Abstract
Exposure of rat cortical neurons to combined oxygen and glucose deprivation results in loss of NAD(P)H autofluorescence that is only partially reversible following restoration of oxygen and glucose, suggesting catabolism of pyridine nucleotides. This study tested the hypothesis that metabolic inhibition caused by cyanide-induced chemical anoxia plus glucose deprivation promotes both release of mitochondrial NAD(H) in response to opening of the permeability transition pore (PTP) and NAD(P)(H) degradation through activation of poly(ADP-ribose) polymerase (PARP). The NAD(P)H autofluorescence of rat neonatal cortical neurons was monitored during and following acute (10 – 30 min) exposure to the respiratory inhibitor, cyanide, in the absence and presence of glucose. Because nitric oxide-derived peroxynitrite is a known activator of PARP, we additionally assessed the effect of a nitric oxide generating agent on the NAD(P)H autofluorescence response to chemical anoxia plus glucose deprivation. Cyanide induced a rapid increase in autofluorescence, followed by a steady decline promoted by the presence of nitric oxide. This decline was primarily due to NAD(H) catabolism, as verified by measurements of total NAD(H) present in cellular extracts. Catabolism was partially blocked by an inhibitor of PARP, by a PTP inhibitor, and by either glucose or pyruvate as a source of reducing power. Overall, data suggest that metabolic, oxidative, and nitrosative stress during in vitro neuronal anoxia and glucose deprivation result in release of mitochondrial pyridine nucleotides in response to PTP opening and rapid, extensive NAD(H) degradation mediated by PARP activation. These events may contribute to the metabolic dysfunction that occurs in vivo during cerebral ischemia and reperfusion and therefore represent prime targets for neuroprotection.
Keywords: Mitochondria, metabolism, oxidative stress, nitric oxide
BACKGROUND
Pyridine nucleotides and their metabolites serve important intracellular energy and signal transducing functions (Pollak et al. 2007; Berger et al. 2004; Ying 2006). For example, a continuous supply of NAD+ is necessary for the glyceraldehyde-3-phosphate dehydrogenase enzyme reaction, necessary for sustaining glycolysis, and for several mitochondrial tricarboxylic acid cycle reactions. Moreover, NADPH is the primary source of reducing power for antioxidant enzyme reactions, e.g., glutathione and thioredoxin reductases. A decline in the levels of either cytosolic or mitochondrial pyridine nucleotides can therefore compromise cell functions and viability through impairment of energy metabolism and promotion of oxidative stress. The net loss of NAD(P)(H) has been strongly implicated in the mechanisms of neurodegeneration associated with both acute injury to the CNS and with chronic neurologic disorders (Xia et al. 2009; Owens et al. 2013).
Intrinsic autofluorescence of reduced pyridine nucleotides in living cells is a convenient non-invasive method for monitoring mitochondrial energy metabolism under both normal and pathological conditions (Shuttleworth et al. 2003; Mayevsky and Rogatsky 2007; Chance et al. 1962b; Chance et al. 1962a). The emission spectra of NADH and NADPH overlap, although the high level of protein-bound NADH that is present in mitochondria contributes to the majority of NAD(P)H autofluorescence in mitochondria-rich cells, e.g., neurons (Li et al. 2008; Shuttleworth 2010).
In a previous study, we demonstrated a loss of NAD(P)H fluorescence during anoxia and reoxygenation in cortical neurons but not astrocytes (Kahraman and Fiskum 2007). There was also a net loss in maximum fluorescence in response to the subsequent inhibition of the electron transfer chain by cyanide after anoxia/reoxygenation in cortical neurons, suggesting either an irreversible oxidation or a net depletion of the mitochondrial pyridine nucleotide pool. Poly (ADP-ribose) polymerase (PARP) isozymes, including PARP-1, are potent NAD+-consuming enzymes under pathological conditions associated with oxidative stress (Berger 1985). PARP-1 contributes to cell death during brain ischemia/reperfusion when extensively activated by DNA damage (Szabo and Dawson 1998). The cell death resulting from PARP-1 activation is linked to NAD+ depletion and energy failure (Berger 1985; Alano et al. 2004). Since mitochondrial membranes are not permeable to NAD+ and NADH, the loss of the mitochondrial pool of pyridine nucleotides could be explained by either the presence of mitochondrial PARP that is activated by damaged mitochondrial DNA (Du et al. 2003) or by the opening of the mitochondrial inner membrane permeability transition pore (PTP) (Dodoni et al. 2004). Pore opening allows for passive release of mitochondrial NAD+ and makes it available to NAD+ consuming PARP enzymes located outside the matrix space in the cytosol and nucleus (Di Lisa et al. 2001). Therefore, we hypothesized that both PTP opening and PARP activation contribute to depletion of neuronal NAD(H) in an in vitro model of cerebral ischemia.
MATERIALS AND METHODS
Materials
All cell culture reagents were from GIBCO-BRL (Grand Island, NY). Potassium cyanide (KCN) was purchased from Fisher Scientific Company L.L.C. (Pittsburgh, PA). Fura-2 AM and hydroethidine (HEt) were from Molecular Probes (Eugene, OR) and NADH oxidase was purchased from Calbiochem (San Diego, CA). Solutions for the Pierce BCA microreagent protein assay were purchased from Pierce (Rockford, IL). Unless otherwise stated, all other chemicals were obtained from Sigma-Aldrich Inc. (St. Louis, MO).
Primary Cultures of Cortical Neurons
Timed-pregnant Sprague Dawley rats were purchased from Charles River Laboratory (Wilmington, MA, USA) for the neuronal cultures. Rats were housed in the animal colony at the University of Maryland School of Medicine until use. All protocols in this study were approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee, and are in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Cortical neurons were isolated from 16th day in utero rats using a modification of the methods of Yavin and Yavin (1980). All dissections were performed in Leibovitz's L-15 with glutamine. After removing the medium, the dissected cerebral cortices were minced and digestedin 0.2% trypsin at 37°C for 2.5 min Trypsin proteolysis was terminated by addition of an equal volume of maintenance medium which contains trypsin inhibitor (Neurobasal medium (NB, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine). The cells were triturated, centrifuged at low speed (800×g), resuspended in fresh medium to a volume of 1 ml per brain, and filtered through a Falcon cell strainer (70 µm pore size). Cortical neurons were cultured on poly-D-lysine-coated 25 mm coverslips for 10 – 14 days in vitro, at a density of ~50,000 cells/coverslip in Neurobasal medium with 2% B27 (Invitrogen) and 2 mM L-glutamine supplements under 95% air / 5% CO2 at 37°C. Glial proliferation was prevented by adding cytosine-arabinofuranoside (5 µM) at in vitro day 4. Fresh media was added at in vitro day 6. Immunocytochemical measurements of glial fibrillary acid protein (GFAP) confirmed that cultures contained <1% astrocytes.
NAD(P)H Autofluorescence Microscopy
On days in vitro 10 – 14, the coverslips were removed and placed in a superfusion chamber in the microscope. The coverslips were constantly superfused (0.5 ml/min) with pH 7.4 artificial cerebrospinal fluid (aCSF) containing 120 mM NaCl, 3.5 mM KCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1 mM MgCl2, 20 mM HEPES, 15 mM glucose and maintained at 37°C. Diethylenetriamine/nitric oxide adduct (DETA-NO, Sigma-Aldrich, St Louis, MO) was added in aCSF 30 min before perfusion to achieve a stable concentration of nitric oxide (NO) in the aCSF. This concentration of DETA-NO was reported to release NO at a rate of ~100 pmol/min (Dranka et al. 2010). When the PARP inhibitor 3,4-Dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ, 30 µM) and cyclosporin A (CsA, 2 µM) were used, the neurons were exposed to these agents under normal culture conditions for 45 min prior to superfusion, when the agents were also present in the perfusate.
The coverslip superfusion chamber was mounted on a Nikon Eclipse TE2000-S inverted microscope (SFluor 20×0.75 N.A., Melville, NY). Single cell fluorescence of NAD(P)H was imaged by excitation at a wavelength of 355 nm (Polychrome IV, Till, Munich, Germany), and emission above 435 nm. Image sequences (10 sec/frame, 120 msec exposure time, 4×4 binning) were acquired by an ORCA-ER cooled digital CCD camera (Hamamatsu Photonics, Hamamatsu, Germany) and imaged with Metafluor 6.3 (Universal Imaging, West Chester, PA) imaging software. The NAD(P)H autofluorescence was expressed as the normalized epifluorescence change (Δf/f0), which is the difference in fluorescence (Δf = f1 - f0) normalized to the initial fluorescence (f0).
Cellular Pyridine Nucleotide Content
At the end of superfusion, each coverslip was placed in lysis buffer containing 0.1 M Tris-HCL pH 8.0, 0.01 M EDTA, 0.05 % Triton X-100 and 30 µM DPQ. Cells were scraped from the coverslips into the buffer and transferred to microfuge tubes. Samples were sonicated on ice for 5 min prior to centrifugation at 4°C for 15 min at 14,000 X g. The supernatant was assayed for total NAD+ plus NADH by using an enzymatic recycling assay (Kristian and Fiskum 2004). The NAD(H) content was determined using a calibration curve with purified NAD+ (Sigma-Aldrich) as the standard. Results were normalized to the protein present in the sonicated cell extract, as measured by the Pierce bicinchoninic acid (BCA) assay (Pierce, Rockford, IL).
Data Analysis
All data are expressed as means ± S.E.M. of n cells. Statistical significance was assessed by one-way ANOVA or a repeated measures ANOVA test followed by the Tukey test for multiple comparisons. For data that were not normally distributed, the Kruskal-Wallis non-parametric ANOVA or Friedman Repeated Measures ANOVA on Ranks with Dunn’s post hoc tests was used. P < 0.05 was considered to be statistically significant.
RESULTS
Loss of NAD(P)H fluorescence induced by chemical anoxia and glucose deprivation
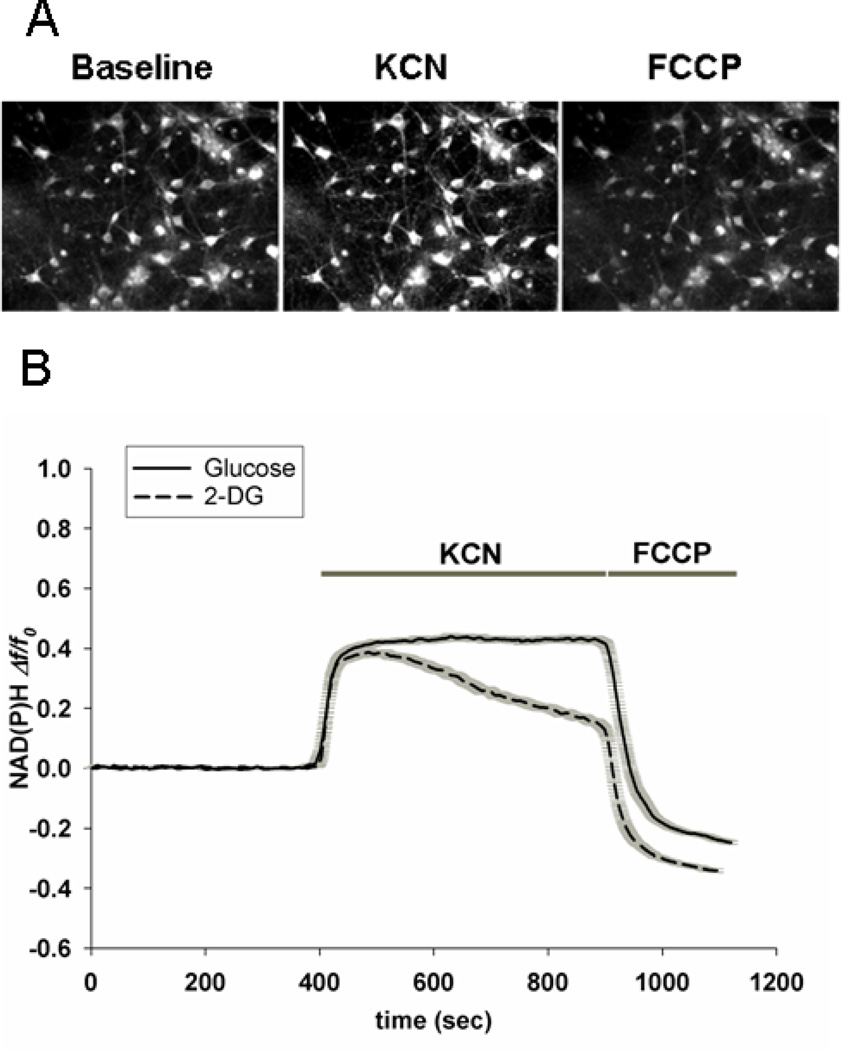
Fig. 1A provides fluorescent images of rat cortical neurons using excitation and emission wavelengths that select for NAD(P)H autofluorescence. Neuronal fluorescence intensity increased when the superfusate medium was changed from artificial CSF (aCSF) to aCSF plus 1 mM potassium cyanide (KCN). The increase in NAD(P)H autofluoresence was consistent with a block of electron flow from NADH to O2 caused by cyanide-mediated inhibition of mitochondrial respiratory chain Complex IV (cytochrome oxidase). Upon washout of KCN and superfusion with aCSF plus the respiratory uncoupler FCCP, the fluorescence intensity decreased to a level lower than baseline. This minimum fluorescence intensity represents the maximally oxidized NAD(P)H oxidation/reduction (redox) state achievable through maximal flow of electrons from NADH to O2 through the respiratory chain.
Figure 1. NAD(P)H autofluorescence within rat cortical neurons in response to respiratory inhibition and respiratory uncoupling.
A. Images of autofluorescence at 355 nm excitation wavelength and emission above 435 nm within rat cortical neurons superfused with artificial CSF (Baseline) and then with aCSF containing 1 mM potassium cyanide (KCN), followed by aCSF containing the protonophore respiratory uncoupling agent FCCP (5.0 µM) in the absenc of KCN. B. Quantification of changes in NAD(P)H autofluorescence in response to respiratory inhibition and uncoupling. Each trace represents the mean change in fluorescence, ± the SEM (gray shadow), compared to the initial fluorescence (Δf/fo) in 40 – 60 neuronal somata per coverslip. After approximately 400 sec of superfusion with aCSF, the perfusate was changed to aCSF plus KCN in the presence of 15 mM glucose or in the absence of glucose. After 500 sec exposure to KCN, the perfusate was changed to aCSF containing 15 mM glucose plus 5.0 µM FCCP.
Quantification of fluorescence intensity within neuronal somata over time in the presence of different superfusates is described in Fig. 1B. These values represent the means (solid lines) ± SEM (gray shadows) for n = 30 – 50 neurons per coverslip. Fluorescence in the presence of aCSF was highly stable during more than 6 min of measurement. Upon superfusion with aCSF plus KCN there was an abrupt increase in fluorescence from virtually all of the neurons, reaching a plateau within less than one min. In the presence of glucose as exogenous reducing power, this maximum fluorescence level remained stable for at least 8 min. When neurons were superfused with aCSF plus KCN in the absence of glucose, the initial rise in fluorescence was very similar to that observed with glucose. However, the fluorescence subsequently declined steadily over the next 8 min toward the initial baseline. Following KCN washout and exposure to aCSF plus glucose and FCCP, the fluorescence intensity rapidly declined to levels well below that of baseline, with the level following perfusion without glucose being lower than that following perfusion with glucose.
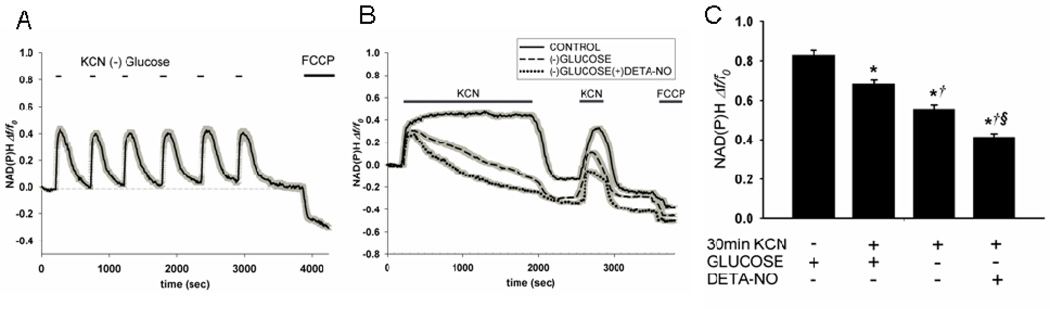
Experiments were then performed to determine the nature and regulation of NAD(P)H fluorescence decay during and after chemical anoxia. To determine if loss of fluorescence intensity (i.e. drop below baseline) is dependent on the duration of anoxia, neurons were exposed alternatively to aCSF plus glucose for 8 min and aCSF minus glucose for 2 min. As shown in Fig. 2A, during six cycles of chemical anoxia and reoxygenation (60 min total), both the maximum increase in NAD(P)H fluorescence and the return to baseline was constant. When neurons were exposed to KCN minus glucose constantly over 30 min, however, the fluorescence declined continuously, eventually dropping to below the original baseline (Fig. 2B; dashed line).
Figure 2. Partial irreversibility of the time-dependent decline in neuronal NAD(P)H fluorescence during respiratory inhibition.
A. Neurons were alternately superfused with aCSF minus glucose plus 1.0 mM KCN for two min and then aCSF plus 15 mM glucose for six min. Following six of these cycles, cells were superfused with aCSF plus glucose plus 5.0 µM FCCP. Traces are mean fluorescence (Δf/fo) ± SEM (gray shadow) of all cell somata in the microscopic field of view (42 cells). B. Neurons were superfused with aCSF containing KCN either with or without glucose for 30 min. In the absence of glucose, 200 µM DETA-NO was either absent or present. Cells were then exposed to normal aCSF plus glucose for 10 min followed by exposure to KCN for 5 min. Neurons were then exposed to normal aCSF plus glucose for another 5 min followed by exposure to FCCP. Each tracing represents the mean ± SEM change in fluorescence for 40 – 50 neuronal somata on an individual coverslip. C. Quantification of maximal NAD(P)H fluorescence in experiments shown in B, as defined by the gradient between the maximal fluorescence observed after the second addition of KCN and the minimum observed following addition of FCCP. Values represent the means ± SEM for n=4 experiments using separate neuronal cultures. *p<0.05 compared to timed control (KCN and FCCP after 40 min superfusion with normal aCSF). †p < 0.05 compared to plus glucose without DETA-NO. § p < 0.05 compared to minus glucose without DETA-NO.
The apparent oxidized shift in NAD(P)H redox state following prolonged chemical anoxia could be due to elevated respiration to reestablish ATP homeostasis. Alternatively, the decay of pyridine nucleotide fluorescence could be due to a net loss, or catabolism of NAD(P)H. Loss of total NAD(P)(H) should be reflected by a drop in maximal fluorescence following exposure to a second addition of cyanide. This possibility was tested by superfusing neurons with KCN plus glucose starting at 10 min following exposure to normal aCSF (i.e. the “reoxygenation” period). When KCN was added during reoxygenation after anoxia plus glucose, fluorescence rose to approximately 80% of that observed during the initial anoxia (Fig. 2B, solid line). When cyanide was added during reoxygenation after anoxia in the absence of glucose, fluorescence rose to a level that was only 50% of that observed during the initial anoxia (Fig. 2B, dashed line). These results strongly suggested that NAD(P)(H) catabolism contributes to loss of NAD(P)H fluorescence, particularly in the presence of KCN and the absence of glucose.
One mechanism known to be responsible for NAD(H) catabolism in other cell stress paradigms is the substrate utilization of NAD+ for poly(ADP-ribosylation) of proteins by poly(ADP-ribose)polymerases, e.g., PARP-1. PARP-1 enzyme activity is activated in response to oxidative DNA modifications caused by reactive oxygen and nitrogen species, e.g., nitric oxide (NO) and its metabolites. Experiments were therefore conducted to determine whether the presence of NO in addition to chemical anoxia and glucose deprivation promotes irreversible loss of NAD(P)H fluorescence during exposure of neurons to KCN. As shown in Fig. 2B, superfusion of neurons with medium pre-equilibrated with the NO donor, DETA-NO (200 µM), accelerated the loss of pyridine nucleotide fluorescence during metabolic inhibition and further lowered the fluorescence level present following the second KCN addition (dotted line). These findings are consistent with the possibility of irreversible NAD(P)H catabolism. Exposure of neurons to this concentration of DETA-NO alone (i.e. in normal aCSF plus glucose) had no effect on NAD(P)H fluorescence (not shown). In order to more accurately compare the effects of the different conditions on loss of total pyridine nucleotide fluorescence, the difference between the maximal signal (obtained after the second exposure to KCN) and the minimum signal (obtained after KCN washout and FCCP exposure) was quantified for the individual neurons (n=140–180) measured under different conditions (Fig. 2C). Compared to the time-controlled exposure to normal aCSF, the total pyridine nucleotide fluorescence values calculated after superfusion with KCN ± glucose and KCN minus glucose plus DETA-NO were all significantly lower (p<0.05). In addition, total NAD(P)H fluorescence after KCN minus glucose was significantly lower than after KCN plus glucose, and further reduced by the presence of DETA-NO (Fig. 2C, each p<0.05).
Inhibitors of PARP and PTP opening partially protect against chemical anoxia-induced loss of neuronal NAD(P)H fluorescence
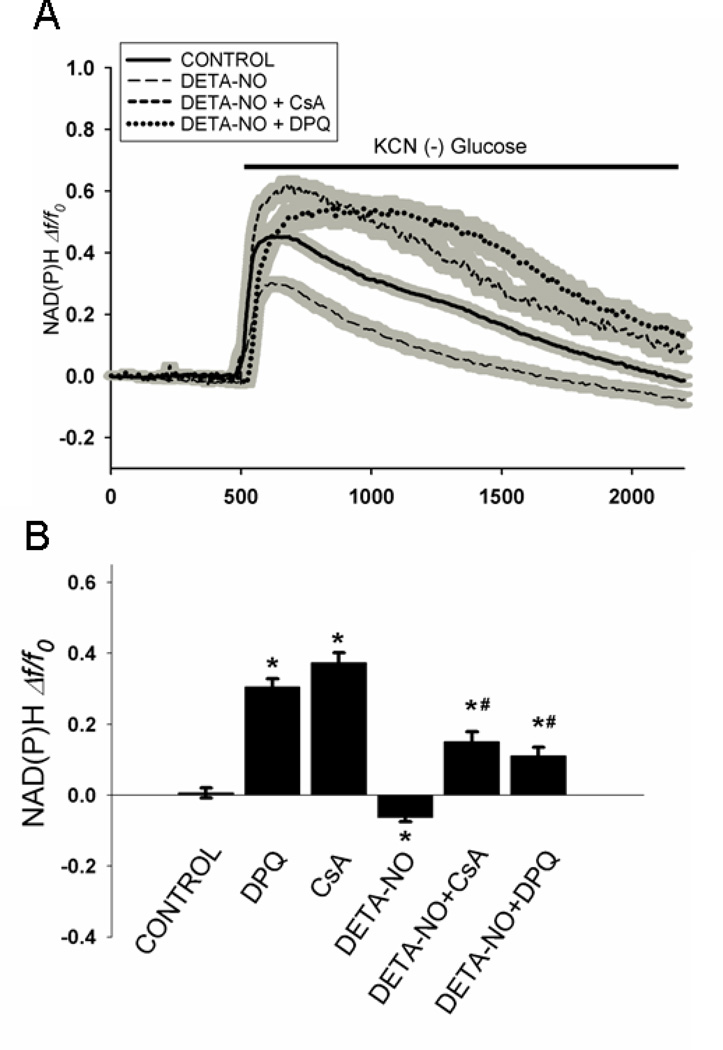
To investigate the involvement of PARP and the mitochondrial permeability transition pore (PTP) in the irreversible loss of NAD(P)H fluorescence, neurons were pre-incubated for 45 min with the PARP inhibitor 3,4-Dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ), or the PTP inhibitor, cyclosporin A (CsA), respectively. As shown in Fig. 3, in the absence of these inhibitors, exposure to KCN in the absence of glucose resulted in an initial rise in fluorescence followed by a steady decline toward baseline (Fig. 3A, solid line). The additional presence of 200 µM DETA-NO elevated both the rate and extent of fluorescence decay (Fig. 3A, dashed line). The presence of either DPQ or CsA inhibited the loss of fluorescence under these conditions (Fig. 3A; dotted lines).
Figure 3. Protection against loss of NAD(P)H fluorescence during metabolic inhibition by inhibitors of PARP and the mitochondrial permeability transition.
A. All coverslips containing neurons were superfused for 8 min with aCSF plus glucose and then exposed to KCN (1 mM) for 30 min in the absence of glucose ± DETA-NO; 200 µM). Cells exposed to KCN plus DETA-NO were superfused with media in the absence or presence of DPQ (30 µM), or CsA (2 µM). Each trace represents the mean fluorescence (Δf/fo) ± SEM (gray shadow) in all cell somata in the microscopic field of view (40 – 50 cells). B. Quantitative comparison of the change in NAD(P)H fluorescence at the end of 30 min exposure to KCN in the absence and presence of DETA-NO, DPQ, and CsA. Values represent the means ± SEM for n=4 – 6 experiments using separate neuronal cultures. *p < 0.001 compared to KCN minus glucose; #p < 0.001 compared to KCN minus glucose plus DETA-NO.
A quantitative comparison was made between the changes in fluorescence immediately prior to exposure to chemical anoxia and at 30 min later, in the absence and presence of DETA-NO, DPQ, and CsA (Fig. 3B). At the end of anoxia alone (Control), NAD(P)H fluorescence of control cells were back to baseline values present prior to anoxia, i.e., Δf/f0 = 0. When DETA-NO was present during anoxia, the final fluorescence level was significantly lower than that of the Control cells by approximately 0.1 arbitrary fluorescent units. The fluorescence observed following anoxia in the presence of either DPQ or CsA was significantly higher than in their absence, whether or not DETA-NO was present during the anoxia. The effects of DPQ and CsA were very similar, with final change in fluorescence after anoxia, ranging from 0.3 – 0.4 fluorescent units in the absence of DETA-NO and from 0.10 – 0.15 fluorescence units in presence of DETA-NO. Neither DPQ nor CsA had an effect on cellular NAD(P)H autofluorescence in the absence of chemical anoxia plus glucose deprivation (not shown). Thus, inhibition of both PARP activity and PTP opening significantly mitigates loss of neuronal NAD(P)H autofluorescence during chemical anoxia either in the absence or presence of DETA-NO.
Chemical anoxia and PARP activation depletes total cellular NAD(H) content
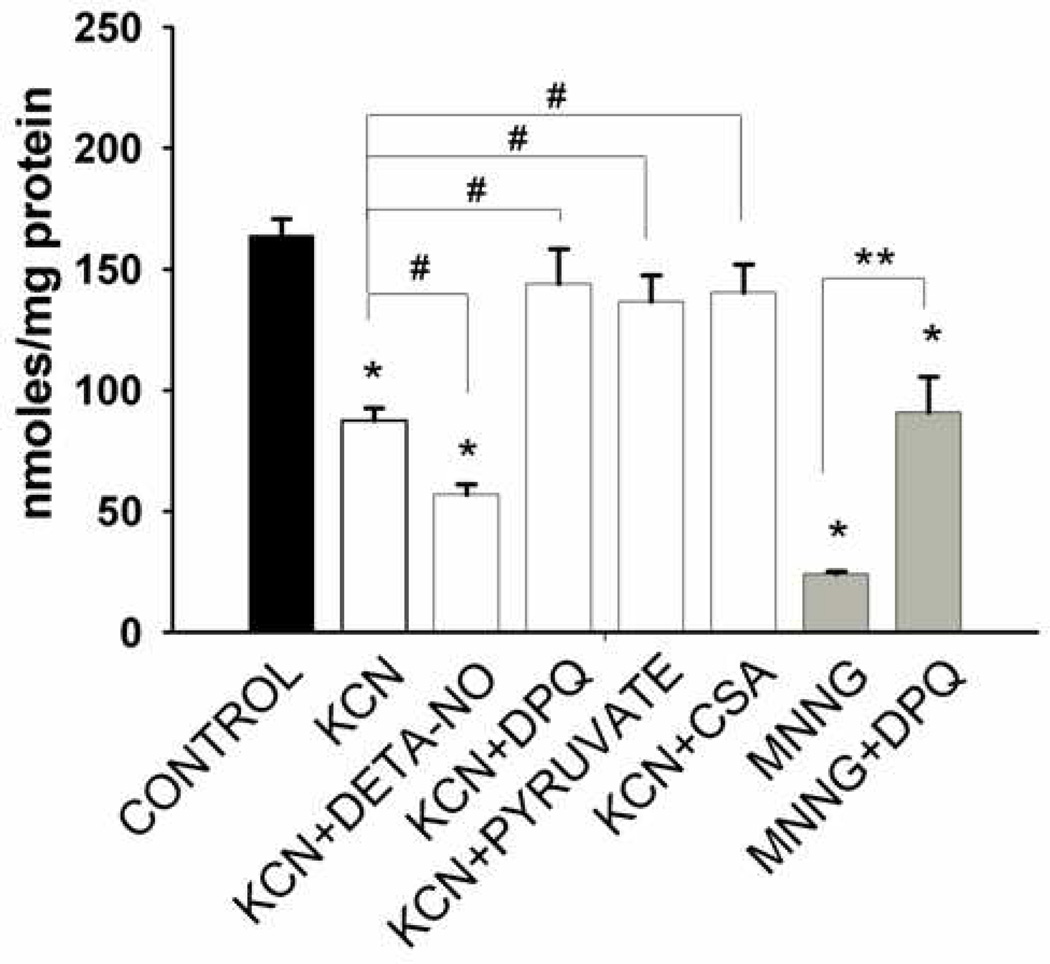
Although the partially irreversible loss of NAD(P)H fluorescence during chemical hypoxia is consistent with pyridine nucleotide catabolism, fluorescence alone does not differentiate between NADH and NADPH. In addition, the loss of fluorescence could be due to irreversible oxidation of NAD(P)(H) or to a shift in the equilibrium between NAD(P)H bound to proteins (high fluorescence) to unbound NAD(P)H (low fluorescence). Validation that chemical anoxia and DETA-NO result in net loss of NAD(H) came from experiments in which these molecules were extracted from neurons and directly measured at the end of the period of chemical anoxia or after the same period under control conditions. Coverslips were placed in lysis buffer containing detergent to disrupt mitochondrial membranes and DPQ to block any PARP-mediated pyridine nucleotide catabolism during processing. As shown in Figure 4, the total NAD(H) present in lysates of neurons superfused for approximately 40 min in aCSF plus glucose was 161 ± 10 nmoles per mg protein (n=8). In contrast, following 30 min superfusion with aCSF minus glucose and plus KCN, total neuronal NAD(H) content was significantly lower (92 ± 8 nmoles/mg; p<0.01, n=6). When the chemical anoxia medium was supplemented with DETA-NO, an even lower level of NAD(H) was observed which was 63% of that obtained in the absence of DETA-NO (p<0.05) and 36% of that obtained under control conditions (p<0.01; n=6). The presence of DPQ during the 30 min anoxic superfusion completed protected against the loss of NAD(H). Complete inhibition of NAD(H) catabolism was also observed after superfusion with either CsA or pyruvate, which is an alternative source of reducing power in the absence of glucose. Finally, as a positive control, we superfused neurons with aCSF plus glucose in the presence of the DNA alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG), which is a chemical activator of PARP activity. MNNG treatment resulted in an 85% reduction in NAD(H) levels compared to Control. This more extensive NAD(H) catabolism was protected, albeit incompletely, when DPQ was present together with MNNG.
Figure 4. Loss of total neuronal NAD(H) during exposure to chemical anoxia.
Cells were incubated with either KCN (1 mM) in glucose free aCSF or in glucose-containing aCSF plus the PARP activator, MNNG (50 µM), for 30 min. The presence of DETA-NO (200 µM) promoted loss of NAD(H) and the presence of either DPQ (30 µM), cyclosporin A (2 µM), or pyruvate (5 mM) significantly protected against loss of NAD(H) during metabolic inhibition. DPQ (30 µM) also partially reversed the loss of NAD(H) induced by MNNG. n = 4 – 12 different neuronal cultures. *p < 0.001 compared to control in normal aCSF, #p = 0.001 compared to KCN in glucose free aCSF, ** p < 0.05 compared to MNNG.
DISCUSSION
This study tested the hypothesis that exposure of neurons to in vitro “ischemia” results in rapid catabolism of NAD(H) due to the enzyme activity of PARP-1, and the release of mitochondrial NAD(H) into the cytosol by opening of the permeability transition pore. The model used to test this hypothesis employed chemical hypoxia induced by superfusion of rat cortical neurons with an artificial CSF solution containing potassium cyanide, which blocks mitochondrial respiration by inhibiting the electron transport chain Complex IV, also known as cytochrome oxidase. Superfusion of neurons with different media was employed to maintain constant concentrations of KCN, protons, and other ions including Ca2+ and Na+. Superfusion also allowed for rapid alternations between different media that have potential effects on cellular redox state, as monitored continuously by NAD(P)H autofluorescence. One limitation of this experimental paradigm is that glutamate released from neurons during KCN-induced deenergization is rapidly removed by superfusion, thus eliminating excitotoxicity as a factor that can influence neuronal responses to in vitro ischemia. This concept was supported by the observation that superfusion with the N-methyl D-aspartate (NMDA) ionotropic glutamate receptor antagonist MK801 had no effect on NAD(P)(H) fluorescence before, during, or after exposure of neurons to cyanide (results not shown). Thus, future experiments could include excitotoxic concentrations of glutamate in the superfusate during chemical anoxia to more closely simulate the conditions to which neurons are exposed during cerebral ischemia in vivo.
Several conditions were tested that significantly protected against loss of pyridine nucleotide autofluorescence and (or) loss of total NAD(H) during chemical anoxia. The first was the presence of DPQ, which inhibits PARP-dependent catabolism of NAD+, (Takahashi et al. 1999), and is also neuroprotective both in vitro and in vivo (Meli et al. 2004; Abramov and Duchen 2008). The presence of 50 µM DPQ during exposure of neurons to KCN plus DETA-NO partially inhibited the decline in NAD(P)(H) autofluorescence (Fig. 3) and completely inhibited the loss of total NAD(H) (Fig. 4). DPQ was somewhat less effective at protecting against loss of pyridine nucleotide fluorescence when tested in the presence of DETA-NO (Fig. 3.B.). This observation is consistent with the promotion of NAD(H) catabolism by DETA-NO and the complete protection against catabolism by DPQ when neurons were exposed to KCN in the absence of DETA-NO.
The majority of neuronal pyridine nucleotides are located within the mitochondrial matrix (Alano et al. 2007). Thus, when we found that at least 50% of the cyanide-re-reducible pool was lost following prolonged exposure to anoxia and glucose deprivation (Fig. 2), we hypothesized that much of this loss was due to release of mitochondrial NAD(P)(H) through opening of the PTP, leading to their degradation. This hypothesis was tested by comparing both the decline in NAD(P)H autofluorescence and the change in total NAD(H) in the absence and presence of the PTP inhibitor CsA. CsA partially attenuated the decline in autofluorescence and completely inhibited the loss of total NAD(H) that occurred during superfusion of neurons with glucose-free medium containing DETA-NO. The finding that CsA was less effective at inhibiting loss of fluorescence than loss of total NAD(H) probably indicates that the fluorescent decline is a combination of both NAD(H) catabolism and net oxidation of NAD(P)H that occurs spontaneously when cells are deprived of reducing power, i.e., metabolic fuel.
The presence of exogenous reducing power was also very effective at inhibiting NAD(H) catabolism, as seen by the stable level of fluorescence observed in the presence of 15 mM glucose (Fig. 1.B), and the maintenance of completely normal levels of NAD(H) in the presence of 5 mM pyruvate (Fig. 4). There are at least two explanations for the protection by glucose or pyruvate against pyridine nucleotide catabolism induced by exposure to KCN. Obviously, glucose can still generate ATP in the presence of cyanide via anaerobic glycolysis, which could inhibit loss of ionic homeostasis and associated oxidative stress, thereby indirectly inhibiting PARP activation. This explanation cannot apply to pyruvate since it can only support TCA cycle metabolism and oxidative phosphorylation, which are both blocked by the cyanide inhibition of the electron transport chain. Pyruvate, however, can provide the reducing power necessary for maintaining a relatively reduced redox state in the mitochondrial matrix, primarily via the pyruvate dehydrogenase reaction, which reduces NAD+ to NADH. In addition, the mitochondrial transhydrogenase reaction transfers hydride ions from NADH to NADP+, forming NADPH. Maintenance of a high NADPH/NADP+ ratio is accompanied by a high reduced/oxidized glutathione ratio, via the glutathione reductase reaction. Constant regeneration of reduced glutathione is necessary to detoxify reactive oxygen species, e.g., peroxides, and is also required for maintaining protein sulfhydryl groups in a reduced redox state. Since oxidation of specific cysteine sulfhydryl groups present on cyclophilin D substantially increase the sensitivity of PTP opening by calcium (Nguyen et al. 2011), the presence of reducing power in the form of glucose or pyruvate likely inhibits PTP opening and subsequent release of mitochondrial pyridine nucleotides into the cytosol where they are subject to PARP-mediated degradation.
Taken together, our results suggest the following series of events responsible for rapid and extensive catabolism of neuronal pyridine nucleotides during chemical anoxia and glucose deprivation. Exposure of cells to cyanide inhibits respiration and oxidative phosphorylation and stimulates mitochondrial ROS production, as has been described for neurons and other cell types (Mills et al. 1996; Niknahad et al. 1995; Roemgens et al. 2011). These ROS react with many molecules throughout the cell, including polyribonucleotides, causing DNA strand breaks that are recognized by PARP-1 (Ali et al. 2012). In response to this recognition, PARP-1 becomes hyperactivated in an effort to poly(ADP-ribosylate) and therefore activate enzymes involved in DNA repair. PARP-1 hyperactivation results in cytosolic NAD(H) catabolism that exceeds the ability of the cell to synthesize NAD(H), particularly under reduced ATP levels, which are necessary for NAD(H) biosynthesis. Increased mitochondrial ROS production also contributes to NADPH and NADH oxidation as they are used in ROS detoxification pathways. The net oxidized shift in the mitochondrial redox state increases the sensitivity of the PTP to open in response to mitochondrial Ca2+ influx. The increased influx is a consequence of elevated cytosolic Ca2+ following a decline in ATP (Costantini et al. 1996; Greco and Fiskum 2010). Once the PTP is opened, pyridine nucleotides are released from the mitochondrial matrix into the cytosol where they are catabolized by PARPs and possibly other enzymes (Kristian et al. 2011). Experiments reported by other labs also indicate that products of PARP-1 enzyme activity, including ADP-ribose and poly(ADP-ribose), might directly increase the permeability of either the inner or outer mitochondrial membranes, contributing to loss of mitochondrial NAD(P)(H) (Cipriani et al. 2005; Baek et al. 2013). It is also possible that poly(ADP-ribosylation) of mitochondrial proteins, e.g., the ATP synthase and the voltage-dependent anion channel, could contribute to PTP opening and release of mitochondrial pyridine nucleotides (Lai et al. 2008). In astrocytes, at least, both NAD(H) depletion and PTP opening are both necessary steps linking PARP-1 activation to cell death . In cells that exhibit a high rate of ATP turnover, e.g., neurons and cardiac myocytes, the PARP-1 mediated death appears primarily necrotic due to the inhibition of both glycolytic and aerobic energy metabolism by NAD(H) depletion. In less energy-demanding cells, death can be due to caspase-independent apoptosis through release of apoptosis initiating factor (AIF) from the mitochondria, translocation to the nucleus, and activation of endonucleases.
The results we obtained with an in vitro model of neuronal ischemia are relevant to many observations made with experimental animals and even patients who experience acute brain injury (Komjati et al. 2005; Kristian et al. 2011). PARP-1 mediated loss on NAD(P)(H) has been strongly implicated in the death of neurons and the neurologic impairment observed in different rodent models of traumatic brain injury (Vagnozzi et al. 1999; Whalen et al. 1999; LaPlaca et al. 2001);(Satchell et al. 2003; Clark et al. 2007; Prieto et al. 2011). In one clinical study, the levels of poly(ADP-ribosylated) proteins present in CSF within four days following severe TBI in children were significantly elevated compared to those in controls and higher in males compared to females (Fink et al. 2008). PARP-1 mediated brain NAD(H) depletion also occurs in rodent and primate stroke models (Yuan et al. 2009; Kaundal et al. 2006; Balan et al. 2010). While similar AIF translocation occurs in male and female mice after ischemic stroke, knockdown of PARP-1 expression reduces ischemic brain damage in males but not in females (Yuan et al. 2009). Accumulation of poly(ADP-ribose) also occurs in human brain autopsy tissue from cardiac arrest patients who die within 48 hr (Love et al. 1999). The finding that a novel PARP-1 inhibitor (MP-124) significantly reduced neurologic impairment and stroke infarct volume when administered to both male and female monkeys at 3 and 6 hr after ischemic stroke provides hope that this approach toward neuroprotection will eventually be translated clinically (Matsuura et al. 2011). Based on the apparent sex differences in the degree to which PARP-1 contributes to cell death and neurologic outcomes after brain injury, interventions designed to mitigate this mechanism might be more effective in males compared to females.
Acknowledgments
Supported by NIH grants R01 NS34152 and P01 HD16596 (GF), NS 07375 (SK), and R01 NS085165 (BMP).
Reference List
- Abramov AY, Duchen MR. Mechanisms underlying the loss of mitochondrial membrane potential in glutamate excitotoxicity. Biochim Biophys Acta. 2008;1777:953–964. doi: 10.1016/j.bbabio.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Alano CC, Tran A, Tao R, Ying W, Karliner JS, Swanson RA. Differences among cell types in NAD(+) compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J Neurosci Res. 2007;85:3378–3385. doi: 10.1002/jnr.21479. [DOI] [PubMed] [Google Scholar]
- Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J Biol Chem. 2004;279:18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Bae ON, Kim EK, Yu SW. Induction of mitochondrial dysfunction by poly(ADP-ribose) polymer: implication for neuronal cell death. Mol Cells. 2013;36:258–266. doi: 10.1007/s10059-013-0172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan IS, Fiskum G, Kristian T. Visualization and quantification of NAD(H) in brain sections by a novel histo-enzymatic nitrotetrazolium blue staining technique. Brain Res. 2010;1316:112–119. doi: 10.1016/j.brainres.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation-reduction states in vivo. Science. 1962a;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- Chance B, Legallais V, Schoener B. Metabolically linked changes in fluorescence emission spectra of cortex of rat brain, kidney and adrenal gland. Nature. 1962b;195:1073–1075. doi: 10.1038/1951073a0. [DOI] [PubMed] [Google Scholar]
- Cipriani G, Rapizzi E, Vannacci A, Rizzuto R, Moroni F, Chiarugi A. Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J Biol Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- Clark RS, Vagni VA, Nathaniel PD, Jenkins LW, Dixon CE, Szabo C. Local administration of the poly(ADP-ribose) polymerase inhibitor INO-1001 prevents NAD+ depletion and improves water maze performance after traumatic brain injury in mice. J Neurotrauma. 2007;24:1399–1405. doi: 10.1089/neu.2007.0305. [DOI] [PubMed] [Google Scholar]
- Costantini P, Chernyak BV, Petronilli V, Bernardi P. Modulation of the mitochondrial permeability transition pore by pyridine nucleotides and dithiol oxidation at two separate sites. J Biol Chem. 1996;271:6746–6751. doi: 10.1074/jbc.271.12.6746. [DOI] [PubMed] [Google Scholar]
- Di Lisa F, Menabo R, Canton M, Barile M, Bernardi P. Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart. J Biol Chem. 2001;276:2571–2575. doi: 10.1074/jbc.M006825200. [DOI] [PubMed] [Google Scholar]
- Dodoni G, Canton M, Petronilli V, Bernardi P, Di LF. Induction of the mitochondrial permeability transition by the DNA alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine. Sorting cause and consequence of mitochondrial dysfunction. Biochim Biophys Acta. 2004;1658:58–63. doi: 10.1016/j.bbabio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med. 2010;48:905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- Fink EL, Lai Y, Zhang X, Janesko-Feldman K, Adelson PD, Szabo C, Berger RP, Sarnaik AA, Kochanek PM, Clark RS. Quantification of poly(ADP-ribose)-modified proteins in cerebrospinal fluid from infants and children after traumatic brain injury. J Cereb Blood Flow Metab. 2008;28:1523–1529. doi: 10.1038/jcbfm.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco T, Fiskum G. Brain mitochondria from rats treated with sulforaphane are resistant to redox-regulated permeability transition. J Bioenerg Biomembr. 2010;42:491–497. doi: 10.1007/s10863-010-9312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahraman S, Fiskum G. Anoxia-induced changes in pyridine nucleotide redox state in cortical neurons and astrocytes. Neurochem Res. 2007;32:799–806. doi: 10.1007/s11064-006-9206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal RK, Shah KK, Sharma SS. Neuroprotective effects of NU1025, a PARP inhibitor in cerebral ischemia are mediated through reduction in NAD depletion and DNA fragmentation. Life Sci. 2006;79:2293–2302. doi: 10.1016/j.lfs.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Komjati K, Besson VC, Szabo C. Poly (adp-ribose) polymerase inhibitors as potential therapeutic agents in stroke and neurotrauma. Curr Drug Targets CNS Neurol Disord. 2005;4:179–194. doi: 10.2174/1568007053544138. [DOI] [PubMed] [Google Scholar]
- Kristian T, Balan I, Schuh R, Onken M. Mitochondrial dysfunction and nicotinamide dinucleotide catabolism as mechanisms of cell death and promising targets for neuroprotection. J Neurosci Res. 2011;89:1946–1955. doi: 10.1002/jnr.22626. [DOI] [PubMed] [Google Scholar]
- Kristian T, Fiskum G. A fluorescence-based technique for screening compounds that protect against damage to brain mitochondria. Brain Res. 2004;13:176–182. doi: 10.1016/j.brainresprot.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, Jenkins LW, Szabo C, Clark RS. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- LaPlaca MC, Zhang J, Raghupathi R, Li JH, Smith F, Bareyre FM, Snyder SH, Graham DI, McIntosh TK. Pharmacologic inhibition of poly(ADP-ribose) polymerase is neuroprotective following traumatic brain injury in rats. J Neurotrauma. 2001;18:369–376. doi: 10.1089/089771501750170912. [DOI] [PubMed] [Google Scholar]
- Li D, Zheng W, Qu JY. Time-resolved spectroscopic imaging reveals the fundamentals of cellular NADH fluorescence. Opt Lett. 2008;33:2365–2367. doi: 10.1364/ol.33.002365. [DOI] [PubMed] [Google Scholar]
- Love S, Barber R, Wilcock GK. Neuronal accumulation of poly(ADP-ribose) after brain ischaemia. Neuropathol Appl Neurobiol. 1999;25:98–103. doi: 10.1046/j.1365-2990.1999.00179.x. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Egi Y, Yuki S, Horikawa T, Satoh H, Akira T. MP-124, a novel poly(ADP-ribose) polymerase-1 (PARP-1) inhibitor, ameliorates ischemic brain damage in a non-human primate model. Brain Res. 2011;1410:122–131. doi: 10.1016/j.brainres.2011.05.069. [DOI] [PubMed] [Google Scholar]
- Mayevsky A, Rogatsky GG. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol. 2007;292:C615–C640. doi: 10.1152/ajpcell.00249.2006. [DOI] [PubMed] [Google Scholar]
- Meli E, Pangallo M, Picca R, Baronti R, Moroni F, Pellegrini-Giampietro DE. Differential role of poly(ADP-ribose) polymerase-1in apoptotic and necrotic neuronal death induced by mild or intense NMDA exposure in vitro. Mol Cell Neurosci. 2004;25:172–180. doi: 10.1016/j.mcn.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Mills EM, Gunasekar PG, Pavlakovic G, Isom GE. Cyanide-induced apoptosis and oxidative stress in differentiated PC12 cells. J Neurochem. 1996;67:1039–1046. doi: 10.1046/j.1471-4159.1996.67031039.x. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Stevens MV, Kohr M, Steenbergen C, Sack MN, Murphy E. Cysteine 203 of cyclophilin D is critical for cyclophilin D activation of the mitochondrial permeability transition pore. J Biol Chem. 2011;286:40184–40192. doi: 10.1074/jbc.M111.243469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknahad H, Khan S, O'Brien PJ. Hepatocyte injury resulting from the inhibition of mitochondrial respiration at low oxygen concentrations involves reductive stress and oxygen activation. Chem Biol Interact. 1995;98:27–44. doi: 10.1016/0009-2797(95)03631-u. [DOI] [PubMed] [Google Scholar]
- Owens K, Park JH, Schuh R, Kristian T. Mitochondrial dysfunction and NAD(+) metabolism alterations in the pathophysiology of acute brain injury. Transl Stroke Res. 2013;4:618–634. doi: 10.1007/s12975-013-0278-x. [DOI] [PubMed] [Google Scholar]
- Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto R, Tavazzi B, Taya K, Barrios L, Amorini AM, Di PV, Pascual JM, Marmarou A, Marmarou CR. Brain energy depletion in a rodent model of diffuse traumatic brain injury is not prevented with administration of sodium lactate. Brain Res. 2011;1404:39–49. doi: 10.1016/j.brainres.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemgens A, Singh S, Beyer C, Arnold S. Inducers of chemical hypoxia act in a gender-and brain region-specific manner on primary astrocyte viability and cytochrome C oxidase. Neurotox Res. 2011;20:1–14. doi: 10.1007/s12640-010-9213-z. [DOI] [PubMed] [Google Scholar]
- Satchell MA, Zhang X, Kochanek PM, Dixon CE, Jenkins LW, Melick J, Szabo C, Clark RS. A dual role for poly-ADP-ribosylation in spatial memory acquisition after traumatic brain injury in mice involving NAD+ depletion and ribosylation of 14-3-3gamma. J Neurochem. 2003;85:697–708. doi: 10.1046/j.1471-4159.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Shuttleworth CW. Use of NAD(P)H and flavoprotein autofluorescence transients to probe neuron and astrocyte responses to synaptic activation. Neurochem Int. 2010;56:379–386. doi: 10.1016/j.neuint.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuttleworth CW, Brennan AM, Connor JA. NAD(P)H fluorescence imaging of postsynaptic neuronal activation in murine hippocampal slices. J Neurosci. 2003;23:3196–3208. doi: 10.1523/JNEUROSCI.23-08-03196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Dawson VL. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Pieper AA, Croul SE, Zhang J, Snyder SH, Greenberg JH. Post-treatment with an inhibitor of poly(ADP-ribose) polymerase attenuates cerebral damage in focal ischemia. Brain Res. 1999;829:46–54. doi: 10.1016/s0006-8993(99)01335-9. [DOI] [PubMed] [Google Scholar]
- Vagnozzi R, Marmarou A, Tavazzi B, Signoretti S, Di Pierro D, del Bolgia F, Amorini AM, Fazzina G, Sherkat S, Lazzarino G. Changes of cerebral energy metabolism and lipid peroxidation in rats leading to mitochondrial dysfunction after diffuse brain injury. J Neurotrauma. 1999;16:903–913. doi: 10.1089/neu.1999.16.903. [DOI] [PubMed] [Google Scholar]
- Whalen MJ, Clark RS, Dixon CE, Robichaud P, Marion DW, Vagni V, Graham SH, Virag L, Hasko G, Stachlewitz R, Szabo C, Kochanek PM. Reduction of cognitive and motor deficits after traumatic brain injury in mice deficient in poly(ADP-ribose) polymerase. J Cereb Blood Flow Metab. 1999;19:835–842. doi: 10.1097/00004647-199908000-00002. [DOI] [PubMed] [Google Scholar]
- Xia W, Wang Z, Wang Q, Han J, Zhao C, Hong Y, Zeng L, Tang L, Ying W. Roles of NAD(+) / NADH and NADP(+) / NADPH in cell death. Curr Pharm Des. 2009;15:12–19. doi: 10.2174/138161209787185832. [DOI] [PubMed] [Google Scholar]
- Yavin Z, Yavin E. Survival and maturation of cerebral neurons on poly(L-lysine) surfaces in the absence of serum. Dev Biol. 1980;75:454–459. doi: 10.1016/0012-1606(80)90176-1. [DOI] [PubMed] [Google Scholar]
- Ying W. NAD+ and NADH in cellular functions and cell death. Front Biosci. 2006;11:3129–3148. doi: 10.2741/2038. [DOI] [PubMed] [Google Scholar]
- Yuan M, Siegel C, Zeng Z, Li J, Liu F, McCullough LD. Sex differences in the response to activation of the poly (ADP-ribose) polymerase pathway after experimental stroke. Exp Neurol. 2009;217:210–218. doi: 10.1016/j.expneurol.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]