ABSTRACT
The inner membrane complex (IMC) of Toxoplasma gondii is a peripheral membrane system that is composed of flattened alveolar sacs that underlie the plasma membrane, coupled to a supporting cytoskeletal network. The IMC plays important roles in parasite replication, motility, and host cell invasion. Despite these central roles in the biology of the parasite, the proteins that constitute the IMC are largely unknown. In this study, we have adapted a technique named proximity-dependent biotin identification (BioID) for use in T. gondii to identify novel components of the IMC. Using IMC proteins in both the alveoli and the cytoskeletal network as bait, we have uncovered a total of 19 new IMC proteins in both of these suborganellar compartments, two of which we functionally evaluate by gene knockout. Importantly, labeling of IMC proteins using this approach has revealed a group of proteins that localize to the sutures of the alveolar sacs that have been seen in their entirety in Toxoplasma species only by freeze fracture electron microscopy. Collectively, our study greatly expands the repertoire of known proteins in the IMC and experimentally validates BioID as a strategy for discovering novel constituents of specific cellular compartments of T. gondii.
IMPORTANCE
The identification of binding partners is critical for determining protein function within cellular compartments. However, discovery of protein-protein interactions within membrane or cytoskeletal compartments is challenging, particularly for transient or unstable interactions that are often disrupted by experimental manipulation of these compartments. To circumvent these problems, we adapted an in vivo biotinylation technique called BioID for Toxoplasma species to identify binding partners and proximal proteins within native cellular environments. We used BioID to identify 19 novel proteins in the parasite IMC, an organelle consisting of fused membrane sacs and an underlying cytoskeleton, whose protein composition is largely unknown. We also demonstrate the power of BioID for targeted discovery of proteins within specific compartments, such as the IMC cytoskeleton. In addition, we uncovered a new group of proteins localizing to the alveolar sutures of the IMC. BioID promises to reveal new insights on protein constituents and interactions within cellular compartments of Toxoplasma.
INTRODUCTION
Apicomplexans are a phylum of obligate intracellular parasites that cause a significant disease burden in humans and animals. The most successful of these is Toxoplasma gondii, which infects approximately one-third of the human population globally and is a threat to immunocompromised individuals and neonates (1). The most notorious apicomplexan parasite is Plasmodium falciparum, which is the causative agent of malaria, which claims ~1 million lives annually (2). A hallmark of apicomplexans is the conservation of a number of unique organelles that serve critical roles in parasite biology and thus are attractive targets for the development of novel therapeutic interventions.
One of these unique organelles is the inner membrane complex (IMC), which consists of two distinct elements: a series of flattened membrane sacs called alveoli underlying the parasite plasma membrane and a supporting rigid cytoskeletal network of intermediate filaments (3, 4). In Toxoplasma species, the alveolar sacs of the IMC are arranged as three rows of fused rectangular membrane plates, sutured together like a quilt, and capped by a single large alveolar plate at the apical end of the parasite (5). While Plasmodium merozoites appear to possess only a single alveolar sac, distinct segmented plates that are sutured together are visible in Plasmodium gametocytes (6, 7). In apicomplexans, the alveolar sacs and underlying cytoskeleton are layered on top of subpellicular microtubules emanating from an apical microtubule-organizing center (8).
The IMC has important functions in parasite motility, host cell invasion, and intracellular replication. The outer leaflet of the IMC membrane acts as the anchor for the actin-myosin motor that powers parasite gliding and invasion (9, 10). In addition, the IMC serves as the structural scaffold for the formation of daughter cells within the mother during asexual reproduction. In Toxoplasma, two daughter buds arise from the maternal parasite, a process called endodyogeny, while in Plasmodium, multiple daughters are formed, termed merogony (11). The nascent IMC appears at the onset of bud formation, and replicated organelles are packaged within this compartment as the buds elongate. The daughter buds complete their maturation by adopting the maternal plasma membrane.
Despite the crucial roles of the IMC, the protein composition of this cellular compartment is still largely unknown. The majority of IMC proteins that have been identified to date belong to a family of articulin-like proteins called alveolins, which are characterized by conserved repeat motifs flanked by variable sequences (12, 13). These proteins are one of the distinguishing hallmarks of the Alveolata infrakingdom that includes apicomplexans, dinoflagellates, and ciliates. In apicomplexans, the alveolins form part of the cytoskeletal network that provides structural stability to the IMC (8).
Even fewer proteins have been localized to the alveolar subcompartment of the IMC. Some of these include components of the parasite actin-myosin motor, such as Gap40, Gap45, and Gap50, which form a complex that recruits the myosin machinery, including MyoA and MLC1, to the IMC (14–16). Another subset of proteins in the membrane sacs is the IMC subcompartment proteins (ISP1, -2, -3, and -4), which are tethered to the membrane bilayers via posttranslational acylations (17, 18). In T. gondii, each ISP localizes to distinct subcompartments of the IMC, which may be delineated by specific subsets of alveoli (17, 18). For example, ISP1 is found exclusively in the apical cap, while ISP2 and -4 are present in the center of the IMC but not the cap or base of the parasite. ISP3, on the other hand, localizes to the center and the base of the IMC. Although their precise functions are unknown, the ISPs appear to be involved in daughter bud formation, as disruption of ISP2 leads to a dysregulation of endodyogeny (17).
The paucity of characterized IMC proteins led us to identify new protein constituents of this organelle. We utilized a technique named BioID that has been developed to uncover novel proteins and elucidate complex networks of protein-protein interactions (19). The BioID approach relies on the proximity-dependent biotinylation of interacting partners and proximal proteins by a promiscuous biotin ligase, BirA*, that is fused to the protein of interest. BirA* contains a mutation that abolishes substrate specificity, allowing it to label proteins interacting with or proximal to the ligase-target protein fusion within its native environment. The biotinylated partners or near neighbors can then be affinity purified for identification by mass spectrometry. The method has proved particularly valuable for cytoskeletal or membrane-bound bait proteins, which are solubilized only under harsh detergent conditions that often disrupt protein-protein interactions in standard immunoprecipitation assays. A further advantage of BioID is the ability to capture transient or weak interactions that could be missed by other techniques. This approach has been successfully used to uncover new components of the nuclear lamina, nuclear pore, and centrioles in mammalian cells, as well as novel bilobe proteins in Trypanosoma brucei (19–22).
We adapted the BioID technique for T. gondii and, as a proof of principle, used the IMC membrane protein ISP3 as bait to identify novel IMC proteins. We demonstrate that the ISP3-BirA* fusion localizes properly to the IMC, is catalytically active, and labels previously described IMC proteins as well as novel targets. A number of these targets were verified by epitope tagging, thereby validating our approach. We further characterized two of these proteins by gene disruption and show that one of them appears to be essential in Toxoplasma. To evaluate labeling in a different suborganellar compartment of the IMC, we then applied the BioID approach to a novel IMC cytoskeletal network protein that is restricted to the apical cap. An array of known and novel targets was again identified, of which a selection of new targets were verified by localization. Of particular interest, we uncovered a group of proteins that localize to the transverse and longitudinal sutures of the rectangular alveolar plates and demonstrate that these novel constituents associate with both the membrane and cytoskeletal elements of the IMC. Together, these studies establish a methodology for identifying novel proteins in Toxoplasma subcellular compartments, substantially increase the repertoire of known IMC proteins, and provide a foundation for evaluating protein-protein interactions in the IMC.
RESULTS
ISP3-BirA* biotinylates proteins in the IMC.
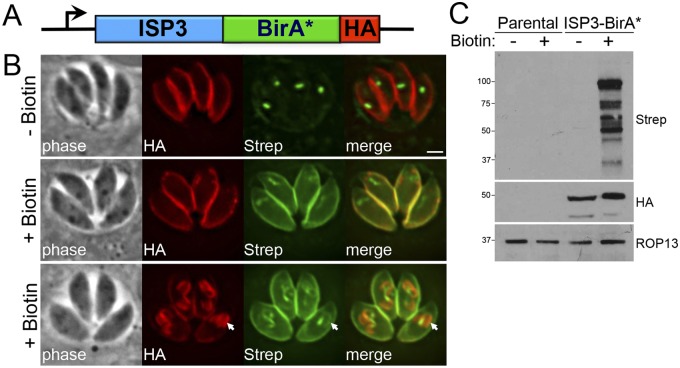
To identify novel IMC components by BioID, we generated a construct containing ISP3 fused to BirA* with a C-terminal hemagglutinin (HA) tag, driven from the ISP3 promoter (Fig. 1A). The ISP3-BirA* fusion was integrated into the genome of RHΔhpt (parental) parasites, and localization of the fusion was assessed by immunofluorescence assay (IFA). As expected, we observed staining in the central and posterior subcompartments of the IMC but not in the apical cap (Fig. 1B), a pattern consistent with endogenous ISP3 (17). Correct IMC localization was confirmed by colocalization with IMC3 (see Fig. S1A in the supplemental material).
FIG 1 .
ISP3-BirA* biotinylates proteins in the parasite IMC. (A) Diagram of the expression cassette encoding ISP3 fused to BirA*, plus a C-terminal 3× HA epitope tag, driven by the ISP3 promoter. (B) IFA of ISP3-BirA*-expressing parasites, grown for 24 h with or without biotin. ISP3-BirA* localizes to the parasite periphery and biotinylates proteins in a biotin-dependent manner. Endogenously biotinylated proteins in the apicoplast are detectable as the background even in the absence of biotin. Bottom panel, ISP3-BirA* biotinylation activity is detectable but not enriched in daughter buds (arrows). Red, mouse anti-HA antibody; green, streptavidin-Alexa 488. Scale bar = 2 µm. (C) Western blot comparing the profile of biotinylated proteins from lysates of parental and ISP3-BirA* parasites.
To determine if ISP3-BirA* could label proximal proteins in the IMC, we grew parasites in medium supplemented with biotin and stained for biotinylated proteins using fluorophore-conjugated streptavidin. Wild-type T. gondii parasites have endogenously biotinylated proteins in the apicoplast (and to a lesser degree in the mitochondria) that are labeled by streptavidin, thus representing the substantial background of this method in Toxoplasma compared with other cell types (Fig. 1B, top) (23). In ISP3-BirA*-expressing parasites, we also detected robust streptavidin staining in the periphery that colocalized with ISP3-BirA*, indicating that the biotin ligase fusion was active and labeled proteins in the IMC (Fig. 1B, middle). Surprisingly, while ISP3-BirA* was enriched in daughter buds during endodyogeny (see Fig. S1A), lower but detectable amounts of biotinylation were seen in daughter buds (Fig. 1B, bottom).
We then assessed the labeling of T. gondii proteins by ISP3-BirA* in whole-cell lysates by Western blotting with a streptavidin-horseradish peroxidase (HRP) probe. Although background biotinylated proteins were detectable in parental lysates when the blot was overexposed (see Fig. S1B), there was a significant increase in biotinylated proteins in lysates of ISP3-BirA* parasites, and only when biotin was present in the growth medium (Fig. 1C). These labeled proteins were unique to ISP3-BirA* parasites and did not appear to correspond to background proteins from the parental line. Collectively, these data demonstrate that ISP3-BirA* is active and biotinylates proteins at the parasite periphery.
Identification of novel IMC proteins by BioID.
To identify targets labeled by ISP3-BirA*, we affinity purified biotinylated proteins from parasite lysates for MuDPIT mass spectrometric analysis. Biotinylated proteins from both parental and ISP3-BirA* lysates were significantly enriched using streptavidin-conjugated magnetic beads (Fig. 2A). Proteins identified by mass spectrometry that were unique to or significantly enriched in ISP3-BirA* samples were scored as hits based on the number of identified spectra and unique peptides (see Table S1 in the supplemental material). The highest-scoring protein in our data set was the ISP3 bait, which is in agreement with previous BioID studies in which the bait-BirA* fusion biotinylated itself and was the top hit (19–22). Importantly, some of the highest-scoring hits in our data set included proteins known to associate with IMC membranes: MLC1, GAP45, GAP50, HSP20, Rab11b, and ISP1 (14–17, 24, 25). In addition, proteins in the IMC cytoskeletal network, such as IMCs 1, 10, and 14, were enriched in our data set (4, 12). These data indicate that ISP3-BirA* biotinylates proteins in both the membrane sacs and the underlying network of the IMC.
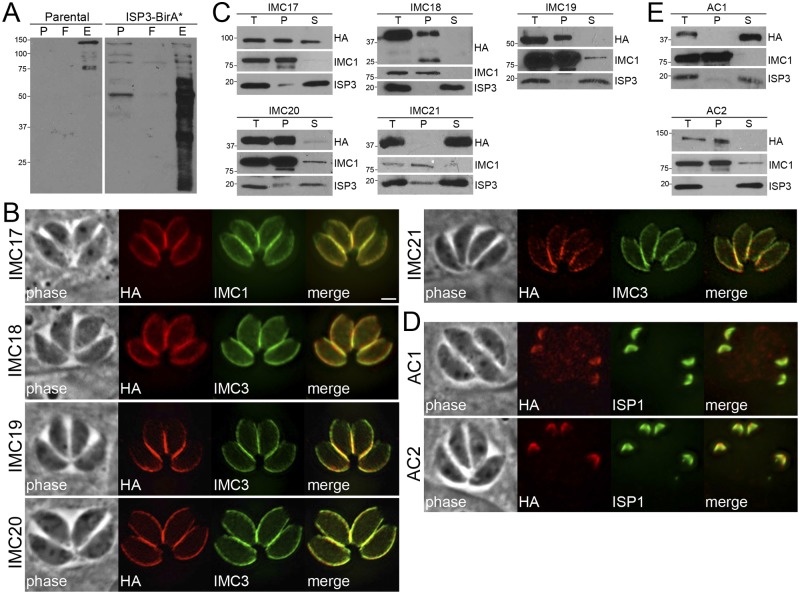
FIG 2 .
Identification of novel IMC proteins by ISP3 BioID. (A) Western blot showing enrichment of biotinylated proteins from parental and ISP3-BirA* lysates by streptavidin magnetic beads, as assessed by monitoring of the precolumn (P), flowthrough (F), and elution (E) fractions. (B) ISP3 BioID hits were localized to the IMC by endogenous tagging, where they colocalize with the IMC markers IMCs 1/3. Similar to ISP3-BirA*, these proteins stain the central and basal subcompartments of the IMC but not the apical cap. Red, mouse or rabbit anti-HA antibodies; green, mouse anti-IMC1 or rat anti-IMC3 antibodies. Scale bar = 2 µm. (C) Western blots showing detergent extraction analysis of ISP3 BioID hits. The total lysate (T) was partitioned into the insoluble pellet (P) or soluble (S) fractions. Fractionation was monitored using IMC1 (insoluble) and ISP3 (soluble) controls. (D) IFA showing two additional ISP3 BioID hits that localize to the apical cap demarked by ISP1. Red, rabbit anti-HA antibody; green, mouse anti-ISP1 antibody. (E) Detergent extractions of ACs 1/2 as described in part C.
Next, we utilized gene expression data to filter hits from our BioID data set for further investigation (summarized in Table 1). IMC proteins are synthesized de novo during endodyogeny, with their gene expression peaking during each round of daughter bud formation at the S and/or M phase (26). We selected a group of hypothetical proteins with this signature cyclical expression pattern similar to known IMC proteins (see Fig. S2A in the supplemental material) and determined their localization in parasites via epitope tagging at their C termini.
TABLE 1 .
Summary of IMC proteins identified in this studya
| Name | TgGT1 gene identifier |
Localization | TX-100 extraction |
Daughters? | P. falciparum PF3D7 identifier | Mol wt | Description/note(s) |
|---|---|---|---|---|---|---|---|
| IMC17 | 286580 | Center/base | Insoluble | No | None | 107 | Contains predicted α/β hydrolase fold |
| IMC18 | 295360 | Center/base | Insoluble | No | None | 30 | Knockout produced no apparent phenotype in vitro; Predicted palmitoylation site at Cys-266 |
| IMC19 | 217510 | Center/base | Insoluble | Yes | None | 42 | Hypothetical protein; predicted palmitoylation site at Cys-362; signal brighter in posterior IMC |
| IMC20 | 271930 | Center/base | Insoluble | No | 1447500 | 26 | Hypothetical protein |
| IMC21 | 232030 | Center/base | Soluble | Yes | None | 23 | Predicted palmitoylation site at Cys-2; signal brighter in posterior IMC |
| IMC22 | 316340 | Center/base | Insoluble | No | None | 26 | Hypothetical protein |
| IMC23 | 304670 | Center/base | Insoluble | Yes | None | 38 | Contains predicted leucine-rich repeats |
| IMC24 | 258470 | Center/base | Insoluble | No | None | 31 | Hypothetical protein |
| AC1 | 311480 | Apical cap | Soluble | Yes | None | 42 | Detected in daughters before ISP1; predicted palmitoylation site at Cys-8 |
| AC2 | 250820 | Apical cap | Insoluble | Yes | None | 121 | Detected in daughters after ISP1 |
| AC3 | 308860 | Apical cap | Insoluble | Yes | None | 66 | Detected in daughters after ISP1 |
| AC4 | 214880 | Apical cap | Insoluble | Yes | None | 98 | Detected in daughters after ISP1 |
| AC5 | 235380 | Apical cap | Insoluble | Yes | None | 65 | Detected primarily in daughters (low signal in maternal IMC) and before ISP1 |
| AC6 | 251850 | Apical cap | Soluble | Yes | 1423300 | 117 | Predicted serine/threonine protein phosphatase; detected in daughters after ISP1 |
| AC7 | 225690 | Apical cap | Insoluble | Yes | None | 194 | Predicted palmitoylation sites at Cys-1255, Cys-1401 |
| ISC1 | 235340 | Sutures | Insoluble | Yes | 1341500 | 77 | Predicted palmitoylation site at Cys-573 |
| ISC2 | 219170 | Sutures | Soluble | Yes | None | 33 | Hypothetical protein |
| ISC3 | 220930 | Sutures | Soluble | Yes | 1431900 | 74 | 8 predicted transmembrane domains; enriched in daughter cells compared to other ISCs |
| ISC4 | 305930 | Sutures | Insoluble | Yes | None | 26 | Predicted palmitoylation site at Cys-3 |
Predicted palmitoylation sites scoring >8.0 in CSS Palm 4.0 (http://csspalm.biocuckoo.org) are listed. No predicted myristoylation sites were detected (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm).
Characterization of novel IMC proteins identified by BioID.
Among our ISP3-BirA* hits, we epitope-tagged six proteins that shared the same localization in parasites as ISP3: TgGT1_286580 (here referred to as IMC17), TgGT1_295360 (IMC18), TgGT_217510 (IMC19), TgGT1_271930 (IMC20), TgGT1_232030 (IMC21), and TgGT1_313380 (Table 1). One of these proteins, TgGT1_313380, had been independently identified from a purification of the MORN1 basal complex, designated ILP1, and characterized elsewhere (27). These proteins have no known functions (Table 1), although IMC17 was annotated as a putative lysophospholipase based on an α/β hydrolase fold predicted by BLAST analysis (28). We localized these hits to the central and basal subcompartments of the IMC but not in the apical cap (Fig. 2B), consistent with proximal biotinylation by ISP3-BirA*. In agreement with the preferential labeling of the maternal IMC by the ISP3-BirA fusion (Fig. 1B, bottom), IMCs 17/18/20 appear to be deposited onto the maternal IMC after endodyogeny and were not detected in daughter buds (see Fig. S2B in the supplemental material). In contrast, IMCs 19/21 were detected in both the maternal IMC and nascent daughter buds, as shown by colocalization with IMC3 during endodyogeny (see Fig. S2C).
Next, we used detergent extraction assays to determine whether these IMC proteins are associated with the membrane sacs or the underlying cytoskeleton (Fig. 2C). Proteins embedded in the IMC network (such as the alveolins) are resistant to extraction by the nonionic detergent Triton X-100, whereas proteins attached to the membrane sacs (such as the ISPs) are solubilized under these conditions (4). IMCs 18/19/20 were entirely resistant to extraction by Triton X-100, indicating these proteins are anchored into the network of the IMC. IMC17 was partially soluble in these extraction assays, suggesting an association with the underlying cytoskeleton that is not as firm as these other novel IMC proteins or alveolins. In contrast, IMC21 appears to localize to the membrane sacs, as it was solubilized in detergent. This result is supported by the presence of a predicted palmitoylation site (a cysteine residue at position 2) that may mediate tethering to the IMC membranes. The identification of hits from both the membrane sacs and the underlying cytoskeleton indicates that ISP3-BirA* labels proteins in both of these IMC suborganellar compartments.
In addition to these proteins in the central/posterior regions of the IMC, we also localized two proteins to the apical cap of the parasite: TgGT1_311840 and TgGT1_250820. TgGT1_311840 was one of the highest-scoring hits on our data set. TgGT1_250820, on the other hand, had a much lower rank but was chosen for analysis based on its IMC-like cyclical expression pattern (see Fig. S2A). Staining of TgGT1_311840 and TgGT1_250820 overlapped perfectly with that of ISP1 in both mature and dividing parasites (Fig. 2D and Fig. S2C in the supplemental material, respectively), demonstrating these proteins also localize to the nascent IMC during replication. We subsequently designated TgGT1_311840 and TgGT1_250820 as apical cap protein 1 (AC1) and AC2. Consistent with extraction data from other ISP3 BioID hits, we found these two apical cap proteins reside in separate subcompartments of the IMC: AC1 associates with the membrane sacs (consistent with a predicted palmitoylation site at Cys-8), while AC2 is anchored into the cytoskeletal network (Fig. 2E). Two additional proteins from the ISP3 BioID data set were localized to a unique compartment of the IMC and are discussed below. As expected, not all proteins chosen for endogenous tagging localized to the IMC: three proteins, TgGT1_240570, TgGT1_320740, and TgGT1_232340, stained punctate vesicular structures throughout the parasite (see Table S1 and Fig. S2D in the supplemental material).
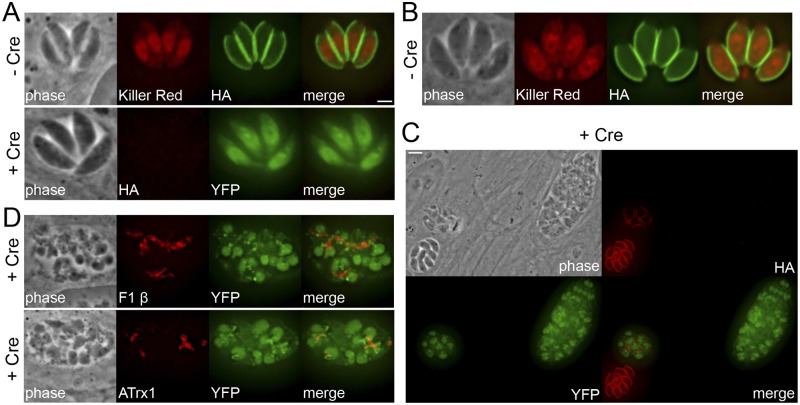
Of these new IMC proteins that we localized, we chose to functionally characterize IMC18 and ILP1 by gene disruption using a combinatorial epitope-tagging/Cre-lox strategy (see the approach in Fig. S3A in the supplemental material). This strategy is a modification of the Cre-lox gene excision system previously established for Toxoplasma (29). We endogenously tagged both the target gene and the upstream gene with an epitope tag followed downstream by an loxP site, thereby flanking the target gene with loxP sites. We performed this genetic engineering in DiCre-ku80::KillerRedfloxYFP parasites that express inducible Cre recombinase and also activate yellow fluorescent protein (YFP) expression upon induction with rapamycin (30). In both target strains, we observed the loss of the 3× HA-tagged target protein at the IMC as well as concomitant expression of YFP upon induction of Cre activity (Fig. 3A and C). Disruption of IMC18 did not have any observable gross effects on parasite morphology, as assessed by IFA (Fig. 3A, bottom), or parasite growth, as measured by plaque assays (data not shown). In contrast, Δilp1 vacuoles were highly disordered and contained morphologically abnormal parasites that appeared enlarged and/or malformed (Fig. 3C), defects that were reminiscent of ILP1-overexpressing parasites (27). In addition, we observed a general mislocalization of organelles, such as mitochondria and apicoplasts, within parasites lacking ILP1 (Fig. 3D). We were unable to successfully clone an ILP1-null, YFP-positive parasite after repeated rounds of limiting dilution from knockout-positive pools, suggesting ablation of this gene is lethal.
FIG 3 .
Gene disruption of selected ISP3 BioID hits using a combinatorial epitope-tagging/Cre-lox strategy. (A) IFA showing parasites with IMC18 endogenously tagged in the DiCre-ku80::KillerRedfloxYFP background (30), grown with or without rapamycin induces Cre recombinase, resulting in excision of the tagged gene. Cre-mediated recombination is monitored by a chromosomal copy of loxP-KillerRed-loxP-YFP under the control of the p5RT70 promoter, which drives YFP expression after the floxed KillerRed gene is excised. Top, untreated parasites. Green, mouse anti-HA antibody. Bottom, ablation of IMC18 had no observable gross effects on parasite morphology as assessed by IFA. Red, mouse anti-HA antibody. Scale bar = 2 µm. (B) IFA showing untreated parasites with ILP1 endogenously tagged in the same background. Green, mouse anti-HA antibody. (C) Disruption of ILP1 leads to disordered vacuoles containing aberrant and misshapen parasites. Red, mouse anti-HA antibody. Scale bar = 5 µm. (D) IFA showing mislocalized mitochondria (top) and apicoplasts (bottom) in ILP1-null parasites. Red, mouse anti-F1 β or mouse anti-ATrx1 antibody.
AC2-BirA* biotinylates proteins in the apical cap of T. gondii.
We also wanted to explore whether the BioID approach was applicable to the cytoskeletal network of the IMC. We simultaneously chose to focus on proteins in the apical cap, and thus we fused BirA* to the cytoskeletal apical cap protein AC2 (Fig. 4A). In this case, we engineered the BirA* sequence into a vector for endogenous tagging and integrated it into the AC2 locus by homologous recombination. The resulting AC2-BirA* fusion localized correctly to the apical cap and also labeled proteins in the apex of mature and dividing parasites upon addition of biotin, as detected by streptavidin staining that colocalized with the bait protein (Fig. 4A).
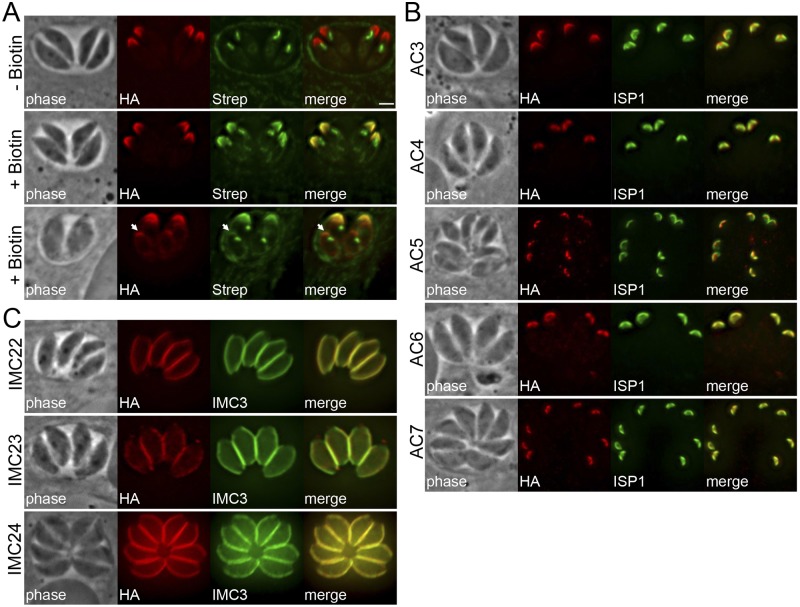
FIG 4 .
Identification of novel IMC proteins by AC2 BioID. (A) IFA of AC2-BirA*-expressing parasites, grown for 24 h with or without biotin. AC2-BirA* biotinylates proteins in the apical cap in mature (middle) and daughter (bottom, arrows) parasites. Red, mouse anti-HA antibody; green, streptavidin-Alexa488. Scale bar = 2 µm. (B) IFA showing AC2 BioID hits localized to the apical cap demarked by ISP1. Red, rabbit anti-HA antibody; green, mouse anti-ISP1 antibody. (C) Three additional AC2 BioID hits localized to the center and posterior of the IMC, as assessed by costaining with IMC3. Red, mouse anti-HA antibody; green, rat anti-IMC3 antibody.
To identify the proteins labeled by AC2-BirA*, we again purified biotinylated proteins from parasites and analyzed them by MuDPIT mass spectrometry (see Table S2 in the supplemental material). Our top hits were enriched for the alveolins IMC1, -3, -4, -6, -7, -10, -12, -13, and -14 (12), indicating that AC2-BirA* preferentially labels proteins in the IMC cytoskeletal network. In addition, several IMC proteins prominent in the apical cap, including ISP1, PhIL1, and IMC11, scored highly in our experiment (12, 17, 31). These hits suggest that the BirA* ligase, when fused to the appropriate bait protein, can be used to selectively biotinylate proteins in specific suborganellar compartments of the parasite.
Identification of cytoskeletal proteins in the apical cap of T. gondii by BioID.
To determine if AC2-BirA* labeled novel targets in the apical cap, we localized several hypothetical proteins with IMC-like cyclical expression patterns from our hits (see Fig. S4A in the supplemental material). By endogenous tagging, we identified 5 novel proteins in the apical cap: TgGT1_308860 (designated AC3), TgGT1_214880 (AC4), TgGT1_235380 (AC5), TgGT1_251850 (AC6), and TgGT1_225690 (AC7). All of these proteins colocalized with the apical cap marker ISP1 in both parental and daughter parasites (Fig. 4B; see also Fig. S4B). In addition, we localized 3 proteins from this data set to the central and posterior regions of the IMC: TgGT1_316340 (IMC22), TgGT1_304670 (IMC23), and TgGT1_258470 (IMC24), all of which colocalized with IMC3 (Fig. 4C). Importantly, we determined using detergent extraction assays that 7 of these 8 proteins reside in the IMC cytoskeleton (see Fig. S5 in the supplemental material), validating BioID as an approach to identify cytoskeletal components in T. gondii parasites. Consistent with our previous BioID experiment, we also tagged two proteins that are not in the IMC: TgGT1_249870 labeled cytoplasmic puncta in the parasite while TgGT1_232130 localized to the subpellicular microtubules (see Table S2 and Fig. S4C). During the course of these studies, AC5 and TgGT1_232130 were detected by mass spectrometric analysis of immunoprecipitations of the subpellicular microtubule-associated protein TrxL1, although their localization was not determined (32).
Identification of proteins that localize to the alveolar sutures of the IMC.
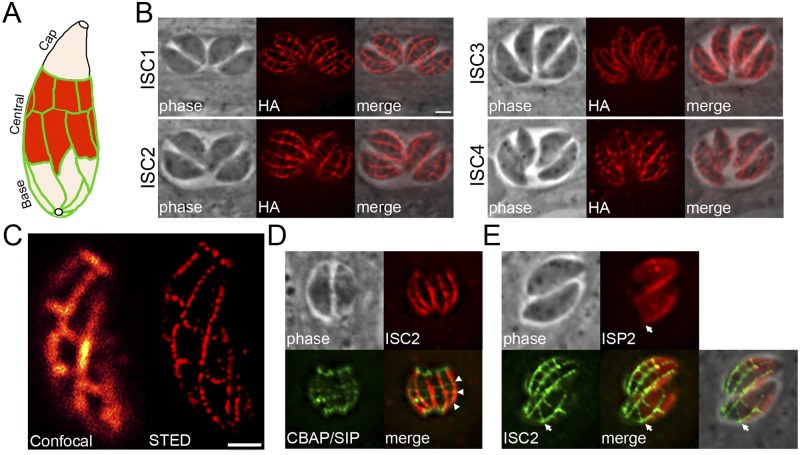
The IMC alveoli are organized as a patchwork of rectangular plates plus the single cone-shaped plate termed the apical cap (Fig. 5A). These plates are sutured together by unknown components to form the IMC. Recently, a protein called CBAP/SIP was identified that appears to label the transverse junctions of the IMC plates, as well as more prominent rings at the apical and basal ends of the parasite (33, 34). One of the most remarkable findings of our IMC BioID approach was the discovery of three proteins (TgGT1_235340 and TgGT1_219170 from the ISP3 BioID data set and TgGT1_220930 from AC2 BioID) that localize to the precise pattern expected for the IMC sutures. This suture staining pattern labels both longitudinal and transverse structures extending along the center and posterior of the IMC (Fig. 5B) but not the apical cap or subpellicular microtubules (see Fig. S6A in the supplemental material) (32). This staining agrees well with the approximate number, size, and shape of the IMC plates shown in freeze fracture electron microscopy studies (5). In independent gene-tagging studies in our lab, we also discovered a fourth IMC suture protein that we also report here (TgGT1_305930). Since TgGT1_235340, TgGT1_219170, TgGT1_220930, and TgGT1_305930 are the first markers that delineate the entirety (e.g., both the longitudinal and transverse) of the sutures between the membrane plates, we designated these proteins IMC suture components (ISCs) 1 to 4, respectively.
FIG 5 .
Identification of proteins that localize to the alveolar sutures of the IMC. (A) Diagram of the IMC membrane sacs subdivided into cap, central, and basal compartments, delineated by the sutures between each membrane sac (green). (B) IFA showing four novel ISC proteins that localize to the IMC sutures. Staining corresponding to both transverse and longitudinal sutures is detectable in parasites. Red, mouse anti-HA antibody. Scale bar = 2 µm. (C) Comparison of ISC3 staining as visualized by confocal (left) and STED microscopy (right). Red, mouse anti-HA antibody. Scale bar = 1 µm. (D) IFA showing that ISC2 and CBAP/SIP colocalize at the transverse sutures of the IMC (arrowheads). Red, rabbit anti-HA antibody; green, mouse anti-Myc antibody. (E) IFA showing that the sutures, labeled by ISC2, form the posterior boundary (arrows) for the central subcompartment of the IMC, demarked by staining of ISP2 in parasites transfected with pISP2-HA-HPT. Red, rabbit anti-HA antibody; green, mouse anti-Myc antibody.
While ISCs 1, 2, and 4 do not contain any apparent functional domains by BLAST analyses, ISC3 contains similarity to choline transporter-like proteins, which may suggest a role in IMC membrane biogenesis (35). Each of the ISCs can be detected on the nascent IMC during endodyogeny, although ISC3 is much more prominent in the budding daughters compared to the other ISCs (see Fig. S6B). ISC3 is also unique in that it contains 8 predicted transmembrane domains that presumably span the alveolar membranes. In agreement with this, ISC3 is released with other IMC membrane proteins upon detergent extraction (see Fig. S6C). ISC2 is also soluble in these assays, although it lacks predicted transmembrane domains or acylation sites, suggesting indirect attachment to the IMC membranes and/or a loose interaction with the cytoskeleton. Surprisingly, ISCs 1/4 appear to be firmly embedded into the IMC cytoskeleton but also have predicted palmitoylation sites that may tether them to the membranes (Table 1). Taken together, these data indicate that there are both membrane and cytoskeletal components of the IMC sutures.
We further examined the ISCs with superresolution microscopy and in relation to other IMC proteins. We visualized the brightest-staining ISC (ISC3) using both confocal and superresolution (STED) microscopy and found that the longitudinal and transverse IMC sutures resolved as puncta rather than continuous lines at higher resolutions (Fig. 5C). We also costained ISC2 with CBAP/SIP, which showed colocalization at the transverse sutures (Fig. 5D), in agreement with their initial discoveries (33, 34). Finally, we had previously hypothesized that the posterior boundary of ISP2, which localizes to the central subcompartment of the IMC, is delineated by discrete alveolar plates (17). To examine this possibility, we colocalized ISP2 with ISC2 and found that the ISP2 subcompartment does indeed terminate at an alveolar junction, as the edges of the ISP2 boundary aligned perfectly with a transverse suture (Fig. 5E). These data provide support for a model in which the IMC subcompartments are defined by specific subsets of alveoli (17).
DISCUSSION
The Toxoplasma IMC consists of distinct membrane and cytoskeletal elements that can be problematic for identifying protein-protein interactions through conventional methods, particularly transient or weaker interactions. In addition, only a small number of IMC proteins (~30) have been identified, and the IMC total protein composition is largely unknown. To circumvent these problems, we adapted the BioID technique for T. gondii and demonstrate that BirA* fusions can biotinylate a robust number of proteins in the IMC. These proteins were identified via MuDPIT despite the inherent background of endogenous biotinylated proteins present in the apicoplast (23). Determining whether these proteins are bona fide interacting partners or merely proximal proteins will need to be addressed using other approaches, such as coimmunoprecipitations, two-hybrid assays, or split green fluorescent protein (GFP). However, we anticipate that the highest-scoring mass spectrometry hits are more likely to represent true binding partners. Additional experimental replicates and reverse BioID experiments with prey proteins will also help to verify interactions and may reveal important interaction networks within the IMC.
We chose the IMC membrane protein ISP3 as bait for the initial BirA* fusion, because ISP3 can be fused to a large tag, such as YFP, and traffic correctly to the IMC (17). Because ISP3 is inserted into the IMC membranes by coordinated acylations (17), we hypothesized that ISP3-BirA* should label proteins in the alveoli and potentially some in the adjacent IMC cytoskeleton. Indeed, our ISP3 BioID data set contains known and novel proteins from both suborganellar compartments (see Table S1 in the supplemental material). Surprisingly, while the ISP3-BirA* fusion targets properly and is enriched in daughter buds like endogenous ISP3 (17), its biotinylation activity appears to be mostly confined to the maternal IMC (Fig. 1B, bottom). This suggests that biotinylation activity of the fusion is altered during endodyogeny or modulated by the surrounding compartment. Alternatively, the lower signal in the nascent IMC could be attributed to the fact that daughter buds have less time to accumulate the biotinylation signal than the maternal IMC. In agreement with this activity, the novel proteins identified by ISP3 BioID are present in, and in some cases restricted to, the maternal IMC. The biotinylation of ISCs by ISP3-BirA* is not surprising, as the sutures appear to be in direct contact with the alveolar membrane sacs. The identification of two new apical cap proteins via ISP3 BioID is less expected but could be attributed to labeling at the junctions between the cap and its neighboring alveolar plates or during early stages of daughter bud formation. It is also clear that ISP3-BirA* does not label the entire subcompartment, as our data set did not include the ISP family members ISP2 and ISP4, whose localizations partially overlap ISP3 in the IMC (17, 18). We anticipate that the extent of biotinylation by BirA* fusions will be dependent on the activity of the particular bait used as well as its proximity to other proteins. It is also important to note that filtering our hits from both data sets described here based on IMC-like gene expression profiles yielded proteins that do not localize to the IMC. Four of these proteins stained punctate vesicular structures in the parasite and one labeled the subpellicular microtubules (see Table S1 and S2 and Fig. S2D and S4C in the supplemental material). Although they were enriched above background proteins from parental parasites, it is not known whether these five hits represent true interactors/near neighbors of our bait proteins.
To begin functional analyses of these novel IMC proteins, we used a modification of the Cre-lox system developed previously to disrupt the genes encoding IMC18 and ILP1 (29). This modified system requires that both the target gene and the upstream gene are arranged in the same orientation in the genome and amenable to endogenous tagging, which results in loxP sites flanking the gene of interest (developed by the M. W. White lab; see Fig. S3A in the supplemental material). This approach has the added benefit of localizing the upstream gene product, if it is unknown (see Fig. S3B and D). Δimc18 parasites displayed no apparent phenotype in vitro, although this result should be taken with caution, as significant compensation has recently been reported in other gene knockouts in T. gondii (36–38). IMC18 does not appear to have paralogs that are detectable by BLAST analysis (data not shown), but we cannot rule out the possibility of other IMC proteins providing functional redundancy. In contrast, disruption of ILP1 resulted in gross morphological defects (Fig. 3C and D) and appears to be lethal to the parasite. These data are in agreement with overexpression studies of ILP1 in Toxoplasma, which resulted in aberrant nuclei and IMC cytoskeletons (27). Similarly, ablation of the ILP1 homolog in Plasmodium berghei produced enlarged, swollen parasites with microtubule and motility defects (39). While the ILP1 homolog is not essential in P. berghei, we were unable to obtain a viable T. gondii Δilp1 mutant in spite of extensive rounds of cloning Cre-induced knockout parasites. It is possible that Plasmodium contains compensatory factors and that their homologs in Toxoplasma are either not present or have lost their ability to suffice in the absence of ILP1. While the tagging/knockout approach used here is rapid and inducible with rapamycin only a subset of the genes in our ISP3 BioID data set have the correct gene orientation for this approach. The development of clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 gene disruption systems in T. gondii will facilitate the functional analysis of large groups of novel genes, such as those identified in this study (40, 41).
The results from our second BioID experiment with AC2 demonstrate the utility of the technique for exploring cytoskeletal networks in T. gondii and for the targeted enrichment of proteins from specific subcompartments of the parasite. In this case, we observed robust labeling of known and novel cytoskeletal proteins in the IMC, many of which are restricted to the apical cap (see Table S2). Interestingly, we found that the 15 novel IMC proteins identified here segregated into two groups of proteins: those that localize to the central and basal subcompartments of the IMC and those that reside in the apical cap (as opposed to IMC1, which stains throughout the IMC). Our results, taken together with other known IMC proteins (12, 17), suggest that the IMC is a highly compartmentalized organelle containing many compartment-specific proteins. As has been previously described for alveolins (12), we observed that some apical cap proteins were detected earlier or later in daughter buds relative to ISP1 (e.g., AC1 is visible on buds before ISP1, while AC4 is detected after [summarized in Table 1]). Analyzing precisely when each of these markers is detected on the nascent IMC will help provide a spatial and temporal understanding of daughter bud formation during endodyogeny.
Perhaps the most intriguing group of proteins uncovered in this study is the ISCs, which appear to specifically label the sutures of the IMC. These proteins localize to both transverse and longitudinal sutures, clearly demarking the rectangular alveolar sacs of the IMC (Fig. 5B). By labeling the entirety of the sutures, the ISCs support the localization of CBAP/SIP to the transverse sutures and indicate that there are differences in protein composition between transverse and longitudinal sutures (33, 34). The staining pattern of MAL13P1.228, a P. falciparum-specific IMC protein reported to localize primarily to rectangular transverse sutures in gametocytes, appears to be more reminiscent of CBAP/SIP than the ISCs (6). Importantly, the ISCs also provide evidence that the IMC subcompartments, delineated by individual ISPs, represent distinct subsets of alveolar plates (17). Although the precise mechanisms targeting ISPs to their subcompartments are unknown, it is possible that these subsets of alveoli express specific enzymes (such as palmitoyl acyltransferases) or receptors that facilitate proper ISP localization.
Interestingly, the ISCs appear to associate with both the membrane and cytoskeletal elements of the IMC. Of the ISCs identified here, ISC3 most closely fits what might be predicted of a membrane suture protein in that it contains 8 predicted transmembrane domains that are presumably anchored in the alveolar sacs and released upon detergent extraction (see Fig. S6C). ISC3 is also similar to choline transporter-like proteins, which import choline for the synthesis of phospholipids for cellular membranes (35). This may suggest a role for ISC3 in choline acquisition for building IMC membranes. In contrast, ISCs 1/4 are embedded in the cytoskeleton and may provide a molecular bridge between the sutured alveolar sacs and the underlying network. ISC2 is released by detergent extraction but lacks apparent transmembrane domains or acylation sites that could tether it to the IMC membranes. It is tempting to speculate that these four ISCs form a complex at the sutures, with ISC2 linking the membrane-bound ISC3 to the ISCs anchored in the cytoskeleton. However, additional studies will be required to elucidate these interactions and determine the function of the ISCs at the sutures. The ISCs also provide new candidate BioID bait proteins that will undoubtedly reveal more constituents of the alveolar sutures and provide clues on how these ISCs interact to form this critical structure within the parasite.
MATERIALS AND METHODS
Toxoplasma and host cell culture.
The T. gondii RHΔhpt, RHΔku80Δhpt, and modified strains were grown on confluent monolayers of human foreskin fibroblast (HFF) host cells in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, as previously described (42).
Antibodies.
The following previously described primary antibodies were used in immunofluorescence (IFA) or Western blot assays: anti-IMC1 (monoclonal antibody [MAb] 45.15) (43), anti-ISP1 (MAb 7E8) (17), mouse anti-ISP3 (17), anti-F1 ATP synthase β subunit (MAb 5F4) (44), anti-ATrx1 (MAb 11G8) (45), anti-ROP7 (MAb 1B10) (46), rabbit anti-ROP13 (47), and rat anti-IMC3 (12). The hemagglutinin (HA) epitope was detected with mouse anti-HA (MAb HA.11) (Covance) or rabbit anti-HA (Invitrogen). The c-Myc epitope was detected with mouse anti-Myc (MAb 9E10) (NeoMarkers) or rabbit anti-Myc (Sigma).
IFA and Western blotting.
For IFA, HFFs were grown to confluence on coverslips and infected with T. gondii parasites. After 18 to 36 h, the coverslips were fixed and processed for indirect immunofluorescence as previously described (48). Primary antibodies were detected by species-specific secondary antibodies conjugated to Alexa 594/488/350. The coverslips were mounted in Vectashield (Vector Labs) and viewed with an Axio Imager.Z1 fluorescence microscope (Zeiss) as previously described (17).
For Western blotting, parasites were lysed in Laemmli sample buffer (50 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 1% 2-mercaptoethanol, 0.1% bromophenol blue), and lysates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Blots were probed with the indicated primary antibodies, followed by secondary antibodies conjugated to horseradish peroxidase (HRP). Target proteins were visualized by chemiluminescence.
Second-copy expression of ISP3-BirA*.
To generate the ISP3-BirA* fusion, the BirA* sequence (19) was PCR amplified (P1/P2; see Table S3 in the supplemental material) and inserted into pISP3-HA-HPT in frame (17), between the 3′ end of ISP3 and the HA epitope tag, to generate pISP3-BirA*-HA-HPT. One hundred micrograms of the construct was linearized with HindIII and transfected into RHΔhpt parasites. The parasites were selected with medium containing 50 µg/ml mycophenolic acid-xanthine, cloned by limiting dilution, and screened by Western blotting and IFA against the HA tag. A clone expressing the correct fusion protein was selected and designated ISP3-BirA*.
Epitope tagging of BioID hits.
For endogenous tagging of proteins, we used the plasmids p3×HA-LIC-DHFR, p3×HA-LIC-HPT, or p3×HA-LIC-CAT (18). For this study, we generated pBirA*-3×HA-LIC-DHFR by inserting the BirA* sequence into p3×HA-LIC-DHFR, in frame between the LIC sites and 3× HA tag. A 3′ portion of each gene was PCR amplified using the designated primers (see Table S3) and inserted into the respective plasmid using a ligation-independent cloning approach to generate a 3× HA epitope tag fusion prior to the stop codon of each gene. One hundred micrograms of each construct was linearized within the amplicon and transfected into RHΔku80Δhpt parasites. Transgenic parasites were selected in the appropriate drug medium (containing either 1 µM pyrimethamine, 50 µg/ml mycophenolic acid-xanthine, or 1 µM chloramphenicol) and cloned by limiting dilution. Clones that had undergone the intended recombination event were screened by IFA and Western blotting against the 3× HA tag. For Cre-lox knockout experiments, we used the plasmids p3×HA-LIC-CAT-loxP and p3×Myc-LIC-HPT-loxP (which contain loxP sites downstream of the drug marker; developed by the M. W. White lab, University of South Florida) for endogenously tagging the target gene and its upstream gene, respectively, in DiCre-ku80::KillerRedfloxYFP parasites (30). Cre-mediated gene excision was performed as previously described (29).
Detergent extractions.
Extracellular parasites were washed in phosphate-buffered saline (PBS), pelleted, and lysed in 1 ml of 1% Triton X-100 lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) supplemented with Complete protease inhibitor cocktail (Roche) for 30 min on ice. Lysates were centrifuged for 15 min at 14,000 × g. Equivalent amounts of total, supernatant, and pellet fractions were separated by SDS-PAGE and analyzed by Western blotting.
Affinity capture of biotinylated proteins.
HFF monolayers infected with parasites expressing BirA* fusions or the respective parental line were grown in medium containing 150 µM biotin for 24 h prior to parasite egress. A total of 150 µM of biotin yielded more robust BirA* labeling within the IMC (which was optimal by 24 h [data not shown]) compared to 50 µM used previously for mammalian cells (19). Extracellular parasites were collected, washed in PBS, and lysed in RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40) supplemented with Complete protease inhibitor cocktail (Roche) for 30 min on ice. Lysates were centrifuged for 15 min at 14,000 × g to pellet insoluble debris and then incubated with PureProteome streptavidin magnetic beads at room temperature for 4 h under gentle agitation. Beads were collected using magnets and washed five times in RIPA buffer, followed by three washes in 8 M urea buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). Ten percent of each sample was boiled in Laemmli sample buffer, and eluted proteins were analyzed by Western blotting by streptavidin-HRP prior to mass spectrometry.
Mass spectrometry of biotinylated proteins.
Purified proteins bound to streptavidin beads were reduced, alkylated, and digested by sequential addition of Lys-C and trypsin proteases (49, 50). The peptide mixture was desalted using C18 tips and fractionated online using a 75-µm inner diameter fritted fused silica capillary column with a 5-µm pulled electrospray tip and packed in house with 15 cm of Luna C18(2) 3 µm reversed-phase particles. The gradient was delivered by an easy-nLC 1000 ultrahigh-pressure liquid chromatography (UHPLC) system (Thermo Scientific). Tandem mass spectrometry (MS/MS) spectra were collected on a Q-Exactive mass spectrometer (Thermo Scientific) (51, 52). Data analysis was performed using the ProLuCID and DTASelect2 implemented in the Integrated Proteomics pipeline IP2 (Integrated Proteomics Applications, Inc., San Diego, CA) (53–55). Protein and peptide identifications were filtered using DTASelect and required a minimum of two unique peptides per protein and a peptide-level false-positive rate of less than 5%, as estimated by a decoy database strategy (56). Normalized spectral abundance factor (NSAF) values were calculated as described (57).
Immunocytochemistry and confocal and superresolution imaging.
Extracellular parasites were fixed, laid onto coverslips, and processed for indirect immunofluorescence as previously described (48). The coverslips were mounted in ProLong Gold (Molecular Probes) and imaged using a TCS SP5 MP-STED confocal microscope (Leica Microsystems) as described (58). In brief, the ATTO 647N-labeled parasites were imaged on an avalanche photodiode (APD; PerkinElmer) using an oil immersion objective (HCX PL APO CS ×100/1.40-numerical-apterature STED), a 640-nm pulsed diode laser (PicoQuant) for excitation, and a 750-nm pulse (Mai Tai Broadband; Spectra-Physics) for STED depletion.
SUPPLEMENTAL MATERIAL
The ISP3-BirA* fusion protein traffics to the IMC. (A) IFA showing ISP3-BirA* localizes to the IMC (top) and is enriched in daughter parasites (bottom), as assessed by costaining with IMC3. Red, mouse anti-HA antibody; green, rat anti-IMC3 antibody. Scale bar = 2 µm. (B) Western blot showing that the background of endogenously biotinylated proteins in parasite lysates is visible after overexposure of the blot. Download
Characterization of ISP3 BioID hits. (A) ISP3 BioID hits were filtered for further characterization based on cell cycle gene expression profiles similar to IMC1. (B) IFA showing that staining of IMCs 17/18/20 is not visible in daughter buds demarked by ISP1 or IMC3. Scale bar = 2 µm. (C) IFA showing that staining of IMCs 19/21 and ACs 1/2 is detected in daughter parasites. Red, mouse or rabbit anti-HA antibodies; green, mouse anti-ISP1 or rat anti-IMC3 antibodies. (D) IFA showing staining of three endogenously tagged proteins from the ISP3 BioID data set that localize to cytoplasmic vesicular structures. Red, mouse anti-HA antibody. Download
Gene disruption of selected ISP3 BioID hits using a combinatorial epitope-tagging/Cre-lox strategy. (A) Strategy for engineering loxP sites flanking the gene of interest by endogenous tagging. Note that this strategy requires the upstream gene and the gene of interest to be in the same orientation. (B) IFA showing mitochondrial localization of TgGT1_295370, encoded by the gene upstream of IMC18, as assessed by costaining with the F1 ATP synthase β subunit. Green, rabbit anti-Myc antibody; blue, mouse anti-F1 β antibody. Scale bar = 2 µm. (C) Western blot showing loss of IMC18 in transgenic parasites after induction of Cre recombinase by rapamycin. (D) IFA showing that TgGT1_313370, encoded by the gene upstream of ILP1, appears to stain punctate vesicular structures in the cytoplasm. Green, mouse anti-Myc antibody. Download
Characterization of AC2 BioID hits. (A) AC2 BioID hits were filtered for further characterization based on cell cycle gene expression profiles similar to IMC1. (B) IFA showing staining of AC2 BioID hits in daughter parasites demarked by ISP1 or IMC3. Note that IMCs 22/24 were not detected in daughter buds. Red, mouse or rabbit anti-HA antibodies; green, mouse anti-ISP1 or rat anti-IMC3 antibodies. Scale bar = 2 µm. (C) IFA showing staining of two endogenously tagged proteins from the AC2 BioID data set that localize to cytoplasmic vesicular structures (top) or the subpellicular microtubules (bottom, colocalized with mEmeraldFP-TrxL2). Red, mouse anti-HA antibody. Download
Almost all of the AC2 BioID hits identified localize to the cytoskeletal compartment of the IMC. Detergent extraction analysis showing 7 of 8 hits from AC2 BioID is embedded in the cytoskeletal network. The total lysate (T), insoluble pellet (P), and soluble (S) fractions were monitored using IMC1/ISP3 controls (as described for Fig. 2C). Download
Characterization of ISCs. (A) IFA showing that staining of ISCs does not colocalize with the apical cap demarked by ISP1 (ISC3, top) or the subpellicular microtubules (ISC1, bottom) visualized by expression of mEmeraldFP-TrxL2. The IFA images of ISC1 from the bottom panel are the same images from Fig. 5B but colocalized with TrxL2. Red, mouse anti-ISP1 or mouse anti-HA antibodies; green, rabbit anti-HA antibody. Scale bar = 2 µm. (B) IFA showing staining of ISCs in daughter parasites demarked by IMC3. Note ISC3 is particularly enriched in daughter buds compared to other ISCs. Red, mouse anti-HA antibody; green, rat anti-IMC3 antibody. (C) Detergent extraction analysis showing that ISCs localize to both the membrane and cytoskeletal subcompartments of the IMC. The total lysate (T), insoluble pellet (P), and soluble (S) fractions were monitored using IMC1/ISP3 controls (as described in Fig. 2C). Download
List of top ISP3 BioID hits identified by mass spectrometry. Endogenously tagged proteins from the ISP3 BioID experiments are shown in blue (tagged proteins not localizing to IMC are denoted by an asterisk). Proteins known to localize to or associate with the IMC (from previous studies) are shown in red. Data are from two experimental replicates.
List of top AC2 BioID hits identified by mass spectrometry. Endogenously tagged proteins from the AC2 BioID experiment are shown in blue (tagged proteins not localizing to IMC are denoted by an asterisk). Proteins known to localize to or associate with the IMC (from previous studies or the ISP3 BioID experiment) are shown in red. Data are from one experiment.
Oligonucleotide primers used in this study. All primer sequences are shown in the 5′ to 3′ orientation.
ACKNOWLEDGMENTS
We thank the following for generously providing reagents: Gary Ward (University of Vermont) for anti-IMC1 antibodies and the pCBAP-3×Myc-LIC-DHFR plasmid, Michael W. White (University of South Florida) for the p3×HA-LIC-CAT-loxP and p3×Myc-LIC-HPT-loxP plasmids, Marc-Jan Gubbels (Boston College) for anti-IMC3 antibodies, Markus Meissner (University Of Glasgow) for providing DiCre-ku80::KillerRedfloxYFP parasites, and Ke Hu (Indiana University) for the pTub-mEmeraldFP-TrxL2 plasmid.
Confocal and superresolution imaging was performed at the California NanoSystems Institute (CNSI) Advanced Light Microscopy/Spectroscopy Shared Resource Facility at UCLA, supported in part by funding from an NSF Major Research Instrumentation grant (CHE-0722519). This work was supported by NIH grant no. AI064616 to P.J.B. and no. GM089778 to J.A.W.
Footnotes
Citation Chen AL, Kim EW, Toh JY, Vashisht AA, Rashoff AQ, Van C, Huang AS, Moon AS, Bell HN, Bentolila LA, Wohlschlegel JA, Bradley PJ. 2015. Novel components of the Toxoplasma inner membrane complex revealed by BioID. mBio 6(1):e02357-14. doi:10.1128/mBio.02357-14
REFERENCES
- 1.Hill DE, Chirukandoth S, Dubey JP. 2005. Biology and epidemiology of Toxoplasma gondii in man and animals. Anim Health Res Rev 6:41–61. doi: 10.1079/AHR2005100. [DOI] [PubMed] [Google Scholar]
- 2.Mackintosh CL, Beeson JG, Marsh K. 2004. Clinical features and pathogenesis of severe malaria. Trends Parasitol 20:597–603. doi: 10.1016/j.pt.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 3.D’Haese J, Mehlhorn H, Peters W. 1977. Comparative electron microscope study of pellicular structures in coccidia (sarcocystis, besnoitia and eimeria). Int J Parasitol 7:505–518. doi: 10.1016/0020-7519(77)90014-5. [DOI] [PubMed] [Google Scholar]
- 4.Mann T, Beckers C. 2001. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol 115:257–268. doi: 10.1016/S0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- 5.Porchet E, Torpier G. 1977. Freeze fracture study of Toxoplasma and Sarcocystis infective stages (author’s transl). Z Parasitenkd 54:101–124. doi: 10.1007/BF00380795 (In French.) [DOI] [PubMed] [Google Scholar]
- 6.Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, Sinha D, Prinz B, Ruch U, Heussler V, Spielmann T, Parkinson J, Gilberger TW. 2012. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol 29:2113–2132. doi: 10.1093/molbev/mss081. [DOI] [PubMed] [Google Scholar]
- 7.Meszoely CA, Erbe EF, Steere RL, Trosper J, Beaudoin RL. 1987. Plasmodium falciparum: freeze-fracture of the gametocyte pellicular complex. Exp Parasitol 64:300–309. doi: 10.1016/0014-4894(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 8.Morrissette NS, Sibley LD. 2002. Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev 66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrowolski JM, Sibley LD. 1996. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84:933–939. doi: 10.1016/S0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 10.Opitz C, Soldati D. 2002. “The glideosome”: a dynamic complex powering gliding motion and host cell invasion by Toxoplasma gondii. Mol Microbiol 45:597–604. doi: 10.1046/j.1365-2958.2002.03056.x. [DOI] [PubMed] [Google Scholar]
- 11.Francia ME, Striepen B. 2014. Cell division in apicomplexan parasites. Nat Rev Microbiol 12:125–136. doi: 10.1038/nrmicro3184. [DOI] [PubMed] [Google Scholar]
- 12.Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, Ferguson DJ, Sahoo N, Gubbels MJ. 2011. A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell Microbiol 13:18–31. doi: 10.1111/j.1462-5822.2010.01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. 2008. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol 25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- 14.Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. 2004. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J Cell Biol 165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson TM, Rajfur Z, Jacobson K, Beckers CJ. 2007. Immobilization of the type XIV myosin complex in Toxoplasma gondii. Mol Biol Cell 18:3039–3046. doi: 10.1091/mbc.E07-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeley A, Soldati D. 2004. The glideosome: a molecular machine powering motility and host-cell invasion by Apicomplexa. Trends Cell Biol 14:528–532. doi: 10.1016/j.tcb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Beck JR, Rodriguez-Fernandez IA, de Leon JC, Huynh MH, Carruthers VB, Morrissette NS, Bradley PJ. 2010. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog 6:e1001094. doi: 10.1371/journal.ppat.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fung C, Beck JR, Robertson SD, Gubbels MJ, Bradley PJ. 2012. Toxoplasma ISP4 is a central IMC sub-compartment protein whose localization depends on palmitoylation but not myristoylation. Mol Biochem Parasitol 184:99–108. doi: 10.1016/j.molbiopara.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firat-Karalar EN, Rauniyar N, Yates JR III, Stearns T. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol 24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. 2014. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A 111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, Anrather D, Kostan J, Djinovic-Carugo K, Roux KJ, Warren G. 2013. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell 12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelenska J, Crawford MJ, Harb OS, Zuther E, Haselkorn R, Roos DS, Gornicki P. 2001. Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc Natl Acad Sci U S A 98:2723–2728. doi: 10.1073/pnas.051629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agop-Nersesian C, Egarter S, Langsley G, Foth BJ, Ferguson DJ, Meissner M. 2010. Biogenesis of the inner membrane complex is dependent on vesicular transport by the alveolate specific GTPase Rab11B. PLoS Pathog 6:e1001029. doi: 10.1371/journal.ppat.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Miguel N, Lebrun M, Heaslip A, Hu K, Beckers CJ, Matrajt M, Dubremetz JF, Angel SO. 2008. Toxoplasma gondii Hsp20 is a stripe-arranged chaperone-like protein associated with the outer leaflet of the inner membrane complex. Biol Cell 100:479–489. doi: 10.1042/BC20080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, Sibley LD, White MW. 2010. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorestani A, Ivey FD, Thirugnanam S, Busby MA, Marth GT, Cheeseman IM, Gubbels MJ. 2012. Targeted proteomic dissection of Toxoplasma cytoskeleton sub-compartments using MORN1. Cytoskeleton 69:1069–1085. doi: 10.1002/cm.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gómez de León CT, Martin D, Mendoza Hernández G, González Pozos S, Ambrosio JR, Mondragón Flores R. 2014. Proteomic characterization of the subpellicular cytoskeleton of Toxoplasma gondii tachyzoites. J Proteomics 111:86–99. doi: 10.1016/j.jprot.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Andenmatten N, Egarter S, Jackson AJ, Jullien N, Herman JP, Meissner M. 2013. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat Methods 10:125–127. doi: 10.1038/nmeth.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieperhoff MS, Pall GS, Jimenez-Ruis E, Das S, Wong EH, Heng J, Mueller S, Blackman MJ, Meissner M. 1 September 2014. Conditional U1 gene silencing in Toxoplasma gondii. bioRxiv. doi: 10.1101/008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilk SD, Raviv Y, Hu K, Murray JM, Beckers CJ, Ward GE. 2006. Identification of PhIL1, a novel cytoskeletal protein of the Toxoplasma gondii pellicle, through photosensitized labeling with 5-[125I]iodonaphthalene-1-azide. Eukaryot Cell 5:1622–1634. doi: 10.1128/EC.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Wetzel L, Zhang Y, Nagayasu E, Ems-McClung S, Florens L, Hu K. 2013. Novel thioredoxin-like proteins are components of a protein complex coating the cortical microtubules of Toxoplasma gondii. Eukaryot Cell 12:1588–1599. doi: 10.1128/EC.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lentini G, Kong-Hap M, El Hajj H, Francia M, Claudet C, Striepen B, Dubremetz JF, Lebrun M. 2015. Identification and characterization of Toxoplasma SIP, a conserved apicomplexan cytoskeleton protein involved in maintaining the shape, motility and virulence of the parasite. Cell Microbiol 17:62–78. doi: 10.1111/cmi.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilley LD, Krishnamurthy S, Westwood NJ, Ward GE. 2014. Identification of TgCBAP, a novel cytoskeletal protein that localizes to three distinct subcompartments of the Toxoplasma gondii pellicle. PLoS One 9:e98492. doi: 10.1371/journal.pone.0098492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel V, Yuan Z, Ramsubir S, Bakovic M. 2006. Choline transport for phospholipid synthesis. Exp Biol Med 231:490–504. [DOI] [PubMed] [Google Scholar]
- 36.Beck JR, Chen AL, Kim EW, Bradley PJ. 2014. RON5 is critical for organization and function of the Toxoplasma moving junction complex. PLoS Pathog 10:e1004025. doi: 10.1371/journal.ppat.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frénal K, Marq JB, Jacot D, Polonais V, Soldati-Favre D. 2014. Plasticity between MyoC- and MyoA-Glideosomes: an example of functional compensation in Toxoplasma gondii invasion. PLoS Pathog 10:e1004504. doi: 10.1371/journal.ppat.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamarque MH, Roques M, Kong-Hap M, Tonkin ML, Rugarabamu G, Marq JB, Penarete-Vargas DM, Boulanger MJ, Soldati-Favre D, Lebrun M. 2014. Plasticity and redundancy among AMA-RON pairs ensure host cell entry of Toxoplasma parasites. Nat Commun 5:4098. doi: 10.1038/ncomms5098. [DOI] [PubMed] [Google Scholar]
- 39.Tremp AZ, Carter V, Saeed S, Dessens JT. 2013. Morphogenesis of Plasmodium zoites is uncoupled from tensile strength. Mol Microbiol 89:552–564. doi: 10.1111/mmi.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen B, Brown KM, Lee TD, Sibley LD. 2014. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5(3):e01114-14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S. 2014. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9:e100450. doi: 10.1371/journal.pone.0100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem 271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 43.Wichroski MJ, Melton JA, Donahue CG, Tweten RK, Ward GE. 2002. Clostridium septicum alpha-toxin is active against the parasitic protozoan Toxoplasma gondii and targets members of the SAG family of glycosylphosphatidylinositol-anchored surface proteins. Infect Immun 70:4353–4361. doi: 10.1128/IAI.70.8.4353-4361.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacot D, Daher W, Soldati-Favre D. 2013. Toxoplasma gondii myosin F, an essential motor for centrosomes positioning and apicoplast inheritance. EMBO J 32:1702–1716. doi: 10.1038/emboj.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeRocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, Bradley PJ, Parsons M. 2008. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot Cell 7:1518–1529. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. 2008. Intervacuolar transport and unique topology of GRA14, a novel dense granule protein in Toxoplasma gondii. Infect Immun 76:4865–4875. doi: 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turetzky JM, Chu DK, Hajagos BE, Bradley PJ. 2010. Processing and secretion of ROP13: a unique Toxoplasma effector protein. Int J Parasitol 40:1037–1044. doi: 10.1016/j.ijpara.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. 2005. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem 280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 49.Kaiser P, Wohlschlegel J. 2005. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol 399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- 50.Wohlschlegel JA. 2009. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol 497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 51.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. 2012. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole Orbitrap mass spectrometer. J Proteome Res 11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 52.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. 2011. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics 10:M111.011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cociorva D, Tabb DL, Yates JR. 2007. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinform Chapter 13:Unit 13.14. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 54.Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu T, Venable JT, Kyu Park S, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR III. 2006. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics 5:S174–S174. [Google Scholar]
- 56.Elias JE, Gygi SP. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 57.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. 2006. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farahani JN, Schibler MJ, Bentolila LA. 2010. Stimulated emission depletion (STED) microscopy: from theory to practice, p 1539–1547. In Mendez-Vilas A, Diaz J (Eds.), Microscopy science, technology, applications and education, vol II, no 4. Formatex Research Center Publishing, Badajoz, Spain. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ISP3-BirA* fusion protein traffics to the IMC. (A) IFA showing ISP3-BirA* localizes to the IMC (top) and is enriched in daughter parasites (bottom), as assessed by costaining with IMC3. Red, mouse anti-HA antibody; green, rat anti-IMC3 antibody. Scale bar = 2 µm. (B) Western blot showing that the background of endogenously biotinylated proteins in parasite lysates is visible after overexposure of the blot. Download
Characterization of ISP3 BioID hits. (A) ISP3 BioID hits were filtered for further characterization based on cell cycle gene expression profiles similar to IMC1. (B) IFA showing that staining of IMCs 17/18/20 is not visible in daughter buds demarked by ISP1 or IMC3. Scale bar = 2 µm. (C) IFA showing that staining of IMCs 19/21 and ACs 1/2 is detected in daughter parasites. Red, mouse or rabbit anti-HA antibodies; green, mouse anti-ISP1 or rat anti-IMC3 antibodies. (D) IFA showing staining of three endogenously tagged proteins from the ISP3 BioID data set that localize to cytoplasmic vesicular structures. Red, mouse anti-HA antibody. Download
Gene disruption of selected ISP3 BioID hits using a combinatorial epitope-tagging/Cre-lox strategy. (A) Strategy for engineering loxP sites flanking the gene of interest by endogenous tagging. Note that this strategy requires the upstream gene and the gene of interest to be in the same orientation. (B) IFA showing mitochondrial localization of TgGT1_295370, encoded by the gene upstream of IMC18, as assessed by costaining with the F1 ATP synthase β subunit. Green, rabbit anti-Myc antibody; blue, mouse anti-F1 β antibody. Scale bar = 2 µm. (C) Western blot showing loss of IMC18 in transgenic parasites after induction of Cre recombinase by rapamycin. (D) IFA showing that TgGT1_313370, encoded by the gene upstream of ILP1, appears to stain punctate vesicular structures in the cytoplasm. Green, mouse anti-Myc antibody. Download
Characterization of AC2 BioID hits. (A) AC2 BioID hits were filtered for further characterization based on cell cycle gene expression profiles similar to IMC1. (B) IFA showing staining of AC2 BioID hits in daughter parasites demarked by ISP1 or IMC3. Note that IMCs 22/24 were not detected in daughter buds. Red, mouse or rabbit anti-HA antibodies; green, mouse anti-ISP1 or rat anti-IMC3 antibodies. Scale bar = 2 µm. (C) IFA showing staining of two endogenously tagged proteins from the AC2 BioID data set that localize to cytoplasmic vesicular structures (top) or the subpellicular microtubules (bottom, colocalized with mEmeraldFP-TrxL2). Red, mouse anti-HA antibody. Download
Almost all of the AC2 BioID hits identified localize to the cytoskeletal compartment of the IMC. Detergent extraction analysis showing 7 of 8 hits from AC2 BioID is embedded in the cytoskeletal network. The total lysate (T), insoluble pellet (P), and soluble (S) fractions were monitored using IMC1/ISP3 controls (as described for Fig. 2C). Download
Characterization of ISCs. (A) IFA showing that staining of ISCs does not colocalize with the apical cap demarked by ISP1 (ISC3, top) or the subpellicular microtubules (ISC1, bottom) visualized by expression of mEmeraldFP-TrxL2. The IFA images of ISC1 from the bottom panel are the same images from Fig. 5B but colocalized with TrxL2. Red, mouse anti-ISP1 or mouse anti-HA antibodies; green, rabbit anti-HA antibody. Scale bar = 2 µm. (B) IFA showing staining of ISCs in daughter parasites demarked by IMC3. Note ISC3 is particularly enriched in daughter buds compared to other ISCs. Red, mouse anti-HA antibody; green, rat anti-IMC3 antibody. (C) Detergent extraction analysis showing that ISCs localize to both the membrane and cytoskeletal subcompartments of the IMC. The total lysate (T), insoluble pellet (P), and soluble (S) fractions were monitored using IMC1/ISP3 controls (as described in Fig. 2C). Download
List of top ISP3 BioID hits identified by mass spectrometry. Endogenously tagged proteins from the ISP3 BioID experiments are shown in blue (tagged proteins not localizing to IMC are denoted by an asterisk). Proteins known to localize to or associate with the IMC (from previous studies) are shown in red. Data are from two experimental replicates.
List of top AC2 BioID hits identified by mass spectrometry. Endogenously tagged proteins from the AC2 BioID experiment are shown in blue (tagged proteins not localizing to IMC are denoted by an asterisk). Proteins known to localize to or associate with the IMC (from previous studies or the ISP3 BioID experiment) are shown in red. Data are from one experiment.
Oligonucleotide primers used in this study. All primer sequences are shown in the 5′ to 3′ orientation.