Abstract
The distal gut harbours ∼1013 bacteria, representing the most densely populated ecosystem known. The functional diversity expressed by these communities is enormous and relatively unexplored. The past decade of research has unveiled the profound influence that the resident microbial populations bestow to host immunity and metabolism. The evolution of these communities from birth generates a highly adapted and highly personalized microbiota that is stable in healthy individuals. Immune homeostasis is achieved and maintained due in part to the extensive interplay between the gut microbiota and host mucosal immune system. Imbalances of gut microbiota may lead to a number of pathologies such as obesity, type I and type II diabetes, inflammatory bowel disease (IBD), colorectal cancer (CRC) and inflammaging/immunosenscence in the elderly. In-depth understanding of the underlying mechanisms that control homeostasis and dysbiosis of the gut microbiota represents an important step in our ability to reliably modulate the gut microbiota with positive clinical outcomes. The potential of microbiome-based therapeutics to treat epidemic human disease is of great interest. New therapeutic paradigms, including second-generation personalized probiotics, prebiotics, narrow spectrum antibiotic treatment and faecal microbiome transplantation, may provide safer and natural alternatives to traditional clinical interventions for chronic diseases. This review discusses host–microbiota homeostasis, consequences of its perturbation and the associated challenges in therapeutic developments that lie ahead.
Keywords: bacterial, host–pathogen interactions, inflammation, mucosa
Introduction
The human gut harbours several hundred species of bacteria featuring a small number of phyla, including Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria and Fusobacteria, highlighting the selectivity of the gut microenvironment 1. The bacterial, archeal, fungal and viral intestinal communities are referred to as the gut microbiota and their collective genomes are referred to as the gut microbiome. Eons of co-evolution have selected those species that bring no harm (commensals) or confer benefit to their host (mutualists) 2–4. These communities feature metabolic specialization, complementarity and co-operation, which results in complex networks of microbe–microbe and microbe–host relationships 5–7. An individual's gut microbiome may encode ∼150 times more genes than our own genomes 8, thus justifying references to the gut microenvironment as a complex bioreactor replete with diverse biochemical activities. The relative balance of specific metabolic activities in the gut and their interaction with the human host may promote both health and disease. The composition and functional capacity of the gut microbiota may modulate risk positively or negatively to a wide variety of health and disease phenotypes. The human gut microbiota displays high interpersonal variation 9,10 but is stable over time, displaying resilience in the face of perturbing influences 11 such as dietary flux 12–15 and exposure to antibiotics 16. Chronic or acute perturbations drive microbiota dysbioses, which has been observed in a number of important human diseases.
Generation of gut microbiota homeostasis
Many studies have identified gut microbiota dysbiosis in association with a large variety of human disease; however, in most cases causality has yet to be established. The study of the gut microbiota over the human lifespan provides important new insights and is crucial to our ability to restore homeostasis through therapeutic interventions intended to correct disease-associated dysbioses (Fig. 1). Studies examining microbial succession communities in early life provide a powerful framework to address how homeostasis and stability are acquired in infants and subsequently lost in elderly people 17. The developing gut microbiota increases in diversity and stability over time. Conversely, gut microbiota of elderly people display communities of reduced diversity and increased interpersonal variability. Human subjects of advanced age represent an opportunity to identify the factors involved in maintaining homeostasis and the relationship to aspects of inflammaging and immunosenescence in elderly people 18–23.
Fig 1.
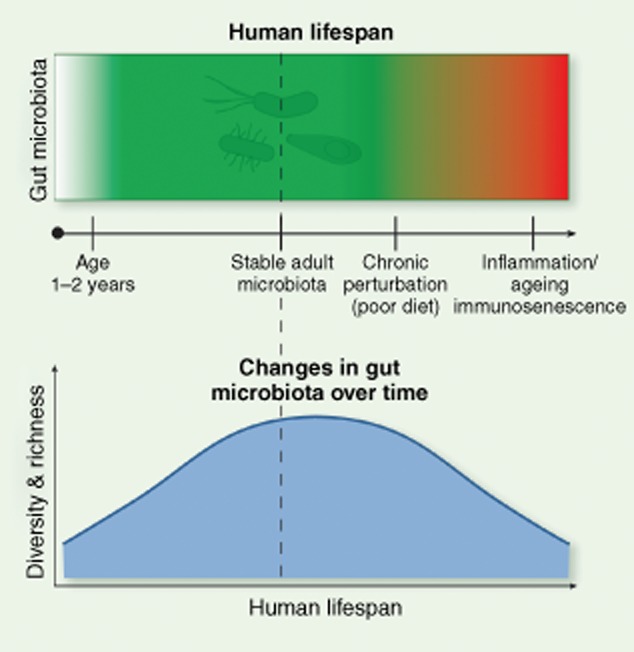
The gut microbiota during the human lifespan. Over the first 2–3 years of life, the gut microbiota undergoes dynamic changes wherein highly adapted communities are established resulting in a healthy microbiome in a state of homeostasis. Environmental factors such as sustained intake of a high-fat, high-carbohydrate diet may drive the gut microbiota into a state of dysbiosis that may influence human diseases such as obesity, diabetes and colorectal cancer. Similarly, the elderly gut microbiota may degenerate into a state of dysbiosis resulting in chronic inflammation (inflammaging) and reduced immune function (immunosenescence).
Successional communities drive the formation of functional networks
Until recently, humans were thought to be born sterile. Accumulating evidence suggests that microbes are detected in amniotic fluid, placenta, meconium and umbilical cord 24,25. The role of these microbes is unclear, but has been suggested to play a role in tolerance to commensal bacteria 26. The microbiota acquired at birth is of low diversity and unstable, evolving in the first years of life, and culminating in a stable configuration with expanded representation of niche-adapted phylotypes. The primary inoculum for vaginally delivered babies is from the mother's vaginal and faecal microbiome, dominated by Lactobacillus spp., Prevotella spp. and Sneathia spp. In caesarean section (C-section) births, the skin of individuals handling the newborns is the primary source of the initial microbiota and features Staphylococcus spp., Corynebacterium spp. and Propionibacterium spp. 27,28. The diversity of adult gut microbiota is higher in vaginally compared to C-section-delivered infants 29–31. Despite the distinct taxonomic representation of the initial microbial inoculums, the evolving communities display increased relatedness over time 30,32,33, suggesting that evolving communities possess a ‘trajectory’ that tends towards stable fitness optima (Fig. 2).
Fig 2.
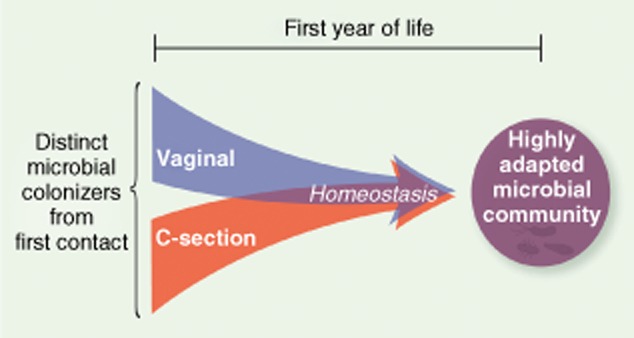
Vaginal versus caesarean-section delivery. The pioneer microbial colonizers acquired vertically from first contact are distinct but microbial succession converges to become more similar over the first years of life. This illustrates that gut microbiota evolves over time to establish a state of homeostasis wherein resident species are highly adapted for survival in the highly competitive gut microenvironment.
Microbial succession is iterative, and driven at least in part by the metabolic activities of the initial pioneer community that necessarily alter the virgin ecosystem, providing novel opportunities for subsequent community succession that broadens the functional complementarity of resident species. The initial colonizers of the gut include many facultative anaerobes owing to the elevated oxygen tension in the newborn gut. The activities of the pioneer colonizers reduce the oxygen tension, aiding succession favouring strict anaerobes. The pH and redox potential along the length of the gut is not uniform and dictates the fitness of individual bacterial species occupying various subdomains. Analysis of gut succession communities in early life bear resemblance to ecological models of punctuated equilibrium, wherein brief periods of transient stability are followed by bursts of change 34–36. In this regard, gut microbiota development also has the property of velocity, wherein rapid change may reflect a maladapted microbiota, e.g. when babies transition from breast milk to a solid food diet.
Microbial succession proceeds with a trend towards increased numbers and interconnectivity of microbe–microbe and host–microbe functional networks 37,38. Many aspects of bacterial metabolism are carried out by co-ordinated and co-operative consortia. These consortia are indirectly discernable by the analysis of metagenomic DNA sequence data to identify broadly co-occurring and co-excluded species 37. The generation of short chain fatty acids (SCFAs) reflects the activities of a network of species that co-operatively degrade resistant starch. The contribution of SCFAs to gut enterocytes is thought to be a significant driving force in host/microbiota interactions 39. The collective fermentative activities of the numerically dominant phyla generate inhibitory quantities of hydrogen (H2). The fermentative consortia are linked functionally to biochemical activities of sulphate-reducing bacteria and methanogens in the community that consume H2. This example illustrates the driving forces of stable and widespread microbial interdependencies. The number and interconnectivity of these networks and the robustness of species representing network hubs may influence the property of community resilience (the resistance to change of community structure in response to perturbation) and elasticity (the rate that communities restore equilibrium following perturbation) (Fig. 3). These traits may be highly personalized and define an individual's stability landscape 11,40. Ecological models suggest that some community topologies or phylotypes will be highly resistant to change, whereas others may be prone to larger change following perturbation (Fig. 4). A major challenge inherent in gut microbiota modulation to correct dysbioses lies in our ability to reliably alter the composition of bacterial communities to achieve desired clinical outcomes while avoiding unintended or poorly perceived negative consequences. An improved understanding of the selective features controlling stable ‘health-promoting’ network formation may define novel therapeutic approaches to achieve restoration of microbiota equilibria and immune homeostasis.
Fig 3.
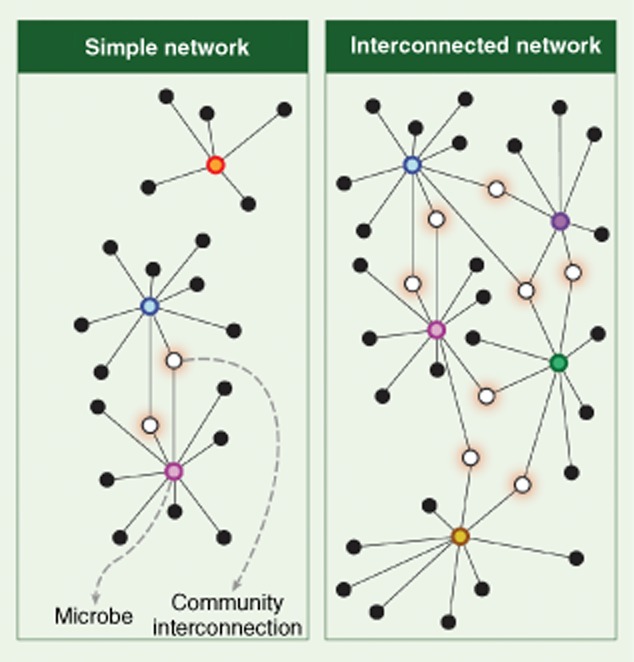
Functional networks link the function of microbes and support community stability and resilience. Functional networks in the gut are extensive and imply that targeted modulation of the gut microbiota may have unexpected impact on the viability of off-target species. Left: low interconnectivity may render gut communities prone to change as the result of relatively small perturbation. Right: highly interconnected networks may be more robust and resistant to change.
Fig 4.
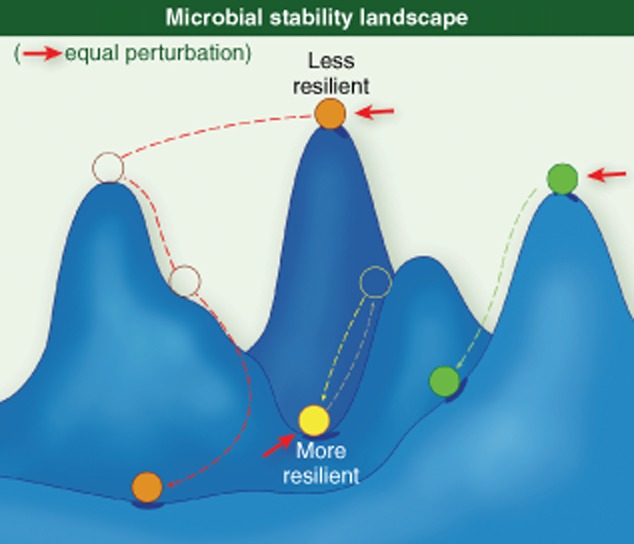
Microbial stability landscape. A theoretical depiction of gut species and their response to perturbation. In some instances, a small perturbation may lead to a large change in the fitness of that species with low resilience, whereas other species may be equally impacted by a similar perturbation but display high resilience.
Gut homeostasis
Interplay of the host immune system and gut microbiota
We are making strong inroads in our understanding of the complex interactions between the gut microbiota and the mucosal immune system of the gastrointestinal (GI) tract. The close physical proximity of dense microbial populations with underlying host tissue facilitates numerous metabolic and immunological opportunities for host benefit while simultaneously posing a constant and proximal threat to human health. The human immune system must establish an appropriate balance between tolerance to the gut commensal microbiota and vigilance to guard against infectious agents and opportunistic pathogens. Gut homeostasis is maintained as an inflammatory tone, allowing a rapid and self-limiting response appropriate to a stress or infectious agent. The cross-talk between the gut microbiota and host is extensive, and involves both innate and adaptive immunity.
Immune surveillance of the gut commensal community involves the recognition of a diversity of pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) and bacterial peptidoglycan cell wall components through Toll-like receptors (TLRs) and nucleotide binding oligomerization domain proteins (NODs). Endogenous and exogenous signals in the gut are recognized by a repertoire of innate immune cell pattern recognition receptors (PRRs) mediating the interaction between bacterial ligands and the host 41,42. TLRs and NODs act in distinct cellular compartments and cell-type specific combinations. Ligand engagement of these receptors on the apical surface (lumen exposed) epithelium promotes tolerance and healthy inflammatory tone; however, the activation of these receptors on the basolateral surface of colonocytes leads to strong proinflammatory responses 41. A variety of microbial ligands stimulate activation of nuclear factor kappa B (NF-κB) and downstream proinflammatory cytokines such as tumour necrosis factor (TNF)-α and interleukin (IL)-1 43.
Bacterial populations are segregated from the gut epithelium by a thick mucin layer produced by goblet cells that is embedded with a number of anti-microbial factors such as immunoglobulin (Ig)A, α and β defensins 44. Despite this segregation, it is evident that components of the gut microbiota play essential roles in immune development, homeostasis and tolerance. Commensal colonization of the gut results in Paneth cell expression of the anti-microbial peptide, regenerating islet-derived 3 gamma (Reg III-γ) 45, whereas NOD2 signalling is required for α-defensin production 46 (Fig. 5). The gut microbiota is involved in maintaining a balance of T-effector cell function. In germ-free mice, natural killer (NK) T cells residing in non-mucosal lymphoid organs could not be primed effectively to mount anti-viral responses, as both macrophages and dendritic cells (DCs) failed to produce type 1 interferon (IFN) 47. The commensal microbiota also contributes to tolerance via TLRs sequestered in the crypts and regulatory T cells (Treg) cells that down-regulate proinflammatory signalling through the production of IL-10 and transforming growth factor (TGF)-β 48. Thymic Treg cells confer tolerance to antigens produced by resident microbiota 49. Additional studies have highlighted additional microbe–host interactions that serve to modulate immune homeostasis. Non-pathogenic Salmonella block the ubiquitination of IκBα, thereby maintaining NF-κB in an inactive state 50. Bacteroides thetaiotamicron, an abundant member of the gut microbiota, increases the nuclear export of the RelA subunit of NF-κB, thereby reducing its activity 51. Lactobacillus casei down-regulates components of the proteasome complex, thereby decreasing the turnover of IκBα 52. B. fragilis increases the production of the anti-inflammatory cytokine IL-10 in gut-associated immune cells 53. Many Clostridium spp. increase TGF-β expression and Treg cell titres 54. Commensally produced ATP is known to stimulate differentiation of Th17 cells 55. Segmented filamentous bacteria (SFB) drive this induction 56. Polysaccharide A (PSA) derived from B. fragilis systemically increases CD4+ T cell numbers in germ-free mice 53 and forkhead box protein 3 (FoxP3) Treg production of IL-10 in mice 57. Taken together, these results make evident that the gut microbiota/host immune interactions are extensive and co-operative. Homeostasis is maintained through what is likely to be a large number of microbial-produced signals that facilitate robust immune surveillance and tolerance.
Fig 5.
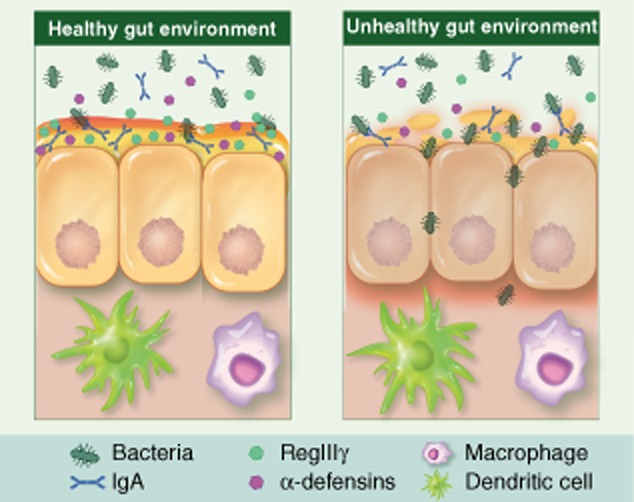
Stratification of the gut epithelium. In a healthy gut environment (left panel), goblet cells secrete mucin to establish a physical barrier excluding direct contact of the gut microbiota from the underlying epithelium. Paneth cells produce a number of anti-microbial defensins that are embedded in the mucin layer. Dendritic cells extend into the lumen to sample the commensal communities potentially as a means of determining self. During pathological conditions (right panel), mucin barrier is compromised which facilitates microbial invasion through the epithelium and leads to inflammation.
Antibiotic treatment reveals gut microbiota immune interactions
Antibiotic treatment modulates the gut microbiota composition (Table 1) along with their immune modulatory ligands, and therefore represents an approach to deepen our knowledge of the interactions between the gut microbiota and host immune function. Antibiotic exposure impacts both innate and adaptive immunity. In mice, antibiotic treatment results in reduction of Paneth cells, goblet cells and enterocytes and production of anti-microbial peptides, including defensins, C-type lectins and cathelicidins 68. Antibiotics that target Gram-negative bacteria impact signalling through TLR-4 and NOD1, whereas those targeting Gram-positive species alter signalling through TLR-2 and NOD2 pathways 69,70. Treatment of mice with amoxicillin and clavulanic acid (broad-spectrum) resulted in reduced serum IgG levels 71. Mice treated with amoxicillin results in the loss of Lactobacillus spp. and an accompanying reduction in α- defensins, matrilysin and phospholipase A2 production. Transcript abundance of major histocompatibility complex (MHC) class II and class Ib genes were also reduced 72. Mice treated with Gram-positive-specific antibiotics (vancomycin or ampicillin) but not Gram-negative-specific treatments (metronidazole and neomycin) resulted in depletion of Th17 cells in the intestine 73 and the identification of SFB as the signal responsible for the Th17 cell collapse 56. The continued presence of the gut microbiota appears to be important for maintaining effector T cell populations in the gut, as treatment of mice with an antibiotic cocktail resulted in the reduced abundance of CD4+ T cells expressing IFN-γ or IL-17A 74. As we begin to improve our understanding of the specific ligands and immune pathways mediating gut homeostasis we will increase our ability to recognize the aberrations associated with microbial dysbioses and how those communities might be altered to restore homeostasis.
Table 1.
Examples of antibiotic-induced alterations of the gut microbiota in humans and mice
| Antibiotic class | Antibiotic and spectrum | Dose and duration | Mechanism of action | n | Method and study | Effect on microbiota | Recovery post–treatment | Reference |
|---|---|---|---|---|---|---|---|---|
| β-lactam | Amoxicillin | 1·5 g/day, 5 days | Inhibition of cell wall synthesis by irreversible binding of β-lactam to Penicillin Binding Protein (PRP) | 6 subjects | 16s rRNA, TTGE was used to monitor test subjects up to 60 days including treatment | Major shift in dominant species after 24 h. Similarity index, an average of 74% day 4 of treatment | Day 30, microbiota largely restored, 88% similarity. Day 60, 4 of 6 subjects had a similarity index >90%. 1 subj. had a similarity index of only 66%. | 58 |
| Narrow, gram-positive | ||||||||
| β-lactam | Aztreonam | 0·1 mg/ml in drinking water 7 days | Inhibition of cell wall synthesis by irreversible binding of β-lactam to PRP | 4–5 mice | 47 selected bacteria were monitored, 16s rDNA analysed by QuantiGene 2·0 Reagent System | ↑Firmicutes (overall increase), ↑Clostridium clostriforme, ↑C. methylpentosum, ↑Eubacterium desmolans, and ↑E. limosum ↓C. celerecrescens. Bacteroidetes: ↑Porphyromonas. Other: ↑Acetovibrio cellulosolvens, Bacillus mycoides, ↑Helicobacter sp, ↑Rumniococcus gnavus and ↑R. schinkii. | Not studied | 59 |
| Narrow, gram-negative | ||||||||
| β-lactam | Amoxicillin | Distributed <1 month of age | Inhibition of cell wall synthesis by irreversible binding of β-lactam to PRP | 28 subjects | RT–PCR was used to monitor selected bacterial species in infants | ↓Bifidobacteria and ↓Bacteroides fragilis group spp. Slightly higher numbers of Bifidobacteria were found in infants with older siblings | Not studied | 29 |
| Narrow, gram-positive | ||||||||
| β-lactam | Amoxicillin and clavulanic acid | 2 × (875 mg amoxicillin, 125 mg clav. acid) per day 10 days | Inhibition of cell wall synthesis by irreversible binding of β-lactam to PRP. | 1 subject | Subj. treated for non-GIT-condition developed | ↑Bacteroides distasonis replaced ↓B. fragilis as the dominant species in the Bacteroides group. ×Clostridium rRNA cluster XIVa (not detected), ↓Clostridium sp. rRNA cluster IV, ×Bifidobacterium spp. were eliminated (not detected) | 2 weeks post-treatment, ↑B. fragilis again the dominant Bacteroides sp., | 60 |
| Narrow, gram-positive | β-lactamamase inhibitor added | Antibiotic Assoc. Diarrhoea. 16s rRNA used to monitor microbiome up to 16 days post-treatment | ↑↑Enterobacteriaceae (34% of all sequences) | ↑Clostridium rRNA cluster XIVa increased compared to pretreatment, ↑Clostridium sp. rRNA cluster IV increased but did not reach pretreatment level,×Bifidobacterium spp. ×Enterobacteriaceae | ||||
| Cephalosporin (β-lactam) | Cefoperazone | 0·5 mg/ml in drinking water, 10 days | Inhibition of cell wall synthesis by irreversible binding of β-lactam to PRP | 15 mice | 16s rRNA analysis of the whole microbiome, Roche 454 GS FLX platform | No information. Sequence tags failed to amplify. Mice killed at end of treatment showed a significant decrease in bacterial load compared to control. | Treated mice co-housed with a wt mouse during a 6-week recovery period, showed greater diversity and higher Bacteroides levels (15% vs 0·33%) compared to mice that recovered alone. | 61 |
| Broad, gram-positive and -negative | ||||||||
| Fluoro-quinolone | Ciprofloxacin | 2 × 500 mg/day, 5 days | Inhibition of DNA topoisomerase enzymes, promotes breakage of DNA | 3 subjects | 16s rRNA tag pyrosequencing | Relative abundance levels of approximately 30% of the identified taxa were affected | 4 weeks post-treatment, the composition had returned to ranges within the temporal variability of pretreatment samples | 62 |
| Broad, gram-positive and -negative | ||||||||
| Fluoro-quinolone | Ciprofloxacin | 30 mg/ml in drinking water 14 days | Inhibition of DNA topoisomerase enzymes, promotes breakage of DNA | 4–5 mice | 47 selected bacteria were monitored, 16s rDNA analysed by QuantiGene 2·0 Reagent System | Firmicutes: ↑Clostridium methylpentosum and ↑Eubacterium desmolans, ↓Lactobacillus murinus and ↓L. salivarius. ↓Bacteroidetes (overall decrease): ↓Bacteroides acidofaciens, ↓B. distasonis, ↓B. forsythus, ↓Bacteroides sp. ASF519 L7-16. Other: ↑Acetovibrio cellulosolvens, ↑Helicobacter sp. L10-17, ↑Klebsiella granulomatis, ↑Rumniococcus gnavus and ↑R. schinkii. × Ralstonia was eliminated (non-detctable level) | Not studied | 59 |
| Broad, gram-positive and -negative | ||||||||
| Fluoro-quinolone | Ciprofloxacin | 2 × 500 mg/day, 5 days Antibiotic course repeated after 6-month antibiotic-free interval | Inhibition of DNA topoisomerase enzymes, promotes breakage of DNA | 3 subjects | 16s rRNA tag pyrosequencing | 25–50% of taxa were eliminated, and species closely related to those removed by antibiotic treatment increased; changes in composition of Bacteroides, Lachnospiraceae, and Ruminococcaceae (excluding Faecalibacterium which was reduced or eliminated) | The Bacteroides spp, Lachnospiraceae and Ruminococcaceae that surged during the antibiotics course, were replaced by original taxa once the treatment had stopped | 16 |
| Broad, gram-positive and -negative | ||||||||
| Glycylcycline | Tigecyclin | 6·25 mg/kg, subcutaneous injection twice daily, 10 days | Inhibition of protein synthesis by binding to 30s ribosomal subunit | 20 mice | 16s rDNA analysis, Roche 454 GS FLX Platform. Mothur 1.27.0, used for comparisons of community structure | ↓Bacteroidetes decreased (phyla consisted largely of Porphyromonadaceae day 3 of treatment), ↓Firmicutes, ↑Enterobacteriaceae and ↑Verrucomicrobia increased | At the family level, microbiota had returned to baseline 2 weeks post-treatment. Overall diversity had still not returned to baseline 5 weeks post-treatment | 63 |
| Broad, gram-positive and -negative, facult. and obligate anaerobes | ||||||||
| Lincosamide | Clindamycin | 4 × 150 mg/day 7 days | Protein synthesis inhibitor, binds to 50S ribosomal subunit | 4 subjects | Long-term impact of treatment was shown using rep-PCR and T-RFLP targeting the genus Bacteroides | ↓Bacteroides spp. | 21 days post-treatment, the composition was changed, but monitored Bacteroides species as a group, had returned to pretreatment level. The composition was still significantly altered 18 months post-treatment | 64 |
| Broad, gram-positive and -negative, anaerobes | ||||||||
| Nitro-imidazole | Metronidazole | 0·1 mg/ml in drinking water 14 days | Inhibition of DNA synthesis, DNA damage by oxidation | 4–5 mice | 47 selected bacteria were monitored, 16s rDNA analysed by QuantiGene 2·0 Reagent System | ↑Firmicutes (overall increase), ↑Eubacterium desmolans, ↑E. limosum, ↓Lactobacillus murinus and ↓L. salivarius. ↓Bacteroidetes (overall decrease), with the exception of ↑Porphyromonas. Other: ↑Acetovibrio cellulosolvens, ↑Bacillus mycoides, ↑Helicobacter sp, ↑Klebsiella granulomatis, ↑Ruminococcus gnavus and ↑R. schinkii | Not studied | 59,65 |
| Narrow, anaerobic bacteria | ||||||||
| Combination (β-lactam/ Amino-glycoside) | Cephalothin+Neomycin | 2 mg/ml per antibiotic in drinking water 7 days | Inhibition of cell wall synthesis by binding to PBP, both β-lactam antibiotics | 4–5 mice | 47 selected bacteria were monitored, 16s rDNA analysed by QuantiGene 2·0 Reagent System | Firmicutes (decreased half of monitored species): ↓Clostridium celerecrescens, ↓C. fusiformis ↓C. methylpentosum, ↓C. polysaccharolyticum, ↓C. scincdens, ↓C. sp ASF502, ↓Eubacterium desmolans, ↓Lactobacillus acidophilus, ↓L. reuteri, ↓L. salivarius. Bacteroidetes: ↑B. acidifaciens and ↑B. distasonis. Other: ↓Acetovibrio cellulosolvens, ↓Helicobacter sp. L10-17 | Not studied | 59 |
| Broad, gram-positive and -negative | ||||||||
| Combination (β-lactam/ nitro-imidazole/ heavy metal) | Amoxicillin + metronidazole + bismuth | 3·0 mg + 0·69 mg + 0·185 mg, in food 5 g/tablet/day/(20 g) mouse 10 days | Inhibition of cell wall synthesis/ inhibition of DNA synhesis/inactivation of proteins | 15 mice | 16s rRNA analysis of the whole microbiome, Roche 454 GS FLX Platform | ↓Bacteroidetes, ↓↓Firmicutes, ↑↑Proteobacteria, ↓other Phyla | 2 weeks post-treatment, Bacteroidetes and Firmicutes returned to dominance and while the Proteobacteria level decreased, it remained higher than in untreated animals (5·8% versus 1·2% of the microbiota) | 61,66 |
| Narrow, gram-positive | ||||||||
| Narrow, anaerobic/Broad, gram-positive and -negative | ||||||||
| Combination (fluoro quinolone/ lincos amide) | Ciprofloxacin + Clindamycin | 2 × 500 mg/day, 7 days + 3 × 500 mg/day for 7 additional days | Inhibition of DNA topoisomerase enzymes/ Protein synthesis inhibitor, binds to 50S ribosomal subunit | 1 subject | DGGE profiles compared pre- and post-treatment | Similarity index of DGGE profiles was 73% post-ciprofloxacin, but was drastically reduced by clindamycin. Ciprofloxacin: ↓Bacteroides vulgatus, ↑B. thetaiotaomicron, →Faecalibacterium prausnitzii (unchanged), ↑↑Ruminococcus productus, Clindamycin: ↓↓Bacteroides vulgatus, ×B. thetaiotaomicron, ↓↓Faecalibacterium prausnitzii, ↓Ruminococcus productus | 16 days post-treatment, Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii had been restored to pretreatment levels. B. vulgatus and Ruminococcus productus had not recovered at all | 67 |
| NBroad, gram-positive and -negative | ||||||||
| Combination (glyco peptide/ β-lactams) | Vancomycin + Imipenem | 0·1 mg/ml per antibiotic in drinking water 14 days | Inhibits proper cross-linking in cell wall/ inhibition of cell wall synthesis by binding to PBP | 4–5 mice | 47 selected individual bacteria were monitored, 16 s rDNA analysed by QuantiGene 2·0 Reagent System | Firmicutes (decreased >half of monitored species): ↓Clostridium celerecrescens, ↓C. clostridioforme, ↓C. fusiformis, ↓C. methylpentosum, ↓C. polysaccharolyticum, ↓C. scincdens, ↓C. sp ASF502, L7-2, ↓Lactobacillus murinus and ↓L. salivarius Bacteroidetes: ↑Bacteroides acidifaciens ↑B. distasonis, ↑B. forsythus, ↑B. sp. ASF19. Other: ↑Bacillus mycoides and ↑Klebsiella granulomatis | Not studied | 59 |
| Narrow, gram-positive/ Broad, gram-positive and -negative |
Gut microbiota dysbiosis
Dysbioses of the gut microbiota have been noted for a large spectrum of human diseases (Table 2). Evaluating the microbiota as a potential causal agent of disease is of great interest. One anticipated challenge in ‘diagnosing’ dysbioses is that signatures of dysbiosis are probably disease-specific and multiple. It will be important to be able to distinguish the disease-associated alterations in microbiota composition that may drive or contribute to disease onset from those that reflect altered selective pressures generated by the disease microenvironment, e.g. chronic or acute inflammation. Systematic characterization of microbial dysbioses that promote disease initiation and the specific host pathways impacted will be critical to improving our understanding of the mechanistic aspects of microbiota as a driver of human disease.
Table 2.
Examples of microbial dysbiosis and/or bacteria associated with disease
| Disease | Dysbiosis and bacterial species | Protective or pathogenic microorganisms, effect on host | Reference |
|---|---|---|---|
| Crohn's disease (CD) | •↑Microbial diversity, ↑mucosal adherent bacteria ↓Bifidobacteria ↑Enteropathogenic bacteria (incl. Eschericia coli), ↓Firmicutes (except colorectal CD ↑Firmicutes increased), ↑Enterobacteriaceae, ↓Lachnospiraceae, ↑Ruminococcus gnavus, ↓Faecalibacterium prausnizii, ↓Roseburia. CD patients have a fivefold increase in risk for developing CRC | F. prausnitzii shows anti-inflammatory effects in colitis mice models, reduction of F. p. is associated with recurrence in postoperative ileal CD. Epsilonproteobacteria and Helicobacteraceae are families that harbour known pathogens, e.g. Campylobacter and H. pylori | 75–79 |
| Ulcerative colitis (UC) | •↓Microbial diversity, ↑mucosal adherent bacteria ↑Enteropathogenic bacteria (incl. Escherichia coli), ↓Clostridium coccoides ↑Epsilon proteobacteria ↓Faecalibacterium prausnizii, ↑Helicobacteraceae, ↓Lachnospiraceae ↑Ruminococcus gnavus. UC patients have a fivefold increase in risk for developing CRC | ||
| Colorectal Cancer (CRC) | •Enterotoxin-producing Bacteroides fragilis (ETBF) increases tumorigenesis in ApcMin/+ mice, stimulates cell proliferation | Enterotoxin increases the ion permeability of epithelial cells | 80,81 |
| •Enterobacteria spp., ↑Citrobacter spp., ↑Shigella spp., ↑Salmonella spp., genera with pathogenic potential have been found at increased levels in normal tissue flanking CRC tumours | Several genera with pathogenic potential, e.g. cell invasive species (Salmonella) | 76 | |
| •Rats colonized with Enterococcus faecalis showed increased DNA damage compared with control rats | Produces reactive oxygen species, which is a possible cause of chromosomal instability (CIN) | 82 | |
| •↑Escherichia coli, pks+, more prevalent in CRC patients than in healthy individuals | Colibactin, causes double-stranded DNA breaks leading to genomic instability | 83–85 | |
| •Three separate studies reported associations of Streptococcus gallolyticus (formerly S. bovis) with CRC (carcinomas or adenomas) | Able to avoid detection by immune system, form biofilms on collagen-rich surfaces | 86,87 | |
| •↑Fusobacterium spp., consistently over-represented in tumour-adherent microbiota | Pathogen causing periodontal infections and Lemierre's syndrome | 88–90 | |
| Obesity | •Ratio of Firmicutes: Bacteroides increase in both obese human subjects and mice Weight loss by diet (1 year) increased ↑Bacteroides and reduced Firmicutes ↑Lactobacillus ↑Faecalibacterium prausnizii (obese children) | Shifts in microbiota composition alter aspects of energy harvest | 91–93 |
| •Numerous studies show that antibiotic treatment leads to weight gain in human subjects (including children) and animals | |||
| •Transplantation of microbiota of obese mice into lean mice led to increased body weight, establishing causality | |||
| Diabetes, type 1 | •Decrease in ratio of Firmicutes: Bacteroides, and ↓microbial diversity was found to correlate with the development of type 1 diabetes-associated autoimmunity (children). ↑Bacteroides ovatus, ↓Clostridiales | 94,95 | |
| •BB-diabetes prone rats (BBDP, diabetes type I model) with a decrease in ↓↓Bacteroides spp., did not develop diabetes. Difference in microbiota composition compared to rats that did develop clinical disease was detectable before clinical onset Treatment with antibiotics decreased incidence and onset | |||
| •BB-diabetes-resistant rats had higher levels of ↑Bifidobacterium and ↑Lactotobacillus compared to BBDP rats | |||
| Immunosenescence | •Decreased microbial diversity, increase in interpersonal variability, ↓Bifidobacterium, ↓Faecalibacterium prausnitzii, ↓Clostridium cluster XIVa, ↑Streptococcus spp., ↑↑Staphylococcus spp. ↓↓Enterococcus spp, ↑Enterobacteria spp. | Increase in genera that often contain pathogenic species | 18,19,22,23 |
The gut microbiota of elderly people
As humans age, a variety of T cells display age-dependent decline 96 that mirror the involution of the thymus which is essentially complete in human subjects >60 years of age 97. In older subjects, CD3+, helper CD4+ and suppressor/cytotoxic CD8+ cell numbers are reduced. These changes are accompanied by an increase in the production of type 1 cytokines: IL-2, IFN-γ, TNF-α and type 2 cytokines, IL-4, IL-6 and IL-10 98. These changes are associated with elevated inflammation in elderly people, a phenomenon known as inflammaging 99. The gut microbiota of elderly people displays decreased diversity and increased interpersonal variability 22,23. The abundance of anti-inflammatory taxa, Bifidobacterium spp., F. prausnitzii and members of Clostridium cluster XIVa are reduced in elderly populations 19. The level of Bifidobacterium is anti-correlated with serum levels of TNF-α and IL-1 19. Streptococcus spp., Staphylococcus spp., Enterococcus spp. and Enterobacteria spp., genera containing pathogenic and inflammatory species are increased. Unique stool microbiota profiles were evident between healthy ‘community-dwelling’ and ‘long-term residential care’ subjects 18. These differences may be due to disparate consumption of fruits and vegetables in the two groups. Alterations in the gut microbiota diversity and richness in elderly people may increase susceptibility to infectious agents by favouring the colonization of pathobionts. The elevated inflammatory status and concomitant reduced mucin production in the aged colon increases the potential for bacteria to adhere to the colonic mucosa. An important unanswered question is whether age-related gut microbiota changes account for the observed increased susceptibility to infection, immunosenescence and inflammaging in elderly people.
Host inflammation in inflammatory bowel disease (IBD)
Among the significant changes associated with IBD is the reduction in epithelial barrier function resulting from increased expression of claudin 2 and the down-regulation and spatial redistribution of claudins 5 and 8, proteins mediating tight junctions between epithelial cells. Pores and discontinuities in epithelial tight junctions increase the likelihood of bacterial breach 100,101. Additional hallmarks of IBD include defective mucin organization and expression, induction of endoplasmic reticulum (ER) stress, autophagy and inflammation 102,103. In human subjects with Crohn's disease (CD), dysregulation of α- and β-defensin is thought to account partially for observed alterations of the gut microbiota composition 104. A study using T-bet (transcription factor)-deficient mice demonstrated that they are colitogenic and fail to develop Th1 cells. These mice harbour an altered gut microbiota. Interestingly, the gut microbiota derived from T-bet-deficient mice conferred colitis to recipient wild-type mice, indicating that the dysbiotic microbiota is sufficient to cause disease 105. Human subjects with IBD display a reduction in community complexity coupled with an overall loss of several prominent commensal species. Among the anti-inflammatory species, F. prausnitzii displays decreased abundance in subjects with IBD 75,106,107. Pathobionts appear to exploit the void and may contribute further to host inflammation. Specific organisms and phyla with either increased mucolytic and/or adherence properties have been identified in subjects with IBD, including Ruminococcus gnavus and R. torques 75,108,109, as well as members of the γ-proteobacteria featuring Escherichia coli strains 106,110,111. Subjects with IBD displayed a significant increase in mucus penetrant bacteria compared to healthy subjects 111. Recent studies have implicated the increased abundance of adherent-invasive E. coli (AIEC) as a potentially significant causal agent in the initiation and/or potentiation of CD 112,113. Defects in autophagy genes NOD2, ATG16L1 and IRGM result in increased prevalence of AIEC 114,115. Another host factor that promotes AIEC colonization is the abnormal expression of CEACAM6, a receptor for AIEC in patients with ileal CD 116. Additional dysbioses have been noted, including the increased abundance of Listeria monocytogenes, Campylobacter spp., Salmonella spp., Yersinia enterolitica and Y. pseudotuberculosis in subjects with CD; however, these associations are not observed uniformly in other studies, suggesting that ‘inflammatory’ species may be considered interchangeable with respect to their ability to drive disease.
Human subjects with IBD are at elevated risk for developing colorectal cancer (CRC). The gut microbiota of colitis-susceptible Il10–/– mice display reduced richness and increased abundance of Verrucomicrobia, Bacteroidetes, Proteobacteria and ∼100-fold increase in E. coli 117. Gnotobiotic azoxymethane (AOM)-treated Il10–/– mice mono-associated with either polyketide synthase (pks+) E. coli (NC101) expressing the colibactin toxin or E. faecalis develop colitis. While both mono-associations induced inflammation, only the E. coli-associated mice develop invasive carcinomas. These results indicate that elevated inflammation was not sufficient for tumour formation and that genotoxic gut bacteria provide additional signals required for tumorigenesis 117. This study also showed that pks+ E. coli was present in ∼67% of subjects with CRC compared to ∼21% in non-IBD/CRC subjects. Mice deficient for components of the NOD-like receptor (NLR) family, pyrin domain containing 6 (NLRP6) inflammasome in a model of IBD-associated inflammation-induced CRC displayed increased inflammation, dysbiotic microbiota composition and increased tumour burden 118. Remarkably, the co-housing of the NLRP6-deficient mice with wild-type mice conferred the colon tumour phenotype to wild-type mice, indicating that dysbiotic microbiota represent a previously unrecognized trigger of CRC initiation and progression. Multiple inflammatory and stress-induced responses are expressed aberrantly in subjects with IBD. Each of these pathways has the potential to alter microbiota composition. Deciphering the role(s) of the microbiota in relation to inflammatory, ER stress and autophagy functions in IBD is likely to be complex and highly challenging.
Modulation of the gut microbiota
Diet as a modulator of the gut microbiota
One of the major advances in our understanding of the gut microbiota is the recognition that it is a metabolically adaptable ‘organ’. Among the numerous factors that modulate the microbial composition of the gut, diet is perhaps the most influential. It has been proposed that the gut microbiota can be grouped into three enterotypes, Bacteroides, Prevotella and Ruminococcus, based upon the relative abundance of the dominant phyla 119. Dietary profiles have been associated with specific enterotypes. Bacteroides is associated with a high-fat diet and Prevotella is associated with a high carbohydrate diet 120. A typical Western diet (high-fat, high-sugar), results in an overall reduction of Bacteroidetes and an increase in Firmicutes 12,121. The microbial communities present in faecal samples derived from vegetarian and vegan controls were distinct from omnivorous control subjects that displayed a reduced abundance of Bacteroides spp., Bifidobacterium spp., E. coli and Enterobactericieae 122,123. Recently, it has been shown that the gut microbiota undergoes rapid change as the result of dietary shifts from an animal-based to plant-based diet 124. These alterations were larger than the interpersonal differences distinguishing gut microbiota, indicating the strong modulating potential of diet. Animal-based diet increased the abundance of bile-tolerant genera including Alistipes, Bilophila and Bacteroides and a reduction of Firmicutes. The shift in microbiota resulted in shifts in the functional composition of the microbiome that featured either amino acid or polysaccharide metabolism. These observations emphasize the potential of dietary intervention to increase human health through modulation of the gut microbiota. The health benefits of fruits and vegetables and the anti-cancer properties of a number of plant-based nutrients are well documented 125,126. However, far less is known regarding how these metabolites are produced and their modes of action. Human intervention studies have been conducted on a number of dietary nutrients. These studies have highlighted something unexpected; that human subjects display variability in their ability to metabolize specific substrates or secondary metabolites, suggesting that the derived health benefits associated with these nutrients may be personalized.
Breast cancer is the most common malignancy and second leading cause of cancer-related death in women worldwide 127. Epidemiological studies have shown a significant disparity in breast cancer incidence when comparing US and Japanese populations 128,129. The elevated consumption of phytoestrogens found in soy-based foods was identified as a potential contributory factor 130. Phytoestrogens, including isoflavones, coumestans, lignans and steroidal phytosterols, possess oestrogenic effects in animals via oestrogen receptor signalling 131. Soy is rich in the compounds genistein and daidzein. While genistein is absorbed readily in the small intestine without further metabolism, daidzein is metabolized in the gut leading to the production of equol and O-desmethylangolensin (O-DMA); however, only 30 and 80% of subjects from the United States are able to convert daidzein to the bioactive compounds, equol and O-DMA, respectively 132–134. Conventional but not germ-free mice produce equol and O-DMA 135,136. Furthermore, incubation of daidzein with faecal microbiota from some individuals but not others results in the production of equol and O-DMA 137, demonstrating that members of the gut microbiota mediate equol and O-DMA production and represent a personal trait 137–140.
The widespread identification of bacterial species/strains capable of carrying out metabolic bioconversions of health-promoting compounds may allow an individual's metabolic phenotype to be defined. This, in turn, may dictate the composition of personalized probiotic formulations as a means of expanding the benefits of a healthy diet. Detailed knowledge of the genes encoding these metabolic enzymes would provide opportunities for the development of diagnostics, synthetic or genetically engineered microorganisms to specifically complement any individual's set of metabolic deficiencies.
Faecal microbiome transplantation
Faecal microbiome transplantation (FMT), generally conducted by colonic enema or by endoscopy, introduces distal gut flora from a healthy donor into an unhealthy recipient, often a family member. This seemingly radical procedure has been performed with remarkable success to treat subjects with recurrent, refractory C. difficile infections. The high level of success of this therapeutic option now reported on more than 200 human subjects with a ∼90% success rate 141 has led to intensified interest to examine whether this approach could be effective in reversing the effects of IBD, IBS, CRC, obesity and other diseases. Further studies will need to be performed to understand more clearly the dynamics of donor and recipient microbiota, following FMT. It will be of interest to determine whether certain bacterial clades or networks are displaced more easily than others. Is there significant variation in the efficacy of FMT across individuals? Are the post-FMT microbiota or therapeutic components stable over time? An analysis of gut communities before and after FMT indicates that successful clinical outcomes are associated with restoration of community diversity that has been reduced as the result of the infection and/or antibiotic treatment 142. The effectiveness of FMT in treating C. difficile infection is consistent with a function of the gut microbiota known as pathogen exclusion, wherein a healthy and diverse commensal flora efficiently colonizes the gut lumen and mucosa, preventing pathogenic organisms to compete or co-exist. The degraded state of the gut microbiota in cases of refractory C. difficile infection may be ideal for efficient ‘regime change’ afforded by FMT. The application to FMT to treat IBD represents an important avenue for future evaluation. It may be predicted that FMT will be an effective treatment option for those diseases involving dysbiotic microbiota.
Probiotics
The use of probiotics has increased sharply in recent years, representing a multi-billion-dollar industry annually. Safety and regulatory concerns have slowed progress in this area in the United States. As a result, there is a growing interest in the development of probiotics that seek to mimic the clinical outcomes observed for FMT. The strong potential of FMT to reverse and cure chronic disease highlights the potential therapeutic direction that seeks to mimic the therapeutic virtues of FMT in a defined probiotic formulation. Probiotics are live microorganisms ingested either through diet, e.g. yogurt or in the form of a probiotic supplement. It has been demonstrated that probiotics containing Bifidobacterium or treatment with inulin reduces the frequency of translocating Enterobacteriaceae in DSS-colitis induced rats and similarly probiotic-treated mice showed decreased mortality following infection with either L. monocytogenes and S. typhimurium 143,144. A recent study revealed the mechanism by which a probiotic strain, E. coli Nissle, confers an anti-infective effect 145. E. coli Nissle competes effectively with pathogenic S. typhimurium for binding of essential and limiting iron in the gut. It remains unclear whether this or another mechanism accounts for the reduced susceptibility to infection observed with other probiotics. An analysis of a human twin pair consuming a probiotic formulation consisting of five probiotic species indicated that the gut microbiota composition was not altered, but that changes in the community gene expression patterns were evident. Nearly 40 metabolites derived from the five probiotic species were noted 146. Reduction in a number of carbohydrates and increased pyrogallol and polyphenols were observed, mirroring the carbohydrate utilization and metabolic capacity of the probiotic species. The transcriptional and metabolic impact was transient and subsided upon cessation of probiotic intake. As researchers begin to characterize the properties of individual gut microbes with potent immunomodulatory potential, it is likely that second-generation probiotic formulations may feature a new species. Future probiotics may target specific human disease states, including effective prophylactics that reduce the incidence of infectious disease in at risk populations, susceptibility to weight gain and alleviation of inflammation and tissue damage associated with IBD. Probiotics may increasingly be prescribed to patients being treated with antibiotics and other chemotherapeutics.
Outlook and conclusions
The rapid progress in our understanding of the complexity of the human microbiome has been remarkable. It has become clear that a wide variety of human diseases and conditions are associated with dysbiosis of the gut microbiota. Investigations of the gut microbiota in the coming years will attempt to evaluate the impact of dysbiosis as a causal or contributing factor in these diseases. This possibility has fuelled heightened interest in identifying strategies to modulate the gut microbiota in order to correct dysbioses and restore immune homeostasis. It appears inevitable that our view of human health and disease will increasingly consider the microbiome as an important component. We are improving our knowledge of beneficial taxa such as Bifidobacterium spp. and Lactobacillus spp., but a complete definition of healthy microbiota is incomplete. Similarly, we need to improve our ability to recognize dysbiotic microbiota in human subject cohorts that are highly variable and understand how specific microbes of groups of microbes influence health and disease. The very large number of species and gene functions present in microbiomes will make this very challenging indeed. The rate that we can apply DNA sequence characterization of the human microbiome will continue to increase. However, new complementary, high-throughput technology platforms are needed to improve our ability to cultivate gut microbes and evaluate their interaction with the human host.
Disclosure
The authors have no competing interests to report.
References
- Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLOS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett. 2006;254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4554–4561. doi: 10.1073/pnas.1000087107. doi: 1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills ME. The CNS and collaborative practice. Clin Nurse Spec. 1990;4:194–195. doi: 10.1097/00002800-199000440-00008. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Toward R, Montandon S, Walton G, Gibson GR. Effect of prebiotics on the human gut microbiota of elderly persons. Gut Microbes. 2012;3:57–60. doi: 10.4161/gmic.19411. doi: 10.4161/gmic.19411. [DOI] [PubMed] [Google Scholar]
- Jirillo E, Jirillo F, Magrone T. Healthy effects exerted by prebiotics, probiotics, and symbiotics with special reference to their impact on the immune system. Int J Vitam Nutr Res. 2012;82:200–208. doi: 10.1024/0300-9831/a000112. doi: 10.1024/0300-9831/a000112. [DOI] [PubMed] [Google Scholar]
- Mueller S, Saunier K, Hanisch C, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinross J, Nicholson JK. Gut microbiota: dietary and social modulation of gut microbiota in the elderly. Nat Rev Gastroenterol Hepatol. 2012;9:563–564. doi: 10.1038/nrgastro.2012.169. doi: 10.1038/nrgastro.2012.169. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4586–4591. doi: 10.1073/pnas.1000097107. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MJ, Conlon B, Landeau M, et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208:226.e1–2267. doi: 10.1016/j.ajog.2013.01.018. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez E, Marin ML, Martin R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Zaura E, Nicu EA, Krom BP, Keijser BJ. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 2014;4:851–858. doi: 10.3389/fcimb.2014.00085. doi: 10.3389/fcimb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueimonde M, Sakata S, Kalliomaki M, Isolauri E, Benno Y, Salminen S. Effect of maternal consumption of lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J Pediatr Gastroenterol Nutr. 2006;42:166–170. doi: 10.1097/01.mpg.0000189346.25172.fd. doi: 10.1097/01.mpg.0000189346.25172.fd. [DOI] [PubMed] [Google Scholar]
- Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelun Barfod M, Magnusson K, Lexner MO, Blomqvist S, Dahlen G, Twetman S. Oral microflora in infants delivered vaginally and by caesarean section. Int J Paediatr Dent. 2011;21:401–406. doi: 10.1111/j.1365-263X.2011.01136.x. doi: 10.1111/j.1365-263X.2011.01136.x. [DOI] [PubMed] [Google Scholar]
- Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl. 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Lif Holgerson P, Harnevik L, Hernell O, Tanner AC, Johansson I. Mode of birth delivery affects oral microbiota in infants. J Dent Res. 2011;90:1183–1188. doi: 10.1177/0022034511418973. doi: 10.1177/0022034511418973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLOS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz MJ, Denef VJ, Costello EK, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4578–4585. doi: 10.1073/pnas.1000081107. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLOS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. The human microbiome: ecosystem resilience and health. Nutr Rev. 2012;70(Suppl. 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Sanderson IR, Walker WA. TLRs in the Gut I. The role of TLRs/NODs in intestinal development and homeostasis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G6–10. doi: 10.1152/ajpgi.00275.2006. doi: 10.1152/ajpgi.00275.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138:416–420. doi: 10.1016/j.cell.2009.07.024. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Izcue A, Coombes JL, Powrie F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol Rev. 2006;212:256–271. doi: 10.1111/j.0105-2896.2006.00423.x. doi: 10.1111/j.0105-2896.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier-Hyams LS, Sloane V, Batten BC, Neish AS. Cutting edge: bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J Immunol. 2005;175:4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Tien MT, Girardin SE, Regnault B, et al. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277:138–145. doi: 10.1016/j.taap.2014.03.009. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLOS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis CM, Theriot CM, Young VB. Alteration of the murine gastrointestinal microbiota by tigecycline leads to increased susceptibility to Clostridium difficile infection. Antimicrob Agents Chemother. 2014;58:2767–2774. doi: 10.1128/AAC.02262-13. doi: 10.1128/AAC.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- Lofmark S, Edlund C, Nord CE. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis. 2010;50(Suppl. 1):S16–23. doi: 10.1086/647939. doi: 10.1086/647939. [DOI] [PubMed] [Google Scholar]
- Marzano IM, Franco MS, Silva PP, et al. Crystal structure, antibacterial and cytotoxic activities of a new complex of bismuth(III) with sulfapyridine. Molecules. 2013;18:1464–1476. doi: 10.3390/molecules18021464. doi: 10.3390/molecules18021464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donskey CJ, Hujer AM, Das SM, Pultz NJ, Bonomo RA, Rice LB. Use of denaturing gradient gel electrophoresis for analysis of the stool microbiota of hospitalized patients. J Microbiol Methods. 2003;54:249–256. doi: 10.1016/s0167-7012(03)00059-9. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U, Hornef MW, Henriques-Normark B, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, et al. Vancomycin-resistant enterococci exploit antibiotic- induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessein R, Gironella M, Vignal C, et al. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58:771–776. doi: 10.1136/gut.2008.168443. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- Dufour V, Millon L, Faucher JF, et al. Effects of a short-course of amoxicillin/clavulanic acid on systemic and mucosal immunity in healthy adult humans. Int Immunopharmacol. 2005;5:917–928. doi: 10.1016/j.intimp.2005.01.007. doi: 10.1016/j.intimp.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Schumann A, Nutten S, Donnicola D, et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17- producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54.e1. doi: 10.1053/j.gastro.2010.08.049. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Phil Soc. 2012;87:701–730. doi: 10.1111/j.1469-185X.2012.00218.x. doi: 10.1111/j.1469-185X.2012.00218.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, Saunders B, Wilkinson K, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126:451–459. doi: 10.1053/j.gastro.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Reinisch W. Intestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases? Best Pract Res Clin Gastroenterol. 2013;27:47–58. doi: 10.1016/j.bpg.2013.03.005. doi: 10.1016/j.bpg.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- Nougayrede JP, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Wang TC. Cancer. Bacteria deliver a genotoxic hit. Science. 2012;338:52–53. doi: 10.1126/science.1229905. doi: 10.1126/science.1229905. [DOI] [PubMed] [Google Scholar]
- Boleij A, Muytjens CM, Bukhari SI, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp. gallolyticus with colorectal cancer. J Infect Dis. 2011;203:1101–1109. doi: 10.1093/infdis/jiq169. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- Boleij A, Schaeps RM, Tjalsma H. Association between Streptococcus bovis and colon cancer. J Clin Microbiol. 2009;47:516. doi: 10.1128/JCM.01755-08. doi: 10.1128/JCM.01755-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- Zheng L, Giri B. Gastrointestinal variant of Lemierre syndrome: Fusobacterium nucleatum bacteremia-associated hepatic vein thrombosis: a case report and literature review. Am J Ther. 2014 doi: 10.1097/MJT.0000000000000084. ; doi: 10.1097/MJT.0000000000000084. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49:2105–2108. doi: 10.1007/s00125-006-0334-0. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S. Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine. 2000;18:1717–1720. doi: 10.1016/s0264-410x(99)00513-7. [DOI] [PubMed] [Google Scholar]
- Sansoni P, Cossarizza A, Brianti V, et al. Lymphocyte subsets and natural killer cell activity in healthy old people and centenarians. Blood. 1993;82:2767–2773. [PubMed] [Google Scholar]
- Zanni F, Vescovini R, Biasini C, et al. Marked increase with age of type 1 cytokines within memory and effector/cytotoxic CD8+ T cells in humans: a contribution to understand the relationship between inflammation and immunosenescence. Exp Gerontol. 2003;38:981–987. doi: 10.1016/s0531-5565(03)00160-8. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann NY Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Gassler N, Rohr C, Schneider A, et al. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am J Physiol Gastrointest Liver Physiol. 2001;281:G216–228. doi: 10.1152/ajpgi.2001.281.1.G216. [DOI] [PubMed] [Google Scholar]
- Zeissig S, Burgel N, Gunzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLOS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Kang S, Furet JP, et al. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis. 2011;17:185–192. doi: 10.1002/ibd.21436. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Andoh A, Sakata S, Koizumi Y, Mitsuyama K, Fujiyama Y, Benno Y. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–962. doi: 10.1002/ibd.20151. doi: 10.1002/ibd.20151. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Theissig F, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56:343–350. doi: 10.1136/gut.2006.098160. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci USA. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–24.e1–2. doi: 10.1053/j.gastro.2009.08.042. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012;66:53–60. doi: 10.1038/ejcn.2011.141. doi: 10.1038/ejcn.2011.141. [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J, Devi RS, Mary RR, Ramakrishna BS. Faecal microbiota composition in vegetarians: comparison with omnivores in a cohort of young women in southern India. Br J Nutr. 2012;108:953–957. doi: 10.1017/S0007114511006362. doi: 10.1017/S0007114511006362. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;23:559–563. doi: 10.1038/nature12820. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manach C, Mazur A, Scalbert A. Polyphenols and prevention of cardiovascular diseases. Curr Opin Lipidol. 2005;16:77–84. doi: 10.1097/00041433-200502000-00013. [DOI] [PubMed] [Google Scholar]
- Arts IC, Hollman PC. Polyphenols and disease risk in epidemiologic studies. Am J Clin Nutr. 2005;81(1 Suppl):317S–325. doi: 10.1093/ajcn/81.1.317S. [DOI] [PubMed] [Google Scholar]
- Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- Dunn JE. Cancer epidemiology in populations of the United States – with emphasis on Hawaii and California – and Japan. Cancer Res. 1975;35:3240–3245. [PubMed] [Google Scholar]
- Gray GE, Pike MC, Henderson BE. Breast-cancer incidence and mortality rates in different countries in relation to known risk factors and dietary practices. Br J Cancer. 1979;39:1–7. doi: 10.1038/bjc.1979.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina MJ, Persky V, Setchell KD, Barnes S. Soy intake and cancer risk: a review of the in vitro and in vivo data. Nutr Cancer. 1994;21:113–131. doi: 10.1080/01635589409514310. doi: 10.1080/01635589409514310. [DOI] [PubMed] [Google Scholar]
- Price KR, Fenwick GR. Naturally occurring oestrogens in foods – a review. Food Addit Contam. 1985;2:73–106. doi: 10.1080/02652038509373531. doi: 10.1080/02652038509373531. [DOI] [PubMed] [Google Scholar]
- Bolca S, Possemiers S, Herregat A, et al. Microbial and dietary factors are associated with the equol producer phenotype in healthy postmenopausal women. J Nutr. 2007;137:2242–2246. doi: 10.1093/jn/137.10.2242. [DOI] [PubMed] [Google Scholar]
- Markiewicz L, Garey J, Adlercreutz H, Gurpide E. In vitro bioassays of non-steroidal phytoestrogens. J Steroid Biochem Mol Biol. 1993;45:399–405. doi: 10.1016/0960-0760(93)90009-l. [DOI] [PubMed] [Google Scholar]
- Muthyala RS, Ju YH, Sheng S, et al. Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors alpha and beta. Bioorg Med Chem. 2004;12:1559–1567. doi: 10.1016/j.bmc.2003.11.035. doi: 10.1016/j.bmc.2003.11.035. [DOI] [PubMed] [Google Scholar]
- Cruz ML, Wong WW, Mimouni F, et al. Effects of infant nutrition on cholesterol synthesis rates. Pediatr Res. 1994;35:135–140. doi: 10.1203/00006450-199402000-00001. doi: 10.1203/00006450-199402000-00001. [DOI] [PubMed] [Google Scholar]
- Rowland I, Wiseman H, Sanders T, Adlercreutz H, Bowey E. Metabolism of oestrogens and phytoestrogens: role of the gut microflora. Biochem Soc Trans. 1999;27:304–308. doi: 10.1042/bst0270304. [DOI] [PubMed] [Google Scholar]
- Atkinson C, Berman S, Humbert O, Lampe JW. In vitro incubation of human feces with daidzein and antibiotics suggests interindividual differences in the bacteria responsible for equol production. J Nutr. 2004;134:596–599. doi: 10.1093/jn/134.3.596. [DOI] [PubMed] [Google Scholar]
- Frankenfeld CL, McTiernan A, Tworoger SS, et al. Serum steroid hormones, sex hormone-binding globulin concentrations, and urinary hydroxylated estrogen metabolites in post- menopausal women in relation to daidzein-metabolizing phenotypes. J Steroid Biochem Mol Biol. 2004;88:399–408. doi: 10.1016/j.jsbmb.2004.01.006. doi: 10.1016/j.jsbmb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Rowland IR, Wiseman H, Sanders TA, Adlercreutz H, Bowey EA. Interindividual variation in metabolism of soy isoflavones and lignans: influence of habitual diet on equol production by the gut microflora. Nutr Cancer. 2000;36:27–32. doi: 10.1207/S15327914NC3601_5. doi: 10.1207/S15327914NC3601_5. [DOI] [PubMed] [Google Scholar]
- Frankenfeld CL, Atkinson C, Thomas WK, et al. Familial correlations, segregation analysis, and nongenetic correlates of soy isoflavone-metabolizing phenotypes. Exp Biol Med. 2004;229:902–913. doi: 10.1177/153537020422900906. [DOI] [PubMed] [Google Scholar]
- Bakken JS. Fecal bacteriotherapy for recurrent Clostridium difficile infection. Anaerobe. 2009;15:285–289. doi: 10.1016/j.anaerobe.2009.09.007. doi: 10.1016/j.anaerobe.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Shahinas D, Silverman M, Sittler T, et al. Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio. 2012;23:e00338–12. doi: 10.1128/mBio.00338-12. : pii:. doi: 10.1128/mBio.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman N, Adawi D, Molin G, Ahrne S, Berggren A, Jeppsson B. Bifidobacterium infantis strains with and without a combination of oligofructose and inulin (OFI) attenuate inflammation in DSS-induced colitis in rats. BMC Gastroenterol. 2006;6:31. doi: 10.1186/1471-230X-6-31. doi: 10.1186/1471-230X-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddington KK, Donahoo JB, Buddington RK. Dietary oligofructose and inulin protect mice from enteric and systemic pathogens and tumor inducers. J Nutr. 2002;132:472–477. doi: 10.1093/jn/132.3.472. [DOI] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra. doi: 10.1126/scitranslmed.3002701. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]


