Abstract
Small, genomically-encoded microRNAs are important factors in the regulation of mRNA translation. Although their biogenesis is relatively well-defined, it is still unclear how they are recruited to their mRNA targets. The fragile X mental retardation protein family members, FMRP, FXR1P and FXR2P are RNA binding proteins that regulate translation of their cargo mRNAs. All three proteins, in addition to the single Drosophila ortholog, dFmrp, associate physically and functionally with the microRNA pathway. In this review, we summarize what is known about the role of the fragile X family members in translation regulation and highlight evidence for their association with the microRNA pathway. In addition, we present a new model for the role of phosphorylation on FMRP function, where phosphorylation of FMRP inhibits Dicer binding, leading to the accumulation of precursor microRNAs and possibly a paucity of activating microRNAs.
Keywords: FMRP, FXR1P, miRNA, translation regulation, phosphorylation, mRNA
Introduction
The Fragile X Mental Retardation Protein (FMRP) is an RNA binding protein that regulates translation of its bound mRNAs (in addition to references 1 and 2, let’s put that new review here on translation regulation that Tory found Costa-Mattioli et al 2009—it reviews quite a lot). FMRP associates with approximately 400 mRNAs in the brain3, 4 and is found nearly ubiquitously throughout the body.5 High expression levels of FMRP in brain6 and its role as a translational regulator1, 2(add Costa-Mattioli et al): suggest an important role in memory, learning, and normal cognition. In fact, loss of FMR1 gene expression causes Fragile X Syndrome, which is characterized by impaired cognitive function and other symptoms.5 Although FMRP is associated with ribosomes7, how FMRP regulates translation of its bound mRNAs at the molecular level is still unclear. The Drosophila ortholog of FMRP, dFmrp, associates with Dicer, Ago1, Ago2, microRNAs (miRNAs) and other components of the miRNA pathway.8–11 Further studies subsequently showed that mammalian FMRP associates with Dicer, miRNAs, Ago2 and miRNA pathway components10, 12, suggesting that FMRP utilizes the miRNA pathway to regulate its target mRNAs. The aim of this review is to highlight recent discoveries implicating an association of the fragile X family of proteins with the miRNA pathway and to examine how phosphorylation of FMRP may influence this association.
Processing of miRNAs
MiRNAs are a class of small RNAs that are 19–25 nucleotides in size and are genomically encoded.13, 14 MiRNAs function by base pairing with sequences in the 3′UTR of their target mRNA sequences. If base-pairing is perfect along their ~22 nucleotide length, the result is mRNA target degradation. In contrast, if base-pairing is imperfect with a bulge in the duplex, the result is translational silencing.15 The most recent Sanger Release (version 12.0 Sept. 2008, http://microrna.sanger.ac.uk/sequences/), lists 948 human microRNAs. Since each miRNA could potentially interact with multiple mRNAs due to incomplete base pairing to each target, it has been bioinformatically predicted that more than 30% of human genes could be regulated by miRNAs.16
In mammals, primary miRNAs are transcribed in the nucleus and processed into 70–80 nt precursor miRNAs by Drosha and DGCR8.17 Precursor miRNAs (pre-miRNAs) are exported to the cytoplasm by exportin 518, where they are processed into short, double stranded duplexes by the Dicer pre-miRNA processing complex.13, 16, 17 The RNA induced silencing complex, RISC, then separates the duplexed strands into a mature miRNA.13 This guide miRNA associates with Argonaute 2 (Ago2), which is the only human Argonaute family protein with endonuclease activity.19, 20 Incomplete complementarity of the miRNA to its target mRNA leads to translational repression (or in some cases activation, as discussed later) while complete complementarity triggers Ago2’s endonuclease activity leading to cleavage of the target mRNA, as observed during RNA silencing. Conversely, in Drosophila, both Ago1 and Ago2 have cleavage activity, with Ago1 mediating miRNA guided cleavage of RNA and Ago2 mediating siRNA cleavage during RNA silencing.19
Although it is unknown exactly how the mature miRNA finds its target mRNA, miRNAs function as part of larger complexes such as RISC (RNA induced silencing complex) and miRNP (miRNA ribonucleoprotein) complexes, suggesting that miRNAs guide associated proteins to target mRNAs to effect degradation, repression, or in some cases, translation activation.20–22 Although both structure and homology studies of Argonaute explain its “slicing role”, there is no similar evidence explaining its role in translation repression.20 Thus, other associated proteins within the miRNP are likely to mediate translational repression or activation. Since FMRP has been found associated with miRNAs and Ago210, 12 and is a known translational regulator which can both activate3, 23 and suppress translation1, 2((add Costa-Mattiola et al):, it likely plays an important role in miRNA mediated translation regulation.
Processing bodies and FMRP
Once mRNAs have been targeted by miRNAs for degradation (because of complete complementarity) or suppression (incomplete complementarity), they move into cytoplasmic foci. These foci were first observed in yeast and subsequently named processing bodies (PBs)24, where mRNPs colocalize with machinery for translation repression and mRNA decay.25 Although the environment of PBs is highly dynamic, decapping machinery including Dcp1p/Dcp2p and activators of Dcp proteins have been found there along with, under certain stress conditions, nonsense-meditated decay pathway proteins.25 PBs require mRNA to form, as shown by their disruption upon RNase treatment and the observation that overexpression of a nontranslating mRNA increases their size in yeast.26
PBs are also called GW bodies because they contain the protein GW182, an RNA binding protein first observed in discrete foci in HeLa cells.27 Knockdown of GW182 with siRNAs (silencing RNAs) results in a loss of GW bodies/PBs, leading to the conclusion that GW182 is a necessary component of PBs, as well as a useful marker for immunofluorescence.28 Whether FMRP localizes to PBs is controversial. One study reported that FMRP localizes to approximately 50% of granules containing the GW182 marker as shown by immunofluoresence in astrocytoma cells, suggesting that it is present in PBs.29 Further, dFmrp also colocalizes with Dcp1p and other known PB proteins.30 However, in a study using HeLa cells, the majority of FMRP was found not to colocalize with Dcp1p, but was instead found in stress granules.31 Thus, whether FMRP is localized to PB may depend on the cell system under study.
Role of FMRP in the microRNA pathway
Although FMRP associates with components of the miRNA pathway, it is also controversial whether it plays an important or essential role in the function of the miRNA pathway itself. As described above, Didiot and colleagues reported that FMRP localizes separately from RISC machinery components and PBs.31 Further, they showed that FMRP was not required for RISC function in transfection studies with reporter constructs bearing miRNA target sequences.31 In contrast, Plante and colleagues showed that FMRP was required for efficient RNA silencing using reporter constructs expressed in murine fibroblast cells.32 They also demonstrated that FMRP directly associates with miRNAs, possibly through the hnRNP K homology domains, to aid assembly of Dicer processed miRNAs onto target mRNAs in vitro.32 Taking this a step further, Xu and colleagues showed that dFmrp was required for assembly of the Dicer-Ago complex, and in the absence of dFmrp, there were fewer complexes, which may explain the reduced levels of the miRNA 122 (miR-122).8 They concluded that dFmrp was required for normal neuronal miRNA levels during development, likely because it was necessary for assembly of Dicer with Ago.8 Differing conclusions about whether FMRP is required for normal function of the miRNA pathway may be due to the use of different cell systems or the evaluation of different miRNAs. Regardless of FMRP’s role in RISC function, it is still possible that FMRP utilizes the miRNA pathway for translation regulation of its cargo mRNAs.
The fragile X family proteins and translation activation
FMRP is part of a family of proteins that includes two autosomal paralogs, Fragile X related protein 1 (FXR1P) and Fragile X related protein 2 (FXR2P).5 Co-expression of the FXR proteins at synapses in patients with Fragile X syndrome demonstrates that the FXRs cannot completely compensate for lack of FMRP33, 34, suggesting different functions for the paralogs. Additionally, although FXR1P can be found throughout the body, it is highly expressed in muscle and heart tissue where FMRP is mostly absent.35–37 Further, inactivation of FXR1P leads to impaired myogenesis in mouse38 and Xenopus (NEW REFERENCE Huot, et al. MBC 2005) and altered expression of muscle specific isoforms of FXR1p may contribute to facioscapulohumeral muscular dystrophy (FSHD).39 Collectively, these studies underscore the important role of FXR1p in normal muscle development.
At the molecular level, recent data show that miRNAs bound to the 3′ untranslated region (3′UTR) of the TNFα gene in quiescent cells recruit both AGO2 and FXR1 proteins, resulting in upregulation of translation.21, 22 The transition between translation upregulation and repression by the AGO2/FXR1 containing miRNPs occurs based on cell cycle.22 Translation activation by fragile X family member FMRP has also been suspected when a subset of mRNA cargoes were found decreased on polysomes in the absence of FMRP (add ref Brown, 2001), suggesting that FMRP was required for their translation. Further, a recent study found increased translation of known mRNA cargo Sod1 through binding by the C terminus of FMRP, which revealed the start codon, perhaps leading to translation initiation.23 Thus, fragile X family members FXR1p and FMRP have both been shown to activate translation of some mRNAs.
Phosphorylated FMRP and regulation of translation
FMRP is phosphorylated on three serines between its nuclear export sequence and the primary RNA binding domain, the RGG box40, in both brain and cell lines. To determine the role of phosphorylation on FMRP function, amino acid substitutions of the primary phosphorylation site were made, mimicking either constitutive phosphorylation or no phosphorylation.40 Constitutively phosphorylated FMRP (P-FMRP) was relatively resistant to ribosomal run-off, suggesting that it was associated with untranslating polyribosomes.40 In contrast, unphosphorylated FMRP was easily run-off, suggesting that it was associated with actively translating polyribosomes.40 Narayanan and colleagues identified a specific cargo mRNA, SAPAP3, whose translation was modulated by phosphorylation of FMRP.3, 41 As predicted by the model, inhibition of phosphatase 2A (PP2A) led to an increase in translation of SAPAP3.41 Consequently, it has been suggested that phosphorylation of FMRP functions as a key step in the regulation of bound target mRNAs through an unknown mechanism. Recent data points toward phosphorylation regulating association of FMRP with the miRNA pathway as a means to regulate translation of target mRNAs.42
The role of phosphorylated FMRP and the miRNA pathway
As described above, FMRP associates with many components of the miRNA pathway and localizes to PBs in some systems.8–11, 30 Morphological data show that loss of FMRP in mouse or Drosophila causes defects in spine and synapse formation.43 Taken together, a role for FMRP in neuronal development mediated through the miRNA pathway seems likely.
Phosphorylation of FMRP provides a rapid and reversible way to modulate FMRP’s association with the miRNA pathway. Since phosphorylation of FMRP and miRNAs are mechanisms for translation regulation of target mRNAs, we investigated whether phosphorylation regulates association of FMRP with the miRNA pathway. Using a phospho-specific antibody that is specific to the three phosphorylated serines (496, 499, and 503) of FMRP’s sole phosphorylation site40, we repeatedly found that P-FMRP associated with a large amount of an 80nt RNA species that appeared to be pre-miRNA.42 Since the Dicer complex is necessary for the processing of pre-miRNAs into mature miRNAs13, our hypothesis is that phosphorylation of FMRP precludes Dicer binding and leads to the abundance of pre-miRNAs observed with P-FMRP (Fig. 1 and 42). Indeed, immunoprecipitations of P-FMRP and total FMRP probed with a Dicer antibody showed that Dicer associates only with FMRP and not P-FMRP.42 RNase treatment had no effect on the FMRP-Dicer interaction, suggesting a protein interaction.42 To further confirm that phosphorylation prevents binding, beads linked to the phospho-peptide sequence of FMRP could not capture Dicer while the same unphosphorylated peptide sequence could capture Dicer.42
Figure 1. Potential regulatory role of P-FMRP in translation regulation.
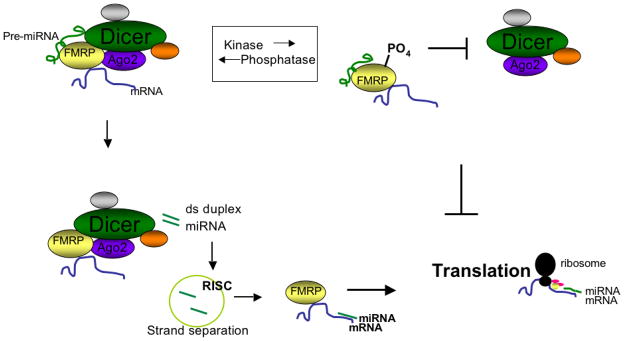
Unphosphorylated FMRP associates with Dicer (top left). The Dicer containing complex then processes pre-miRNAs into mature, double-stranded duplex miRNAs (bottom left). After RISC separates the duplex strands, the single-stranded mature miRNA binds FMRP’s target mRNA to induce translation. Conversely, when FMRP is phosphorylated, Dicer cannot bind and pre-miRNAs are not processed into mature miRNAs (top right). Without activating miRNAs, translation of the target mRNA cannot occur and the result is indirect suppression of translation by FMRP due to loss of Dicer binding.
We conclude that Dicer-FMRP association requires FMRP’s unphosphorylated region 496-503 and that phosphorylation of FMRP abolishes this interaction. This is a new role for phosphorylation in regulating FMRP function by modulating association with Dicer and the miRNA pathway. If miRNAs are required for translation activation21, 22 and FMRP is involved in translation activation23, phosphorylation of FMRP would indirectly suppress translation by decreasing miRNA production through loss of Dicer binding (Fig. 1). Future studies will determine whether there is translation activation of specific mRNAs by miRNAs associated with non-phosphorylated FMRP, similar to the reported translational activation function of FXR1.21,22 Ultimately, the multiple layers of regulation associated with FMRP and the miRNA pathway will need to be unraveled to fully comprehend the complexity of Fragile X syndrome and translation regulation via the miRNA pathway.
Acknowledgments
We would like to thank fellow Ceman lab members for critically reading this manuscript. This work was supported by University of Illinois startup funds and in part by Public Health Service grant HD41591 from the National Institute of Child Health and Human Development and by the Spastic Paralysis Research Foundation of the Illinois-Eastern Iowa District of Kiwanis International to S. Ceman.
References
- 1.Li Z, Zhang Y, Ku L, Wilkinson K, Warren S, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laggerbauer B, Ostareck D, Keidel E, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–38. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 3.Brown V, Jin P, Ceman S, Darnell J, O’Donnell W, Tenebaum S, et al. Microarray Identification of FMRP-Associated Brain mRNAs and Altered mRNA Translational Profiles in Fragile X Syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 4.Miyashiro K, Beckel-Mitchener A, Purk T, Becker K, Barret T, Liu L. RNA cargoes associating with FMRP reveal deficits in cellular functioning Fmr1 null mice. Neuron. 2003;37:417–31. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 5.Terracciano A, Chiurazzi P, Neri G. Fragile X Syndrome. Semin Med Genet. 2005;137C:32–7. doi: 10.1002/ajmg.c.30062. [DOI] [PubMed] [Google Scholar]
- 6.Weiler I, Irwin S, Klintsova A, Spencer C, Brazelton A, Miyashiro K, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. PNAS. 1997;94:5395–400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khandjian E, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–3. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Li Y, Wang F, Gao F. The Steady-State Level of the Nervous-System-Specific MicroRNA-124a Is Regulated by dFMR1 in Drosophila. J Neuro. 2008;28:11883–9. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishizuka A, Siomi M, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin P, Zarnescu D, Ceman S, Nakamoto M, Mowrey J, Jongens T, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neuro. 2004;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 11.Caudy A, Myers M, Hannon G, Hammond S. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–6. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P, Alisch R, Warren S. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–53. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D. MircroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Ross J, Carlson J, Brock G. miRNA: the new gene silencer. Am J Clin Pathol. 2007;128:830–6. doi: 10.1309/2JK279BU2G743MWJ. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Choi H, Kim J, Yim J, Lee J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–19. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Yi R, Qin Y, Macara I, Cullen B. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hock J, Meister G. The Argonaute protein family. Genome Biology. 2008;9:210.1–.8. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fillipowicz W, Jaskiewicz L, Kolb F, Pillai R. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struc Bio. 2005;15:331–41. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Vasudevan S, Steitz J. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–18. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasudevan S, Tong Y, Steitz J. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007 doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 23.Bechara E, Didiot M, Melko M, Davidovic L, Bensaid M, Martin P, et al. A Novel Function for Fragile X Mental Retardation Protein in Translational Activation. PLoS. 2009;7:0133–45. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kedersha N, Anderson P. Mammalian Stress Granules and Processing Bodies. Methods Enzym. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 25.Parker R, Sheth U. P Bodies and the Control of mRNA Translation and Degradation. Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing Bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eystathioy T, Chan E, Tenebaum S, Keene J, Griffith K, Fritzler M. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–51. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang A, Jakymiw A, Wood M, Eystathioy T, Rubin R, Fritzler M, et al. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–78. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 29.Moser J, Eystathioy T, Chan E, Fritzler M. Markers of mRNA stabilization and degradation, and RNAi within astrocytoma GW bodies. J Neuro Res. 2007;85:3619–31. doi: 10.1002/jnr.21439. [DOI] [PubMed] [Google Scholar]
- 30.Barbee S, Estes P, Cziko A, Hillebrand J, Luedeman R, Coller J, et al. Staufen- and FMRP-Containing Neuronal RNPs Are Structurally and Functionally Related to Somatic P Bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didiot M, Subramanian M, Flatter E, Mandel J, Moine H. Cells Lacking the Fragile X Mental Retardation Protein (FMRP) have Normal RISC Activity but Exhibit Altered Stress Granule Assembly. Mol Biol Cell. 2008;20:428–37. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plante I, Davidovic L, Ouellet D, Gobeil L, Tremblay S, Khandjian E, et al. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed and Biotech. 2006;2006:1–12. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiler I, Greenough W. Synaptic synthesis of the Fragile X protein: possible involvement in synapse maturation and elimination. Am J Med Genet. 1999;83:248–52. doi: 10.1002/(sici)1096-8628(19990402)83:4<248::aid-ajmg3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 34.Hoogeveen A, Willemsen R, Oostra B. Fragile X Syndrome, The Fragile X Related Proteins, and Animal Models. Micros Res Tech. 2002;57:148–55. doi: 10.1002/jemt.10064. [DOI] [PubMed] [Google Scholar]
- 35.Devys D, Lutz Y, Rouyer N, Bellocq J, Mandel J. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of fragile X premutation. Nat Genet. 1993;4:335–40. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 36.Coy J, Sedlacek Z, Bachner D, Hameister H, Joos S, Lichter P, et al. Highly conserved 3′UTR and expression pattern of FXR1 points to a divergent gene regulation of FXR1 and FMR1. Hum Mol Genet. 1995;4:2209–18. doi: 10.1093/hmg/4.12.2209. [DOI] [PubMed] [Google Scholar]
- 37.Khandjian E, Fortin A, Thibodeau A, Tremblay S, Cote F, Devys D, et al. A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum Mol Genet. 1995;4:783–9. doi: 10.1093/hmg/4.5.783. [DOI] [PubMed] [Google Scholar]
- 38.Mientjes E, Willemsen R, Kirkpatrick L, Nieuwenhuizen I, Hoogeveen-Westerveld M, Verweij M, et al. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum Mol Genet. 2004;13:1291–302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 39.Davidovic L, Sacconi S, Bechara E, Delplace S, Allegra M, Desnuelle C, et al. Alteration of expression of muscle specific isoforms of the fragile X related protein (FXR1P) in facioscapulohumeral muscular dystrophy patients. J Med Genet. 2008;45:679–85. doi: 10.1136/jmg.2008.060541. [DOI] [PubMed] [Google Scholar]
- 40.Ceman S, O’Donnell W, Reed M, Patton S, Pohl J, Warren S. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 41.Narayanan U, Nalavadi V, Nakamoto M, Pallas D, Ceman S, Bassell G, et al. FMRP Phosphorylation Reveals an Immediate-Early Signaling Pathway Triggered by Group I mGluR and Mediated by PP2A. J Neuro. 2007;27:14349–57. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA. 2009;15:1–5. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao F. Posttranscriptional control of neuronal development by microRNA networks. TRENDS Neuro. 2007;31:20–6. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


