ABSTRACT
Reef-building corals form essential, mutualistic endosymbiotic associations with photosynthetic Symbiodinium dinoflagellates, providing their animal host partner with photosynthetically derived nutrients that allow the coral to thrive in oligotrophic waters. However, little is known about the dynamics of these nutritional interactions at the (sub)cellular level. Here, we visualize with submicrometer spatial resolution the carbon and nitrogen fluxes in the intact coral-dinoflagellate association from the reef coral Pocillopora damicornis by combining nanoscale secondary ion mass spectrometry (NanoSIMS) and transmission electron microscopy with pulse-chase isotopic labeling using [13C]bicarbonate and [15N]nitrate. This allows us to observe that (i) through light-driven photosynthesis, dinoflagellates rapidly assimilate inorganic bicarbonate and nitrate, temporarily storing carbon within lipid droplets and starch granules for remobilization in nighttime, along with carbon and nitrogen incorporation into other subcellular compartments for dinoflagellate growth and maintenance, (ii) carbon-containing photosynthates are translocated to all four coral tissue layers, where they accumulate after only 15 min in coral lipid droplets from the oral gastroderm and within 6 h in glycogen granules from the oral epiderm, and (iii) the translocation of nitrogen-containing photosynthates is delayed by 3 h.
IMPORTANCE
Our results provide detailed in situ subcellular visualization of the fate of photosynthesis-derived carbon and nitrogen in the coral-dinoflagellate endosymbiosis. We directly demonstrate that lipid droplets and glycogen granules in the coral tissue are sinks for translocated carbon photosynthates by dinoflagellates and confirm their key role in the trophic interactions within the coral-dinoflagellate association.
INTRODUCTION
Photosynthesis plays a central role in many aquatic animals symbiotically associated with microalgae or cyanobacteria (1). Shallow-water reef-building scleractinian corals hosting photosynthetic dinoflagellates of the genus Symbiodinium (“zooxanthellae”) represent an emblematic example of such a stable mutualistic endosymbiotic relationship, which is critical for the development and health of coastal coral reef ecosystems in (sub)tropical oceans. The dinoflagellate endosymbionts, located within the coral gastrodermal cells (see Fig. S1 in the supplemental material), significantly contribute to the nutrition of their animal host partner by transferring a large fraction (up to 90%) of their photosynthetically assimilated carbon (C) and nitrogen (N) to support growth, respiration, reproduction, and biocalcification of the coral in nutrient-poor marine environments (2, 3). These photosynthates are produced by dinoflagellates through the fixation of dissolved inorganic carbon (DIC) via the Calvin-Benson “C3” photosynthetic pathway (4) and through the photosynthesis-dependent acquisition of dissolved inorganic nitrogen (DIN), ultimately via the glutamine synthetase-glutamate synthase (GS-GOGAT) enzymatic cycle (5, 6). The nature of translocated photosynthates (“mobile compounds”) ranges from soluble low-molecular-weight compounds, such as glycerol, glucose, amino acids, and organic acids (7–9), to more complex molecules, such as free fatty acids (10) or glycoconjugates (11). However, the detailed pathway of this nutritional autotrophic flux from the dinoflagellate endosymbionts to the different cellular layers composing the coral host tissue, as well as the precise fate and turnover of photosynthates in the symbiotic system, remain poorly documented at the (sub)cellular level.
Symbiotic reef-building corals are regarded as “fat organisms” because they contain 9 to 47% (dry weight) lipids in their tissue, mostly in the form of neutral lipids (triglycerides, wax esters, and sterols) packed into lipid droplets (LDs), which are hypothesized to be a main sink for C-rich photosynthates translocated by dinoflagellates to the coral tissue (12–15). In support of this view, most previous bulk-level studies using radioactive (14C) or stable (13C) isotope labeling found preferential incorporation of translocated photosynthates into a chemically extracted lipid fraction, as well as structural polymeric compounds such as proteins (16–21). Additionally, recent observations indicate morphological and compositional changes of coral LDs upon coral bleaching (i.e., loss of dinoflagellates or their pigmentation) and a positive correlation between abundance of coral LDs and dinoflagellate density or light intensity (22, 23). Nevertheless, despite their supposed key role in the trophic interactions within the coral-dinoflagellate endosymbiosis, a direct demonstration that coral LD biosynthesis is linked with the release of photosynthates by dinoflagellates is still lacking.
Glycogen is another potentially important C reserve pool in the endosymbiosis, previously detected in stony corals both biochemically and ultrastructurally (24, 25). Gene expression for glycogen synthase and glycogen phosphorylase enzymes, which regulate the production and mobilization of glycogen stores, was detected in the reef coral Acropora aspera transcriptome (26). However, the possible incorporation of photosynthates such as glucose (9) into coral glycogen has not been investigated. Furthermore, little attention has been paid to the allocation and turnover of photosynthates within the dinoflagellate subcellular compartments, especially in their C storage structures, which are LDs and starch granules (27, 28).
Nanoscale secondary ion mass spectrometry (NanoSIMS) ion microprobe imaging is a powerful tool to simultaneously image and quantify the distribution and turnover of stable isotopic tracers (e.g., 13C and 15N) inside cells, especially when correlated with ultrastructural transmission electron microscopy (TEM) imaging (29–33). Here, we used this methodological approach on microcolonies (nubbins) of the common Indo-Pacific symbiotic reef-building coral Pocillopora damicornis, which were pulse-labeled in an aquarium for 6 h simultaneously with [13C]bicarbonate and [15N]nitrate (15NO3−), followed by an extended chase of 186 h under either normal light/dark cycling (12 h/12 h) or prolonged darkness. We used [15N]nitrate to unambiguously track the flow of N in the endosymbiotic system because nitrate is assimilated by the dinoflagellates only, in contrast to ammonium, the preferred source of DIN for most reef corals, which is simultaneously assimilated by both dinoflagellate and coral cells (32, 33).
The two main objectives of this study were to visualize and measure in situ, with subcellular resolution, the photosynthesis-dependent incorporation, fate, and turnover of inorganic C in the dinoflagellate endosymbionts and to track the translocation of dinoflagellate photosynthates toward the coral host tissue layers (see Fig. S1 in the supplemental material), especially their incorporation and turnover in coral LDs and glycogen granules.
RESULTS
Assimilation and turnover of carbon and nitrogen in dinoflagellates.
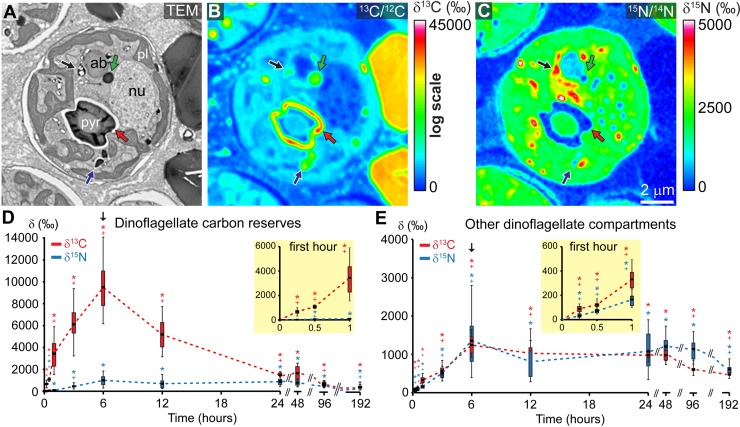
NanoSIMS 13C/12C and 15N/14N isotopic images of dinoflagellate endosymbionts indicate spatially heterogeneous intracellular distribution of 13C and 15N incorporated during the 6-h labeling pulse under light (a representative dinoflagellate cell is illustrated in Fig. 1A to C). Regarding incorporated 13C, preferential accumulation was systematically recorded on isotopic images in the following dinoflagellate compartments, identified on corresponding ultrastructural TEM micrographs and collectively termed “C reserves”: (i) large, “primary” starch granules (terminology of Doyle and Doyle [27]) forming a cap around the plastidial pyrenoid (red arrows in Fig. 1A and B), (ii) small “secondary” starch granules distributed in the cytosol (blue arrows in Fig. 1A and B), and (iii) highly osmiophilic electron-dense LDs (green arrows in Fig. 1A and B).
FIG 1 .
Photosynthesis-dependent carbon and nitrogen assimilation and turnover in dinoflagellate endosymbionts. (A) TEM micrograph of a representative dinoflagellate cell within the coral oral gastroderm after 6 h in the pulse of dual isotopic labeling under light with [13C]bicarbonate (2 mM) and [15N]nitrate (30 µM). (B and C) Corresponding NanoSIMS 13C/12C (B) and 15N/14N (C) isotopic images. (D and E) NanoSIMS quantified 13C/12C (in red) and 15N/14N (in blue) isotope ratios in the dinoflagellate C reserves (including lipid droplets and starch granules) and in the remaining dinoflagellate compartments, respectively, during the pulse-chase experiment conducted for 8 days under light/dark cycling (12 h/12 h). The black arrow on graphs indicates the end of the 6-h pulse of labeling under light. Significant differences (pairwise Wilcoxon rank sum test or pairwise t test, P < 0.05) are indicated between labeled and unlabeled control corals (*) and between samples from two consecutive time points (+); the number of replicate ROIs and P values of statistical analyses are given in Data Set S1 in the supplemental material; the box-whisker plot separates data into quartiles, with the top of the box defining the 75th percentile, the bottom the 25th percentile, the middle line the average value, the upper “whisker” the 95th percentile, and the lower “whisker” the 5th percentile. ab, accumulation body; nu, nucleus; pl, plastid; pyr, pyrenoid; red arrows, primary starch; blue arrows, secondary starch; green arrows, dinoflagellate LDs; black arrows, vesicles containing uric acid crystals.
NanoSIMS isotopic measurements from regions of interest (ROIs) defined in the dinoflagellate population (as illustrated in Fig. S2 in the supplemental material) show that the average 13C enrichment of the dinoflagellate C reserves increased rapidly, after 15 min, during the 6-h labeling pulse under light, but then it declined by ~80% of its peak value, during the first 18 h of the chase under light/dark cycling, over a period that includes the first 12-h dark phase (Fig. 1D). (Summary data tables and P values from statistical analyses are provided in Data Set S1 in the supplemental material). In parallel, incorporated 13C started to accumulate after 15 min into the pulse in the various other dinoflagellate compartments (e.g., nucleus and plastid) (Fig. 1A, B, and E). In contrast to C reserves, the 13C enrichment of other compartments in the dinoflagellates only slowly decreased throughout the 186-h chase period, by about 60% of its peak value (Fig. 1E; see Data Set S1).
Regarding 15N incorporated by the endosymbionts, this tracer was detected to accumulate in the dinoflagellate C reserves after 30 min into the pulse of labeling, but the 15N enrichment in these compartments remained essentially stable during the chase (Fig. 1D). In the various other dinoflagellate compartments, 15N was incorporated after 15 min into the pulse with a pattern of isotopic depletion throughout the chase almost identical to that of 13C (Fig. 1E). Note that preferential incorporation of 15N into vesicles containing uric acid crystals was confirmed (33, 34) (black arrows in Fig. 1A and C). Figure S3 in the supplemental material illustrates the turnover of both C and N during the pulse-chase experiment in representative dinoflagellates.
In corals maintained for 186 h in constant darkness, following the 6-h labeling pulse under light, the pattern of rapid 13C decrease in the C reserves of dinoflagellates was confirmed (see Fig. S4 in the supplemental material). In addition, the observed ultrastructural changes indicate that both dinoflagellate LDs and starch granules were progressively depleted in the absence of photosynthetic carbon replenishment, an observation concomitant with the gradual appearance of ultrastructural symptoms of dinoflagellate cell degeneration (see Fig. S4). Severe paling of coral microcolonies was also noticed after this prolonged dark treatment (8 days), strongly suggesting that coral bleaching had occurred.
No significant incorporation of 13C and 15N was observed in dinoflagellate and coral cells pulse-labeled for 6 h in darkness following a 24-h pretreatment in darkness (see Fig. S5 in the supplemental material). This result confirms that C and N assimilation by dinoflagellates is light dependent (i.e., related to photosynthesis) and that the contribution of nonphotosynthetic “heterotrophic” C fixation by carboxylation reactions was negligible in both dinoflagellate and coral cells.
Translocation of carbon and nitrogen to the coral host tissue.
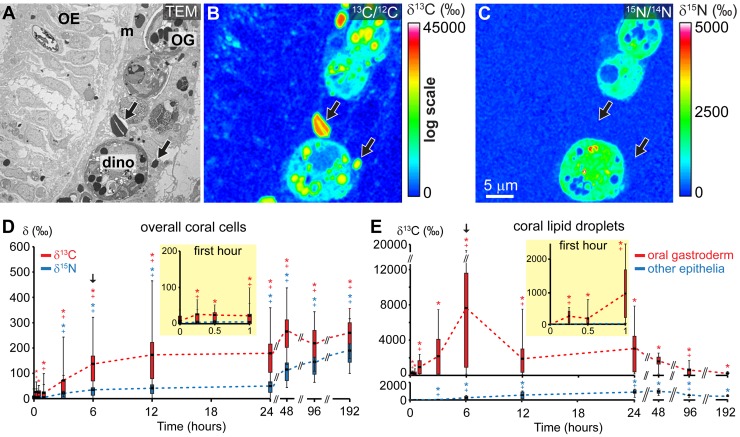
At the overall coral tissue level, NanoSIMS isotopic measurements from ROIs (as illustrated in Fig. S2 in the supplemental material) demonstrate the gradual translocation of 13C from the dinoflagellate endosymbionts to all four coral epithelia, starting after only 15 min and stabilizing after 48 h in the pulse-chase experiment with light/dark cycling (Fig. 2A, B, and D). (Summary data tables and P values of statistical analyses are given in Data Set S1 in the supplemental material, and Fig. S6 in the supplemental material illustrates a sequence of representative TEM and NanoSIMS isotopic images of the coral oral tissue.) Among the coral tissue layers, 13C-labeled photosynthates transferred by dinoflagellates during the pulse were observed to rapidly (within 15 min) accumulate in the coral LDs from the oral gastroderm. In contrast, incorporation into coral LDs from the three other epithelia was much less efficient and occurred with a 3-h delay (Fig. 2A, B, and E; see Data Set S1 and Fig. S6). In the 186-h chase, coral LDs from the oral gastroderm were then strongly depleted in 13C, with more than a 90% decrease in average 13C enrichment compared to the peak value at the end of the pulse (Fig. 2E). Note the very high level of variability in 13C enrichments among individual coral LDs from the oral gastroderm, especially after 6 h in the pulse (Fig. 2E).
FIG 2 .
Photosynthate translocation from dinoflagellates into the coral host tissue and their lipid droplets. (A) Representative TEM micrograph of the coral oral tissue after 6 h in the pulse of dual isotopic labeling under light. (B and C) Corresponding NanoSIMS 13C/12C (B) and 15N/14N (C) isotopic images. (D) NanoSIMS quantified 13C/12C (in red) and 15N/14N (in blue) isotopic ratios in the whole coral tissue (including all four epithelia) during the pulse-chase experiment under light/dark cycling. (E) NanoSIMS quantified 13C/12C isotopic ratio in coral lipid droplets from the oral gastroderm (in red) and from the three other epithelia (oral epiderm, aboral gastroderm, and calicoderm [in blue]). Results and their statistical significance are reported as described in the legend to Fig. 1. OE, oral epiderm; OG, oral gastroderm; m, mesoglea; dino, dinoflagellate cell; black arrows, coral lipid droplets.
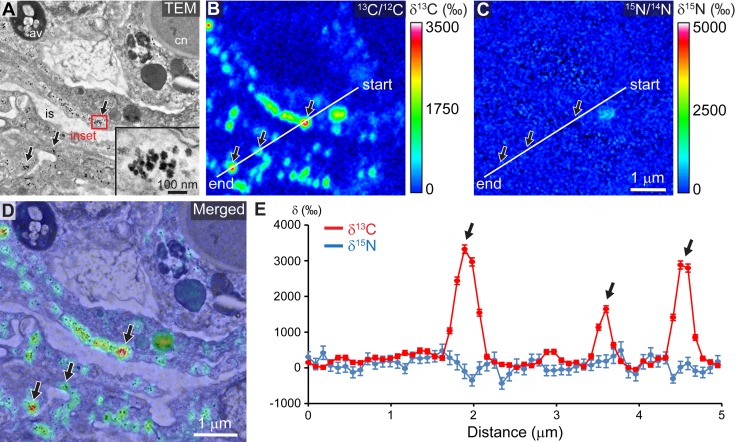
At the end of the 6-h pulse, 13C-labeled photosynthates translocated by dinoflagellates were also systematically found associated with numerous ~50-nm-diameter glycogen granules, located in the apical region of coral cells from the oral epiderm (Fig. 3). (An enlarged view of Fig. 3D is provided in Fig. S7 in the supplemental material.) In these areas rich in glycogen granules, the 13C labeling was found from NanoSIMS line profiles to gradually decrease over the 186-h chase under light/dark cycling, reaching ~85% 13C depletion compared to the peak value at the end of the pulse (see Fig. S8 in the supplemental material).
FIG 3 .
Photosynthate accumulation into coral glycogen granules. (A) Representative TEM micrograph of the coral oral epiderm with a higher-magnification view of glycogen granules (inset). (B and C) Corresponding NanoSIMS 13C/12C (B) and 15N/14N (C) isotopic images. (D) Merged image between the TEM micrograph and the NanoSIMS 13C/12C isotopic map. An enlarged view is provided in Fig. S7 in the supplemental material. (E) Fluctuations of both 13C (in red) and 15N (in blue) enrichments along the NanoSIMS profile depicted in panels B and C. Standard deviations of the mean are based on Poisson statistics. Black arrows point to areas rich in glycogen granules. av, autophagic vacuole; cn, cnidocyte; is, intercellular space.
Translocation by dinoflagellates of 15N-labeled photosynthates to the coral tissue (overall coral cells) was recorded starting after 3 h into the pulse—i.e., with a much longer delay in comparison to 13C (Fig. 2D). Moreover, in contrast to translocated 13C, the spatial distribution of translocated 15N was found to be relatively homogeneous in coral cells, with no specific subcellular compartments in the host tissue benefitting preferentially from the dinoflagellate supply of 15N-labeled materials (Fig. 2A and C; see Fig. S6 in the supplemental material). Major spatial patterns and temporal time scales of 13C and 15N fluxes in the symbiosis are summarized in Table 1.
TABLE 1 .
Summary of the main events and time scales traced in this study
| Symbiotic partner | Major event traced | Time scale |
|---|---|---|
| Dinoflagellates | Photosynthesis-dependent assimilation of bicarbonate and nitrate | 15 min |
| C allocation to C reserves (lipid droplets and starch) | 15 min | |
| C turnover in C reserves related to diurnal light cycle | ~80% 13C depletion in 18 h | |
| C and N turnover in other compartments | ~60% 13C and 15N depletion in 186 h | |
| Coral | C translocation by dinoflagellates | 15 min |
| N translocation by dinoflagellates | 3 h | |
| Allocation of translocated C to lipid droplets (oral gastroderm) | 15 min | |
| Allocation of translocated C to glycogen (oral epiderm) | 6 h | |
| C turnover in lipid droplets (oral gastroderm) | >90% 13C depletion in 186 h | |
| C turnover in glycogen (oral epiderm) | ~85% 13C depletion in 186 h |
DISCUSSION
Subcellular imaging of photosynthetic C and N assimilation and utilization by the dinoflagellate endosymbionts.
This study demonstrates that in the tropical reef-building coral P. damicornis, a substantial fraction of photosynthetically assimilated inorganic C and N was retained in the dinoflagellate cells during the pulse-chase experiments. C-containing photosynthates rapidly (within 15 min) accumulated into dinoflagellate lipid droplets (LDs) and starch granules (both primary and secondary), with subsequent rapid turnover. Most accumulated 13C was depleted from dinoflagellate C reserves within the first 18 h of the chase under light/dark cycling, over a period that includes the first 12-h dark phase. This result strongly suggests a diurnal rhythmicity in the formation (under light) and utilization (under dark) of LDs and starches by dinoflagellates. The 15N labeling observed in the 13C-enriched dinoflagellate LDs and starch granules most likely reflects the incorporation of 15N-labeled proteins, possibly enzymes involved in the synthesis and further catabolism of neutral lipids or carbohydrates, onto the surface or into the internal matrix of these compartments (35, 36). In addition, assimilated 13C and 15N were allocated to the various other dinoflagellate compartments (including, e.g., the nucleus and plastid), albeit for C with a lower efficiency and a slower turnover than for the C reserves, most likely reflecting the utilization of C and N for dinoflagellate maintenance, growth, and division.
These NanoSIMS results obtained in situ (i.e., in the intact coral-dinoflagellate association) are in agreement with data from previous bulk-level isotopic incubation analyses with 14C- or 13C-labeled bicarbonate, which report a rapid loss of 14C or 13C enrichment in the dinoflagellate fraction within the first hours of the chase, especially during the first night (17, 21, 37–39). In particular, by labeling the reef coral Acropora pulchra simultaneously with [13C]bicarbonate and [15N]nitrate, Tanaka et al. (21) found a dramatic nighttime decrease in the dinoflagellate fraction of the C:N ratio of light-produced compounds, suggesting rapid consumption of photosynthates with high C content (i.e., lipids and carbohydrates) by dinoflagellate respiration. Conversely, in Stylophora pistillata colonies, the rapid decrease in 14C or 13C labeling of the dinoflagellate fraction, observed over a chase period of 24 to 48 h, was mainly ascribed to delayed translocation of 14C- or 13C-labeled photosynthates to the coral host partner (17, 39).
Here, we observed that the rapid 13C decrease in dinoflagellate endosymbionts of P. damicornis is occurring via isotopic depletion of their intracellular LDs and starch granules and not through depletion of their other cell compartments. This result supports the hypothesis that lipids and carbohydrates stored by dinoflagellates during daytime are quickly remobilized, especially during nighttime, via mitochondrial respiration (releasing 13CO2) to sustain dinoflagellate metabolism. Nevertheless, the following additional mechanisms for such rapid 13C depletion in dinoflagellate C reserves cannot be excluded: (i) the effect of an isotopic dilution due to the additional storage of newly produced [12C]photosynthates during the light periods of the chase, (ii) the translocation of 13C-labeled compounds toward the coral host tissue (during light and dark periods of the chase), and/or (iii) the remobilization of stored 13C as building blocks for dinoflagellate maintenance, growth, and division.
Interestingly, by maintaining P. damicornis microcolonies under constant darkness, a treatment known to trigger coral bleaching after about 4 days (40, 41), the ultrastructural disappearance of dinoflagellate LDs and starch granules was systematically accompanied by features of cell vacuolization and damage of organelles, indicative of in situ degradation of the endosymbionts. These observations are additional evidence that photosynthates stored in dinoflagellates under light are essential to further sustain their respiration and metabolism, especially during nighttime.
In marine microalgae, photosynthates produced and stored under light might provide C skeletons and energy to support DIN (ammonium and nitrate) uptake and assimilation in the dark (42). The existence of an internal C reservoir in dinoflagellate endosymbionts, metabolized during nighttime to sustain N acquisition, is suggested by the extended time of darkness (at least 15 h) needed to efficiently inhibit ammonium incorporation in symbiotic reef corals (5, 33). Similarly, we demonstrate that prolonged dark pretreatment (24 h) fully repressed nitrate assimilation in dinoflagellates of the reef coral P. damicornis. Interestingly, in coastal marine environments, migrating free-living dinoflagellates reach the illuminated sea surface, poor in DIN, during the day, to accumulate excess photosynthates not channeled toward protein synthesis, whereas they descend during the night to a depth enriched in nitrate, which is then efficiently assimilated through the remobilization of light-produced C reserves (43). Hence, the storage of lipids and carbohydrates by dinoflagellates during daytime might constitute a C reserve helping reef corals to efficiently sustain DIN acquisition during nighttime, representing an adaption to nutrient-poor environments.
The fate of C and N translocated to the coral host.
The present study reveals the subcellular pathways of photosynthetic C translocation from the dinoflagellate cells toward all four epithelia composing the coral host tissue, extending our previous NanoSIMS observations of transepithelial movements of nitrogenous compounds translocated by the endosymbionts (32, 33). In early autoradiographic investigations, transepithelial metabolite fluxes have been reported in marine cnidarian tissue, albeit at light microscopy level in thick histological sections (44, 45). Moreover, using bulk-level isotopic measurements of separated fractions of dinoflagellate-containing oral gastroderm and dinoflagellate-free oral epiderm prepared from tentacles of a sea anemone, Trench (18) previously found that 18 to 31% of total net photosynthates moved toward the oral epiderm within 10 h of labeling in light with [14C]bicarbonate.
Here, we observed that coral LDs from the oral gastroderm epithelium constitute a major accumulation site for translocated photosynthetic 13C, providing direct validation of previous hypotheses (12, 13, 15, 22, 23). The rapid C translocation toward gastrodermal coral LDs, which we visualized already at 15 min from the onset of the pulse, is consistent with results from bulk-level isotopic investigations of symbiotic sea anemones, which reported rapid translocation of photosynthates to the host fraction, within a few minutes following their production (9, 46, 47). Interestingly, we frequently observed 13C-enriched LD-like structures located in the symbiosomal space between the dinoflagellate endosymbiont and the coral gastrodermal host cell (white arrows in Fig. S3 in the supplemental material). These structures have previously been interpreted as “extra-algal” LDs produced by dinoflagellates, in the process of exocytosis toward the host gastrodermal cells (48, 49). However, ultrastructural evidence for such a potential exocytotic process is still lacking. Moreover, the occurrence of LDs within coral cells of dinoflagellate-free epithelia (oral epiderm and calicoderm) implies the existence of other, still unknown, mechanisms of coral LD formation.
This study also provides evidence that coral glycogen granules in the coral oral tissue constitute another major sink for 13C-photosynthates translocated by dinoflagellates. In bulk isotopic analyses of sea anemones, glucose was found to be a major metabolite translocated within minutes from the dinoflagellate endosymbionts to the host fraction (7–9). Moreover, transcriptome analyses have revealed that scleractinian (Acropora genus) corals have the enzymatic machinery for synthesis and remobilization of glycogen from and to glucose (26). Consistent with these reports, combined TEM and NanoSIMS observations show incorporation (within 6 h) of external [13C6]glucose (30 µM) into glycogen granules of the coral oral epidermal cells (see Fig. S9 in the supplemental material). Thus, our results provide direct evidence of the functional mechanisms of storage of photosynthetic C, translocated by the dinoflagellate endosymbionts (probably in the form of glucose) to coral glycogen in the oral tissue.
Similar to dinoflagellate C reserves, the 13C depletion observed for coral LDs in the oral gastroderm and for the glycogen granules in the oral tissue most likely reflects the breakdown of neutral lipids and carbohydrates to sustain coral cell respiration. Nevertheless, we cannot exclude the potential contribution of (i) an isotopic dilution effect resulting from the translocation of photosynthates with normal C isotopic composition to coral LDs and glycogen and (ii) the reallocation of stored C toward coral cell maintenance, growth, and division.
Translocation of N-containing compounds by dinoflagellates was not recorded immediately in the coral host partner but was observed with a delay of 3 h following the onset of the pulse-chase experiment under light/dark cycling, confirming previous observations (33). These results suggest a temporal separation between the translocation by dinoflagellates of C- and N-containing photosynthetic assimilates. Compounds bearing C are released within a few minutes after their production, compared to a time scale of hours for N-bearing compounds (summarized in Table 1). It is also possible that early translocation of N to the coral tissue has been partly masked by a potentially high rate of N recycling by the dinoflagellates (5) or by extraction of low-molecular-weight soluble nitrogenous compounds (e.g., free amino acids) during sample preparation. Alternative NanoSIMS sample preparation methods (e.g., cryofixation and cryosubstitution) might improve tracking of low-molecular-weight soluble photosynthates.
Conclusion.
By combining pulse-chase double stable-isotopic labeling (13C and 15N) with TEM ultrastructural and NanoSIMS isotopic imaging, we have visualized and quantified at subcellular levels the incorporation and turnover of C and N in the symbiotic reef-building coral P. damicornis. These results provide a qualitative baseline of subcellular allocation and turnover of C- and N-containing photosynthates in the coral-dinoflagellate symbiosis. In the future, more precise quantitative C and N budgets for symbiotic reef corals could be constructed, using additional respiration and photosynthesis measurements. Moreover, alterations in the pattern of C and N utilization by symbiotic corals might be more precisely characterized in response to heterotrophic feeding and to global environmental changes.
MATERIALS AND METHODS
Design of 13C and 15N dual isotopic labeling experiments.
Experiments were carried out on microcolonies (~5 cm tall) of the reef-building coral Pocillopora damicornis (Linnaeus, 1758) prepared from one large colony grown at the Aquarium Tropical, Palais de la Porte Dorée, Paris, France. Coral microcolonies were acclimatized for 4 weeks prior to experimental manipulation in a large tank equilibrated with fish and benthic organisms, filled with artificial seawater (Instant Ocean Salts) and containing low nutrient concentrations (NH4+, <1 µM; NO2−, <1 µM; NO3− <5 µM; PO43−, <1 µM [Salifert tests]). The temperature was 25 ± 1°C, the salinity was 35 ± 1‰, and the pH was 8.1 ± 0.1. Light irradiance of 100 µmol · m−2 · s−1 in the photosynthetically active radiation range was provided by 8 fluorescent T5 tubes of 39 W (6 × 10,000 K, 2 × 20,000 K) with 12-h/12-h light/dark cycling. During the acclimatization period of 4 weeks and subsequent experiments, coral microcolonies were not fed with plankton and therefore mostly relied on the uptake and assimilation of particulate and dissolved organic matter and on the autotrophic input from their photosynthetic dinoflagellate symbionts.
Isotopic dual labeling pulse-chase experiments were conducted in closed-system 20-liter glass tanks. In a first step, 17 coral microcolonies were incubated for 6 h in light in 0.2-µm-pore-filtered artificial seawater (adapted from Harrison et al. [50]), initially free of bicarbonate and nitrate, and supplemented with 2 mM [13C]bicarbonate (NaH13CO3, 99 atom% [Sigma-Aldrich]) and 30 µM [15N]nitrate (K15NO3, 98 atom% [Sigma-Aldrich]). This pulse of isotopic dual labeling started about 2 h after the onset of the light period. Then 5 coral microcolonies were transferred for a 186-h (8-day) chase period with light/dark cycling (12 h/12 h) in a tank filled with 20 liters artificial seawater sampled from the large acclimatization tank with normal C and N isotopic composition. The effect of coral autotrophic starvation on C and N remobilization pattern was investigated in a parallel 186-h chase experiment with 5 coral microcolonies exposed to similar conditions but maintained under constant darkness. During the chase phase, 25% of the incubation volume was renewed daily with artificial seawater from the large acclimatization tank. For TEM ultrastructural and NanoSIMS isotopic imaging, apexes of coral branches were sampled with cutting pliers during the pulse-chase experiments at 0, 0.25, 0.5, 1, 3, 6, 12, 24, 48, 96, and 192 h, respectively. At each time step, a different coral microcolony (nubbin) was sampled, except at 6 h, where 3 replicate nubbins were sampled. Throughout the pulse-chase experiment under light/dark cycling, coral microcolonies displayed no macromorphological indications of stress (i.e., no unusual tentacle retraction, extensive mucus secretion, or paling). During the chase period, ammonium and nitrate concentrations fluctuated mildly between values of 1 to 4 µM and 5 to 10 µM, respectively, corresponding to closed-system aquarium conditions.
Contribution of nonphotosynthetic carboxylation reactions (“heterotrophic” C fixation) to [13C]bicarbonate incorporation by the symbiotic system and the effect of dark inhibition on [15N]nitrate assimilation by symbiotic dinoflagellates were assessed as follows: 3 coral microcolonies were pretreated for 24 h under constant darkness and then incubated for 6 h in darkness in a tank filled with 2 liters of 0.2-µm-pore-filtered artificial seawater (adapted from Harrison et al. [50]), initially free of bicarbonate and nitrate and supplemented with 2 mM [13C]bicarbonate (NaH13CO3, 99 atom% [Sigma-Aldrich]) and 30 µM [15N]nitrate (K15NO3, 98 atom% [Sigma-Aldrich]). Such extended pretreatment under darkness is required to effectively inhibit DIN assimilation in symbiotic reef corals (5, 33). Apexes of coral branches were sampled with cutting pliers at 0, 3, and 6 h during the labeling pulse.
To limit spatial fluctuations due to metabolic heterogeneities along coral branches or between different coral tissue areas (51–53), TEM ultrastructural observations and NanoSIMS isotopic quantitative imaging were systematically performed on coral tissue sampled from the subapical area of light-exposed branches, within the coenosarcal connective tissue linking polyp units together (see Fig. S1 in the supplemental material)
TEM ultrastructural observations.
Coral samples were chemically fixed for 24 h at room temperature in Sörensen-sucrose phosphate buffer (0.1 M phosphate at pH 7.5, 0.6 M sucrose, 1 mM CaCl2) containing both 2.5% glutaraldehyde and 1% formaldehyde. They were then decalcified for 4 to 5 days in 0.1 M Sörensen phosphate buffer containing 0.5 M EDTA at 4°C. Tissue samples were dissected under the stereomicroscope into small pieces containing one or two polyps, postfixed 1 h at room temperature with 1% OsO4 in 0.1 M Sörensen phosphate buffer, dehydrated in graded series of ethanol (50, 70, 90, and 100%), and embedded in Spurr’s resin. Tissue was preferentially oriented to obtain longitudinal sections parallel to the vertical growth direction of polyps. Sections were cut with a Diatome 45° diamond knife. Semithin sections (~0.5 µm) were stained with methylene blue-Azur II and observed with a Zeiss Axio Imager Z2 light microscope equipped with a Zeiss AxioCamMRc 5 digital camera. Ultrathin sections (~70 to 90 nm) were mounted on Formvar carbon-coated alphanumeric grids counterstained with 4% uranyl acetate and Reynold’s lead citrate solution. Ultrastructural observations were carried out at 80 kV with a Philips CM 100 transmission electron microscope within the Electron Microscopy Facility (EMF) at the University of Lausanne (Switzerland).
Quantitative NanoSIMS isotopic imaging and ROI definition.
In order to image and quantify in situ the subcellular distribution of 13C and 15N enrichment within endosymbiotic dinoflagellate cells and coral host tissue, the exact same areas in the coral tissue imaged by TEM were analyzed with the NanoSIMS 50L ion microprobe in the Laboratory for Biological Geochemistry (EPFL, Lausanne, Switzerland), enabling direct correlation of ultrastructural (TEM) and isotopic (NanoSIMS) images.
TEM grids were mounted on 10-mm aluminum stubs with double-stick Cu tape and coated with about a 10-nm thickness of gold. They were bombarded with a 16-keV primary ion beam of Cs+ (1 to 3 pA) focused to a spot size of about 100 to 150 nm on the sample surface. Secondary molecular ions 12C2−, 13C12C−, 12C14N−, and 12C15N− were simultaneously collected in electron multipliers at a mass resolution sufficient to avoid potentially problematic isobaric interferences on 13C12C− and 12C15N−. Charge compensation was not necessary. Isotopic images ranging between 5 × 5 µm2 and 50 × 50 µm2 with 256 by 256 pixels were obtained by rastering the primary beam across the sample surface with a dwell time of 5 ms. 13C/12C and 15N/14N ratio distribution maps were obtained by taking the ratio between the drift-corrected 13C12C− and 12C2− images and 12C15N− and 12C14N− images, respectively. 13C and 15N enrichments were expressed in the following delta notations:
| (1) |
where Cmes is the measured 13C/12C ratio and Cnat is the average natural 13C/12C ratio measured several times per day in nonlabeled, identically prepared coral samples throughout the period of NanoSIMS analyses and
| (2) |
where Nmes is the measured 15N/14N ratio and Nnat is the average natural 15N/14N ratio measured several times per day in nonlabeled, identically prepared coral samples throughout the period of NanoSIMS analyses.
For each ultrathin section analyzed (for each coral microcolony sampled at each time step of the pulse-chase experiments), ~12 NanoSIMS isotopic maps were acquired to obtain a representative view of both isotopic enrichment and spatial distribution within the coral-dinoflagellate system. All analyzed dinoflagellates in this study were located in the oral gastroderm epithelium, which contains by far the highest density of symbiotic cells compared to the aboral gastroderm (see Fig. S1 in the supplemental material).
Data were processed using the L’IMAGE software. A smooth width of 3 pixels was applied for NanoSIMS 13C/12C and 15N/14N isotopic images, and a line width of 1 pixel was applied when illustrative line scans were defined from these smoothed images. NanoSIMS quantification of both 13C and 15N enrichments was carried out by defining regions of interest (ROIs), as illustrated in Fig. S2 in the supplemental material. For the whole dinoflagellates and their C reserves (i.e., starch granules and LDs), the ROIs were obtained by drawing their contours. Quantification of the remaining cell compartments (i.e., the whole dinoflagellate cell minus the C reserves) was obtained by subtracting corresponding ROIs. In the coral host tissue, ROIs were defined as circles of 2 to 3 µm covering each of the four coral epithelia, avoiding the mesoglea and intercellular spaces. For coral LDs, ROIs were obtained by drawing their contours. Because their small size (~50 nm in diameter) prevented accurate drawing of ROIs around each glycogen granule, their accumulation and turnover were assessed qualitatively from NanoSIMS line scans, and the spatial correlation between isotopic labeling and glycogen granules was confirmed in merged TEM and NanoSIMS images.
Importantly, conventional TEM sample preparation (i.e., chemical fixation with aldehydes and osmium tetroxide postfixation, followed by decalcification and ethanol dehydration) extracts most low-molecular-weight soluble compounds and diffusible ions located in coral and dinoflagellate cells. Consequently, we imaged with NanoSIMS 13C and 15N incorporation into macromolecules (e.g., proteins, unsaturated lipids, and carbohydrates, such as glycogen and starch) stabilized by the sample preparation procedure.
Statistical analyses.
Data were statistically analyzed with the R software. Shapiro-Wilk and Bartlett tests were used to assess data normality and homoscedasticity. In the case of non-Gaussian distributions, the nonparametric Kruskal-Wallis test was applied combined with a pairwise Wilcoxon rank sum test, instead of the one-way analysis of variance (ANOVA) combined with a pairwise t test. Holm’s correction was systematically employed when doing pairwise multiple-comparison tests. Results were considered significant at the 5% level.
SUPPLEMENTAL MATERIAL
Histological organization of the symbiotic reef-building coral P. damicornis. (A) Schematic representation of a coral colony composed of numerous polyps linked together by coenosarc tissue and their calcareous exoskeleton (aragonite). (B) Optical micrograph of a semithin section (0.5 µm) stained with methylene blue-Azur II from the connective coenosarcal tissue, which is composed of two oral cellular layers (epithelia) facing the seawater (oral epiderm and oral gastroderm) and two aboral epithelia facing the decalcified skeleton (aboral gastroderm and skeletogenic calicoderm). The gastroderm lines a gastro-vascular cavity, named the coelenteron, and contains the Symbiodinium dinoflagellate endosymbionts (red arrow), primarily abundant in the oral gastroderm. OE, oral epiderm; OG, oral gastroderm; coel, coelenteron; AG, aboral gastroderm; cal, calicoderm. Download
Definition of regions of interest (ROIs) for NanoSIMS isotopic quantification of dinoflagellate and coral cells. (A) TEM micrograph (left panel) of an endosymbiotic dinoflagellate cell (displayed in Fig. 1A to C) and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images. (B) TEM micrograph (left panel) of the coral tissue (oral epithelia [displayed in Fig. 2A to C]) and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images. (C) Table reporting 13C and 15N enrichments measured from individual ROIs indicated in panels A and B. Standard deviations of the mean for NanoSIMS data sampled within each individual ROI surface are based on Poisson statistics. Download
Visualization of C and N incorporation and turnover in dinoflagellates during the pulse-chase experiment under light/dark cycling. (A to F) Each sequence includes, from left to right, a representative TEM micrograph of a dinoflagellate cell and its corresponding NanoSIMS 13C/12C and 15N/14N isotopic images at 15 min and 3, 6, 24, 96, and 192 h in the pulse-chase experiment, respectively. ab, accumulation body; nu, nucleus; pl, plastid; pyr, pyrenoid; red arrows, primary starch; blue arrows, secondary starch; green arrows, dinoflagellate LDs; white arrows, extra-algal LDs; black arrows, vesicles containing uric acid crystals. Download
Turnover of C and N in dinoflagellates during the chase under prolonged darkness. (A to D) Each sequence includes a representative TEM micrograph (left panel) of a dinoflagellate cell and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images at 6, 18, 90, and 186 h into the chase period under constant darkness. nu, nucleus; pl, plastid; pyr, pyrenoid; red arrows, primary starch; blue arrows, secondary starch; green arrows, dinoflagellate LDs. Download
Dark control. (A) TEM micrograph of the coral oral epithelia after 6 h of dual isotopic labeling under dark with [13C]bicarbonate (2 mM) and [15N]nitrate (30 µM), following 24 h of pretreatment under constant darkness to inhibit photosynthetic processes. (B and C) Corresponding NanoSIMS 13C/12C (B) and 15N/14N (C) isotopic images. (D) Fluctuations of both 13C and 15N enrichments along the profile depicted in panels B and C. The yellow band indicates the statistical fluctuations of 13C and 15N enrichments measured from similar NanoSIMS profiles in unlabeled control corals (δ13C and δ15N, 0 ± 150‰ [3 SD]). Note the complete lack of isotopic enrichment in the dark. Standard deviations of the mean are based on Poisson statistics. OE, oral epiderm; OG, oral gastroderm; m, mesoglea; dino, dinoflagellate cell; mu, mucocyte. Download
Visualization of the translocation of C and N from dinoflagellates to the coral tissue. (A to E) Each sequence during the pulse-chase experiment under light/dark cycling includes, from left to right, a representative TEM micrograph of the coral oral tissue and its corresponding NanoSIMS 13C/12C and 15N/14N isotopic images. OE, oral epiderm; OG, oral gastroderm; m, mesoglea; dino, dinoflagellate cell; black arrows, coral lipid droplets. Download
Enlarged view of the merged image in Fig. 3D. Download
Carbon turnover in coral glycogen granules. (A to F) Each sequence during the pulse-chase experiment under light/dark cycling includes, from left to right, a representative TEM micrograph of the coral oral epiderm, its corresponding NanoSIMS 13C/12C isotopic map, the merged image between the TEM micrograph and the NanoSIMS isotopic map, and the 13C fluctuations along the depicted NanoSIMS profile. Black arrows point to areas rich in glycogen granules. Standard deviations of the mean are based on Poisson statistics. Download
Incorporation of [13C]glucose into coral glycogen granules. (A) Representative TEM micrograph of the coral oral epiderm after a 6-h incubation under light with [13C6]glucose (30 µM) and (B) its corresponding NanoSIMS 13C/12C isotopic image. (C) Merged image from panels A and B. (D) 13C fluctuations along the profile depicted in panel B. Black arrows point to areas rich in glycogen granules. Standard deviations of the mean are based on Poisson statistics. Download
Excel file reporting summary data tables and P values of statistical analyses from Fig. 1 and 2. Download
ACKNOWLEDGMENTS
This work was supported by European Research Council advanced grant 246749 (BIOCARB), Fonds National Suisse grant CR2312-141048, and the École Polytechnique Fédérale de Lausanne to A.M., CNRS grant Interface 2010 (NanoSIMS and symbiosis) to I.D.C., and the Faculty of Biology and Medicine of the University of Lausanne.
We thank Jean Daraspe, Chakib Djediat, Céline Loussert, and Antonio Mucciolo for providing help with TEM preparations. We are grateful for access to and expert help with aquarium facilities at ATPD, in particular to Jean-Daniel Galois and Sylvain Joumier. Mathieu Pernice, Stephanie Cohen, and Thomas Krueger are thanked for discussions. Two anonymous reviewers are thanked for highly constructive comments and suggestions that helped improve the manuscript.
Footnotes
Citation Kopp C, Domart-Coulon I, Escrig S, Humbel BM, Hignette M, Meibom A. 2015. Subcellular investigation of photosynthesis-driven carbon assimilation in the symbiotic reef coral Pocillopora Damicornis. mBio 6(1):e02299-14. doi:10.1128/mBio.02299-14.
REFERENCES
- 1.Venn AA, Loram JE, Douglas AE. 2008. Photosynthetic symbioses in animals. J Exp Bot 59:1069–1080. doi: 10.1093/jxb/erm328. [DOI] [PubMed] [Google Scholar]
- 2.Yellowlees D, Rees TA, Leggat W. 2008. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ 31:679–694. doi: 10.1111/j.1365-3040.2008.01802.x. [DOI] [PubMed] [Google Scholar]
- 3.Davy SK, Allemand D, Weis VM. 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trench R. 1993. Microalgal-invertebrate symbioses—a review. Resources 9:135–175. [Google Scholar]
- 5.Muscatine L, D’Elia CF. 1978. The uptake, retention, and release of ammonium by reef corals. Limnol Oceangr 23:725–734. doi: 10.4319/lo.1978.23.4.0725. [DOI] [Google Scholar]
- 6.Rahav O, Dubinsky Z, Achituv Y, Falkowski PG. 1989. Ammonium metabolism in the zooxanthellate coral, Stylophora pistillata. Proc R Soc Lond B Biol Sci 236:325–337. doi: 10.1098/rspb.1989.0026. [DOI] [Google Scholar]
- 7.Muscatine L, Cernichiari E. 1969. Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol Bull 137:506–523. doi: 10.2307/1540172. [DOI] [PubMed] [Google Scholar]
- 8.Whitehead LF, Douglas AE. 2003. Metabolite comparisons and the identity of nutrients translocated from symbiotic algae to an animal host. J Exp Biol 206:3149–3157. doi: 10.1242/jeb.00539. [DOI] [PubMed] [Google Scholar]
- 9.Burriesci MS, Raab TK, Pringle JR. 2012. Evidence that glucose is the major transferred metabolite in dinoflagellate-cnidarian symbiosis. J Exp Biol 215:3467–3477. doi: 10.1242/jeb.070946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papina M, Meziane T, Van Woesik R. 2003. Symbiotic zooxanthellae provide the host-coral Montipora digitata with polyunsaturated fatty acids. Comp Biochem Physiol B Biochem Mol Biol 135:533–537. doi: 10.1016/S1096-4959(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 11.Markell DA, Trench RK. 1993. Macromolecules exuded by symbiotic dinoflagellates in culture: amino acid and sugar composition. J Phycol 29:64–68. doi: 10.1111/j.1529-8817.1993.tb00280.x. [DOI] [Google Scholar]
- 12.Patton JS, Abraham S, Benson AA. 1977. Lipogenesis in the intact coral Pocillopora capitata and its isolated zooxanthellae: evidence for a light-driven carbon cycle between symbiont and host. Mar Biol 44:235–247. doi: 10.1007/BF00387705. [DOI] [Google Scholar]
- 13.Stimson JS. 1987. Location, quantity and rate of change in quantity of lipids in tissue of Hawaiian hermatypic corals. Bull Mar Sci 41:889–904. [Google Scholar]
- 14.Harland AD, Navarro JC, Spencer Davies P, Fixter LM. 1993. Lipids of some Caribbean and Red Sea corals: total lipid, wax esters, triglycerides and fatty acids. Mar Biol 117:113–117. doi: 10.1007/BF00346432. [DOI] [Google Scholar]
- 15.Peng SE, Chen WN, Chen HK, Lu CY, Mayfield AB, Fang LS, Chen CS. 2011. Lipid bodies in coral-dinoflagellate endosymbiosis: proteomic and ultrastructural studies. Proteomics 11:3540–3555. doi: 10.1002/pmic.201000552. [DOI] [PubMed] [Google Scholar]
- 16.Von Holt C, Von Holt M. 1968. Transfer of photosynthetic products from zooxanthellae to coelenterate hosts. Comp Biochem Physiol 24:73–81. doi: 10.1016/0010-406X(68)90959-6. [DOI] [PubMed] [Google Scholar]
- 17.Muscatine L, Falkowski PG, Porter JW, Dubinsky Z. 1984. Fate of photosynthetic fixed carbon in light- and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc R Soc Lond B Biol Sci 222:181–202. doi: 10.1098/rspb.1984.0058. [DOI] [Google Scholar]
- 18.Trench R. 1971. The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. I. The assimilation of photosynthetic products of zooxanthellae by two marine coelenterates. Proc R Soc Lond B Biol Sci 177:225–235. [Google Scholar]
- 19.Cooksey KE, Cooksey B. 1972. Turnover of photosynthetically fixed carbon in reef corals. Mar Biol 15:289–292. doi: 10.1007/BF00401387. [DOI] [Google Scholar]
- 20.Crossland CJ, Barnes DJ, Cox T, Devereux M. 1980. Compartmentation and turnover of organic carbon in the staghorn coral Acropora formosa. Mar Biol 59:181–187. doi: 10.1007/BF00396866. [DOI] [Google Scholar]
- 21.Tanaka Y, Miyajima T, Koike I, Hayashibara T, Ogawa H. 2006. Translocation and conservation of organic nitrogen within the coral-zooxanthella symbiotic system of Acropora pulchra, as demonstrated by dual isotope-labeling techniques. J Exp Mar Biol Ecol 336:110–119. doi: 10.1016/j.jembe.2006.04.011. [DOI] [Google Scholar]
- 22.Luo Y-J, Wang L-H, Chen W-NU, Peng S-E, Tzen JT-C, Hsiao Y-Y, Huang H-J, Fang L-S, Chen C-S. 2009. Ratiometric imaging of gastrodermal lipid bodies in coral-dinoflagellate endosymbiosis. Coral Reefs 28:289–301. doi: 10.1007/s00338-008-0462-8. [DOI] [Google Scholar]
- 23.Chen W-NU, Kang H-J, Weis VM, Mayfield AB, Jiang P-L, Fang L-S, Chen C-S. 2012. Diel rhythmicity of lipid-body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs 31:521–534. doi: 10.1007/s00338-011-0868-6. [DOI] [Google Scholar]
- 24.Hosoi K. 1938. Contribution to the biochemistry of the coral. 1. On the occurrence of glycogen and its content in the polyp of Fungia actiniformis var. palawensis Döderlein Palao. Trop Biol Stn 3:447–451. [Google Scholar]
- 25.Hayes RL, Goreau NI. 1977. Intracellular crystal-bearing vesicles in the epidermis of scleractinian corals, Astrangia danae (Agassiz) and Porites porites (Pallas). Biol Bull 152:26–40. doi: 10.2307/1540724. [DOI] [PubMed] [Google Scholar]
- 26.Leggat W, Seneca F, Wasmund K, Ukani L, Yellowlees D, Ainsworth TD. 2011. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6(10):e26687. doi: 10.1371/journal.pone.0026687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle WL, Doyle MM. 1940. The structure of zooxanthellae. Tortugas Lab 32:129–142. [Google Scholar]
- 28.Taylor DL. 1968. In situ studies on the cytochemistry and ultrastructure of a symbiotic marine dinoflagellate. J Mar Biol Assoc 48:349–366. doi: 10.1017/S0025315400034548. [DOI] [Google Scholar]
- 29.Lechene, Francois H, Douglas B, Patrick KJ, Daniel D, Yvette L, Joseph B, Kwon P, Susumu I, Gilles B, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. 2006. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol 5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clode PL, Stern RA, Marshall AT. 2007. Subcellular imaging of isotopically labeled carbon compounds in a biological sample by ion microprobe (NanoSIMS). Microsc Res Tech 70:220–229. doi: 10.1002/jemt.20409. [DOI] [PubMed] [Google Scholar]
- 31.Hoppe P, Cohen S, Meibom A. 2013. NanoSIMS: technical aspects and applications in cosmochemistry and biological geochemistry. Geostand Geoanal Res 37:111–154. doi: 10.1111/j.1751-908X.2013.00239.x. [DOI] [Google Scholar]
- 32.Pernice M, Meibom A, Van Den Heuvel A, Kopp C, Domart-Coulon I, Hoegh-Guldberg O, Dove S, Wang D, Huang R, Chang X, Chain PS, Xie G, Ling J, Xu J. 2012. A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J 6:1314–1324. doi: 10.1038/ismej.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp C, Pernice M, Domart-Coulon I, Djediat C, Spangenberg JE, Alexander DT, Hignette M, Meziane T, Meibom A. 2013. Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4(3):e00052-13. doi: 10.1128/mBio.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clode PL, Saunders M, Maker G, Ludwig M, Atkins CA. 2009. Uric acid deposits in symbiotic marine algae. Plant Cell Environ 32:170–177. doi: 10.1111/j.1365-3040.2008.01909.x. [DOI] [PubMed] [Google Scholar]
- 35.Walther TC, Farese RV. 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldwin PM. 2001. Starch granule-associated proteins and polypeptides: a review. Starch 53:475–503. doi:. [DOI] [Google Scholar]
- 37.Patton JS, Battey JF, Rigler MW, Porter JW, Black CC, Burris JE. 1983. A comparison of the metabolism of bicarbonate 14C and acetate 1-14C and the variability of species lipid compositions in reef corals. Mar Biol 75:121–130. doi: 10.1007/BF00405994. [DOI] [Google Scholar]
- 38.Hughes A, Grottoli A, Pease T, Matsui Y. 2010. Acquisition and assimilation of carbon in non-bleached and bleached corals. Mar Ecol Prog Ser 420:91–101. doi: 10.3354/meps08866. [DOI] [Google Scholar]
- 39.Tremblay P, Grover R, Maguer JF, Legendre L, Ferrier-Pagès C. 2012. Autotrophic carbon budget in coral tissue: a new 13C-based model of photosynthate translocation. J Exp Biol 215:1384–1393. doi: 10.1242/jeb.065201. [DOI] [PubMed] [Google Scholar]
- 40.Downs CA, Kramarsky-Winter E, Martinez J, Kushmaro A, Woodley CM, Loya Y, Ostrander GK. 2009. Symbiophagy as a cellular mechanism for coral bleaching. Autophagy 5:211–216. doi: 10.4161/auto.5.2.7405. [DOI] [PubMed] [Google Scholar]
- 41.DeSalvo MK, Estrada A, Sunagawa S, Medina M. 2012. Transcriptomic responses to darkness stress point to common coral bleaching mechanisms. Coral Reefs 31:215–228. doi: 10.1007/s00338-011-0833-4. [DOI] [Google Scholar]
- 42.Huppe HC, Turpin DH. 1994. Integration of carbon and nitrogen metabolism in plant and algal cells. Annu Rev Plant Physiol Plant Mol Biol 45:577–607. doi: 10.1146/annurev.pp.45.060194.003045. [DOI] [Google Scholar]
- 43.Cullen JJ. 1985. Diel vertical migration by dinoflagellates: roles of carbohydrate metabolism and behavioral flexibility. Contrib Mar Sci 27:135–152. [Google Scholar]
- 44.Muscatine L, Hand C. 1958. Direct evidence for the transfer of materials from symbiotic algae to the tissues of a coelenterate. Proc Natl Acad Sci U S A 44:1259–1263. doi: 10.1073/pnas.44.12.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goreau TF, Goreau NI. 1960. Distribution of labeled carbon in reef-building corals with and without zooxanthellae. Science 131:668–669. doi: 10.1126/science.131.3401.668. [DOI] [PubMed] [Google Scholar]
- 46.Battey JF, Patton JS. 1984. A reevaluation of the role of glycerol in carbon translocation in zooxanthellae-coelenterate symbiosis. Mar Biol 79:27–38. doi: 10.1007/BF00404982. [DOI] [Google Scholar]
- 47.Swanson R, Hoegh-Guldberg O. 1998. Amino acid synthesis in the symbiotic sea anemone Aiptasia pulchella. Mar Biol 131:83–93. doi: 10.1007/s002270050299. [DOI] [Google Scholar]
- 48.Kellogg RB, Patton JS. 1983. Lipid droplets, medium of energy exchange in the symbiotic anemone Condylactis gigantea: a model coral polyp. Mar Biol 75:137–149. doi: 10.1007/BF00405996. [DOI] [Google Scholar]
- 49.Patton JS, Burris JE. 1983. Lipid synthesis and extrusion by freshly isolated zooxanthellae (symbiotic algae). Mar Biol 75:131–136. doi: 10.1007/BF00405995. [DOI] [Google Scholar]
- 50.Harrison PJ, Waters RE, Taylor F. 1980. A broad spectrum, artificial sea water medium for coastal and open ocean phytoplankton. J Phycol 16:28–35. [Google Scholar]
- 51.Gladfelter EH, Michel G, Sanfelici A. 1989. Metabolic gradients along a branch of the reef coral Acropora palmata. Bull Mar Sci 44:1166–1173. [Google Scholar]
- 52.Hill R, Schreiber U, Gademann R, Larkum AWD, Kühl M, Ralph PJ. 2004. Spatial heterogeneity of photosynthesis and the effect of temperature-induced bleaching conditions in three species of corals. Mar Biol 144:633–640. doi: 10.1007/s00227-003-1226-1. [DOI] [Google Scholar]
- 53.Ralph PJ, Schreiber U, Gademann R, Kühl M, Larkum AWD. 2005. Coral photobiology studied with a new imaging pulse amplitude modulated fluorometer. J Phycol 41:335–342. doi: 10.1111/j.1529-8817.2005.04034.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histological organization of the symbiotic reef-building coral P. damicornis. (A) Schematic representation of a coral colony composed of numerous polyps linked together by coenosarc tissue and their calcareous exoskeleton (aragonite). (B) Optical micrograph of a semithin section (0.5 µm) stained with methylene blue-Azur II from the connective coenosarcal tissue, which is composed of two oral cellular layers (epithelia) facing the seawater (oral epiderm and oral gastroderm) and two aboral epithelia facing the decalcified skeleton (aboral gastroderm and skeletogenic calicoderm). The gastroderm lines a gastro-vascular cavity, named the coelenteron, and contains the Symbiodinium dinoflagellate endosymbionts (red arrow), primarily abundant in the oral gastroderm. OE, oral epiderm; OG, oral gastroderm; coel, coelenteron; AG, aboral gastroderm; cal, calicoderm. Download
Definition of regions of interest (ROIs) for NanoSIMS isotopic quantification of dinoflagellate and coral cells. (A) TEM micrograph (left panel) of an endosymbiotic dinoflagellate cell (displayed in Fig. 1A to C) and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images. (B) TEM micrograph (left panel) of the coral tissue (oral epithelia [displayed in Fig. 2A to C]) and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images. (C) Table reporting 13C and 15N enrichments measured from individual ROIs indicated in panels A and B. Standard deviations of the mean for NanoSIMS data sampled within each individual ROI surface are based on Poisson statistics. Download
Visualization of C and N incorporation and turnover in dinoflagellates during the pulse-chase experiment under light/dark cycling. (A to F) Each sequence includes, from left to right, a representative TEM micrograph of a dinoflagellate cell and its corresponding NanoSIMS 13C/12C and 15N/14N isotopic images at 15 min and 3, 6, 24, 96, and 192 h in the pulse-chase experiment, respectively. ab, accumulation body; nu, nucleus; pl, plastid; pyr, pyrenoid; red arrows, primary starch; blue arrows, secondary starch; green arrows, dinoflagellate LDs; white arrows, extra-algal LDs; black arrows, vesicles containing uric acid crystals. Download
Turnover of C and N in dinoflagellates during the chase under prolonged darkness. (A to D) Each sequence includes a representative TEM micrograph (left panel) of a dinoflagellate cell and its corresponding NanoSIMS 13C/12C (middle panel) and 15N/14N (right panel) isotopic images at 6, 18, 90, and 186 h into the chase period under constant darkness. nu, nucleus; pl, plastid; pyr, pyrenoid; red arrows, primary starch; blue arrows, secondary starch; green arrows, dinoflagellate LDs. Download
Dark control. (A) TEM micrograph of the coral oral epithelia after 6 h of dual isotopic labeling under dark with [13C]bicarbonate (2 mM) and [15N]nitrate (30 µM), following 24 h of pretreatment under constant darkness to inhibit photosynthetic processes. (B and C) Corresponding NanoSIMS 13C/12C (B) and 15N/14N (C) isotopic images. (D) Fluctuations of both 13C and 15N enrichments along the profile depicted in panels B and C. The yellow band indicates the statistical fluctuations of 13C and 15N enrichments measured from similar NanoSIMS profiles in unlabeled control corals (δ13C and δ15N, 0 ± 150‰ [3 SD]). Note the complete lack of isotopic enrichment in the dark. Standard deviations of the mean are based on Poisson statistics. OE, oral epiderm; OG, oral gastroderm; m, mesoglea; dino, dinoflagellate cell; mu, mucocyte. Download
Visualization of the translocation of C and N from dinoflagellates to the coral tissue. (A to E) Each sequence during the pulse-chase experiment under light/dark cycling includes, from left to right, a representative TEM micrograph of the coral oral tissue and its corresponding NanoSIMS 13C/12C and 15N/14N isotopic images. OE, oral epiderm; OG, oral gastroderm; m, mesoglea; dino, dinoflagellate cell; black arrows, coral lipid droplets. Download
Enlarged view of the merged image in Fig. 3D. Download
Carbon turnover in coral glycogen granules. (A to F) Each sequence during the pulse-chase experiment under light/dark cycling includes, from left to right, a representative TEM micrograph of the coral oral epiderm, its corresponding NanoSIMS 13C/12C isotopic map, the merged image between the TEM micrograph and the NanoSIMS isotopic map, and the 13C fluctuations along the depicted NanoSIMS profile. Black arrows point to areas rich in glycogen granules. Standard deviations of the mean are based on Poisson statistics. Download
Incorporation of [13C]glucose into coral glycogen granules. (A) Representative TEM micrograph of the coral oral epiderm after a 6-h incubation under light with [13C6]glucose (30 µM) and (B) its corresponding NanoSIMS 13C/12C isotopic image. (C) Merged image from panels A and B. (D) 13C fluctuations along the profile depicted in panel B. Black arrows point to areas rich in glycogen granules. Standard deviations of the mean are based on Poisson statistics. Download
Excel file reporting summary data tables and P values of statistical analyses from Fig. 1 and 2. Download