ABSTRACT
Type III secretion systems (T3SS) translocate effector proteins into target cells in order to disrupt or modulate host cell signaling pathways and establish replicative niches. However, recognition of T3SS activity by cytosolic pattern recognition receptors (PRRs) of the nucleotide-binding domain leucine rich repeat (NLR) family, either through detection of translocated products or membrane disruption, induces assembly of multiprotein complexes known as inflammasomes. Macrophages infected with Yersinia pseudotuberculosis strains lacking all known effectors or lacking the translocation regulator YopK induce rapid activation of both the canonical NLRP3 and noncanonical caspase-11 inflammasomes. While this inflammasome activation requires a functional T3SS, the precise signal that triggers inflammasome activation in response to Yersinia T3SS activity remains unclear. Effectorless strains of Yersinia as well as ΔyopK strains translocate elevated levels of T3SS substrates into infected cells. To dissect the contribution of pore formation and translocation to inflammasome activation, we took advantage of variants of YopD and LcrH that separate these functions of the T3SS. Notably, YopD variants that abrogated translocation but not pore-forming activity failed to induce inflammasome activation. Furthermore, analysis of individual infected cells revealed that inflammasome activation at the single-cell level correlated with translocated levels of YopB and YopD themselves. Intriguingly, LcrH mutants that are fully competent for effector translocation but produce and translocate lower levels of YopB and YopD also fail to trigger inflammasome activation. Our findings therefore suggest that hypertranslocation of YopD and YopB is linked to inflammasome activation in response to the Yersinia T3SS.
IMPORTANCE
The innate immune response is critical to effective clearance of pathogens. Recognition of conserved virulence structures and activities by innate immune receptors such as NLRs constitute one of the first steps in mounting the innate immune response. However, pathogens such as Yersinia actively evade or subvert components of host defense, such as inflammasomes. The T3SS-secreted protein YopK is an essential virulence factor that limits translocation of other Yops, thereby limiting T3SS-induced inflammasome activation. However, what triggers inflammasome activation in cells infected by YopK-deficient Yersinia is not clear. Our findings indicate that hypertranslocation of pore complex proteins promotes inflammasome activation and that YopK prevents inflammasome activation by the T3SS by limiting translocation of YopD and YopB themselves.
INTRODUCTION
The innate immune system plays a crucial role in host defense against pathogens. Pattern recognition receptors (PRRs) recognize conserved microbial structures expressed by both pathogenic and nonpathogenic bacteria, commonly termed pathogen-associated molecular patterns (PAMPs) (1, 2). Cytosolic receptors detect both virulence activities as well as bacterial molecules within the cytosol, which generally occurs as a direct consequence of pathogen virulence machinery activities, provide an additional layer of sensing of pathogenic bacteria (3).
Virulence activities, such as pore formation by toxins or secretion systems and delivery of bacterial products into the cytosol of target cells, trigger the activation of a cytosolic immune surveillance pathway that culminates in assembly of multiprotein complexes termed inflammasomes (4, 5). Inflammasome assembly typically requires a nucleotide binding domain leucine-rich repeat protein (NLR), which responds to particular pathogen-associated stimuli and induces oligomerization of apoptosis-associated speck-like protein (ASC) and the cysteine protease caspase-1 (5, 6). Inflammasome activation results in pyroptosis, a programmed cell death characterized by caspase-1 processing and the release of interleukin-1 (IL-1) family cytokines IL-1α, IL-1β, and IL-18 (7). Additionally, a recently described noncanonical inflammasome also engages caspase-11 to induce cytotoxicity and IL-1α and contributes to maximal IL-1β processing and secretion by caspase-1 (8–11).
Different inflammasomes respond to distinct signals. The NLRP3 inflammasome recognizes endogenous indicators of tissue damage and tissue stress, such as extracellular ATP and sodium urate crystals, as well as microbial products, such as viral and bacterial pore-forming proteins (5, 12–15), whereas NLRC4 in conjunction with the NLR family apoptosis inhibitory protein (NAIP) detects the presence of cytosolic flagellin and structurally similar products of the type III secretion system (T3SS) itself, such as PrgJ (16–22). Recognition of pathogen-specific signals by inflammasomes promotes antimicrobial defense either via the immunological effects of IL-1 family cytokines (23, 24) or through induction of pyroptosis (25).
The pathogenic yersiniae Yersinia pseudotuberculosis, Yersinia enterocolitica, and Yersinia pestis express a conserved T3SS that injects Yersinia outer proteins (Yops) into target cells (26). Once inside the cell, the Yops disrupt a number of cellular processes, including actin cytoskeleton rearrangement, protein phosphorylation, and NF-κB and mitogen-activated protein kinase (MAPK) signaling (27, 28). Two Yops, YopK and YopM, modulate inflammasome activation, thereby interfering with host innate immune defense and promoting Yersinia infection (29–31).
Once translocated into the cell, YopK interacts with integral membrane components of the translocation complex, YopD and YopB, to prevent hypertranslocation of other Yops (29, 32, 33). YopK may therefore limit translocation of other Yops by physically blocking or modulating the size of the translocon pore. YopK may also limit translocation by interacting with the host scaffolding protein, RACK1, or potentially other cellular factors (33, 34). In addition to a hypertranslocation of effectors, yopK mutants exhibit inflammasome-dependent attenuation in animal infections, indicating that inappropriate translocation of other Yops or additional factors triggers inflammasome activation, and leads to bacterial clearance in vivo (29). Interestingly, NLRP3, not NLRC4 is the primary driver of inflammasome activation in response to the Yersinia T3SS, in contrast to several other bacterial pathogens, such as Salmonella, likely due to inverse regulation of flagellin and T3SS expression in Yersinia (17, 29, 35).
The YopB and YopD proteins are integral membrane proteins that form the translocon pore through which other Yops are delivered into the host cell. Thus, complete deletion of either YopB or YopD results in a nonfunctional T3SS (36–39). However, specific in-frame deletions in YopD result in loss of translocation ability while maintaining pore-forming activity (40). Conversely, mutations in the YopB and YopD chaperone, LcrH, impact pore-forming capacity due to reduced steady-state expression levels of YopB and YopD, yet maintain wild-type (WT) levels of translocation, implying that pore formation and translocation are separable biological features of the T3SS (41).
Intriguingly, in addition to forming the translocation pore through which the classical effector Yops are injected into host cells, YopB and YopD themselves are also translocated into the host cell, albeit at relatively low levels in WT Yersinia (34, 42). The functional consequences of this translocation of components of the T3SS pore complex are not well understood but appear to be a general feature of the T3SS as Salmonella SipC and SipB pore complex proteins are also translocated (43).
Critically, the mechanism by which the Yersinia T3SS induces NLRP3 inflammasome activation remains unclear. As both pore-forming toxins and translocated bacterial products can cause inflammasome activation (13, 44–46), inflammasome activation could potentially be the result of pore formation generated by the insertion of the pore complex or due to translocation of a molecule by the T3SS.
Here we employed the YopD and LcrH mutants that separate pore formation and translocation activities and find that pore formation is not sufficient to activate the inflammasome, consistent with recent observations (47). Strikingly, we also note that while translocation competence is required for inflammasome activation, it is not sufficient, as LcrH mutants with a functional translocon that display wild-type (WT) levels of effector Yop translocation but do not translocate detectable levels of YopB or YopD fail to trigger inflammasome activation. Moreover, hypertranslocation of YopB and YopD correlated with an increased likelihood of caspase-1 activation at the single-cell level. These findings suggest that the translocation of YopB and/or YopD may mediate inflammasome activation in response to the Yersinia T3SS and that YopK functions to prevent excessive translocation of these proteins.
RESULTS
Translocation is required for inflammasome activation in response to Yersinia T3SS.
Translocation of currently undefined T3SS client proteins may be responsible for NLRP3 inflammasome activation in macrophages by the Yersinia T3SS (29, 47). Alternatively, alteration in the stoichiometry, conformation, or pore-forming properties of the translocon components YopB and YopD could be responsible. In order to dissect the relative contributions of translocation and pore formation of the T3SS to inflammasome activation, we generated in-frame yopD deletion mutations in a Y. pseudotuberculosis strain lacking Yops E, J, and K (ΔyopEJK), which induces robust T3SS-dependent NLRP3 inflammasome activation in murine macrophages. These in-frame deletion mutations in yopD separate the pore-forming and translocation activities of YopD in wild-type Yersinia (40).
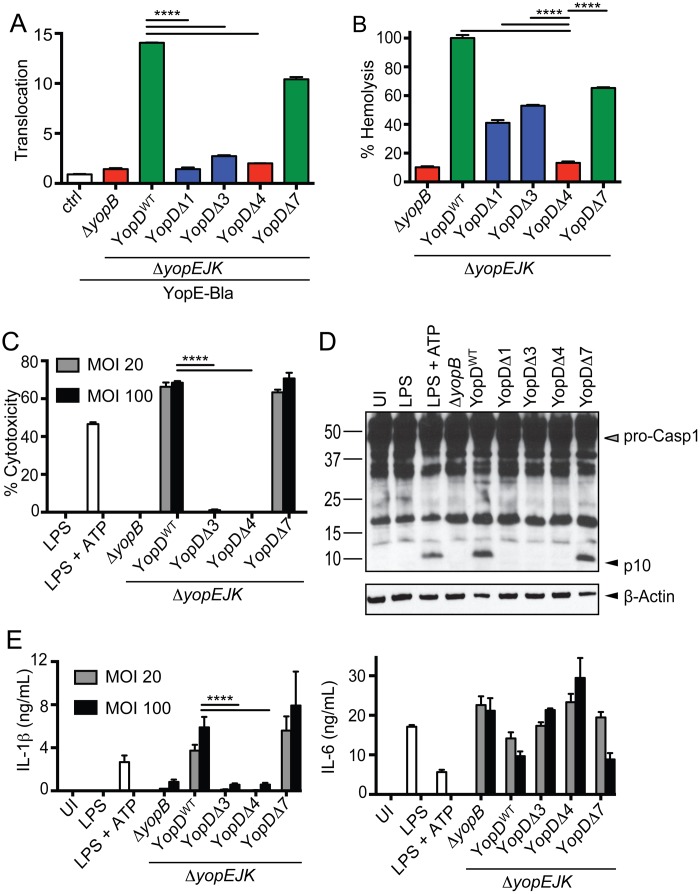
To accurately determine bacterially induced pore formation and differences in translocation, we performed translocation and pore formation assays with HeLa cells and sheep red blood cells (RBCs) that do not undergo inflammasome-mediated cell death in response to infection. We observed that deletion of amino acids (aa) 4 to 20 or 53 to 68 (termed YopDΔ1 and YopDΔ3, respectively) completely abrogated translocation of a YopE-β-lactamase reporter construct similarly to what is seen with ΔyopB or YopDΔ4 (aa 73 to 90), which have been previously demonstrated to generate nonfunctional T3SS. In contrast, YopDΔ7 (deletion of aa 150 to 170) maintained translocation, although at a reduced level compared to wild-type YopD (YopDWT) (Fig. 1A). Pore formation as measured by hemolysis in sheep red blood cells was significantly reduced in YopDΔ1, YopDΔ3, and YopDΔ7 in comparison to YopDWT. However, these variants still caused significant levels of pore formation relative to the ΔyopB and YopDΔ4 variants, which eliminated the hemolytic capacity of the T3SS (Fig. 1B). To address potential threshold effects resulting from the reduced pore-forming activity of YopDΔ1 and YopDΔ3, we performed our macrophage infections at both a multiplicity of infection (MOI) of 20 and an MOI of 100. Critically, at both MOIs, YopDΔ1 and YopDΔ3 failed to induce IL-1β secretion, lactate dehydrogenase (LDH) release, or cleavage of caspase-1, suggesting that eliminating translocation abrogated the ability of the Yersinia T3SS to induce inflammasome activation (Fig. 1C to E). Differential secretion of IL-1β in response to different YopD variants was not due to effects of YopD on Toll-like receptor (TLR) signaling per se, as secretion of IL-6 was unaffected in response to infection with different YopD variants (Fig. 1E). Interestingly, while the YopDΔ1, -Δ3, and -Δ7 variants were all capable of inducing hemolysis in RBCs, they did not induce propidium iodide (PI) uptake in HeLa cells, another measure of “pore formation,” implying that this pore formation, at least in nucleated cells, is distinct from the T3SS pore itself (see Fig. S1 in the supplemental material). Together, these data suggest that the pore-forming capacity of the Yersinia T3SS is insufficient to induce inflammasome activation: rather, translocation of an unknown molecule induces NLRP3 inflammasome activation.
FIG 1 .
Translocation is required for inflammasome activation by Yersinia T3SS. (A) HeLa cells were infected with indicated strains expressing a YopE–β-lactamase fusion protein (YopE-Bla) or ΔyopEJK mutant lacking the β-lactamase construct (ctrl). Cells were loaded with CCF4-AM dye, and the ratio of blue to green signal (translocation) was calculated as described in Materials and Methods. Results are representative of 3 independent experiments. (B) Sheep red blood cells were infected with indicated YopD deletion mutants or the ΔyopB and ΔyopEJK controls. Supernatants were assayed for release of hemoglobin as described in Materials and Methods. The graph is representative of two to four independent experiments with 6 independent replicates per sample. (C) BMDMs were infected with indicated YopD deletion mutants (ΔyopB or ΔyopEJK) or treated with LPS or LPS plus ATP, and cytotoxicity was determined by LDH release. The graph is of a representative experiment from one of five independent experiments (MOI, 20) or 2 independent experiments (MOI, 100) with 3 replicates per sample. (D) BMDMs were infected with the indicated bacterial strains or treated with LPS or LPS plus ATP. Cell lysates were assayed for p10 antibody label, processed caspase-1 (pro-Casp1), and actin as indicated. (E) Supernatants from BMDMs infected with indicated bacterial strains were assayed for levels of secreted IL-1β and IL-6 by ELISA as described in Materials and Methods. UI, uninfected. ****, P < 0.0001.
Deletion of yopK results in hypertranslocation of YopD and YopB.
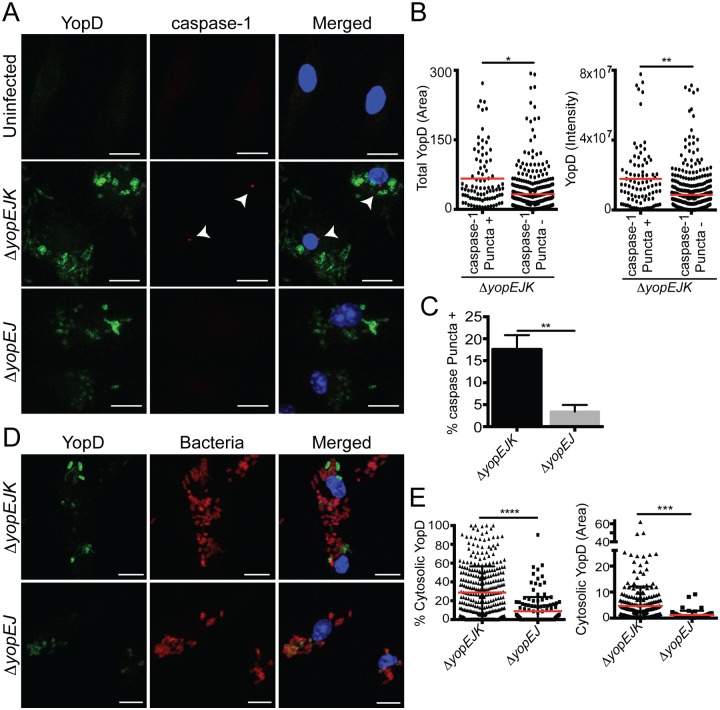
Our data and previously published work (29, 32, 34, 40, 48) suggest that YopK normally functions to regulate translocation by the T3SS and that the hypertranslocation taking place in the absence of YopK triggers inflammasome activation. The integral translocon components YopB and YopD are also translocated during infection, raising the possibility that excess translocation of YopD or YopB themselves might be responsible for T3SS-induced inflammasome activation (34, 42). To determine whether YopB and YopD translocation might be correlated with inflammasome activation, we performed confocal microscopy on Yersinia-infected macrophages and analyzed both the translocation of T3SS pore complex proteins and formation of caspase-1 puncta. Macrophages were treated with N-benzyloxycarbonyl–Tyr-Val-Ala-Asp(OMe)-fluoromethylketone (Z-YVAD-FMK) to enhance visualization of puncta by preventing death of the macrophages or escape of caspase-1 from the puncta. Formation of caspase-1 puncta indicates oligomerization of caspase-1 and is an indicator of inflammasome formation (49). Confocal fluorescence microscopy analysis of YopD translocation revealed a significant correlation between the degree of YopD translocation, defined by the intensity and area of anti-YopD staining, and the formation of caspase-1 puncta (Fig. 2). A similar correlation was found for YopB (see Fig. S2 in the supplemental material). Infection with the ΔyopEJK mutant resulted in a significantly elevated percentage of punctum-positive cells compared to infection with the ΔyopEJ mutant, consistent with previous studies that showed YopK prevents inflammasome activation (29). Interestingly, YopB and YopD accumulated in the cytosol in large punctate or vesicular structures that were devoid of bacteria (Fig. 2D and E). Both the extent (area) and percentage of cytosolic YopD were significantly elevated in the absence of YopK. We did not observe colocalization of either YopD or YopB with the caspase-1 puncta themselves, implying that any role of YopB or YopD in inflammasome activation was not the result of direct recognition by NLRP3.
FIG 2 .
Increased YopD translocation correlates with activation of caspase-1. (A) Combined z-stacks of confocal microscopy images of BMDMs that were pretreated with Z-YVAD-FMK and left uninfected or infected with the ΔyopEJK or ΔyopEJ mutant for 2 h. Cells were stained for YopD (green), caspase-1 (red), and DNA (blue). Arrows denote caspase-1 puncta. (B) Total area of YopD (μm2) in caspase-1 punctum-positive cells versus caspase-1 punctum-negative cells in ΔyopEJK mutant infection. Shown is the total intensity of YopD staining for caspase-1 punctum-positive cells versus caspase-1 punctum-negative cells infected with the ΔyopEJK mutant. Each point represents a single cell. (C) Percentage of YopD-staining cells positive for caspase-1 puncta. (D) Combined z-stacks of confocal microscopy images of cells that were uninfected or infected with the ΔyopEJK or ΔyopEJ mutant for 2 h. Cells were stained with antibodies against YopD (green), Yersinia (red), and Hoechst to visualize DNA (blue). Amount of cytosolic YopD was determined as described in Materials and Methods. (E) Percentage and total area of cytosolic YopD (µm2) obtained from analyzing microscopy images as in panel D. (B, C, and E) Each point represents a single cell; data are pooled from 3 independent experiments. Red lines represent means of data. Scale bars are 10 µm. (B and C) A total of 477 individual cells were analyzed. (E) A total of 321 individual cells were analyzed. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Translocation is not sufficient for T3SS-induced inflammasome activation.
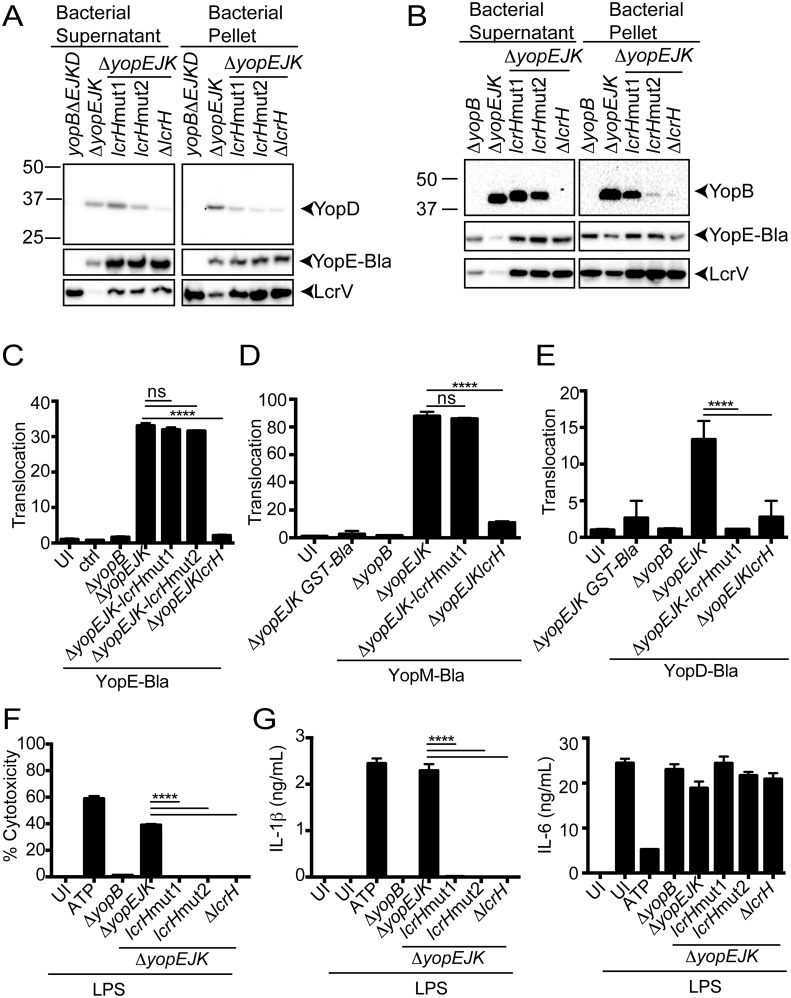
To further test the possibility that YopD or YopB translocation might be responsible for inflammasome activation, we generated two separate LcrH mutations, designated lcrHmut1 (encoding LcrHY40C I101M K102R) and lcrHmut2 (encoding LcrHQ46R F72S Q81R H162R), as well as the lcrH null mutant (ΔlcrH) in the ΔyopEJK strain background. LcrH is a chaperone for both YopB and YopD, and specific amino acid mutations in LcrH lead to reduced steady-state levels of stable YopB and YopD without affecting the translocation of other Yops (41). Consistent with established work that these amino acid changes in LcrH affect the T3SS, both lcrHmut1 and lcrHmut2 expressed and secreted lower levels of YopB and YopD (Fig. 3A and B) but did not affect the translocation of a YopE-β-lactamase reporter (Fig. 3D). Furthermore, lcrHmut1 also had no effect on translocation of a YopM-β-lactamase reporter, demonstrating that these LcrH mutants do not impact translocation of secreted effector Yops. Importantly, another structural component of the T3SS, LcrV, as well as YopE–β-lactamase, was expressed to an equal extent in the bacteria themselves but exhibited a hypersecretory phenotype in the lcrH mutant strain backgrounds, consistent with previous observations that LcrH mutations or translocon mutations that reduce expression of YopD or YopB result in hypersecretion (50–53) (Fig. 3A and B). Critically, the lcrHmut1 and ΔlcrH strains were unable to translocate YopD–β-lactamase (Fig. 3E). Despite translocating effector Yops in a manner that was indistinguishable from wild-type bacteria, both lcrHmut1 and lcrHmut2 strains failed to trigger macrophage cytotoxicity and IL-1β secretion (Fig. 3E and F). Collectively these data suggest that hypertranslocation of LcrH-dependent pore-complex proteins is required for inflammasome activation.
FIG 3 .
Minimal translocon is insufficient for inflammasome activation by Yersinia T3SS. (A) Trichloroacetic acid (TCA)-precipitated bacterial supernatants and bacterial pellet from inducing cultures were probed for YopD, YopE–β-lactamase (YopE-Bla), and LcrV. The ΔyopEJKBD strain lacking the YopE-Bla construct was used as a control. (B) YopB and LcrV. (C) HeLa cells were infected with the lcrH wild-type, point mutant, or deletion strains, as indicated, or ΔyopB control strains expressing YopE-Bla. The ΔyopEJK mutant lacking the YopE-Bla plasmid was used as a negative control (ctrl). Translocation was determined as described in Materials and Methods. The graph is of a representative experiment from one of three independent experiments with 3 replicates per treatment per experiment. UI, uninfected. (D and E) HeLa cells were infected with the lcrH wild-type, point mutant, or deletion strains, as indicated, or ΔyopB control strains expressing YopM-Bla (D) and YopD-Bla (E). Translocation was determined as described in Materials and Methods. The graph is a representative experiment from one of three independent experiments with 3 replicates per treatment per experiment. (F) The percentage of cytotoxicity was determined by assaying LDH in supernatants following infection of BMDMs with the indicated bacterial strains, as in panel C, or treatment with LPS or LPS plus ATP. (G) ELISA for IL-1β (left) or IL-6 (right) from LPS-primed BMDMs infected with the indicated bacterial mutant (ΔyopB or ΔyopEJK) strains or treated with ATP or the medium control alone. Shown is a representative graph from one of three independent experiments with 3 replicates per treatment. *, P < 0.05; ****, P < 0.0001.
Hypertranslocation of YopD is necessary for formation of caspase-1 puncta.
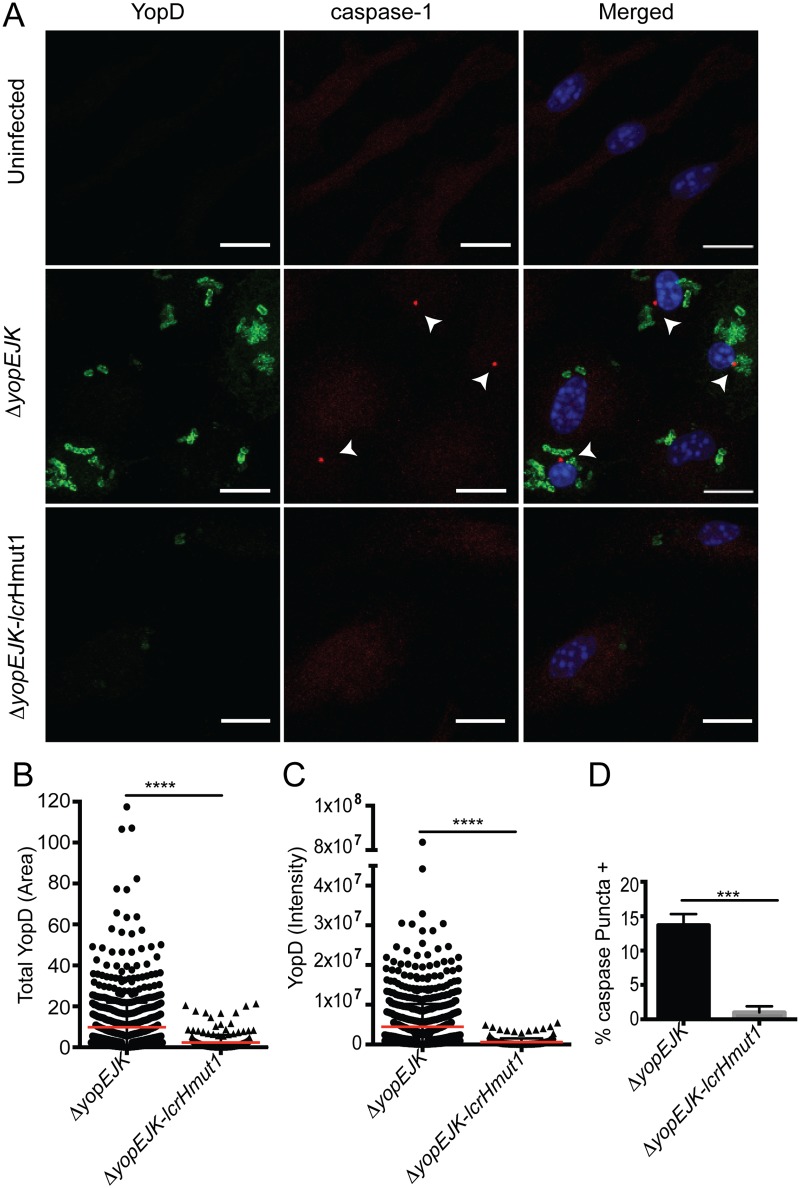
To test the contribution of translocated YopD and YopB to inflammasome activation, we examined inflammasome activation in the context of lcrHmut1 as the lcrHmut1 mutant had reduced steady-state levels of YopB and YopD but the reduction was not as great as in lcrHmut2 (Fig. 3A). Notably, confocal microscopy analysis of individual Yersinia-infected cells demonstrated that lcrHmut1 Yersinia strains translocated significantly lower levels of YopD than wild-type Yersinia, as quantified by intensity and area of antibody staining, in accordance with our findings using the YopD–β-lactamase reporter (Fig. 4A to C). Critically, this reduced translocation of YopD was linked to loss of inflammasome activation, as a significantly higher percentage of ΔyopEJK Yersinia-infected cells exhibited evidence of caspase-1 puncta in comparison with lcrHmut1 mutant-infected cells (Fig. 4D). Translocation of YopB was similarly reduced in lcrHmut1 mutant-infected cells, and no caspase-1 puncta were observed (see Fig. S2 in the supplemental material). Altogether, our data support a model in which excess translocation of YopD and/or YopB triggers NLRP3 inflammasome activation. Although an additional T3SS-translocated product that is inappropriately translocated in the absence of YopK may also lead to NLRP3 inflammasome activation, the lack of inflammasome activation by specific LcrH mutants that limit translocation of Yops D and B without impacting translocation of other T3SS substrates indicates that YopD and/or B plays a critical role in triggering this response.
FIG 4 .
Hypertranslocation of pore complex proteins is necessary for inflammasome activation. (A) Combined z-stacks of confocal microscopy images of Z-YVAD-FMK-pretreated BMDMs that were left uninfected or infected with the ΔyopEJK or ΔyopEJ mutant for 2 h. Cells were stained for YopD (green), caspase-1 (red), and DNA (blue). White arrowheads denote active caspase-1 puncta. (B) Total area of YopD staining (square µm) in cells infected with the ΔyopEJK (circles) or ΔyopEJK-lcrHmut1 (triangles) mutant. (C) Total intensity of YopD staining in cells infected with the ΔyopEJK (circles) or ΔyopEJK-lcrHmut1 (triangles) mutant. Each point represents a single cell. A total of 1,242 individual cells were analyzed. All data were pooled from 3 independent experiments. Scale bars are 10 µm. (D) Percentage of cells with YopD staining that were positive for caspase-1 puncta. ***, P < 0.0005; ****, P < 0.0001.
DISCUSSION
Inflammasome activation plays a key role in inflammatory responses during infection or disruption of tissue homeostasis, and distinct NLR proteins respond to different inflammatory triggers, such as the presence of microbial products within the cytosol of target cells. The NLRP3 inflammasome is triggered by structurally diverse microbial and stress-associated signals (13, 14, 24, 44, 54–60), and previous studies demonstrated that the Yersinia type III secretion system can trigger NLRP3 inflammasome activation in the absence of all known secreted effectors (29). Several studies have demonstrated that in the absence of YopK, a secreted Yersinia effector protein that negatively regulates the translocation activity of the T3SS, NLRP3 inflammasome activation is significantly enhanced (29, 31, 47). However, the mechanism by which the NLRP3 inflammasome is triggered by the Yersinia T3SS is unknown.
Interactions between the Yersinia virulence machinery and inflammasome activation involve an intricate interplay between Yersinia effector proteins, Toll-like receptor (TLR) signaling, and innate immune pathways. In the context of NF-κB and mitogen-activated protein kinase (MAPK) blockade by YopJ, cells such as naive macrophages trigger a distinct form of caspase-1 activation that engages the cell extrinsic death pathway and promotes anti-Yersinia immune defense independently of known inflammasome components (61, 62). Interestingly, lipopolysaccharide (LPS)-primed macrophages do not undergo YopJ-induced caspase-1 activation, possibly as a result of inhibition of caspase-1 activation by YopM (30, 31). However, in the absence of all known effector Yops or in the absence of Yops J and K alone, the Yersinia T3SS triggers a robust NLRP3 inflammasome activation that involves both canonical and noncanonical inflammasomes in both LPS-primed and unprimed macrophages (23, 29, 63, 64). The mechanism by which the Yersinia T3SS induces this NLRP3 inflammasome activation in the absence of all known secreted effectors remains unclear.
The various activities of the T3SS have been extensively probed by taking advantage of bacterial genetic tools to generate mutants deficient in key components of the translocon and associated proteins (40, 41). In particular, in-frame deletions in the integral translocon component YopD make it possible to separate the pore-forming ability of the translocon from the capacity of the T3SS to translocate effector proteins (40). Specifically, deletion of amino acids 4 to 20 or 53 to 68 in YopD compromises the ability of the T3SS to translocate effectors but has a minimal impact on the ability of the T3SS to induce hemolysis in sheep RBCs, one measure of pore-forming ability (40) (Fig. 1A). Interestingly, we found that these YopD deletion mutants failed to induce inflammasome activation, despite being able to lyse RBCs, consistent with recent findings by Kwuan et al., that pore formation is insufficient to induce inflammasome activation (47). Thus, some structural feature of the translocation process and/or a particular translocated molecule(s) is the cue for inflammasome activation.
Indeed, we found that translocation is necessary but insufficient for NLRP3 inflammasome activation, as mutations in the T3SS chaperone gene lcrH that do not affect YopE or YopM translocation, but are not capable of forming pores in RBCs, are also incapable of triggering inflammasome activation. Critically, our data show that LcrH mutant Yersinia that produce a functional translocon in the presence of limited YopB and YopD proteins, have a specific defect in translocation of YopD and YopB. Single-cell-based analyses of YopD and YopB translocation revealed a significant correlation between the extent of translocation of these proteins and the formation of an active inflammasome complex.
Taken together, these studies suggest that hypertranslocation of YopD or YopB in the absence of YopK is responsible for NLRP3 inflammasome activation. An alternative possibility is that structural differences in the translocon resulting from mutation of LcrH or YopD alters the membrane-perturbing properties of the translocon. While the YopDΔ1, -Δ3, and -Δ7 mutants are all capable of inducing RBC lysis, we did not detect measurable PI uptake in HeLa cells infected with these mutants (see Fig. S1 in the supplemental material). These data suggest that membrane perturbation or lysis in different cell types involves distinct features of the translocon. Indeed, neither of the LcrH mutants used in this study induces RBC lysis, despite being fully competent for translocation (41) (Fig. 3). These data suggest that pore formation measured by leakage of dyes across eukaryotic cell membranes during Yersinia infection detects the presence of a host-cell-derived pore that is assembled or activated in response to the activity of the T3SS. Whether this pore is itself dependent on caspase-1 activation (65) or is responsible for mediating caspase-1 activation, or whether this system constitutes a self-amplifying positive-feedback loop, remains to be determined.
How translocation of YopD or YopB is linked to NLRP3 inflammasome activation is currently unclear. YopD and YopB are both amphipathic and membrane active, raising the possibility that at elevated concentrations in the target cell, insertion of one or both of these proteins into membranes of cellular organelles may induce inflammasome activation. Disruption of lysosomal membranes or Golgi membranes has been linked to NLRP3 inflammasome activation (12, 66, 67), raising the possibility that insertion of YopD or YopB into these membranes triggers NLRP3. We observed large aggregates or superstructures of YopD and YopB in cells infected by YopK-deficient Yersinia (Fig. 2 and 4; see Fig. S2 in the supplemental material). These structures were not simply the presence of YopD or YopB staining around the Yersinia, as a significant percentage of these structures were not associated with intact bacteria (Fig. 2D–E). Protein aggregates or crystals are known to induce NLRP3 inflammasome activation; large aggregates of YopB or YopD could therefore lead to NLRP3 inflammasome activation. Future studies will dissect the molecular mechanism for NLRP3 activation by translocated YopD and/or YopB.
It is possible that YopD and YopB translocation serves as a surrogate for elevated translocation of an unknown factor or protein that is more directly responsible for T3SS-induced inflammasome activation. While it remains possible that another protein or bacterial molecule is translocated by the Yersinia T3SS and induces NLRP3 activation, our data suggest that YopD and/or YopB translocation into the cytosol is responsible and that YopK prevents the excessive translocation of YopD and YopB into target cells in order to avoid triggering this innate inflammatory response. Interestingly, initial studies of inflammasome activation by Salmonella suggested that microinjected SipB, the Salmonella homolog of YopB, induced caspase-1 activation (68). Thus, activation of the NLRP3 inflammasome by the bacterial T3SS machinery may be a broadly conserved innate response to the cytosolic presence of T3SS pore complex proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Yersinia strains are described in Table S1 in the supplemental material. Yersiniae were grown overnight with aeration in 2× yeast extract-tryptone (YT) broth at 26°C. The bacteria were diluted into fresh 2× YT containing 20 mM sodium oxalate and 20 mM MgCl2. Bacteria were grown with aeration for 1 h at 26°C followed by 2 h at 37°C prior to infection.
Hemolysis assay.
Pore formation was determined by hemolysis assay, as previously described with minor modifications (40). Briefly: sheep erythrocytes (RBCs) were infected with T3SS-induced Yersinia at an MOI of 0.05 to 0.1 for 3 h at 37°C. RBCs were then resuspended in 150 µl of cold PBS and spun down at 4°C for 15 min. One hundred microliters of supernatant was transferred to a flat-bottom plate and read for absorbance at 545 nm. The percentage of hemolysis was normalized to the parent strain.
PI uptake assay.
HeLa cells were seeded at 1 × 104/well overnight in a 12-well chamber slide. Cells were loaded for 45 min with CellTracker Green (1:2,000 [Life Technologies]) and then washed with PBS. PI was added to the medium (1:200), and cells were infected at an MOI of 100. Gentamicin was added 2 h postinfection. Cells were imaged 4 h postinfection using Leica DMI4000 with a Yokagawa CSU-X1 spinning disk confocal attachment (10× objective). Images were then analyzed using Metamorph 7.6 to quantify PI-positive cells.
Translocation assay.
HeLa cells were plated in a clear-bottom, black 96-well tissue culture plate (Greiner Bio One) overnight. Cells were infected at a MOI of 5 with Yersinia expressing β-lactamase fused to YopE (69), YopM, or YopD (34) for 2 h. Gentamicin was added. CCF4-AM was loaded into cells using LiveBLAzer-FRET B/G Loading kit (Life Technologies) for 1.5 to 2 h. The fluorescence was read using BioTek Synergy plate reader. The response ratio was then calculated using the formula (blue/green ratio)/(average negative blue/green ratio).
Cell death assay.
Cytotoxicity was assayed using a lactate dehydrogenase (LDH) release assay kit (Clontech). In brief, C57BL/6 mouse bone marrow-derived macrophages (BMDMs) were differentiated from bone marrow with 30% L929 supernatant medium for 7 days before being plated. They were then infected with an MOI of 10, 20, or 100 for 4 h. In some cases, cells were primed for 3 to 4 h with 500 ng/ml of LPS. ATP (2.5 mM) was added to primed cells as a control for NLRP3 inflammasome activation. After 1 h, gentamicin was added. Four hours postinfection, the plate was spun down and assayed for LDH release.
Cytokine production.
Cells were primed for 3 to 4 h with 500 ng/ml of LPS. The cells were then infected as in the cytotoxicity assay. The supernatants and recombinant cytokine standards (R&D Systems) were collected and added to enzyme-linked immunosorbent assay (ELISA) plates that had been coated overnight with purified IL-1β or IL-6 purified antibodies (eBioscience). The cytokines were then detected with the corresponding biotin antibodies (eBioscience) for 1 h at room temperature. Streptavidin-horseradish peroxidase (HRP) (Fisher Scientific) was then added. The plates were developed with citric acid buffer with O-phenylenediamine (Sigma Aldrich). Three molar sulfuric acid was used to stop the reaction. Absorbance was read at 490 nm.
Western blotting.
Western blots were performed as described previously (3). In brief, cell lysates were run on a 4% to 12% gradient gel (Life Technologies) and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blotted with the primary antibodies rabbit anti-mouse caspase-1 p10 antibody (SC-514 [Santa Cruz]) and mouse anti-actin (Sigma) or anti-YopD, anti-YopB (30, 70), anti-β-lactamase (QED Bioscience, Inc.), and anti-LcrV (a kind gift from Matthew Nilles, University of North Dakota) antibodies. The secondary antibodies were HRP-conjugated goat anti-rabbit (Jackson ImmunoResearch) or horse anti-mouse (Cell Signaling). Blots were developed with the Pierce ECL enhanced chemiluminescence Western blotting substrate (Fisher Scientific).
Immunofluorescence staining.
BMDMs were plated at 2 × 105/well on glass coverslips in 24-well plates overnight. Two hours prior to infection, cells were treated with Z-YVAD-FMK (R&D Systems) to a final concentration of 20 µM. BMDMs were then infected with an MOI of 5 for 2 h. Cells were fixed in 4% paraformaldehyde prepared from 16% paraformaldehyde (Electron Microscopy Sciences) for 20 min at room temperature and then washed twice in PBS. Cells were permeabilized for 10 min at room temperature in 0.2% Triton X-100 (Sigma) and washed once in PBS. Cells were blocked for 1 h at room temperature in 10% bovine serum albumin (BSA)–PBS. Coverslips were then inverted onto primary antibody cocktails on Parafilm and incubated for 1 h at 37°C. Mouse monoclonal antibodies against YopD (1:500) and YopB (1:500) and rabbit polyclonal antibody against the total Yersinia cell have been described previously (30, 70, 71). Rabbit anti-caspase-1 p10 antibody (1:100) was from Santa Cruz. Coverslips were washed and stained with goat anti-mouse–Alexa 488 (Life Technologies) and goat anti-rabbit–Alexa 568 (Life Technologies). Coverslips were washed and stained with a cocktail of Hoechst (1:1,000 [Life Technologies]) and phalloidin-Alexa 647 (1:40 [Life Technologies]). Coverslips were incubated for 1 h at 37°C and then washed 4 times. Coverslips were mounted on glass slides with Fluoromount-G (Southern Biotech). Cells were imaged with a Leica SP5 inverted confocal microscope and analyzed using Metamorph 7.6.
Metamorph analysis.
Analysis of translocation in single cells was performed by first compressing all individual z-plane stacks into a single “maximum projection.” This enables analysis of the total area in the cell occupied by translocated protein. The final image was segmented into individual cells based on analysis of nuclear and actin staining. Each defined cell was converted into a region and transferred onto images of YopD and caspase-1 staining. Thresholds for positive and negative signals were set based on the presence of no primary antibody, uninfected, and ΔyopB- or ΔyopD mutant-infected control samples. The images were then analyzed for the intensity and area of staining in the regions. To quantify the amount of YopD not associated with bacteria, the area defined by YopD and/or bacterial staining was separated into whether pixels in that specific region were positive or negative for staining, and areas of YopD and bacterial overlap were determined. Areas of the cell determined to contain both YopD and bacterial staining were subtracted from total YopD staining. The degree of YopD staining was determined in pre- and postsubtraction images, and the percentage of bacterium-free YopD was calculated on a per cell basis by the area occupied by YopD postsubtraction by the area occupied by YopD before subtraction.
Statistical analysis.
Statistical analysis was performed using Prism 6 (GraphPad Software, Inc.). One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used for the analysis of data requiring multiple comparisons. Otherwise, unpaired Student’s t tests were used for the analysis with Welch’s correction in cases of unequal standard deviation.
SUPPLEMENTAL MATERIAL
In-frame YopD deletions eliminate pore formation in HeLa cells independent of translocation and hemolysis. HeLa cells were infected at an MOI of 100 with the indicated YopD mutants or the ΔyopEJKD and ΔyopEJK controls in the presence of PI for 2 h prior to the addition of gentamicin. Images were taken 2 h post-gentamicin addition. Images were analyzed for the percentage of HeLa cells containing PI. Data were pooled from three independent experiments. ****, P < 0.0001. Download
Increased translocation of YopB correlates with caspase-1 activation. (A) Combined z-stacks of confocal microscopy images of Z-YVAD-FMK-pretreated BMDMs that were left uninfected or were infected with the ΔyopEJK or ΔyopEJK-lcrHmut1 mutant for 2 h. Cells were stained for YopB (green), caspase-1 (red), and DNA (blue). White arrowheads indicate active caspase-1 puncta. (B) Total area (µm2) and total intensity of YopB staining in cells infected with the ΔyopEJK (circles) or ΔyopEJK-lcrHmut1 (squares) mutant. Data were pooled from at least three independent experiments, respectively. Each point represents a single cell. A total of 646 individual cells were analyzed. Red bars indicate means of data. (C) Percentage of YopB-staining cells that contain active caspase-1 puncta. Scale bars are 10 µm. (B) One-way ANOVA with Tukey multiple comparison tests. **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001. (C) Student’s t test. ****, P < 0.0001. Download
Yersinia strains used in this study
ACKNOWLEDGMENTS
This work was supported by the Microbial pathogenesis and Genomics NIH T32 training grant T32 AI060516 (E.E.Z.) and NIH R01AI103062 (I.E.B.). We thank the Zhu lab for technical assistance. We thank the Penn Vet Imaging Core (supported by NIH S10RR027128, the School of Veterinary Medicine, the University of Pennsylvania, and the Commonwealth of Pennsylvania) for technical assistance. We thank the Hunter and Weiss labs for reagents. We thank Dieter Schifferli for critical reading. We thank Baofeng Hu, Lance Peterson, Daniel Beiting, and the Shin, Hunter, Lopez, and Scott labs for scientific discussion.
Footnotes
Citation Zwack EE, Snyder AG, Wynosky-Dolfi MA, Ruthel G, Philip NH, Marketon MM, Francis MS, Bliska JB, Brodsky IE. 2015. Inflammasome activation in response to the Yersinia type III secretion system requires hyperinjection of translocon proteins YopB and YopD. mBio 6(1):e02095-14. doi:10.1128/mBio.02095-14.
Contributor Information
Joan Mecsas, Tufts University.
Arturo Zychlinsky, Max Planck Institute for Infection Biology.
REFERENCES
- 1.Janeway CA., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Vance RE, Isberg RR, Portnoy DA. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky IE, Monack D. 2009. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin Immunol 21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Lamkanfi M, Dixit VM. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol Rev 227:95–105. doi: 10.1111/j.1600-065X.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 6.Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 7.Cookson BT, Brennan MA. 2001. Pro-inflammatory programmed cell death. Trends Microbiol 9:113–114. doi: 10.1016/S0966-842X(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 8.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. 2012. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. 2012. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem 287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. 2011. Non-canonical inflammasome activation targets caspase-11. Nature 479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 11.Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. 2012. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell 150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanneganti TD, Ozören N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Núñez G. 2006. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 14.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 15.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 16.Gong YN, Shao F. 2012. Sensing bacterial infections by NAIP receptors in NLRC4 inflammasome activation. Protein Cell 3:98–105. doi: 10.1007/s13238-012-2028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 18.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. 2008. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A 105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy CR, Zamboni DS. 2006. Cytosolic detection of flagellin: a deadly twist. Nat Immunol 7:549–551. doi: 10.1038/ni0606-549. [DOI] [PubMed] [Google Scholar]
- 21.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. 2014. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol Cell 54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Zhao Y, Shi J, Shao F. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, Shin S. 2013. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog 9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornelis GR, Wolf-Watz H. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol 23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 27.Viboud GI, Bliska JB. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol 59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 28.Palmer LE, Hobbie S, Galán JE, Bliska JB. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol 27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 29.Brodsky IE, Palm NW, Sadanand S, Ryndak MB, Sutterwala FS, Flavell RA, Bliska JB, Medzhitov R. 2010. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung LK, Philip NH, Schmidt VA, Koller A, Strowig T, Flavell RA, Brodsky IE, Bliska JB. 2014. IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM. mBio 5(4):e01402-14. doi: 10.1128/mBio.01402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaRock CN, Cookson BT. 2012. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 12:799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmström A, Petterson J, Rosqvist R, Håkansson S, Tafazoli F, Fällman M, Magnusson KE, Wolf-Watz H, Forsberg A. 1997. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol 24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 33.Thorslund SE, Edgren T, Pettersson J, Nordfelth R, Sellin ME, Ivanova E, Francis MS, Isaksson EL, Wolf-Watz H, Fällman M. 2011. The RACK1 signaling scaffold protein selectively interacts with Yersinia pseudotuberculosis virulence function. PLoS One 6:e16784. doi: 10.1371/journal.pone.0016784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dewoody R, Merritt PM, Marketon MM. 2013. YopK controls both rate and fidelity of Yop translocation. Mol Microbiol 87:301–317. doi: 10.1111/mmi.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minnich SA, Rohde HN. 2007. A rationale for repression and/or loss of motility by pathogenic Yersinia in the mammalian host. Adv Exp Med Biol 603:298–310. doi: 10.1007/978-0-387-72124-8_27. [DOI] [PubMed] [Google Scholar]
- 36.Neyt C, Cornelis GR. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol 33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 37.Nordfelth R, Wolf-Watz H. 2001. YopB of Yersinia enterocolitica is essential for YopE translocation. Infect Immun 69:3516–3518. doi: 10.1128/IAI.69.5.3516-3518.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosqvist R, Persson C, Håkansson S, Nordfeldt R, Wolf-Watz H. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib Microbiol Immunol 13:230–234. [PubMed] [Google Scholar]
- 39.Sory MP, Cornelis GR. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 40.Olsson J, Edqvist PJ, Bröms JE, Forsberg A, Wolf-Watz H, Francis MS. 2004. The YopD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J Bacteriol 186:4110–4123. doi: 10.1128/JB.186.13.4110-4123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edqvist PJ, Aili M, Liu J, Francis MS. 2007. Minimal YopB and YopD translocator secretion by Yersinia is sufficient for Yop-effector delivery into target cells. Microbes Infect 9:224–233. doi: 10.1016/j.micinf.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Francis MS, Wolf-Watz H. 1998. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol Microbiol 29:799–813. doi: 10.1046/j.1365-2958.1998.00973.x. [DOI] [PubMed] [Google Scholar]
- 43.Collazo CM, Galán JE. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol 24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 44.McNeela EA, Burke A, Neill DR, Baxter C, Fernandes VE, Ferreira D, Smeaton S, El-Rachkidy R, McLoughlin RM, Mori A, Moran B, Fitzgerald KA, Tschopp J, Pétrilli V, Andrew PW, Kadioglu A, Lavelle EC. 2010. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog 6:e1001191. doi: 10.1371/journal.ppat.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ. 2013. The dhx33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craven RR, Gao X, Allen IC, Gris D, Bubeck Wardenburg J, McElvania-Tekippe E, Ting JP, Duncan JA. 2009. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS One 4:e7446. doi: 10.1371/journal.pone.0007446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwuan L, Adams W, Auerbuch V. 2013. Impact of host membrane pore formation by the Yersinia pseudotuberculosis type III secretion system on the macrophage innate immune response. Infect Immun 81:905–914. doi: 10.1128/IAI.01014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dewoody R, Merritt PM, Houppert AS, Marketon MM. 2011. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol 79:1445–1461. doi: 10.1111/j.1365-2958.2011.07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. 2010. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson DM, Ramamurthi KS, Tam C, Schneewind O. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol 184:1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edqvist PJ, Bröms JE, Betts HJ, Forsberg A, Pallen MJ, Francis MS. 2006. Tetratricopeptide repeats in the type III secretion chaperone, LcrH: their role in substrate binding and secretion. Mol Microbiol 59:31–44. doi: 10.1111/j.1365-2958.2005.04923.x. [DOI] [PubMed] [Google Scholar]
- 52.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis GR. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci U S A 91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams AW, Straley SC. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J Bacteriol 180:350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL, Sutterwala FS. 2013. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S-, Joe Y, Jeong SO, Zheng M, Back SH, Park SW, Ryter SW, Chung HT, Lee K, Jang Y. 2013. Endoplasmic reticulum stress is sufficient for the induction of IL-1beta production via activation of the NF-kappaB and inflammasome pathways. Int Immunol 25:623–632. doi: 10.1093/intimm/dxt029. [DOI] [PubMed] [Google Scholar]
- 57.Menu P, Mayor A, Zhou R, Tardivel A, Ichijo H, Mori K, Tschopp J. 2012. ER stress activates the NLRP3 inflammasome via an UPR-independent pathway. Cell Death Dis 3:e261. doi: 10.1038/cddis.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. 2012. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shio MT, Eisenbarth SC, Savaria M, Vinet AF, Bellemare MJ, Harder KW, Sutterwala FS, Bohle DS, Descoteaux A, Flavell RA, Olivier M. 2009. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog 5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou R, Yazdi AS, Menu P, Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 61.Monack DM, Mecsas J, Ghori N, Falkow S. 1997. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci U S A 94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Philip NH, Dillon CP, Snyder AG, Fitzgerald P, Wynosky-Dolfi MA, Zwack EE, Hu B, Fitzgerald L, Mauldin EA, Copenhaver AM, Shin S, Wei L, Parker M, Zhang J, Oberst A, Green DR, Brodsky IE. 2014. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-kappaB and MAPK signaling. Proc Natl Acad Sci U S A 111:7385–7390. doi: 10.1073/pnas.1403252111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergsbaken T, Cookson BT. 2007. Macrophage activation redirects Yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog 3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin H, Cornelis GR. 2007. Type III secretion translocation pores of Yersinia enterocolitica trigger maturation and release of pro-inflammatory IL-1beta. Cell Microbiol 9:2893–2902. doi: 10.1111/j.1462-5822.2007.01004.x. [DOI] [PubMed] [Google Scholar]
- 65.Fink SL, Cookson BT. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol 8:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 66.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito M, Yanagi Y, Ichinohe T. 2012. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog 8:e1002857. doi: 10.1371/journal.ppat.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A 96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brodsky IE, Medzhitov R. 2008. Reduced secretion of YopJ by Yersinia limits in vivo cell death but enhances bacterial virulence. PLoS Pathog 4:e1000067. doi: 10.1371/journal.ppat.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noel BL, Lilo S, Capurso D, Hill J, Bliska JB. 2009. Yersinia pestis can bypass protective antibodies to LcrV and activation with gamma interferon to survive and induce apoptosis in murine macrophages. Clin Vaccine Immunol 16:1457–1466. doi: 10.1128/CVI.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grabenstein JP, Marceau M, Pujol C, Simonet M, Bliska JB. 2004. The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect Immun 72:4973–4984. doi: 10.1128/IAI.72.9.4973-4984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In-frame YopD deletions eliminate pore formation in HeLa cells independent of translocation and hemolysis. HeLa cells were infected at an MOI of 100 with the indicated YopD mutants or the ΔyopEJKD and ΔyopEJK controls in the presence of PI for 2 h prior to the addition of gentamicin. Images were taken 2 h post-gentamicin addition. Images were analyzed for the percentage of HeLa cells containing PI. Data were pooled from three independent experiments. ****, P < 0.0001. Download
Increased translocation of YopB correlates with caspase-1 activation. (A) Combined z-stacks of confocal microscopy images of Z-YVAD-FMK-pretreated BMDMs that were left uninfected or were infected with the ΔyopEJK or ΔyopEJK-lcrHmut1 mutant for 2 h. Cells were stained for YopB (green), caspase-1 (red), and DNA (blue). White arrowheads indicate active caspase-1 puncta. (B) Total area (µm2) and total intensity of YopB staining in cells infected with the ΔyopEJK (circles) or ΔyopEJK-lcrHmut1 (squares) mutant. Data were pooled from at least three independent experiments, respectively. Each point represents a single cell. A total of 646 individual cells were analyzed. Red bars indicate means of data. (C) Percentage of YopB-staining cells that contain active caspase-1 puncta. Scale bars are 10 µm. (B) One-way ANOVA with Tukey multiple comparison tests. **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001. (C) Student’s t test. ****, P < 0.0001. Download
Yersinia strains used in this study