Abstract
To study the evolutionary dynamics of regulatory DNA, we mapped >1.3 million DNase I hypersensitive sites (DHSs) in 45 mouse cell and tissue types, and systematically compared these with human DHS maps from orthologous compartments. The mouse and human genomes have undergone extensive cis-regulatory rewiring that combines branch-specific evolutionary innovation and loss with widespread repurposing of conserved DHSs to alternative cell fates mediated by turnover of transcription factor (TF) recognition elements. Despite pervasive evolutionary remodeling of the location and content of individual cis-regulatory regions, within orthologous mouse and human cell types the global fraction of regulatory DNA bases encoding recognition sites for each TF has been strictly conserved. Our findings provide new insights into the evolutionary forces shaping mammalian regulatory DNA landscapes.
The laboratory mouse Mus musculus is the major model organism for mammalian biology and has provided extensive insights into human developmental and disease processes (1), At 2.7 Gb, the mouse genome is of comparable size, structure, and sequence composition with the 3.3 Gb human genome (2, 3), and >80% of mouse genes have clear human orthologs (1, 4). Human-to-mouse transgenic experiments have collectively demonstrated that the mouse is capable of recapitulating salient features of human gene regulation, often with striking precision, even in the case of human genes that lack mouse orthologs (5). By contrast, comparative analyses of regulatory regions governing individual gene systems (6), as well as the occupancy patterns of several transcription factors (7), have highlighted the potential for cis-regulatory divergence. However, broader efforts to identify and quantify the major forces shaping the evolution of the mammalian cis-regulatory landscape have been hampered by the lack of expansive and highly detailed regulatory DNA maps from diverse cell fates that can be directly compared between mouse and human.
DNase I hypersensitive sites (DHSs) mark all major classes of cis-regulatory elements in their cognate cellular context, and systematic delineation of DHSs across many human cell types and states has provided fundamental insights into many aspects of genome control (8). In conjunction with the Mouse ENCODE Project (9), we undertook comprehensive mapping of DHSs in diverse mouse cell and tissue types and systematically compared the resulting maps to those from orthologous and non-orthologous human cells and tissues.
We mapped DHSs in 45 mouse cell and tissue types including adult primary tissues (n=19); purified adult and primitive primary cells (n=10); primary embryonic tissues (n=4); embryonic stem cell (ESC) lines (n=4); and model immortalized primary (n=3) and malignant cell lines (n=5) (Fig. 1A, fig. S1A and table S1). We identified between 74,386 and 218,597 DHSs per cell type at a false discovery rate (FDR) threshold of 1%, and collectively delineated 1,334,703 distinct ~150bp DHSs, each of which was detected in one or more mouse cell/tissue types. The genomic distribution of DHSs relative to annotated genes and transcripts was similar to that observed in human (8) (Fig. S1B). On average, 13.5% of DHSs marked promoters, with the remaining 86.5% distributed across the intronic and intergenic compartments in roughly equal proportions, and the vast majority located within 250 kb of the nearest annotated transcriptional start site (TSS) (Fig. S1C). However, average intergenic DHS-to-TSS distances in the mouse genome were markedly compressed (median 48.7 kb vs. 91.6 kb for human) relative to genome size (2.7 Gb vs. 3.3 Gb) indicating differential rates of genome remodeling within DHS-rich regions (Fig. S1D), with a pronounced difference in both size and density of distal elements (Fig. S2A,B).
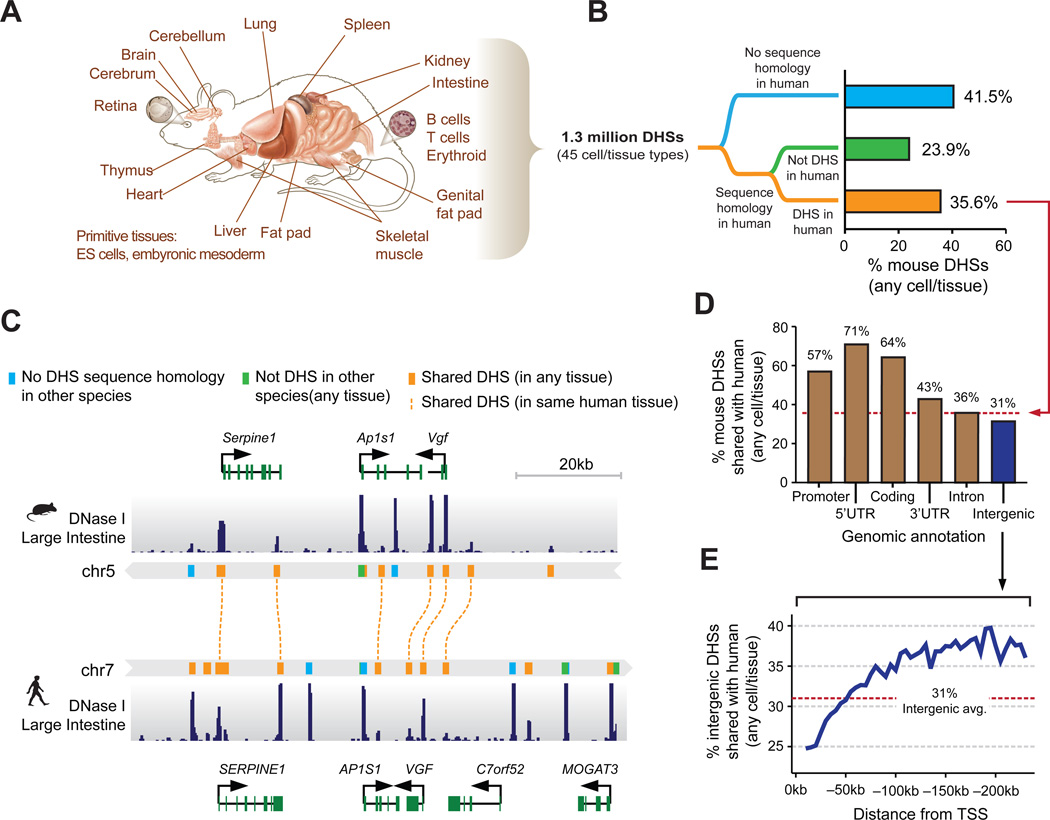
Figure 1. Conservation of mouse regulatory DNA in humans.
(A) The accessible landscape of the mouse was derived from 45 tissues and cell types (B) Proportions of the mouse regulatory DNA landscape with sequence homology and functional conservation with human. (C) Example of the conservation of the cis-regulatory elements surrounding within the Vgf/VGF locus in mouse and human intestine. (D) Gene proximal DHSs are more likely to be conserved than distal DHSs. Dashed red line indicates the average conservation of DHSs. (E) The rate of intergenic DHS conservation vs. distance to nearest TSS indicates a rapidly evolving cis-regulatory domain.
To gain insight into the evolution of mammalian regulatory DNA, we comprehensively integrated the mouse DHS maps with human maps generated using the same methods derived from 232 cell/tissues types from the ENCODE Project (n=103) (8) and the Roadmap Epigenomics Project (n=126) (10). These human maps collectively encompass ~3 million distinct DHSs from primary cells, adult and fetal tissues, immortalized and malignant lines, and ESCs (Table S2). We projected the genomic sequence underlying all mouse and human DHSs to the other species using high-quality pairwise alignments and a conservative reciprocal mapping and filtering strategy (Figs. 1B,C, fig. S3A). Collectively 59.5% of mouse DHSs (52.5–78.8% per cell type) could be aligned with high confidence to the human genome, of which 35.6% (38.6–60% per cell type) coincided with a human DHS (Fig. 1B and table S3). The remaining 23.9% (13–22.7% per cell type) may correspond either with yet-to-be defined human DHSs, or with human lineage-specific extinction of an ancestral element. In support of the latter, mouse DHSs aligning outside of human DHSs show excess sequence divergence evidenced by fewer alignable or identical nucleotides than mouse DHSs aligning to human DHSs (Fig. S3B,C). A lower proportion of human DHSs align with a mouse DHS (17.3%, fig. S3A and table S4); however, this is largely a reflection of the >2-fold greater number human DHSs. Given the breadth of mouse and human tissues analyzed, these values suggest upper and lower limits of regulatory DNA conservation between mouse and human.
To trace the evolutionary origins and dynamics of individual regulatory regions, we aligned all mouse and human DHS sequences to >30 vertebrate genomes spanning ~550 million years of evolutionary distance (Figs. S4A,B). Despite the deep sequence conservation of many DHSs, turnover of individual regulatory regions within different branches of the evolutionary tree appears frequently. Of the 80% of mouse DHS sequences that predate the divergence of humans from a common ancestor, only 58.5% are detectable in human, and comparison of mouse DHSs aligning to a human DHS or to a non-DHS region yields nearly identical evolutionary profiles (Fig. S4A,B). Overall, the proportion of DHSs that encompass evolutionarily conserved sequence elements increases with alignability and conservation of DNase I hypersensitivity (Fig. S4B). Unexpectedly, however, ~40% of mouse-human shared DHSs lack conserved elements.
The aforementioned trends are also reflected in patterns of human variation. Analysis of nucleotide diversity (π) within DHSs revealed graded constraint depending on the extent of sequence and DHS conservation (Fig. S5A). Notably, mean π within human-specific DHSs approximated that of four-fold synonymous sites within coding regions, compatible with relaxed (but not absent) nucleotide-level constraint. Despite decreased constraint (both evolutionary and recent), human-specific DHSs are significantly enriched (vs. all DHSs) in disease- and trait-associated variants identified by genome-wide association studies (Fig. S5B, Permutation test, Pnull < 0.005). The above results indicate that while mouse-human shared DHSs are collectively under selection over evolutionary timescales and within human populations, the sequence information with the cis-regulatory compartment is rapidly evolving in both mice and humans.
Whereas the overall density of mouse-human shared DHSs was higher in gene-proximal regions such as promoters, exons and UTRs (Fig. 1D), the relative proportion of shared DHSs (to all DHSs) increased markedly with distance from the TSS (Fig. 1E and Fig. S6). From 10kb to 50kb upstream of the TSS, the proportion of DHSs that are shared with human (avg. 27%) is lower than the average for intergenic regions (avg. 31%, fig. 1E), while in far distal regions this proportion increases substantially to a plateau of ~38%. These data suggest that regulatory elements functioning over long range (11) comprise a genomic compartment that may be functionally distinct from a more rapidly evolving gene-proximal region, and less buffered against evolutionary alteration.
Genesis of novel regulatory DNA sequences appears to have played a substantial role in shaping the DHS landscape in both mouse and human (Fig. 1B and fig. S2A). Over 50% of the mouse and human genomes comprise repetitive DNA (2, 3), which is proportionately reflected in their respective DHS compartments (Figs. S7A,B). Species-specific DHSs were enriched (relative to all DHSs) for nearly all classes of repetitive elements (Fig. S7C), and 5–10% of shared DHSs overlap ancient repeats that predate mouse/human divergence (Fig. S7D), compatible with an important role for transposons in the evolution of mammalian regulatory genomes.
Transposable elements have recently been implicated in the rapid expansion of a TF recognition elements (12, 13). To test the generality of this phenomenon, we estimated the total proportion of TF recognition sequences residing within species-specific DHSs that arose from transposon expansion during mouse and human evolution, which revealed substantial asymmetries (Fig. S8A–C). For example, the recognition motif for the pluripotency factor OCT4 (and other POU family TFs), has been greatly expanded in the murine lineage on a LTR/ERVL element (12), accounting for >25% of mouse-specific sites vs. <5% in humans with a similar class of retroelement (Fig. S8A). By contrast, expansions of CTCF (12) and retinoic acid receptor recognition elements (14) have been driven chiefly by SINE elements in both mouse and human (Figs. S8B,C). These results suggest that expansion of TF recognition sequences by repetitive elements is a general feature shaping mammalian cis-regulatory landscapes.
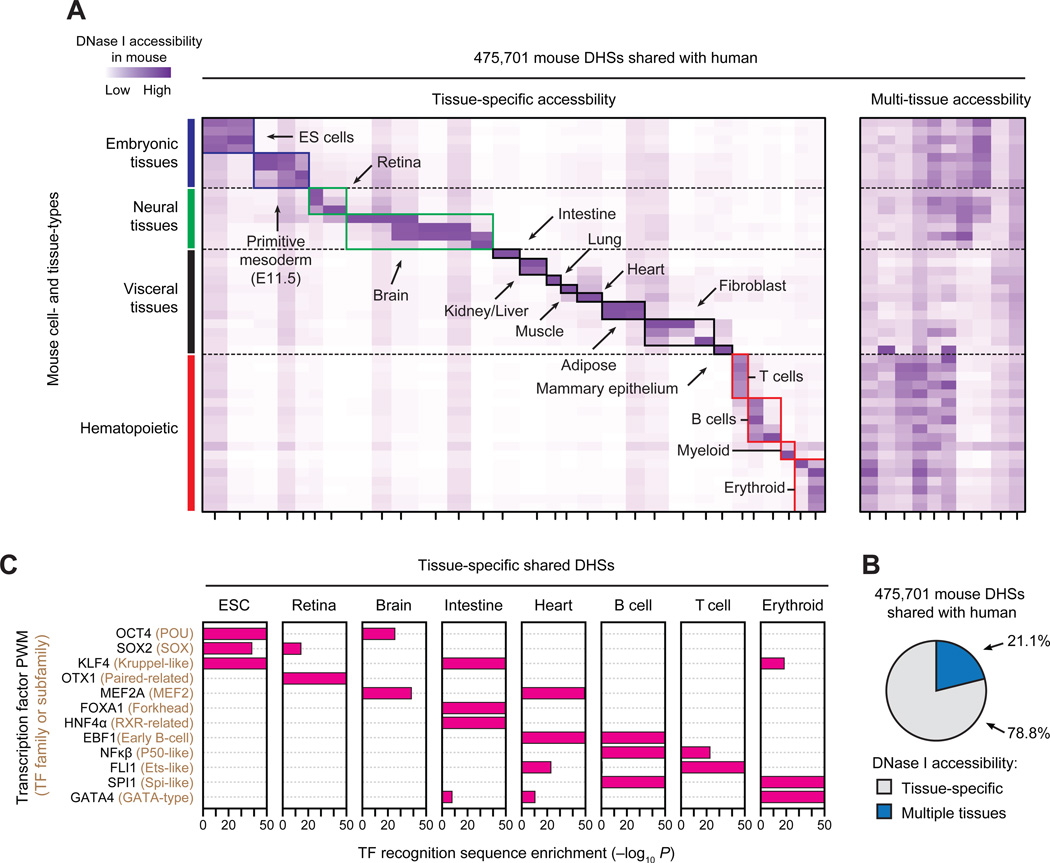
DHS patterns encode cellular fate and identity in a manner that reflects both current and future regulatory potential and informs developmental trajectory (15). To visualize cell- and tissue-selective activity patterns, we clustered shared DHS by normalized DNase I cleavage measured in each of the 45 mouse cell- and tissue-types (Fig. 2A). The vast majority of shared DHSs (78.8%) evinced tissue-selective accessibility, and were readily organized into distinct cohorts. A minority (21.2%) exhibited high accessibility across multiple tissue types, and <5% was constitutive (Fig. 2B). Tissue-selective shared DHSs were enriched in distal regions (Fig. S9) and reflected both tissue organization and anatomic or functional compartments within tissues. For example, the 91,951 shared brain-selective DHSs in turn comprised four sub-clusters corresponding to distinct anatomical and developmental partitions (Fig. 2A, green box). Similarly, shared blood-selective DHSs were sub-compartmentalized into major hematopoietic lineages, including T, B, myeloid, and erythroid cell cohorts (Fig. 2A, red boxes). Across all compartments, cell/tissue-selective shared DHSs were preferentially localized around genes critical for the development and maintenance of their respective cell or tissue type (Fig. S10).
Figure 2. Cell and tissue lineage encoding within shared regulatory elements.
(A) k-means clustering of DHSs by accessibility at each of the 475,701 mouse DHSs shared with human. Columns correspond clusters of mouse DHSs that are also accessible in human and rows correspond to the 45 mouse cell/tissue types. Colors (axes and boxes) distinguish tissue groupings. Left, tissue-selective clusters. Right, clusters containing DHSs active in multiple tissues. (B) Proportion of shared DHSs that are tissue-selective or active in multiple tissues. (C) Enrichment of TF recognition sequences within tissue-selective DHSs computed using the cumulative hypergeometric distribution.
We hypothesized that tissue-selective shared DHSs should encompass elements critical for basic mammalian regulatory processes such as development and differentiation, and that this would be reflected in their TF recognition sequence content. We thus computed, for each TF, the number of DHSs within each cluster that contained its recognition sequence, and compared this value to the overall distribution of recognition sequences within all shared DHSs. Tissue-selective DHSs showed pronounced enrichment for nearly all known lineage- or cell identity-specifying regulators, which were further organized combinatorially into their respective functional compartments (Fig. 2C and fig. S11). For example, OCT4, SOX2, and KLF4 recognition sites were collectively concentrated within ES-selective shared DHS landscapes, consistent with coordinated expression of their cognate factor in ES cells. KFL4 recognition sites were also enriched within intestine- and erythroid-specific DHSs, consistent with the known role of Krüppel-like TFs (many of which share the KLF4 recognition sequence) in intestinal epitheliogenesis (16) and in erythropoiesis (17). Analogously, sequence elements recognized by the cardiac regulators MEF2A, EBF1 FLI1 and GATA4 (18–20) are enriched within heart-selective shared DHSs, compatible with important functions for these TFs or their cognates in defining their respective cell fates (18, 21, 22). Notably, the tissue-selective enrichments we observed are nonetheless consistent with the known cell-selective activity of TFs even after recognition sequences are systematically grouped by similarity (Fig. S11). Together, the above results indicate that mouse-human shared DHSs densely encode regulatory information fundamental to diverse cell and tissue specification programs, and thus collectively define a core mammalian regulon.
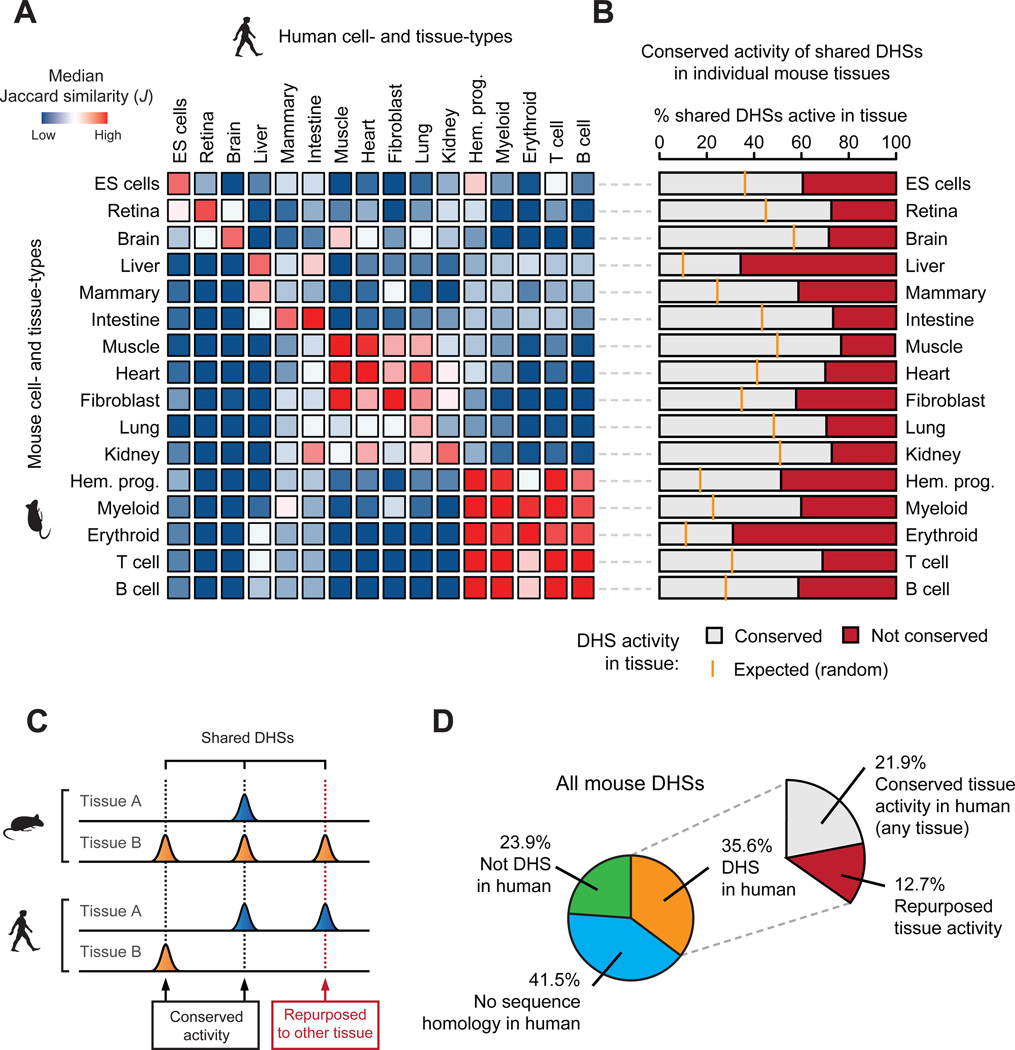
Since most shared DHSs showed strong cell/tissue-selectivity in mouse, we next asked to what degree these patterns were preserved in human. Computing the Jaccard similarity index over all possible combinations of mouse and human cell types revealed surprisingly limited similarity in the tissue-selective usage of shared DHSs (Figs. S12A–C), even when accounting for variability in DNase I cleavage density and peak identification parameters (Fig. S13). Unsupervised hierarchical clustering loosely grouped shared DHSs by cells or tissues derived from the same progenitor or developmental lineage (Fig. 3A).
Figure 3. Conservation and repurposing of regulatory DNA accessibility.
(A) Pairwise comparison (median Jaccard distance) of shared DHS landscape usage between all mouse (rows) and human (columns) tissues largely mirrors their conserved morphological and embryological origins. (B) The conservation of mouse cis-regulatory DNA accessibility in human for individual tissue types. Orange ticks indicate the expected overlap of randomly selected DHSs. (C) The activity patterns of individual shared DHSs during mouse and human evolution may have been conserved (activity in at least one similar tissue) or repurposed to another tissue. (D) Overall conservation of tissue-level accessibility patterns of mouse DHSs shared with human.
Weak correspondence between orthologous tissues suggested that a substantial fraction of shared DHSs had undergone functional ‘repurposing’ via alteration of tissue activity patterns from one cell/tissue type in mouse to a different one in the human (Figs. 3B,C). Indeed, analysis of well-matched mouse and human tissue pairs confirmed substantial repurposing ranging from 22.9–69% of shared DHSs, depending on the tissue (Fig. 3B). For example, of the 77,060 shared DHSs active in mouse muscle, 59,658 (77.4%) were also DHSs in human muscle, while the remaining 17,402 (22.6%) were DHSs in a different human tissue (Fig. 3B, 7th from top). Overall at least 35.7% of shared DHSs (12.7% of mouse DHSs overall) have undergone repurposing (Fig. 3D), chiefly affecting distal elements (Fig. S14). Facile repurposing of regulatory DNA from one tissue context to another thus emerges as an important evolutionary mechanism shaping the mammalian cis-regulatory landscape.
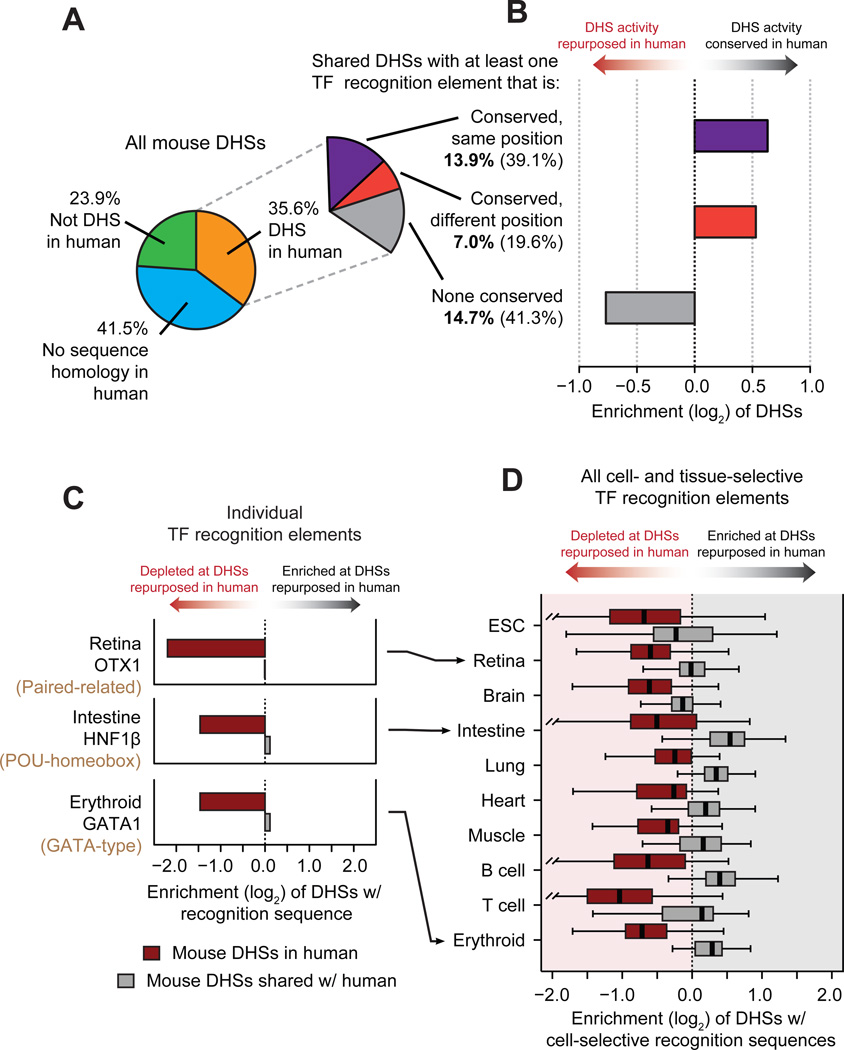
To examine the conservation of individual TF recognition elements within the shared DHS compartment, we distinguished between elements that were positionally conserved vs. those that were operationally conserved – i.e., present arose independently at a different position within the DHS (Fig. S15A). In shared DHSs, 39.1% of TF recognition sequences were positionally conserved, and 19.6% operationally conserved (Fig. 4A). Both positional and operational conservation were significantly concentrated (χ2 test, P < 10−15) within shared DHSs that maintained their tissue activity profile (Fig. 4B and fig. S15B). Surprisingly, 41.3% of shared DHSs (chiefly repurposed DHSs) lacked any positionally or operationally conserved TF recognition elements, (Figs. 4A,B and figs. S15C,D). Additionally, the overall density of TF recognition elements did not differ significantly between shared DHSs with positionally, operationally, or non-conserved TFs (Fig. S15E). This indicates that new regulatory features are continuously evolving within the same ancestral DNA segment.
Figure 4. Evolutionary dynamics of transcription factor recognition sequences.
(A) Conservation of TF recognition sequences within shared DHSs. (B) Positional and operational conservation of TF recognition sequences are enriched within DHSs that have conserved tissue activity patterns. (C) Recognition sequences for cell-selective transcription factors are preferentially lost at mouse DHSs that are repurposed in human, while maintained in or gained in human. Representative examples of individual TF regulators in retina, intestine and erythroid tissues. (D) Same as C for recognition sequences of all cell-selective TF regulators (identified in fig. 2C) within mouse DHSs repurposed in human.
We next elaborated the relationship between conservation of TF recognition sites and the maintenance of tissue accessibility patterns. Reasoning that known regulators of cell fate would play an outsized role in repurposing, we hypothesized that recognition sequences for such TFs would be preferentially maintained (or gained) in DHSs with conserved tissue activity spectra, but preferentially lost at repurposed DHSs (Fig. S16). We found this to be the case across a spectrum of lineage-regulating TFs. For example, recognition sites for the retinal master regulator OTX1 (and other paired-related homeodomain transcription factors), were >4-fold depleted within mouse retinal DHSs that had undergone repurposing in human compared with orthologous DHSs that had conserved retinal activity (Fig. 4C). Analogously, sequence elements recognized by the intestinal master regulator HNF1β (and other POU-homoebox transcription factors), were selectively depleted in repurposed intestinal DHSs, and those recognized by the major erythroid regulator GATA1 (and other GATA-type factors), were selectively depleted in repurposed erythroid DHSs (Fig. 4C). Overall recognition sites for cell fate-modifying TFs were consistently depleted within repurposed DHSs (Fig. 4D), linking the conservation and repurposing of DHSs to preservation vs. turnover of specific TF recognition sequences. These results also suggest an incremental process whereby the composition of TFs within a given DHS is remodeled over evolutionary time via sequential small mutations (23) that could ultimately affect function and phenotype (24). The presence of a substantial population of shared DHSs without conserved TF recognition sites yet preserved tissue-selectivity patterns highlights the plasticity of individual cis-regulatory templates, and indicates that the same higher-level regulatory outcome may be encoded by many different combinations of instructive TF recognition events.
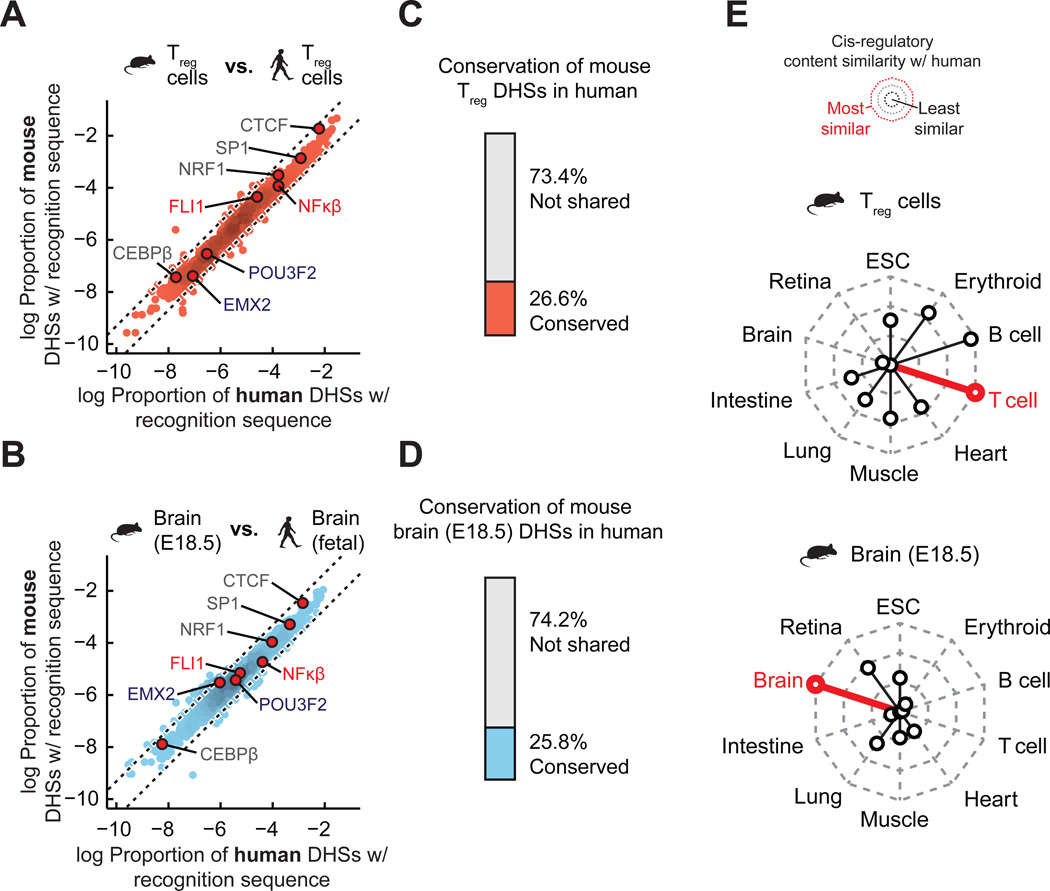
To investigate how the marked plasticity of TF recognition elements within the evolving cis-regulatory landscape was reflected in global patterns in the types and quantities of such elements, we computed the global density of recognition sequences for each of 744 TFs within all mouse and human DHSs (separately, and irrespective of conservation status) from each cell/tissue type. This analysis revealed striking conservation of the proportion of the regulatory DNA landscape of each cell type devoted to recognition sites of each TF. Figs. 5A,B show examples for mouse vs. human regulatory T cell DHSs, and mouse brain vs. human fetal brain; in each case, a linear relationship is observed indicating that the proportion of the DHS compartment devoted to recognition sequences of each of the 744 TFs has been strictly conserved (Fig. 5A). It is particularly notable that this finding obtains across a wide spectrum of TFs that encompasses diverse functional roles and biophysical mechanisms of DNA recognition. These findings markedly contrast with the weak conservation (~25%) of individual mouse regulatory T cell and brain DHSs (Figs. 5C,D). TF recognition sequence content varied between cell/tissue types, with effector TFs selectively enriched within their cognate cell type (Fig. S17), and TF recognition sequence density was consistently most similar between orthologous cell/tissue pairs vs. non-orthologous cells/tissues (Fig. 5E and fig. S18). It has been proposed that in large genomes such as mouse and human, maximization of the occupancy of any given TF demands an excess of its recognition sites in order to ensure high occupancy of sites with critical regulatory roles across a range of TF concentrations (25). Consistent with this model, the majority of DHSs in both the mouse and human genome show relaxed sequence constraint over evolutionary distances (Fig. S4C) and within human populations (Fig. S5A). This model also predicts that the cis-regulatory programs of TF genes themselves should be more highly conserved than other gene classes. Comparing DHSs within 50 kb of the TSSs of TF genes (n=911) relative to those of all orthologous genes (n=14,666 with at least 10 identified DHSs in mouse) revealed an overall increase in the conservation of TF-linked DHSs (Wilcoxon rank-sum test, P < 10−15) (Fig. S19), particularly for DHSs surrounding the TSSs of genes within canonical TF families, such Hox- and Sox-factors. As such, TFs are distinguished from other trans-acting regulators in that their activity appears to directly shape their cis-regulatory landscape.
Figure 5. Conservation of cis-regulatory content dominates over the conservation of individual regulatory elements.
(A) Density of individual TF recognition sequences in both human (x-axis) and mouse (y-axis) regulatory T cells. Dotted black lines demarcate a 2-fold difference in density between mouse and human. (B) Same as A for human and mouse brain. (C–D) Proportion of mouse DHSs that are conserved in a matched human tissue. Top, mouse regulatory T cells DHSs that are conserved in human regulatory T cells. Bottom, mouse embryonic brain DHSs that are conserved in human fetal brain. (E) Radar plots showing the median similarity (Euclidean distance between the distributions of TF recognition sequence densities) of the cis-regulatory content between mouse and human tissues.
Taken together, the results reported herein have important implications for understanding the major mechanisms and forces governing the evolution of mammalian regulatory DNA. Performing genomic footprinting on 25 of the cell and tissue samples analyzed herein reveals that the effective in vivo recognition repertoires of human and mouse TFs are nearly identical, and that the high turnover of individual TF occupancy sites within regulatory DNA is accompanied by striking evolutionary stability at the level of regulatory networks (26). As such, the combination of a highly conserved trans-regulatory environment with a large genome (under weakened selection) may function to potentiate both the de novo creation and the cis-migration of operational TF binding elements. We speculate that high cis-regulatory plasticity may be a key facilitator of mammalian evolution by increasing the potential for innovation of novel functions in the context of an evolutionarily inflexible trans-regulatory environment.
Supplementary Material
Acknowledgments
This work was supported by NIH grants U54HG007010 to J.A.S. and 1RC2HG005654 to J.A.S. and M.G. Additional support was provided by NIH grants R37DK44746 to M.G. and M.A.B., and 2R01HD04399709 to L.S. J.V. is supported by a National Science Foundation Graduate Research Fellowship under grant no. DGE-071824. E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., SynapDx Corp., and DNAnexus, Inc. J.V. and J.A.S. designed the experiments and analysis. E.R., R.S. and R.E.T. aided in data analysis and management. All other authors participated in data generation and sample collection. J.V. and J.A.S. wrote the manuscript with help from E.R. We would like to thank H. Wang and E.K. Salinas for help with figures. All sequence data generated in this study can be accessed with GEO accession numbers found within tables S1 and S2.
Footnotes
Supplementary Materials
Materials and Methods
Figures S1 to S19
Tables S1 to S4
References and Notes (27–52)
References
- 1.Hardouin SN, Nagy A. Mouse models for human disease. Clin. Genet. 2000;57:237–244. doi: 10.1034/j.1399-0004.2000.570401.x. [DOI] [PubMed] [Google Scholar]
- 2.Mouse Genome Sequencing Consortium et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Guénet JL. The mouse genome. Genom. Res. 2005;15:1729–1740. doi: 10.1101/gr.3728305. [DOI] [PubMed] [Google Scholar]
- 5.Peterson KR, Stamatoyannopoulos G. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol. Cell. Biol. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Cell. Biol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium MEP, et al. An Integrated and Comparative Encyclopedia of DNA Elements in the Mouse Genome. Nature. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 12.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genom. Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacques PÉ, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Biol. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laperriere D, Wang T-T, White JH, Mader S. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007;8:23. doi: 10.1186/1471-2164-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stergachis AB, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Q, et al. Comprehensive characterization of erythroid-specific enhancers in the genomic regions of human Kruppel-like factors. BMC Genomics. 2013;14:587. doi: 10.1186/1471-2164-14-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garel S, Marin F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- 19.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 20.Schachterle W, Rojas A, Xu S-M, Black BL. ETS-dependent regulation of a distal Gata4 cardiac enhancer. Dev. Biol. 2012;361:439–449. doi: 10.1016/j.ydbio.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 22.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne JL, Wagner A. The robustness and evolvability of transcription factor binding sites. Science. 2014;343:875–877. doi: 10.1126/science.1249046. [DOI] [PubMed] [Google Scholar]
- 24.Prud'homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, Riggs AD. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975;4:107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- 26.Stergachis AB, Neph S, Sandstrom R, Haugen E. Conservation of trans-acting networks during mammalian regulatory evolution. Nature. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John S, et al. Genome-scale mapping of DNase I hypersensitivity. Curr Protoc Mol Biol. 2013 doi: 10.1002/0471142727.mb2127s103. Chapter 27, Unit 21.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genom. Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genom. Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedges SB. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 36.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genom. Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 38.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genom. Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 40.Vernot B, et al. Personal and population genomics of human regulatory variation. Genom. Res. 2012;22:1689–1697. doi: 10.1101/gr.134890.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit A, Hubley R, Green P. RepeatMasker OpenB3.0 Software Package. (available at http://www.repeatmasker.org).
- 42.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 43.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arthur D, Vassilvitskii S. k-means++: the advantages of careful seeding. Society for Industrial and Applied Mathematics; 2007. pp. 1027–1035. [Google Scholar]
- 46.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genom. Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.