
Relative abundances of 16S rRNA sequences belonging to marine Acidimicrobiales and “Ca. Actinomarinales” in metagenomic data sets. (a) Global Ocean Sampling data set. Open ocean data sets and coastal data sets are shown separately. (b) Southern Ocean (Antarctic) transect, an Arctic metagenome, and two metagenomes from the deep ocean: Marmara (1,000 m) and Puerto Rico Trench (4,000 m). Color keys are at top right. Download