ABSTRACT
Granulomas sit at the center of tuberculosis (TB) immunopathogenesis. Progress in biomarkers and treatment specific to the human granuloma environment is hindered by the lack of a relevant and tractable infection model that better accounts for the complexity of the host immune response as well as pathogen counterresponses that subvert host immunity in granulomas. Here we developed and characterized an in vitro granuloma model derived from human peripheral blood mononuclear cells (PBMCs) and autologous serum. Importantly, we interrogated this model for its ability to discriminate between host and bacterial determinants in individuals with and without latent TB infection (LTBI). By the use of this model, we provide the first evidence that granuloma formation, bacterial survival, lymphocyte proliferation, pro- and anti-inflammatory cytokines, and lipid body accumulation are significantly altered in LTBI individuals. Moreover, we show a specific transcriptional signature of Mycobacterium tuberculosis associated with survival within human granuloma structures depending on the host immune status. Our report provides fundamentally new information on how the human host immune status and bacterial transcriptional signature may dictate early granuloma formation and outcome and provides evidence for the validity of the granuloma model and its potential applications.
IMPORTANCE
In 2012, approximately 1.3 million people died from tuberculosis (TB), the highest rate for any single bacterial pathogen. The long-term control of TB requires a better understanding of Mycobacterium tuberculosis pathogenesis in appropriate research models. Granulomas represent the characteristic host tissue response to TB, controlling the bacilli while concentrating the immune response to a limited area. However, complete eradication of bacteria does not occur, since M. tuberculosis has its own strategies to adapt and persist. Thus, the M. tuberculosis-containing granuloma represents a unique environment for dictating both the host immune response and the bacterial response. Here we developed and characterized an in vitro granuloma model derived from blood cells of individuals with latent TB infection that more accurately defines the human immune response and metabolic profiles of M. tuberculosis within this uniquely regulated immune environment. This model may also prove beneficial for understanding other granulomatous diseases.
INTRODUCTION
The World Health Organization estimates that 8.6 million people were infected with tuberculosis (TB) in 2013, including 1.1 million cases among people living with human immunodeficiency virus (HIV) (1). TB ranks as the second leading cause of death from a single infectious agent, after HIV infection (2). The long-term control of TB requires a better understanding of bacterial virulence properties and host response in appropriate research models.
Granulomas in humans sit at the center of TB immunopathogenesis. Active TB can result from either early progression of a primary granuloma during the infection process (rare) or reactivation of Mycobacterium tuberculosis from an established granuloma in a latently infected person (10% lifetime risk in immunocompetent individuals and 10% annual risk in untreated HIV-infected individuals). Such individuals infected with M. tuberculosis are latent carriers of the bacterium and, although they exhibit no signs of disease, represent reservoirs for later reactivation and transmission. The cellular and biochemical factors that control granuloma formation, maintenance, function, and resolution in humans are multifaceted, involving a complex interplay between the host immune system and survival strategies employed by the bacilli. These factors remain poorly understood since current in vitro, in silico, and in vivo models do not fully recapitulate the true microenvironment found in human granulomas.
Granulomas are organized structures of macrophages, including blood-derived macrophages, epithelioid cells (uniquely differentiated macrophages), and multinucleated giant cells (Langerhans cells, fused macrophages), surrounded by a ring of lymphocytes (3–6). The granuloma’s main function is to localize and contain M. tuberculosis infection while concentrating the immune response to a limited area (7). Although it is presumed that bacterial killing occurs within the granuloma, complete eradication does not occur, since M. tuberculosis has its own strategies to persist within the granuloma and to reactivate and escape under certain circumstances. Thus, the M. tuberculosis-containing granuloma represents a unique battlefield that dictates both the host immune and bacterial responses (8).
Progress on therapies that are active in human granulomas is hindered by the lack of a relevant and tractable infection model that better recapitulates the complexity of the host response as well as M. tuberculosis counterresponses that subvert host immunity. In vitro studies using human peripheral blood mononuclear cells (PBMCs) have shown that the host immune status impacts mycobacterial growth (9–12). Recently, human PBMC-related in vitro models that mimic the microenvironment encountered by M. tuberculosis within human granulomas have been developed (13–18). However, none of these models have focused on how the granulomatous response may be affected by the human host immune status. Here we addressed this gap in knowledge by interrogating a human in vitro granuloma model for identifying potential differences in the host-M. tuberculosis interplay in individuals with and without latent tuberculosis infection (LTBI). We posited that a dynamic model representing the human TB granulomatous response, comprising multiple interactive cells/molecules, in which changes over time can be studied, is necessary to advance our understanding of the mechanisms regulating the biological events involved as well as their impact on bacterial gene expression. Such a model would be able to identify new biomarker and therapeutic targets uniquely expressed in this tissue microenvironment.
By assaying for multiple cellular and immunologic parameters, we demonstrated that individuals with LTBI exhibit a more robust granulomatous response than naive individuals. Further, we demonstrate that M. tuberculosis expresses enzymes representing fundamentally different metabolic pathways during early granuloma formation in LTBI individuals. These findings provide new insight regarding the mechanisms underlying the earliest phases of the host immune response to M. tuberculosis and related pathogen counterdefenses. Such findings will aid in the identification of host-pathogen molecules and pathways that are active in granulomas. Moreover, this granuloma model may prove to be useful for studying other infectious and noninfectious granulomatous diseases.
RESULTS
In vitro granuloma-like structures are formed by infection of PBMCs with M. tuberculosis.
Granuloma-like structures were achieved by infecting PBMCs with M. tuberculosis H37Rv in the presence of 10% autologous serum at a multiplicity of infection (MOI) of 1 bacillus to 1 macrophage as shown in Fig. 1a and b. Cell cultures were maintained for up to 12 days postinfection, and the stage of granuloma formation was determined semiquantitatively daily. Cellular aggregation started around day 4 to day 6 postinfection, depending on interindividual variability. Between days 7 and 12 postinfection, a multicellular, multilayered structure was observed as shown in Fig. 1d and f (see also Movie S1 in the supplemental material). Uninfected cell cultures from the same donors were used as negative controls, and no cellular aggregation was observed throughout the course of the studies (Fig. 1c).
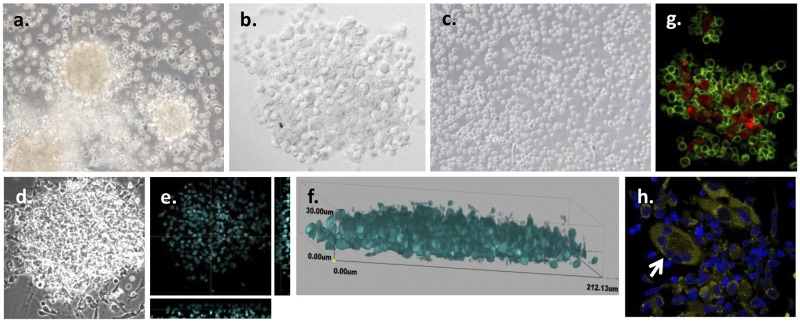
FIG 1 .
In vitro granuloma-like structures are formed by M. tuberculosis infection of PBMCs from LTBI individuals. (a) PBMCs obtained from LTBI individuals were infected with M. tuberculosis H37Rv (MOI 1:1), resulting in the formation of granulomas by day 7 postinfection. (b) Higher magnification of the granulomas by confocal microscopy. (c) No formation of granulomas was observed in uninfected PBMCs obtained from LTBI individuals for up to 12 days postinfection. (d to f) Confocal microscopy images of the granulomas at day 7 postinfection revealed multicellular, multilayered structures containing approximately 4 to 8 cell layers. (d) Differential inference contrast (DIC) image. (e) Image of transverse and straight sections by orthogonal view. (f) Three-dimensional (3D) view image. Nuclei were stained with DAPI (cyan). (g) Granuloma-like structures include macrophages (CD11b+, red) and T cells (CD3+, green). (h) Confocal microscopy images of the granulomas at day 7 postinfection revealed the presence of multinucleated giant cells (CD11b++, yellow; white arrow). Nuclei were stained with DAPI (dark blue). Representative images from n = 12 experiments are shown. The images in panels a and c are shown with ×40 magnification; the remaining images are ×60.
To determine the cellular components of granuloma-like structures that formed early, immunofluorescence studies were performed. The expression of several host cell markers was studied to determine cell type composition. Granuloma-like structures were composed of CD11b+ mononuclear phagocytes generally surrounded by CD3+ lymphocytes (Fig. 1g; see also Movie S1 in the supplemental material). Interestingly, during the maturation process, monocytes differentiated into macrophages, aggregated, and fused into multinucleated giant cells (Fig. 1h; see also Fig. S2). The presence of CD4+ and CD8+ T cells was also determined in granuloma-like structures (see Fig. S4). Finally, B cells were observed in 30% to 50% of these structures which varied according to the donor (see Fig. S4).
In vitro granuloma formation is accelerated in cells from individuals with LTBI.
We hypothesized that the generation of granuloma-like structures would be more robust using cells from individuals with LTBI than using cells from naive individuals as a consequence of immunologic memory. To test this hypothesis, we recruited a total of 28 individuals, comprising 14 otherwise healthy LTBI and 14 uninfected individuals, between 18 and 45 years of age, including males and females, and representatives of American Indian/Alaska native, Asian, African American, and white races and Hispanic/Latino ethnicity (see Table S1 in the supplemental material).
Based on published work (16, 17, 19, 20), we established a scoring system to quantify aggregation, defined as a progressive increase in the size and specific distribution of cells (see Fig. S1 in the supplemental material) in both groups (LTBI and naive) studied. The scoring system was applied at day 7 postinfection since both groups consistently showed cellular aggregation at this time point. The index score of granuloma-like structures generated from LTBI individuals was significantly higher than that seen with naive individuals (Fig. 2b). As shown in Fig. 2a, differences in the speed of cellular recruitment were observed between granulomas derived from LTBI and naive individuals. LTBI individuals showed cellular aggregation as early as day 4 postinfection, while naive individuals started this process no earlier than day 6 after infection. LTBI individuals also had larger granuloma-like structures. Finally, despite interindividual variability, these differences remained throughout the course of the studies for up to 12 days.
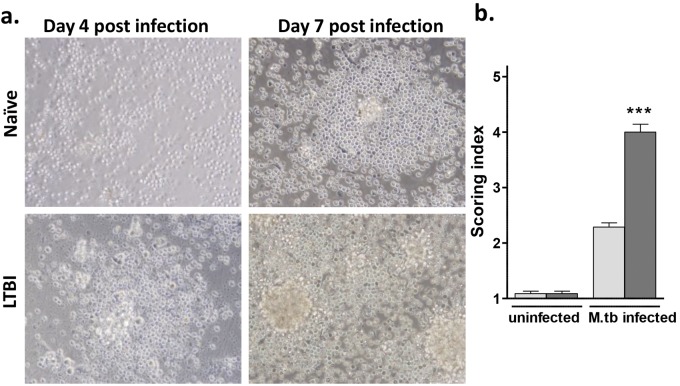
FIG 2 .
Granuloma-like structure formation is accelerated in cells from individuals with LTBI. (a) PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis H37Rv (MOI 1:1) for up to 12 days. While naive individuals typically demonstrated no evidence of cell aggregation until day 6 postinfection, on average, the LTBI cell cultures exhibited phase 3 to 4. (b) The scoring system was applied on day 7 postinfection, and 15 high-power fields/sample were evaluated. Naive individuals, light grey bars; LTBI individuals, dark grey bars. All images are ×40. Uninfected cells were used as controls. n = 12 in triplicate. ***, P < 0.001 (by t test). M.tb, M. tuberculosis.
Granuloma-like structures from LTBI individuals have better control of the bacillary load over time.
There were no previous studies that provided evidence on whether the host immune status has an impact on the intracellular growth of M. tuberculosis within in vitro granulomas. Previous in vitro studies using PBMCs have shown that the immune status of the host can impact bacillary control and that direct contact between T cells and macrophages is required for mycobacterial growth-inhibitory effects (9–12). Due to the presence of immunologic memory in LTBI individuals and on the basis of the above-mentioned studies, we hypothesized that there would be a more effective anti-M. tuberculosis host response within in vitro granuloma-like structures.
To assess whether the host immune status had an impact on the intracellular growth of bacteria within the granuloma-like structures, we performed several assays, including CFU, relative light unit (RLU), and confocal microscopy analyses, following staining of bacteria. Granuloma-like structures were developed as previously described, and bacterial growth was studied over time.
Results showed that intracellular bacterial survival rates between days 0 and 3 postinfection were similar. However, after day 4 postinfection in the CFU assay and day 6 postinfection in the RLU assay, there was significantly greater control of the bacillary load in the granuloma-like structures obtained from LTBI individuals than was seen with naive individuals (Fig. 3a and b). Immunofluorescence studies performed at day 7 postinfection, where bacteria were stained with auramine-O, showed similar results (Fig. 3c; see also Movies S2 [naïve] and S3 [LTBI]). Although the bacterial load continued to increase over time, the difference between the two groups existed throughout the course of the studies. These results provide evidence for a growth-inhibitory effect present in the granuloma-like structures obtained from LTBI individuals compared with naive individuals. Importantly, sterility was never achieved.
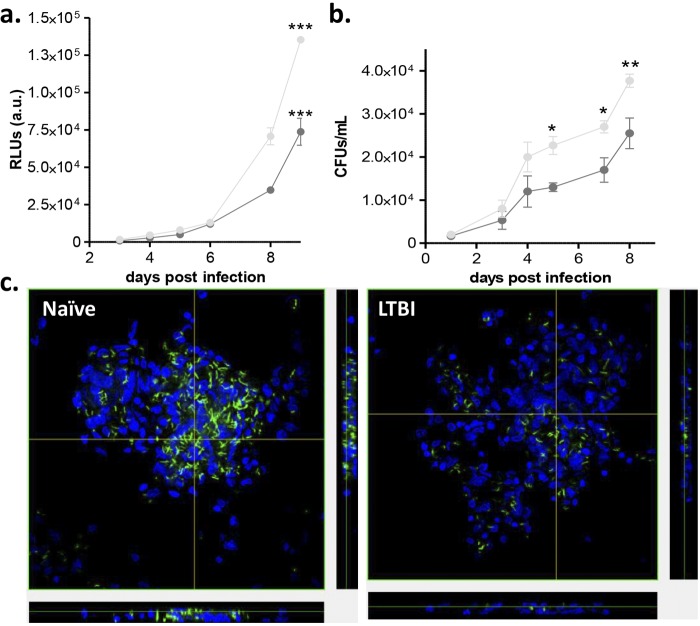
FIG 3 .
Granuloma-like structures from LTBI individuals have better control of M. tuberculosis growth over time. PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis Rv-lux (MOI 1:1) for up to 9 days. (a and b) RLUs (a) and CFU (b) were determined at several time points after infection. (c) Confocal microscopy images of the granulomas at day 7 postinfection revealed more bacilli (green) in granuloma-like structures from naive individuals than in those from LTBI individuals. Naive individuals, light grey bars; LTBI individuals, dark grey bars. Uninfected cells were used as controls. All images are ×60. n = 3 in triplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by two-way ANOVA with Bonferroni posttest).
The proliferative activity of host cell in vitro granulomas from LTBI individuals is significantly greater than the activity seen with naive individuals.
Trafficking of cells is the main mechanism of cellular accumulation in the granuloma (20), although proliferation may also play a role (21). The presence of clonal expansion in mycobacterium-induced granulomas suggests that local proliferation of T cells might occur and contribute to the granuloma composition (22, 23). To determine whether host cell proliferative activity occurred within the granuloma-like structures, two assays were performed: a total cell count assay and an EdU assay. Granuloma-like structures were generated as previously described, and host cell proliferation was studied over time.
First, total cell counts of LTBI and naive groups were performed. Results revealed an increasing number of cells in the LTBI group (2.43-fold change, n = 2) compared to the naive group (1.46-fold change, n = 2) at day 7 postinfection. Since macrophages are terminally differentiated, these data suggested a T cell proliferation process. Therefore, an Edu labeling index analysis (an optimized proliferation assay where the thymidine analogue EdU is efficiently incorporated into newly synthesized DNA and fluorescently labeled with an Alexa Fluor dye) was performed. The labeling index was calculated as the mean fluorescence intensity (MFI) of M. tuberculosis-infected cells minus the basal proliferation of uninfected cells within granuloma-like structures. The Edu labeling index values were compared between granulomas from LTBI individuals and naive individuals. As shown in Fig. 4a and b, LTBI individuals showed a significantly higher EdU labeling index (1.8-fold change, n = 3).
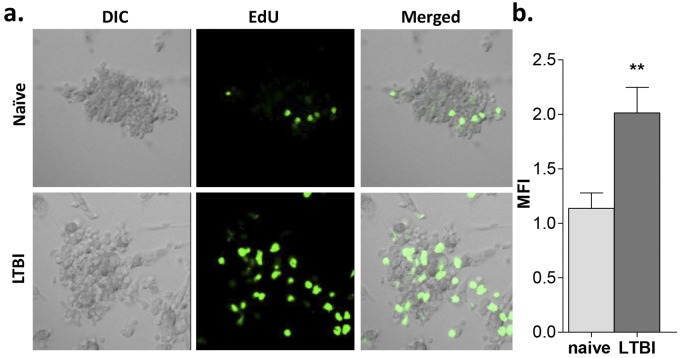
FIG 4 .
The proliferative activity of granuloma-like structures from LTBI individuals is significantly greater than that seen with naive individuals. PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis H37Rv (MOI 1:1) for up to 7 days, and cell proliferation was determined by EdU assay. (a) Confocal microscopy images of the granulomas at day 7 postinfection revealed greater proliferation (green) in granulomas from LTBI individuals than in those from naive individuals. (b) Quantification of the cellular fluorescence shown by calculation of MFI values. Naive individuals, light grey bars; LTBI individuals, dark grey bars. Uninfected cells were used as controls. All images are ×40. n = 3. **, P < 0.01 (by t test).
Granuloma-like structures from LTBI individuals produce greater inflammatory cytokines.
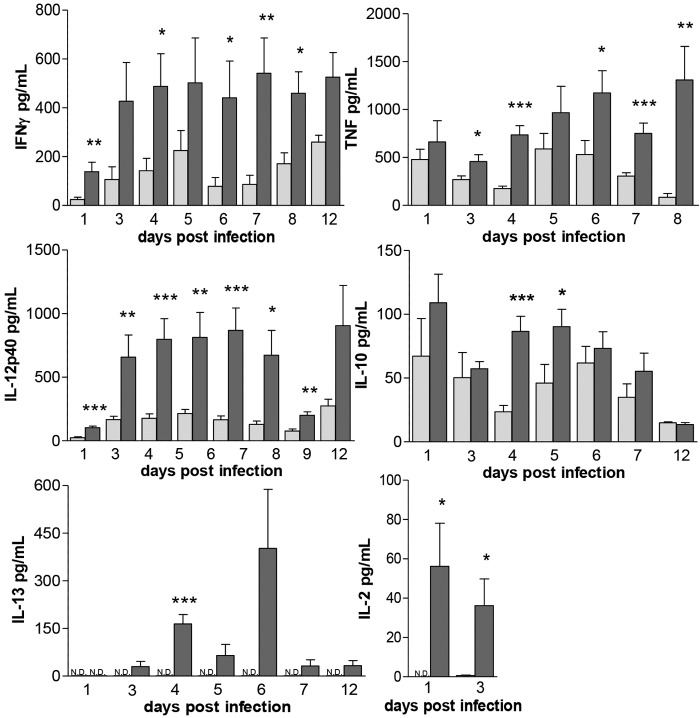
Cytokines were determined in in vitro granuloma cultures and culture supernatants collected from LTBI and naive individuals over time. The release of gamma interferon (IFN-γ), tumor necrosis factor (TNF), interleukin-12 p40 (IL-12p40), IL-2, IL-10, IL-4, and IL-13 was evaluated by enzyme-linked immunosorbent assay (ELISA) (Fig. 5) and mRNA expression by quantitative real-time PCR (qRT-PCR) (data not shown). Expression levels of the various cytokines followed similar trends in the two studied groups, although the levels were always significantly higher in the granuloma-like structures from LTBI individuals. IFN-γ levels peaked between days 3 and 12 postinfection and remained constant. IL-12p40 peaked at day 3 postinfection until day 8 postinfection. There was a decrease in the Il-12p40 level at day 9 postinfection, and it peaked again at day 12. There was a trend toward an increase in TNF levels over time. IL-2 could be detected only at very early time points postinfection and was barely detectable in naive individuals compared to LTBI individuals. There was an increase in the IL-10 level after infection, and it peaked again around day 5 postinfection. IL-4 was not detected in any of the studied groups. Finally, the IL-13 level peaked around day 4 postinfection in the LTBI and was undetectable in the naive individuals. Overall, our results demonstrate that the granulomas from LTBI individuals contained a complex cytokine environment, with proinflammatory, anti-inflammatory, and immunoregulatory cytokines all present.
FIG 5 .
Granuloma-like structures from LTBI individuals produce more robust inflammatory cytokines. PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis H37Rv (MOI 1:1) for up to 12 days, and cytokine levels in cell culture supernatants collected at several time points after infection were determined by ELISA. Naive individuals, light grey bars; LTBI individuals, dark grey bars. Uninfected cells were used as controls. ND, not detected. n = 3 (in triplicate). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by t test).
LTBI individuals have greater lipid body (LB) accumulation in macrophages within granuloma-like structures.
Since there is an increase in the intracellular accumulation of LBs in macrophages after M. tuberculosis infection (24–29), we next assessed whether LB formation was induced within the granuloma-like structures and, specifically, whether the host immune status had an impact on LB accumulation. Granuloma-like structures were generated, and staining using Nile red (a red fluorescent probe for intracellular lipids) was performed. Nile red can also stain lipids within M. tuberculosis (30). Our results showed that, although LBs in macrophages were present in the granulomas of both groups, granuloma-like structures from LTBI individuals had much greater LB accumulation than those from naive individuals at day 7 postinfection that was distinct from bacterial results (see Fig. S3 in the supplemental material [arrows]). In stark contrast, Nile red staining was primarily associated with bacteria in the TB-naive group. Auramine-O staining tended to decrease in both groups with time (although the staining results differed from donor to donor), previously considered to be consistent with a stress-related phenotype (31, 32). There were fewer bacteria in the LTBI group than in the naive group, and they were less elongated. Taken together, these results indicate significant differences in the accumulation of host and bacterial lipids that are influenced by the host immune status.
Differences occur in the expression levels of M. tuberculosis H37Rv cell wall mannosylated lipoglycan and glycolipid biosynthetic enzymes within granuloma-like structures from LTBI individuals compared with naive individuals.
Several studies, including those from our laboratory, have shown that M. tuberculosis surface mannosylation plays an important role in the host-pathogen interaction, resulting in a highly regulated immune response that dictates M. tuberculosis survival within the host (33, 34, 35–37). M. tuberculosis-containing granulomas represent a unique battlefield, dictating both the host immune response and the bacterial response. The precise metabolic status of M. tuberculosis within human granulomas, particularly during early granuloma formation, remains unclear (38). In this regard, it is completely unknown what bacterial biosynthetic pathways are operating within granulomas for the production of major M. tuberculosis cell wall determinants, such as the mannosylated lipoglycans and glycolipids. The apparent redundancies in these biosynthetic enzyme pathways suggest that a subset may be particularly important for bacterial adaptation and persistence within granulomas. We initially focused on the transcript levels of selected genes that are involved in the mannose donor biosynthesis pathway (manA, manB, pmmB, mrsA, pgmA, manC, ppm1, Rv3779, and ppgS) and within the lipoarabinomannan (LAM) biosynthesis pathway (pimA, pimB, and pimF). We have previously shown that there are differences in the expression profiles of mannose donor biosynthetic genes in M. tuberculosis grown in broth culture as well as within human macrophages, suggesting that the level of mannose donors may vary during the course of infection and thereby impact the biosynthesis of mannose-containing cell wall molecules (39) (see Table S2 in the supplemental material for primer sequences). To evaluate the expression profile of this set of genes within granuloma-like structures from LTBI individuals and naive individuals, granulomas were generated, bacterial RNA was isolated, and the expression profiles were analyzed and compared for the two groups.
As shown in Table 1, most of the genes located within the mannose donor biosynthesis pathway (manA, manB, pmmB, and manC) were highly expressed within granuloma-like structures from the LTBI individuals. Similarly, there was increased expression of some genes located within the LAM biosynthesis pathway (pimA, pimB, and pimF) in granulomas from LTBI individuals compared to naive individuals. These data reveal significant differences in the levels of bacterial cell wall biosynthesis-related gene expression within the granuloma-like structures that are dependent on the host immune status.
TABLE 1 .
Expression of M. tuberculosis H37Rv cell wall mannosylated lipoglycan and glycolipid biosynthetic enzymes within granuloma-like structures from LTBI individuals compared with naive individuals at day 7 postinfectiona
| Gene name/gene product (Rv no.) | Encoded protein function | Metabolic pathway | LTBI versus naive fold difference (mean ± SD) |
|---|---|---|---|
| manA/mannose isomerase (Rv3255c) | Fructose 6P to mannose 6P | Mannose donor biosynthesis pathway | 2.271 ± 1.977** |
| manB/phosphomannomutase (Rv3257c) | Mannose 6P to mannose 1P | Mannose donor biosynthesis pathway | 9.3 ± 9.6* |
| pmmB/phosphomannomutase (Rv3308) | Mannose 6P to mannose 1P | Mannose donor biosynthesis pathway | 0.95 ± 1.012** |
| mrsA/phosphomannomutase (Rv3441c) | Mannose 6P to mannose 1P | Mannose donor biosynthesis pathway | 0.91 ± 1.18 |
| pgmA/phosphomannomutase (Rv3068c) | Mannose 6P to mannose 1P | Mannose donor biosynthesis pathway | 1.08 ± 1.5 |
| manC/GDP-mannose pyrophosphorilase (Rv3264c) | Mannose 1P to GDP-mannose | Mannose donor biosynthesis pathway | 2.84 ± 1.79* |
| ppm1/polyprenol phosphomannose synthase (Rv2051c) | Transfer mannose molecule | Mannose donor biosynthesis pathway | 3.9 ± 3.4 |
| Polyprenol phosphomannose synthase (Rv3779) | Transfer mannose molecule | Mannose donor biosynthesis pathway | 1.23 ± 0.63 |
| ppgS/ polyprenol phosphomannose synthase (Rv3631) | Transfer mannose molecule | Mannose donor biosynthesis pathway | 1.36 ± 0.71 |
| pimA/mannosyltransferase (Rv2610c) | Lipoarabinomannan biosynthesis | Lipoarabinomannan biosynthesis pathway | 1.3 ± 0 |
| pimB/mannosyltransferase (Rv2188c) | Lipoarabinomannan biosynthesis | Lipoarabinomannan biosynthesis pathway | 1.1 ± 0 |
| pimF/mannosyltransferase (Rv1500) | Lipoarabinomannan biosynthesis | Lipoarabinomannan biosynthesis pathway | 0.88 ± 0.7 |
Each gene was normalized to the rpoB housekeeping gene, and relative expression levels were determined by the 2−ΔΔCT method. The fold change was determined by comparing levels of expression within granulomas between LTBI individuals and naive individuals. n = 8 in triplicate. *, P < 0.05; **, P < 0.01 (Student’s t test).
Expression levels of key metabolic enzymes in M. tuberculosis H37Rv within granuloma-like structures from LTBI individuals differ from those from naive individuals.
Previous studies have shown that M. tuberculosis, which is metabolically flexible (40, 41), preferentially utilizes fatty acids as carbon and energy sources during chronic infection in the murine model (42). This preferential utilization is hypothesized to be due to increased availability of lipids in the infected host cell (24). Moreover, genes involved in fatty acid metabolism are required during growth and persistency in vivo (43–47). These observations provide evidence that M. tuberculosis is able to switch its carbon source from sugars to fatty acids during the persistent phase of infection or during latency. In this regard, it is completely unknown when the switch to these biosynthetic pathways involving fatty acid metabolism begins during granuloma formation. We questioned whether fatty acid metabolism is upregulated in M. tuberculosis early within granuloma-like structures from LTBI individuals as a result of a more robust inflammatory response staged against the bacilli.
We examined the transcript levels in day 7 granulomas of selected key genes that are involved in glycolysis (pfkA and ppgK) and the Krebs cycle (icd1 and sdhC) as well as in fatty acid degradation (fadB, fadE5, and fadE23) and utilization (glyoxylate shunt, icl and aceA; gluconeogenesis, pckA) (see Table S2 in the supplemental material for primer sequences). As shown in Table 2, fatty acid degradation and utilization genes and Krebs cycle genes were more highly expressed during M. tuberculosis growth within granuloma-like structures from LTBI individuals than from naive individuals. This was also true for the icl glyoxylate shunt gene. In contrast, transcription of glycolysis genes (pfkA and ppgK) was more highly expressed during M. tuberculosis growth within granuloma-like structures from naive individuals.
TABLE 2 .
Expression of M. tuberculosis H37Rv key metabolic enzymes within granuloma-like structures from LTBI individuals compared with naive individuals at day 7 postinfectiona
| Gene name/gene product (Rv no.) | Encoded protein function | Metabolic pathway | LTBI versus naive fold difference (mean ± SD) |
|---|---|---|---|
| pckA/phophoenolpyruvate carboxylase (Rv0211) | Gluconeogenic enzyme | Gluconeogenesis | 6.460 ± 5.523 |
| aceA/isocitrate lyase (Rv1915) | Glyoxylate bypass (at the first step) | Glyoxylate shunt cycle | 5.624 ± 1.963** |
| icl/isocitrate lyase (Rv0467) | Glyoxylate bypass (at the first step) | Glyoxylate shunt cycle | 3.07 ± 2.49 |
| fadE5/macyl-CoA dehydrogenase (Rv0244c) | Involved in lipid degradation | Fatty acid beta-oxidation | 4.875 ± 4.066 |
| fadE23/acyl-CoA dehydrogenase (Rv3140) | Involved in lipid degradation | Fatty acid beta-oxidation | 0.400 ± 0.0 |
| fadB/fatty acid oxidation protein (Rv0860) | Involved in fatty acid degradation | Fatty acid beta-oxidation | 3.125 ± 3.0 |
| icd1/isocitrate dehydrogenase (Rv3339c) | Involved in the Krebs cycle | Krebs cycle | 5.333 ± 4.163 |
| sdhC/cytochrome B-556 of succinate dehydrogenase (Rv3316) | Involved in the Krebs cycle | Krebs cycle | 4.873 ± 4.4 |
| ppgk/glucokinase (Rv2702) | Catalyzes the phosphorylation of glucose | Glycolysis | 0.53 ± 0.0 |
| pfkA/phosphofructose kinase (Rv3010c) | Involved in glycolysis | Glycolysis | 0.1 ± 0.0 |
Each gene was normalized to the rpoB housekeeping gene, and relative expression levels were determined by the 2−ΔΔCT method. The fold change was determined by comparing expression within granulomas between LTBI individuals and naïve individuals. n = 8 (in triplicate). **, P < 0.01 (Student’s t test). CoA, coenzyme A.
Our results are consistent with the notion that M. tuberculosis upregulates metabolic pathways that utilize fatty acids as carbon and energy sources while downregulating metabolic pathways that utilize sugars within granulomas from LTBI individuals. The opposite trend was observed in granulomas from naive individuals, where there was higher expression of glycolysis-related genes at the same time point studied. These results suggest that, even at this early time point during granuloma development in our model, there are differences in nutrient and carbon source availability that depend on the host immune status and that this has a direct impact on M. tuberculosis metabolism over time.
DISCUSSION
Here we have developed and interrogated an in vitro human TB granuloma model representing the complex dynamics of early granuloma formation involving host and bacterial responses that may dictate the subsequent events within this critical tissue environment. Our model shares features with human TB granulomas, including cell aggregation and formation of multinucleated giant cells, lymphocytes, a robust and diverse cytokine profile, and altered bacterial growth. Importantly, we demonstrate that LTBI individuals exhibit a granulomatous response that is significantly different from that exhibited by naive individuals. We further demonstrate that M. tuberculosis changes its expression of metabolic pathway genes during early granuloma formation in LTBI individuals in a manner different from that seen with naive individuals. Overall, our results present clear-cut evidence that host immune status directly impacts the early granulomatous response to M. tuberculosis, providing fundamentally new information about the potential mechanisms involved during the earliest phases of granuloma formation and establishment. This information will be useful in considering new strategies to control M. tuberculosis within granulomas, specifically targeting those host and bacterial parameters that are uniquely regulated in this environment. Moreover, this model may prove to be useful for the study of other infectious and noninfectious granulomatous diseases.
Recent studies have highlighted the importance of studying the local tissue microenvironment in order to gain a better understanding of the cellular and molecular details underlying M. tuberculosis-host interactions during infection (17, 48, 49). It is generally thought that M. tuberculosis must adapt to a highly dynamic environment within the granuloma, with various levels of cell death, oxygen tension, acidity, inflammatory cytokines, oxidants, and hydrolytic enzymes. While the significance of some of these features for M. tuberculosis survival and growth remains unclear, it is predictable that M. tuberculosis must adapt to this environment to enable its persistence.
A substantial barrier to our understanding of the granulomatous response has been the inability to experimentally mimic the conditions encountered by M. tuberculosis within human granulomas. Studies have used various in vivo and in vitro models (8), e.g., the nonhuman primate (50) and zebrafish (20) models. Although it lacks lung structure and thus the full tissue microenvironmental condition (e.g., parenchymal elements), the human in vitro granuloma model presented here offers several advantages, notably the ability to analyze early bacterial and host responses during the development of granuloma structures, the ability to characterize each individual’s response by studying that individual’s own PBMCs and sera (thereby allowing the retention of each individual’s undefined biological parameters), and sample availability for repeated measures. The most impactful element of the model, however, is its ability to account for host immune status, a crucial parameter in dictating the nature of the granulomatous response.
There are no previous studies on whether the host immune status has an impact on M. tuberculosis intracellular growth within in vitro granulomas. We observe that the control of bacterial growth starts after in vitro granulomas have achieved phase 4 in the defined scoring index, a level achieved only in individuals with LTBI, and that this control is maintained thereafter. To generate protective immunity against M. tuberculosis, a specific tissue milieu where activated lymphocytes and macrophages work together in controlling bacterial growth was used that led to local robust inflammation and host cell proliferation. Future studies are now enabled to perform more refined analyses on cellular type, antigen (Ag) presentation, etc. The present report emphasizes the importance of cell kinetic analyses in granulomatous processes, one aspect that is gaining more attention in TB studies (51, 52).
One of the most important factors required for the establishment of infection and bacterial control is the relative abundance of local pro- and anti-inflammatory cytokines produced. For example, TNF and IFN-γ are particularly important in promoting the formation and function of granulomas as well as in bacterial control, whereas IL-10 acts as negative regulator of this response. It is notable that most animal and human studies have detected both cytokines and IL-10 in the immune system host response (53). In concert, we found that granuloma structures from LTBI individuals express various “protective” cytokines, including IFN-γ, TNF, IL-12p40, and IL-2 but also IL-13 and IL-10. Thus, previous studies and ours support the concept that optimum protection (for a healthy individual with LTBI) will be defined as a cytokine equilibrium generated by a number of cell types in the innate and adaptive immune systems. Locally produced IL-10 found in granulomas can explain the presence of multinucleated giant cells (54).
Formation of macrophage LBs is dependent on, among other factors, the activation of Toll-like receptors (TLRs) by pathogen-derived agonists and the presence of proinflammatory signals, such as TNF and CCL2 (26), as well as hypoxia (27). Moreover, there is an increase in the intracellular accumulation of macrophage LBs after M. tuberculosis infection (24–28). In our study, granuloma-like structures from individuals with LTBI had a much greater accumulation of macrophage LBs than naive individuals (see Fig. S3 in the supplemental material). Based on a recent publication demonstrating that M. tuberculosis can utilize host triacylglycerols to accumulate lipid droplets (27), our finding suggests that bacilli have greater access to lipids in macrophages within the in vitro LTBI granuloma environment which could impact their growth rate and lipid metabolism as discussed below.
The ability of M. tuberculosis to survive within human granulomas requires a specific transcriptional signature with the coordinated expression of numerous bacterial determinants, including metabolism genes, persistence genes, and virulence genes among others (55). The identification of the M. tuberculosis transcriptome within human granulomas is of particular interest, as it will allow the development of new therapeutic strategies that target M. tuberculosis factors highly expressed within these structures. Several large-scale studies profiling the mycobacterial transcriptome in in vitro and in vivo models have been performed (55–58). However, none have focused on M. tuberculosis gene expression profiling within the early stages of human granuloma development or the importance of the host immune response. Human biopsy specimens from active TB patients give a static, typically late-stage view of this process and may or may not be pertinent.
In this study, we were specifically interested in determining the expression of several genes encoding enzymes for major M. tuberculosis cell wall mannosylated lipoglycans, including phosphatidyl-myo-inositol mannosides (PIMs), lipomannan (LM), and mannose-capped LAM (ManLAM), within the human in vitro granuloma model given their importance in pathogenesis (35). PIMs, LM, and ManLAM are synthesized via specific mannosyltransferases that use the donors GDP-mannose and polyprenol-phosphate mannose (PPM) (59). The putative genes that build these donors in M. tuberculosis include manA (an isomerase) (60), manB, pmmB, and mrsA (a phosphomannomutase) (61), manC (a GDP-mannose pyrophosphorylase) (62), and Ppm, ppgS, and Rv3779 (polyprenol-phosphate mannose synthase) (59, 63, 64). Studies have shown that mannose biosynthesis plays a pivotal role in cell wall synthesis, protein glycosylation (60), and mannosylation of phospholipids and is essential for viability of mycobacteria in the host (65). Since our results indicate that several genes involved in the mannose donor and LAM biosynthesis pathways are upregulated in granuloma-like structures from LTBI individuals compared to naive individuals, we speculate that one of the survival strategies M. tuberculosis uses in this “immune pressured” environment is to more heavily decorate its cell wall with mannosylated lipoglycans.
We also examined the transcriptional response of M. tuberculosis genes predicted to be involved in the stress encountered during infection with a specific focus on fatty acid degradation and utilization, glycolysis, and the Krebs cycle. During persistence, M. tuberculosis shows a metabolic shift in which bacterial glycolysis is decreased and glyoxylate shunt activity is upregulated, allowing the bacteria to use fatty acids as a carbon and energy source when the availability of primary carbon sources is limited. icl (encoding isocytrate lyase) was one of the first M. tuberculosis genes shown to be required for persistency in the TB murine model (66), and the encoded enzyme is the initial enzyme in the glyoxylate shunt. Genes involved in fatty acid utilization (glyoxylate shunt, icl; gluconeogenesis, pckA) are required for growth and persistence in vivo (43–47). Our results demonstrate that as early as 7 days postinfection during granuloma development in LTBI individuals, M. tuberculosis shifts metabolically to upregulate expression of genes involved in the Krebs cycle, fatty acid degradation, glyoxylate shunt, and gluconeogenesis and to downregulate expression of genes involved in glycolysis. This trend is opposite that seen in the granulomas from naive individuals. Thus, we provide evidence that host immune responses can dictate differences in nutrient and carbon source availability within the granuloma environment during the earliest stages of granuloma development which may impact subsequent events during latency and reactivation. Our data suggest that, during LTBI, M. tuberculosis’s shift to lipid metabolic pathways positions it well to access the increased LBs seen in macrophages within the granulomas. We are beginning a more comprehensive study involving whole-transcriptome sequencing analysis of M. tuberculosis isolated from in vitro granulomas, comparing LTBI and naive individuals. Ultimately, it will be critical to more carefully assess the structures of these complex molecules within the granulomas using newer, more sensitive biochemical techniques, although this will be challenging.
The host-pathogen interplay during infection with M. tuberculosis is incredibly complex and, despite accelerating progress in research, remains poorly understood, particularly with regard to the human processes involved. At the heart of these complex interactions is the granuloma, the hallmark pathology of TB but also of other diseases, such as Crohn’s disease, leprosy, and sarcoidosis to name a few (67–70). Our report characterizes an in vitro granuloma research model that we reason more accurately represents the complex interactions that take place inside human TB granulomas. Clinical correlations between this model and biopsy studies in humans and/or nonhuman primates will be important to pursue in parallel in future work. The development of a “real-time,” tractable in vitro granuloma model will allow researchers to explore genetic and immunologic variables contributing to TB immunopathogenesis, including the metabolic state of mycobacteria contained inside human granulomas. In doing so, our understanding of the pathophysiology of the human M. tuberculosis granulomatous response will be greatly improved, thus enabling new ideas for biomarkers, therapy, and vaccine development.
MATERIALS AND METHODS
Chemicals, reagents, and bacterial strains.
All chemicals used were from Sigma-Aldrich (St. Louis, Mo), unless otherwise specified, and were of the highest purity available. Dulbecco’s phosphate-buffered saline (PBS) (no Ca2+, no Mg2+) and RPMI 1640 with l-glutamine were purchased from Invitrogen Life Technologies (Carlsbad, CA). HyClone Standard heat-inactivated fetal bovine serum (hi-FBS) was purchased from Thermo Fisher Scientific (Waltham, MA).
Lyophilized M. tuberculosis H37Rv (ATCC 25618) was obtained from the American Tissue Culture Collection (Manassas, VA), reconstituted, and used as described previously (71). For each experiment, aliquots of M. tuberculosis frozen stocks were plated on Difco Mycobacteria 7H11 agar plates (BD/Difco) with 0.5% glycerol and 5% Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment and bacteria were grown for 9 to 14 days at 37°C in 5% CO2. M. tuberculosis Rv-lux strain was created by introducing the luciferase-expressing plasmid construct pmv306hsp + Lux (kindly provided by Brian Robertson of London Imperial College) (72) into M. tuberculosis H37Rv. The M. tuberculosis Rv-lux strain generated contains the entire bacterial Lux operon cloned in a mycobacterial integrative expression vector.
Single-cell suspensions of each M. tuberculosis strain were prepared as we previously described (71). Briefly, bacteria were scraped from agar plates, suspended in cell culture media, briefly (five 1-s pulses) subjected to vortex mixing with two glass beads (3-mm diameter), and allowed to settle for 30 min. The upper bacterial suspension (devoid of clumps) was then transferred to a second tube and let rest for an additional 5 min to obtain the final single-cell suspension. Bacterial concentrations were determined by counting using a Petroff-Hausser chamber and subsequent plating for CFU determinations. The concentration of bacteria was 1 × 108 to 2 × 108 bacteria/ml, and the degree of clumping was ≤10%. Bacteria prepared in this fashion are ≥90% viable by CFU assay.
Ethics statement.
Human blood samples were collected and processed from otherwise healthy LTBI and uninfected individuals following signed informed consent using an approved institutional review board (IRB) protocol. Subjects were limited to healthy adults (18 to 45 years of age), and both genders were included without discrimination with respect to race or ethnicity (see demographics in Table S1 in the supplemental material). Individuals with LTBI had had a positive test result for M. tuberculosis latent infection by the Mantoux screening test and/or interferon-γ release assay (IGRA) within the previous 12 months.
PBMC isolation from human blood and granuloma-like structure formation.
PBMCs were separated on a Ficoll-Paque Plus (GE Healthcare) cushion, retrieved, and cultured in RPMI 1640 medium containing 10% autologous serum in 100-by-15-mm or 24-well tissue culture dishes at 37°C in a 5% CO2 atmosphere (33). A total of 2 × 106 PBMCs/ml (containing approximately 2 × 105 monocytes), freshly isolated as described above, were immediately infected with M. tuberculosis at a MOI of 1:1 in the presence of 10% autologous serum and incubated for up to 12 days, during which time granulomas were developed and studied. Media and serum were replenished every 4 days. Use of autologous serum allowed for retention of the undefined characteristics that were unique to each individual.
Measuring in vitro granuloma-like formation.
The stage of granuloma formation was determined semiquantitatively daily (for 12 days in total) for each experimental group using the following approach. Each sample was assessed by light microscopy (Olympus IX71 DP71 microscope digital camera). At least 15 separate high-power fields per sample were evaluated, and at least 3 replicates were used to establish a scoring index. The score was calculated as the mean of the sum of granuloma scores for each sample. The scoring system is described in Fig. S1 in the supplemental material and is based on published literature and established models (16, 19, 20), where there is a progressive increase in the size and specific cell distribution of granulomas.
M. tuberculosis intracellular growth.
To assess intracellular bacterial growth, three different approaches were used. (i) For CFU assays, infected granuloma-like structures were lysed at different time periods from 0 to 9 days postinfection, as described previously (73). Lysates were serially diluted, and several dilutions for each group were plated on Middlebrook 7H11 agar plates and incubated at 37°C for determining CFU at 21 days (73). (ii) For RLU assays, M. tuberculosis Rv-lux-infected granuloma-like structures were lysed as described above at different time periods from 0 to 9 days postinfection. Lysate (500 μl) from each sample was dispensed in a 12-mm-by-50-mm polystyrene tube and read in a single-tube Sirius luminometer (Berthold). Values are expressed in RLUs. (iii) Acid-fast staining was performed on infected granuloma-like structures using a Mycobacteria fluorescent stain kit (Fluka Analytical/Sigma-Aldrich). Briefly, granulomas were formed on glass coverslips in 24-well tissue culture plates and fixed with 4% paraformaldehyde for 20 min, and bacteria were stained with 0.3% fluorescent auramine-O (in a 3% phenol solution), incubated in the dark for 15 min, washed three times with distilled water, and treated with decolorizing solution for 2 min. Then, coverslips were washed three times with distilled water and 0.1 µg/ml of DAPI DNA stain (4′,6-diamidino-2-phenylindole; Molecular Probes, Carlsbad, CA)–PBS was added for 5 min. Finally, the coverslips were washed three times with distilled water and mounted on glass slides using ProLong Gold Antifade (Invitrogen Life Technologies, Carlsbad, CA).
Analysis of cell surface antigens by microscopy.
Granuloma-like structures were formed on glass coverslips in 24-well tissue culture plates. Cells were fixed with 4% paraformaldehyde for 20 min and washed three times with PBS. When permeabilization was required, cells were treated with methanol for 5 min and washed three times with PBS. Finally, cells were incubated with confocal buffer (5 mg/ml bovine serum albumin [BSA]–10% hi-FBS–PBS) overnight at 4°C. For fluorescence microscopy analysis, confocal buffer was carefully removed and coverslips were washed three times with distilled water. Mouse monoclonal antibody (H5A4) against CD11b was obtained from the Developmental Studies Hybridoma Bank (The University of Iowa, Iowa City, IA). Mouse monoclonal antibody (UCHT1) against CD3 was obtained from BioLegend (BioLegend Inc., San Diego, CA). Rabbit monoclonal antibody (EPR7276) against CD4 was obtained from Abcam (Cambridge, MA). Goat polyclonal antibody (C-19) against CD8 and rabbit polyclonal antibody (H-300) against CD19 were obtained from Santa Cruz (Santa Cruz Biotechnology Inc., Dallas, TX). Briefly, cells were blocked with 5% goat serum–0.5% BSA–PBS for 1 h, incubated with CD11b antibody for 3 h, washed 3 times with PBS, and incubated with an anti-mouse AF488 (Invitrogen) secondary antibody for 1 h, all at room temperature. AF647 anti-human CD3 antibody was incubated for 30 min at 37°C. The nuclei were stained with 0.1 µg/ml DAPI DNA stain (Molecular Probes, Carlsbad, CA). Finally, the coverslips were washed three times with distilled water and mounted on glass slides using ProLong Gold Antifade (Invitrogen Life Technologies, Carlsbad, CA).
Measurement of cell proliferation. (i) Cell counts.
To count the total number of cells present per well, a cell enumeration assay was performed as previously described (34, 74). Briefly, granuloma-like structures were developed as described above. At different time points, cells were washed once with PBS and lysed with 1% cetavlon–0.1 M citric acid–0.05% naphthol blue black (pH 2.2) for 15 min at room temperature. Cell lysates were loaded on a hemocytometer, and stained nuclei representing cells were enumerated using phase-contrast microscopy.
(ii) EdU assay.
Granuloma-like structures were formed on glass coverslips in 24-well tissue culture plates. Cell proliferation was measured using a Click-iT EdU Alexa Fluor 488 imaging kit (Invitrogen Life Technologies, Carlsbad, CA) following the instructions of the manufacturer. Cells were treated with 10-µM EdU component A for 36 h. Media was carefully removed, and cells were fixed with 4% paraformaldehyde for 20 min. Cells were washed with 3% BSA–PBS and permeabilized with 0.5% Triton X-100–PBS for 20 min. Cells were washed twice with 3% BSA–PBS, and the Click-iT reaction cocktail was added to each well and incubated for 30 min at room temperature. Finally, cells were washed with 3% BSA–PBS and then with PBS and mounted on glass slides using ProLong Gold Antifade (Invitrogen Life Technologies, Carlsbad, CA).
Cytokine ELISAs.
The cytokine content of cell culture supernatants was measured by ELISA (R&D Systems, Minneapolis, MN) following the instructions of the manufacturer. The following kits were used: for IFN-γ, DY285; for TNF, DY210; for IL-12p40, DY1240; for IL-2, DY202; for IL-10, DY217B; for IL-4, DY204; and for IL-13, DY213.
Cytokine gene expression studies using TaqMan qRT-PCR.
Host cellular RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) following the instructions of the manufacturer and established methods (75). RNA purity and quantity were analyzed using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Total RNA (100 ng) was reverse transcribed to cDNA by using SuperScript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) following the recommendations of the manufacturer and established methods (75). Quantitative real-time PCR (qRT-PCR) was performed using human (IFN-γ, TNF, IL-12, IL-10, IL-4, and IL-13) TaqMan gene expression systems (Applied Biosystems). Negative controls included no-reverse transcriptase and no-template reactions. All samples were run in triplicate using a cfx96 real-time system (Bio-Rad) and analyzed by the threshold cycle (2−ΔΔCT) method (76). Expression of each gene was normalized to β-actin as a housekeeping gene (ΔCT).
Lipid body analysis.
Granuloma-like structures were formed on glass coverslips in 24-well tissue culture plates. Cells were stained with Nile red dye (Sigma-Aldrich, St. Louis, Mo), incubated in the dark for 15 min, and washed three times with distilled water. Counterstaining was performed with 0.1% potassium permanganate, and coverslips were washed three times with distilled water. Finally, 0.1 µg/ml of DAPI DNA stain (Molecular Probes, Carlsbad, CA)–PBS was added for 5 min. The coverslips were then washed three times with distilled water and mounted on glass slides using ProLong Gold Antifade (Invitrogen, Carlsbad, CA). Each sample was assessed by confocal microscopy (Olympus Fluoview 10i). At least 15 separate high-power fields per sample were evaluated, and at least 3 replicates were analyzed.
Gene expression analysis of M. tuberculosis within granuloma-like structures. (i) RNA isolation.
Isolation of bacterial RNA from within granuloma-like structures was achieved by using guanidine thiocyanate (GTC)-based differential lysis solution as previously published (77) and established methods using lysing matrix B tubes containing 0.1-mm-diameter silica beads and a FastPrep 24 instrument (both from MPBio) (39). Total RNA was purified by using an RNeasy Mini Column kit and treated with DNase I (both from Qiagen, Valencia, CA) to avoid genomic DNA contamination. RNA purity and quantity were analyzed by using a NanoDrop 1000 spectrophotometer (Thermo Scientific).
(ii) qRT-PCR.
Mycobacterial RNA was reverse transcribed to cDNA using SuperScript II reverse transcriptase, 10 mM deoxynucleoside triphosphate (dNTP) mix, 3 µg of random hexamers, and 10 U RNase inhibitor (all from Invitrogen, Carlsbad, CA) following the recommendations of the manufacturer and established methods (39). Control reactions were performed in parallel without reverse transcriptase to verify the absence of genomic DNA contamination. qRT-PCR was performed on the resulting cDNA using 300 nM custom-made primers with iQ SYBR green master mix (Bio-Rad) and 4% dimethyl sulfoxide (DMSO). All samples were run in triplicate using a cfx96 real-time system (Bio-Rad) and analyzed by the 2−ΔΔCT method (76). The mycobacterial rpoB housekeeping gene was used as the reference gene to normalize the CT values of the target genes, and input bacteria were used as the calibrator. The data presented are cumulative values (± standard errors of the means [SEM]). Bacterial gene-specific custom primers are listed in Table S2 in the supplemental material.
Statistics.
All experiments were performed at least 3 times in triplicate. Twenty-eight adult individuals (18 to 45 years of age), comprising 14 otherwise healthy LTBI and 14 uninfected individuals, including both genders without discrimination of race or ethnicity (see demographics in Table S1 in the supplemental material), were recruited. For cytokine ELISAs and qRT-PCR analyses, the magnitudes of response from each independent experiment differed among the donors; however, the pattern of experimental results was the same from donor to donor. To account for this variability, we normalized the data to an internal control (uninfected cells) in each experiment. A t test was used for comparing the means of the results for two groups, and one-way analysis of variance (ANOVA) followed by a Bonferroni posttest was used to test the significance of group differences between more than two groups. Statistical significance was defined as follows: *, P < 0.05, **; P < 0.01; and ***, P < 0.001.
SUPPLEMENTAL MATERIAL
Total enrollment report by ethnicity, race, and gender.
List of specific custom primers used for qRT-PCR.
Evolution of human granuloma-like structures after exposure to M. tuberculosis H37Rv. phase I (score 1), no cellular aggregation. Phase II (score 2), cellular aggregation starts, with the recruitment of mostly monocytes and some lymphocytes (approximate diameter of the base, <25 µm). Phase III (score 3), lymphocyte-like cell recruitment increases (approximate diameter of the base, 25 to 100 µm). Phase IV (score 4), the recruited cells form multilayers, comprising macrophages and lymphocytes (approximate diameter of the base, 100 to 200 µm). Phase V (score 5), the cellular structure shows signs of cellular differentiation, such as multinucleated giant cells, with large, differentiated macrophage-like cells surrounded by lymphocytes (approximate diameter of the base, >200 µm). All images are ×40. Representative images from n = 28 experiments are shown. Download
Multinucleated giant cells are present in granuloma-like structures. PBMCs obtained from LTBI individuals were infected with M. tuberculosis H37Rv (MOI 1:1), which resulted in the formation of granuloma-like structures by day 7 postinfection. Confocal microscopy images of granulomas at day 7 postinfection revealed the presence of multinucleated giant cells (CD11b+, red; white arrows). Nuclei were stained with DAPI (dark blue). Each image in the sequence (images 1 to 9) represents the view obtained from steps of 5 µm each. Representative images from n = 12 experiments are shown. All images are ×60. Download
Granuloma-like structures from LTBI individuals have greater lipid body (LB) accumulation in macrophages. PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis H37Rv (MOI 1:1) for up to 7 days. LB accumulation was determined by Nile red staining. Confocal microscopy images of the granulomas at day 7 postinfection revealed increased LBs (apart from bacteria) in macrophages (red; arrows indicate a couple of examples) in granuloma-like structures from LTBI individuals compared to naive individuals. Uninfected cells were used as controls. All images are ×60. n = 3. Download
T and B lymphocytes are present in granuloma-like structures. PBMCs obtained from LTBI individuals were infected with M. tuberculosis H37Rv (MOI 1:1), which resulted in the formation of granuloma-like structures by day 7 postinfection. Confocal microscopy images of granulomas at day 7 postinfection revealed the presence of T lymphocytes (CD4+ and CD8+, green) and B lymphocytes (CD19+, green). Nuclei were stained with DAPI (dark blue). Representative images from n = 2 experiments are shown. All images are ×60. Download
Movie S1. Download
Movie S2. Download
Movie S3. Download
ACKNOWLEDGMENTS
Images in this report were generated using instruments and services at the Campus Microscopy and Imaging Facility, The Ohio State University. The data presented were obtained at the biosafety level 3 (BSL3) laboratory facilities managed by The Ohio State University BSL3 Program. We thank Columbus Public Health for providing a clinical site for individuals with latent TB infection.
Footnotes
Citation Guirado E, Mbawuike U, Keiser TL, Arcos J, Azad AK, Wang S, Schlesinger LS. 2015. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 6(1):e02537-14. doi:10.1128/mBio.02537-14.
REFERENCES
- 1.WHO. 2013. WHO global tuberculosis report 2013 factsheet. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Zumla A, Raviglione M, Hafner R, Von Reyn CF. 2013. Tuberculosis. N Engl J Med 368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 3.Sandor M, Weinstock JV, Wynn TA. 2003. Granulomas in schistosome and mycobacterial infections: a model of local immune responses. Trends Immunol 24:44–52. doi: 10.1016/S1471-4906(02)00006-6. [DOI] [PubMed] [Google Scholar]
- 4.Saunders BM, Cooper AM. 2000. Restraining mycobacteria: role of granulomas in mycobacterial infections. Immunol Cell Biol 78:334–341. doi: 10.1046/j.1440-1711.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- 5.Flynn JL, Chan J, Lin PL. 2011. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol 4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paige C, Bishai WR. 2010. Penitentiary or penthouse condo: the tuberculous granuloma from the microbe’s point of view. Cell Microbiol 12:301–309. doi: 10.1111/j.1462-5822.2009.01424.x. [DOI] [PubMed] [Google Scholar]
- 7.Saunders BM, Frank AA, Orme IM. 1999. Granuloma formation is required to contain bacillus growth and delay mortality in mice chronically infected with Mycobacterium tuberculosis. Immunology 98:324–328. doi: 10.1046/j.1365-2567.1999.00877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guirado E, Schlesinger LS. 2013. Modeling the Mycobacterium tuberculosis granuloma—the critical battlefield in host immunity and disease. Front Immunol 4:98. doi: 10.3389/fimmu.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silver RF, Li Q, Boom WH, Ellner JJ. 1998. Lymphocyte-dependent inhibition of growth of virulent Mycobacterium tuberculosis H37Rv within human monocytes: requirement for CD4+ T cells in purified protein derivative-positive, but not in purified protein derivative-negative subjects. J Immunol 160:2408-2417. doi: 10.3389/fimmu.2013.00098. [DOI] [PubMed] [Google Scholar]
- 10.Cheon SH, Kampmann B, Hise AG, Phillips M, Song HY, Landen K, Li Q, Larkin R, Ellner JJ, Silver RF, Hoft DF, Wallis RS. 2002. Bactericidal activity in whole blood as a potential surrogate marker of immunity after vaccination against tuberculosis. Clin Diagn Lab Immunol 9:901–907. doi: 10.1128/CDLI.9.4.901-907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoft DF, Worku S, Kampmann B, Whalen CC, Ellner JJ, Hirsch CS, Brown RB, Larkin R, Li Q, Yun H, Silver RF. 2002. Investigation of the relationships between immune-mediated inhibition of mycobacterial growth and other potential surrogate markers of protective Mycobacterium tuberculosis immunity. J Infect Dis 186:1448–1457. doi: 10.1086/344359. [DOI] [PubMed] [Google Scholar]
- 12.Worku S, Hoft DF. 2003. Differential effects of control and antigen-specific T cells on intracellular mycobacterial growth. Infect Immun 71:1763–1773. doi: 10.1128/IAI.71.4.1763-1773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birkness KA, Guarner J, Sable SB, Tripp RA, Kellar KL, Bartlett J, Quinn FD. 2007. An in vitro model of the leukocyte interactions associated with granuloma formation in Mycobacterium tuberculosis infection. Immunol Cell Biol 85:160–168. doi: 10.1038/sj.icb.7100019. [DOI] [PubMed] [Google Scholar]
- 14.Franklin GF, Coghill G, McIntosh L, Cree IA. 1995. Monocyte aggregation around agarose beads in collagen gels: a 3-dimensional model of early granuloma formation. J Immunol Methods 186:285–291. doi: 10.1016/0022-1759(95)00153-2. [DOI] [PubMed] [Google Scholar]
- 15.Seitzer U, Gerdes J. 2003. Generation and characterization of multicellular heterospheroids formed by human peripheral blood mononuclear cells. Cells Tissues Organs 174:110–116. [DOI] [PubMed] [Google Scholar]
- 16.Puissegur MP, Botanch C, Duteyrat JL, Delsol G, Caratero C, Altare F. 2004. An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell Microbiol 6:423–433. doi: 10.1111/j.1462-5822.2004.00371.x. [DOI] [PubMed] [Google Scholar]
- 17.Kapoor N, Pawar S, Sirakova TD, Deb C, Warren WL, Kolattukudy PE. 2013. Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS One 8:e53657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parasa VR, Rahman MJ, Ngyuen Hoang AT, Svensson M, Brighenti S, Lerm M. 2014. Modeling Mycobacterium tuberculosis early granuloma formation in experimental human lung tissue. Dis Model Mech 7:281–288. doi: 10.1371/journal.pone.0053657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vordermeier HM, Chambers MA, Cockle PJ, Whelan AO, Simmons J, Hewinson RG. 2002. Correlation of ESAT-6-specific gamma interferon production with pathology in cattle following Mycobacterium bovis BCG vaccination against experimental bovine tuberculosis. Infect Immun 70:3026–3032. doi: 10.1128/IAI.70.6.3026-3032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis JM, Ramakrishnan L. 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani H. 2013. Granuloma cells in chronic inflammation express CD205 (DEC205) antigen and harbor proliferating T lymphocytes: similarity to antigen-presenting cells. Pathol Int 63:85–93. doi: 10.1111/pin.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan LH, Macvilay K, Barger B, Co D, Malkovska I, Fennelly G, Sandor M. 2001. Mycobacterium bovis strain bacillus Calmette-Guerin-induced liver granulomas contain a diverse TCR repertoire, but a monoclonal T cell population is sufficient for protective granuloma formation. J Immunol 166:6367–6375. doi: 10.4049/jimmunol.166.10.6367. [DOI] [PubMed] [Google Scholar]
- 23.Hogan LH, Wang M, Suresh M, Co DO, Weinstock JV, Sandor M. 2002. CD4+ TCR repertoire heterogeneity in Schistosoma mansoni-induced granulomas. J Immunol 169:6386–6393. doi: 10.4049/jimmunol.169.11.6386. [DOI] [PubMed] [Google Scholar]
- 24.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. 2009. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol 10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh V, Jamwal S, Jain R, Verma P, Gokhale R, Rao KV. 2012. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe 12:669–681. doi: 10.1016/j.chom.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 26.D’Avila H, Melo RC, Parreira GG, Werneck-Barroso E, Castro-Faria-Neto HC, Bozza PT. 2006. Mycobacterium bovis bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: intracellular domains for eicosanoid synthesis in vivo. J Immunol 176:3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- 27.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLOS Pathog 7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. 2008. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog 4:e1000204. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehr M, Elamin AA, Singh M. 2013. Lipid inclusions in mycobacterial infections, p 31–56. In Tuberculosis—current issues in diagnosis and management. Intech, Rijeka, Croatia. [Google Scholar]
- 30.Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. 2002. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology 148:2951–2958. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds HY. 1987. Bronchoalveolar lavage. Am Rev Respir Dis 135:250–263. [DOI] [PubMed] [Google Scholar]
- 32.Deb C, Lee CM, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlesinger LS. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol 150:2920–2930. doi: 10.1371/journal.pone.0010777. [DOI] [PubMed] [Google Scholar]
- 34.Rajaram MV, Brooks MN, Morris JD, Torrelles JB, Azad AK, Schlesinger LS. 2010. Mycobacterium tuberculosis activates human macrophage peroxisome proliferator-activated receptor gamma linking mannose receptor recognition to regulation of immune responses. J Immunol 185:929–942. doi: 10.4049/jimmunol.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrelles JB, Schlesinger LS. 2010. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis 90:84–93. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlesinger LS, Azad AK, Torrelles JB, Roberts E, Vergne I, Deretic V. 2008. Determinants of phagocytosis, phagosome biogenesis and autophagy for Mycobacterium tuberculosis, p 1–22. Wiley Interscience, Hoboken, NJ: http://www.wiley-vch.de/books/sample/3527318879_c01.pdf. [Google Scholar]
- 37.Kang PB, Azad AK, Torrelles JB, Kaufman TM, Beharka A, Tibesar E, Desjardin LE, Schlesinger LS. 2005. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med 202:987–999. doi: 10.1084/jem.20051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voskuil MI. 2004. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis 84:138–143. doi: 10.1016/j.tube.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Keiser TL, Azad AK, Guirado E, Bonacci R, Schlesinger LS. 2011. Comparative transcriptional study of the putative mannose donor biosynthesis genes in virulent Mycobacterium tuberculosis and attenuated Mycobacterium bovis BCG strains. Infect Immun 79:4668–4673. doi: 10.1128/IAI.05635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler PR, Ratledge C. 1994. Metabolism of Mycobacterium tuberculosis. In Bloom BR (ed), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, DC. [Google Scholar]
- 41.Shi L, Sohaskey CD, Pfeiffer C, Datta P, Parks M, McFadden J, North RJ, Gennaro ML. 2010. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol Microbiol 78:1199–1215. doi: 10.1111/j.1365-2958.2010.07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloch H, Segal W. 1956. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J Bacteriol 72:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins DM, Wilson T, Campbell S, Buddle BM, Wards BJ, Hotter G, De Lisle GW. 2002. Production of avirulent mutants of Mycobacterium bovis with vaccine properties by the use of illegitimate recombination and screening of stationary-phase cultures. Microbiology 148:3019–3027. [DOI] [PubMed] [Google Scholar]
- 44.Liu K, Yu J, Russell DG. 2003. pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology 149:1829–1835. doi: 10.1099/mic.0.26234-0. [DOI] [PubMed] [Google Scholar]
- 45.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. 2010. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A 107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stout RD, Watkins SK, Suttles J. 2009. Functional plasticity of macrophages: in situ reprogramming of tumor-associated macrophages. J Leukoc Biol 86:1105–1109. doi: 10.1189/jlb.0209073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henning LN, Azad AK, Parsa KV, Crowther JE, Tridandapani S, Schlesinger LS. 2008. Pulmonary surfactant protein A regulates TLR expression and activity in human macrophages. J Immunol 180:7847–7858. doi: 10.4049/jimmunol.180.12.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capuano SV III, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL. 2003. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun 71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fallahi-Sichani M, Marino S, Flynn JAL, Linderman J, Kirschner DE. 2013. A systems biology approach for understanding granuloma formation and function in tuberculosis, p 127–155. In McFadden J, Beste DJV, Kierzek AM (ed), Systems biology of tuberculosis. Springer, New York, NY. [Google Scholar]
- 52.Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, Sacchettini J, Fortune SM, Flynn JL. 2014. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med 20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saunders BM, Britton WJ. 2007. Life and death in the granuloma: immunopathology of tuberculosis. Immunol Cell Biol 85:103–111. doi: 10.1038/sj.icb.7100027. [DOI] [PubMed] [Google Scholar]
- 54.Shrivastava P, Bagchi T. 2013. IL-10 modulates in vitro multinucleate giant cell formation in human tuberculosis. PLoS One 8:e77680. doi: 10.1371/journal.pone.0077680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward SK, Abomoelak B, Marcus SA, Talaat AM. 2010. Transcriptional profiling of Mycobacterium tuberculosis during infection: lessons learned. Front Microbiol 1:121. doi: 10.1371/journal.pone.0077680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danelishvili L, McGarvey J, Li YJ, Bermudez LE. 2003. Mycobacterium tuberculosis infection causes different levels of apoptosis and necrosis in human macrophages and alveolar epithelial cells. Cell Microbiol 5:649–660. doi: 10.1046/j.1462-5822.2003.00312.x. [DOI] [PubMed] [Google Scholar]
- 57.McGarvey JA, Wagner D, Bermudez LE. 2004. Differential gene expression in mononuclear phagocytes infected with pathogenic and non-pathogenic mycobacteria. Clin Exp Immunol 136:490–500. doi: 10.1111/j.1365-2249.2004.02490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy DJ, Brown JR. 2007. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis 7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guy MR, Illarionov PA, Gurcha SS, Dover LG, Gibson KJ, Smith PW, Minnikin DE, Besra GS. 2004. Novel prenyl-linked benzophenone substrate analogues of mycobacterial mannosyltransferases. Biochem J 382:905–912. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patterson JH, Waller RF, Jeevarajah D, Billman-Jacobe H, McConville MJ. 2003. Mannose metabolism is required for mycobacterial growth. Biochem J 372:77–86. doi: 10.1042/BJ20021700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy TR, Torrelles JB, Macfarlane AS, Katawczik M, Kutzbach B, Desjardin LE, Clegg S, Goldberg JB, Schlesinger LS. 2005. Overexpression of Mycobacterium tuberculosis manB, a phosphomannomutase that increases phosphatidylinositol mannoside biosynthesis in Mycobacterium smegmatis and mycobacterial association with human macrophages. Mol Microbiol 58:774–790. doi: 10.1111/j.1365-2958.2005.04862.x. [DOI] [PubMed] [Google Scholar]
- 62.Ning B, Elbein AD. 1999. Purification and properties of mycobacterial GDP-mannose pyrophosphorylase. Arch Biochem Biophys 362:339–345. doi: 10.1006/abbi.1998.1053. [DOI] [PubMed] [Google Scholar]
- 63.Gurcha SS, Baulard AR, Kremer L, Locht C, Moody DB, Muhlecker W, Costello CE, Crick DC, Brennan PJ, Besra GS. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem J 365:441–450. doi: 10.1042/BJ20020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rana AK, Singh A, Gurcha SS, Cox LR, Bhatt A, Besra GS. 2012. Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PLoS One 7:e48211. doi: 10.1371/journal.pone.0048211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishra AK, Krumbach K, Rittmann D, Batt SM, Lee OY, De S, Frunzke J, Besra GS, Eggeling L. 2012. Deletion of manC in Corynebacterium glutamicum results in a phospho-myo-inositol mannoside- and lipoglycan-deficient mutant. Microbiology 158:1908–1917. doi: 10.1371/journal.pone.0048211. [DOI] [PubMed] [Google Scholar]
- 66.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR Jr, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 67.Chambers TJ, Morson BC. 1979. The granuloma in Crohn’s disease. Gut 20:269–274. doi: 10.1136/gut.20.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narayanan RB. 1988. Immunopathology of leprosy granulomas—current status: a review. Lepr Rev 59:75–82. [PubMed] [Google Scholar]
- 69.Wheat J, Sarosi G, McKinsey D, Hamill R, Bradsher R, Johnson P, Loyd J, Kauffman C. 2000. Practice guidelines for the management of patients with histoplasmosis. Clin Infect Dis 30:688–695. doi: 10.1086/313752. [DOI] [PubMed] [Google Scholar]
- 70.Wallis RS, Broder MS, Wong JY, Hanson ME, Beenhouwer DO. 2004. Granulomatous infectious diseases associated with tumor necrosis factor antagonists. Clin Infect Dis 38:1261–1265. [DOI] [PubMed] [Google Scholar]
- 71.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol 144:2771–2780. [PubMed] [Google Scholar]
- 72.Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. 2010. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777. doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olakanmi O, Britigan BE, Schlesinger LS. 2000. Gallium disrupts iron metabolism of mycobacteria residing within human macrophages. Infect Immun 68:5619–5627. doi: 10.1128/IAI.68.10.5619-5627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drowart A, Selleslagls J, Huygen K, De Bruyn J, Van Vooren J-P. 1993. Absence of IgM and IgG antibody response to the three components of the Mycobacterium bovis BCG 85 complex antigen after BCG vaccination. Chest 103:320–321. doi: 10.1378/chest.103.1.320-b. [DOI] [PubMed] [Google Scholar]
- 75.Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. 2011. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A 108:17408–17413. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 77.Monahan IM, Mangan JA, Butcher PD. 2001. Extraction of RNA from intracellular Mycobacterium tuberculosis: methods, considerations and applications, p 31–42. In Parish T, Stoker NG (ed), Mycobacterium tuberculosis protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total enrollment report by ethnicity, race, and gender.
List of specific custom primers used for qRT-PCR.
Evolution of human granuloma-like structures after exposure to M. tuberculosis H37Rv. phase I (score 1), no cellular aggregation. Phase II (score 2), cellular aggregation starts, with the recruitment of mostly monocytes and some lymphocytes (approximate diameter of the base, <25 µm). Phase III (score 3), lymphocyte-like cell recruitment increases (approximate diameter of the base, 25 to 100 µm). Phase IV (score 4), the recruited cells form multilayers, comprising macrophages and lymphocytes (approximate diameter of the base, 100 to 200 µm). Phase V (score 5), the cellular structure shows signs of cellular differentiation, such as multinucleated giant cells, with large, differentiated macrophage-like cells surrounded by lymphocytes (approximate diameter of the base, >200 µm). All images are ×40. Representative images from n = 28 experiments are shown. Download
Multinucleated giant cells are present in granuloma-like structures. PBMCs obtained from LTBI individuals were infected with M. tuberculosis H37Rv (MOI 1:1), which resulted in the formation of granuloma-like structures by day 7 postinfection. Confocal microscopy images of granulomas at day 7 postinfection revealed the presence of multinucleated giant cells (CD11b+, red; white arrows). Nuclei were stained with DAPI (dark blue). Each image in the sequence (images 1 to 9) represents the view obtained from steps of 5 µm each. Representative images from n = 12 experiments are shown. All images are ×60. Download
Granuloma-like structures from LTBI individuals have greater lipid body (LB) accumulation in macrophages. PBMCs obtained from LTBI and naive individuals were infected with M. tuberculosis H37Rv (MOI 1:1) for up to 7 days. LB accumulation was determined by Nile red staining. Confocal microscopy images of the granulomas at day 7 postinfection revealed increased LBs (apart from bacteria) in macrophages (red; arrows indicate a couple of examples) in granuloma-like structures from LTBI individuals compared to naive individuals. Uninfected cells were used as controls. All images are ×60. n = 3. Download
T and B lymphocytes are present in granuloma-like structures. PBMCs obtained from LTBI individuals were infected with M. tuberculosis H37Rv (MOI 1:1), which resulted in the formation of granuloma-like structures by day 7 postinfection. Confocal microscopy images of granulomas at day 7 postinfection revealed the presence of T lymphocytes (CD4+ and CD8+, green) and B lymphocytes (CD19+, green). Nuclei were stained with DAPI (dark blue). Representative images from n = 2 experiments are shown. All images are ×60. Download
Movie S1. Download
Movie S2. Download
Movie S3. Download