Abstract
Background
The H,K-ATPase, consisting of α and β subunits, belongs to the P-type ATPase family. There are two isoforms of the α subunit, HKα1 and HKα2 encoded by different genes. The ouabain-resistant gastric HKα1-H,K-ATPase is Sch28080-sensitive. However, the colonic HKα2-H,K-ATPase from different species shows poor primary structure conservation of the HKα2 subunit between species and diverse pharmacological sensitivity to ouabain and Sch28080. This study sought to determine the contribution of each gene to functional activity and its pharmacological profile using mouse models with targeted disruption of HKα1, HKα2, or HKβ genes.
Methods
Membrane vesicles from gastric mucosa and distal colon in wild type (WT), HKα1, HKα2 or HKβ knockout (KO) mice were extracted. K-ATPase activity and pharmacological profiles were examined.
Results
The colonic H,K-ATPase demonstrated slightly greater affinity for K+ than the gastric H,K-ATPase. This K-ATPase activity was not detected in the colon of HKα2 KO, but was observed in HKβ KO with properties indistinguishable from WT. Neither ouabain nor Sch28080 had a significant effect on the WT colonic K-ATPase activity, but orthovanadate abolished this activity. Amiloride and its analogues benzamil and 5-N-ethyl-N-isopropylamiloride inhibited K-ATPase activity of HKα1-containing H,K-ATPase; the dose dependence of inhibition was similar for all three inhibitors. In contrast, the colonic HKα2-H,K-ATPase was not inhibited by these compounds.
Conclusions
These data demonstrated that the mouse colonic H,K-ATPase exhibits a ouabain- and Sch28080-insensitive, orthovanadate-sensitive K-ATPase activity. Interestingly, pharmacological studies suggested that the mouse gastric H,K-ATPase is sensitive to amiloride.
Keywords: targeted gene disruption, potassium transport, proton transport, P-type ATPase, distal colon, amiloride
1. Introduction
The H,K-ATPase is an integral membrane protein that actively transports protons (H+) and potassium (K+) ions across the plasma membrane and is important for acid-base balance and potassium homeostasis [1]. The H,K-ATPase belongs to the P-type ATPase family and consists of two essential subunits, α and β. The α subunit contains ATP and cation binding sites responsible for ATP hydrolysis and electroneutral exchange of H+ and K+. The highly glycosylated β subunit is involved in the correct packing, plasma membrane localization, and stabilization of the holoenzyme [2]. There are two isoforms of the α subunit (HKα1 and HKα2) and each exhibits a unique tissue distribution. The HKα1 isoform is expressed in the stomach, kidney and other tissues [3, 4]. The HKα1 isoform in the gastric parietal cells is the motor of the ion transport system responsible for acid secretion. The gastric H,K-ATPase is sensitive to the P-type ATPase inhibitor orthovanadate, and to the more specific inhibitors Sch28080 and omeprazole, but is insensitive to ouabain, a Na,K-ATPase inhibitor [5]. The HKα2 isoform is expressed in the distal colon, skin, kidney, prostate and uterus [6], principally expressed in distal colon epithelial cells and renal collecting duct. Colonic H,K-ATPase was initially characterized as a ouabain-insensitive activity in the rabbit descending colon epithelium membranes [7]. Biochemical studies found that the apical membranes from rat distal colon epithelial cells contain both ouabain-sensitive and ouabain-insensitive K-ATPase activities [8, 9].
HKα2 has been cloned from several species [10–13]. The mammalian interspecies difference in amino acid composition of the α2-subunit of the colonic H,K-ATPase is about 15%, in contrast to only 1–2% difference in amino acid sequence of gastric H,K-ATPase and Na,K-ATPase from different species. The properties of the colonic H,K-ATPase have been studied in several functional expression systems including Xenopus oocytes [13–18], HEK 293 cells [19–21] and baculovirus expression system [22–24]. These studies yield rather diverse results, especially regarding the pharmacological sensitivity to ouabain and Sch28080. This might be due to the different species, and functional expression systems used. Most functional expression studies have been performed with rat colonic H,K-ATPase [15, 16, 21–23]. Expression of rat HKα2 in Xenopus oocytes resulted in Sch28080-insensitive and poor ouabain-sensitive K+-activated rubidium uptake and K-ATPase activity, with IC50 values at 0.4–0.6 mM or 1 mM respectively [15, 16]. Rat HKα2 expressed in HEK293 cells resulted in a ouabain- and Sch28080-insensitive, orthovanadate-sensitive K-ATPase activity and rubidium uptake [21]. Similar result was observed with rat HKα2 expressed in baculovirus expression system [23]. However, rat HKα2 without pairing β-subunit expressed in insect Sf9 cells displayed a ouabain-resistant, Sch28080-sensitive K-ATPase activity with 18% inhibition at 0.1 mM Sch28080 [22]. Overall, the functionally expressed rat colonic H,K-ATPase is rather insensitive to the inhibition of Sch28080 and ouabain. Bufo marinus bladder H,K-ATPase expressed in Xenopus oocytes displays intermediate sensitivity to ouabain and Sch28080 with Ki of 25 μM and 230 μM respectively [13]. Expression of human HKα2 in Xenopus oocytes, HEK 293 cells and baculovirus expression system resulted in similar Sch28080 and ouabain-sensitive rubidium uptake and ATPase activity [18, 20, 24]. However, guinea pig HKα2 expressed in HEK 293 cells shows intermediate ouabain-sensitivity (IC50 value of 52 μM) but resistance to Sch28080 [19]. Thus, there is a need to characterize the pharmacological profile of the colonic H,K-ATPase in mouse, given the ability to examine the impact of single gene disruption in the intact animal.
In recent years, mouse models with targeted disruption of the HKα1, HKα2, or HKβ genes [25–27] have been generated, allowing the contribution of the specific gene to functional activity to be unambiguously deciphered. In the present study, we measured K-ATPase activity in the native tissues of wild type mice compared to the HKα1, HKα2 or HKβ knockout (KO) mice. Use of the KO mice allows the wild type enzymatic activities to be specifically linked to each gene in question. Results from our study of HKα1 knockout mice were consistent with the reported properties of the enzyme, thereby validating the assay conditions. Comparison of the K-ATPase activity between wild type and HKα2 knockout mice demonstrated that the colonic H,K-ATPase exhibits a ouabain- and Sch28080-insensitive, orthovanadate-sensitive K-ATPase activity. Interestingly, our pharmacological studies suggested that the gastric H,K-ATPase is sensitive to amiloride.
2. Materials and Methods
2.1 Animals
All animal studies were performed in accordance with the VAMC IACUC. All mice were housed and bred in the VAMC animal facility with free access to standard lab chow (Harlan) and water. Mice between 10–20 weeks were used for the experiments. For HKα1 knockout mice [27], heterozygous 129 Black Swiss mice were bred for wild type and knockout, and siblings were used for each experiment. The genotypes of the experimental mice were confirmed by polymerase chain reaction (PCR) analysis of genomic DNA extracted from the mouse tail. PCR reactions used two gene-specific primers and one neomycin resistance gene-specific primer. Primers a1.1 (5’-GCC TGT CAC TGA CAG CAA AGA GG-3’) and a1.2 (5’-GGT CTT CTG TGG TGT CCG CC-3’) amplified a 175-bp fragment from the HKα1 wild type allele. Primer a1.1 and neo-specific primer a1.3 (5’-CTG ACT AGG GGA GGA GTA GAA GG-3’) amplified a 310-bp fragment from the mutant allele. For HKα2 C57BL/6 Black 6 knockout mice [25], primers a2.1 (5’-CTG GAA TGG ACA GGC TCA ACG-3’) and a2.2 (5’-GTA CCT GAA GAG CCC CTG CTG-3’) amplified a 154-bp fragment of exon 20 from wild type allele. Primer a2.1 and neo-specific primer a1.3 amplified a 298-bp fragment from mutant allele containing portions of exon 20 and the neomycin resistance gene. For H,K-ATPase β BALB/c knockout mice [26], primers b.1 (5’-CCT CAC ACA GAG GAG ACT A-3’) and b.2 (5’-TGC CCA GTG TCC GGG TTC CA-3’) amplified a 134-bp fragment from wild type allele. Primer b.2 and the neo-specific primer b.3 (5’-ATA TTG CTG AAG AGC TTG GCG GC-3’) amplified a 650-bp fragment from the mutant allele that contains portions of exon 1 and neomycin resistance gene.
2.2 Isolation of gastric and distal colon membrane protein
Mice were anesthetized by intraperitoneal injection of sodium pentobarbital (200 mg/kg body weight) then euthanized by cervical dislocation. Epithelial cells from the stomach were removed by scraping with a glass slide then homogenized in buffer (250 mM sucrose, 2 mM MgCl2, 1 mM EGTA·Tris, 25 mM Tris·HCl, pH 7.2) containing protease inhibitors (2 μg/ml aprotinin, 2 μg/ml leupeptin, and 0.5 μM phenylmethyl sulfonyl fluoride) [28]. The homogenate was centrifuged at 1,000 × g for 10 min to remove fragmented cells and debris. The supernatant was centrifuged 15 min at 13,000 × g to pellet organelles. The resulting supernatant was further centrifuged at 100,000 × g for 1 h to pellet the plasma membrane fraction. The final pellet was homogenized in suspension buffer (2 mM MgCl2, 1 mM EGTA·Tris, 25 mM Tris·HCl, pH 7.2) containing protease inhibitors. Resuspended gastric membrane vesicles were quickly frozen by dry ice/acetone and stored at −70 °C. The concentration of isolated membrane protein was measured using the BCA Protein Assay Kit from Pierce Biotechnology, Inc. (Rockford, IL) using bovine serum albumin as a standard.
For collection of distal colonic membrane protein, the epithelial cells of the distal colon were scraped and the membrane vesicles were isolated by differential centrifugation as described above. The resuspended membrane vesicles were then permeabilized according to Forbush [29], mixed with 100 μl treatment solution (1.0 mg/ml lauryl sulfate Tris, 1% BSA, in suspension solution), incubated at 22°C for 10 min, mixed with 500 μl ice-cold suspension solution with 0.3% BSA, then stored on ice until assays were performed.
2.3 K-ATPase activity assays
K-ATPase activity was determined by measuring inorganic phosphate hydrolyzed from (γ-32P) ATP according to the protocol of Codina et al. [9] with some modifications. Briefly, in a total reaction volume of 100 μl, 0.5–1 μg membrane protein was used in the reaction solution containing 30 mM Tris·HCl, pH 7.2, 1 mM EDTA·Tris, 0.1 mM EGTA·Tris, 4 mM MgCl2, 3 mM ATP·Tris, 4.0 μg/ml oligomycin without or with 2.5 mM KCl. Gastric K-ATPase activity was measured in the presence of 1.0 mM ouabain. The reaction was initiated by the addition of 3.0 mM ATP with 1–10 × 106 cpm of (γ-32P) ATP, incubated at 37°C for 30 min, then stopped by 500 μl ice-cold 15% (W/V) charcoal. The supernatant was filtered (0.45 μM, Whatman) then analyzed on a Beckman scintillation counter. K-ATPase activity was defined as the difference between ATPase activity measured in the presence and absence of 2.5 mM KCl and expressed as micromoles of inorganic phosphate librated per milligram protein per hour.
2.4 Statistical analysis
Results were expressed as mean values ± se (standard error) from the indicated number of animals (n). Comparison of the K-ATPase activities between groups was performed by Student’s t test. A P value < 0.05 was considered statistically significant.
3. Results
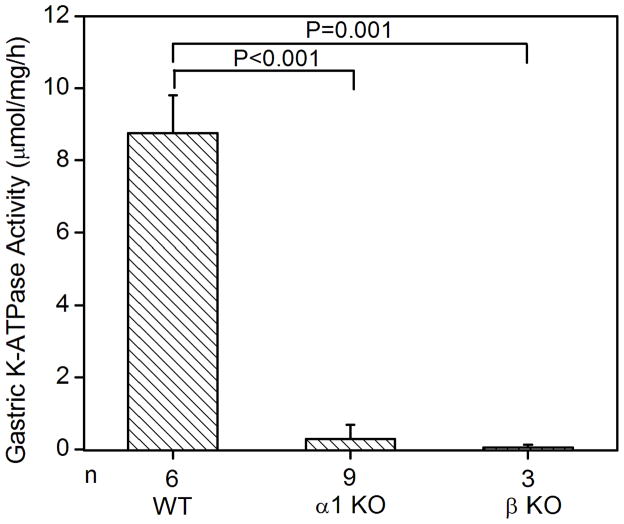
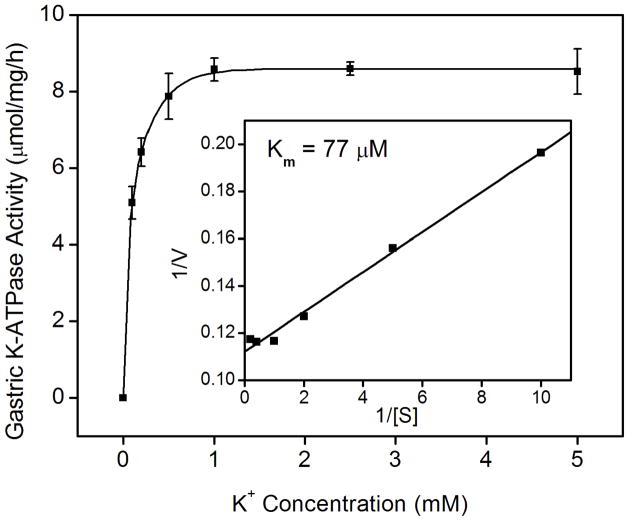
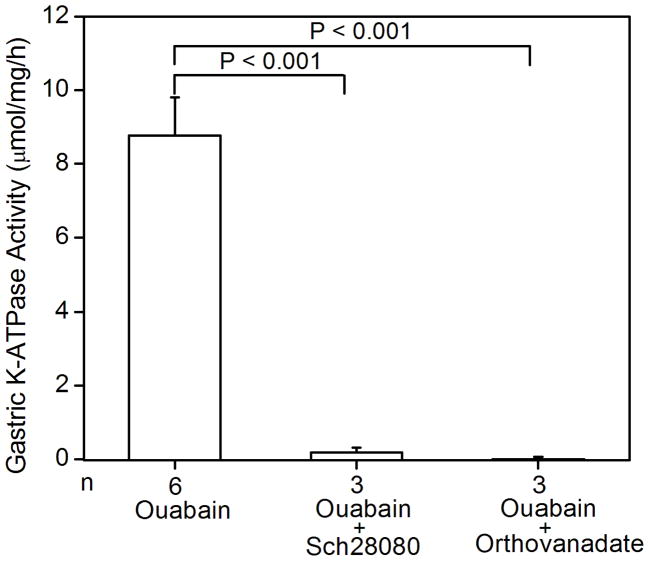
Since the K+-dependent ATP hydrolysis activity of the gastric enzyme has been well-established, wild type and HKα1 knockout mice were used to validate the K-ATPase assay. As expected, wild type murine gastric membrane vesicles exhibited ouabain-resistant, Sch28080- and orthovanadate-sensitive K-ATPase activity. The specific activity was 8.8 ± 1.0 μmol/mg/h (n = 6). Gastric membranes from HKα1 knockout animals were devoid of this K-ATPase activity. As well, the K-ATPase activity was measured in the gastric membrane vesicles from BALB/c WT (HKβ KO strain), yielding similar K-ATPase activity to that of the gastric membranes of 129 Black Swiss HKα1 WT. However, measurable K-ATPase activity was absent in the gastric membranes of HKβ knockouts (Fig. 1A). The apparent Km for K+ of the gastric H,K-ATPase was 77 μM (Fig. 1B). Additionally, we confirmed that the ouabain-resistant gastric K-ATPase activity was inhibited by both Sch28080 and orthovanadate (Fig. 1C).
Figure 1.
K-ATPase activity in the gastric membrane vesicles of mice (A) Gastric K-ATPase activity was determined in the gastric membrane vesicles of wild type (WT), HKα1 knockout (α1 KO), and HKβ knockout (β KO) mice in the presence of 1 mM ouabain. Values (in micromole ATP hydrolyzed per mg membrane protein per hour) are means ± se from the indicated number of animals (n). (B) K-dependent ATPase activity was measured in the gastric membrane vesicles extracted from HKα1 wild type mice. K-ATPase activity of mouse gastric mucosa was fully stimulated by 1.0 mM K+. Inset, double-reciprocal plot of 1/V versus 1/[S] indicates apparent Km of 77 μM for the gastric K-ATPase activity. (C) Gastric K-ATPase activity was measured in the presence of 1 mM ouabain or in the presence of ouabain plus 100 μM Sch28080, or ouabain plus 100 μM orthovanadate. Values are means ± se from the indicated number of animals (n). Comparisons between groups were performed by Student’s t-test.
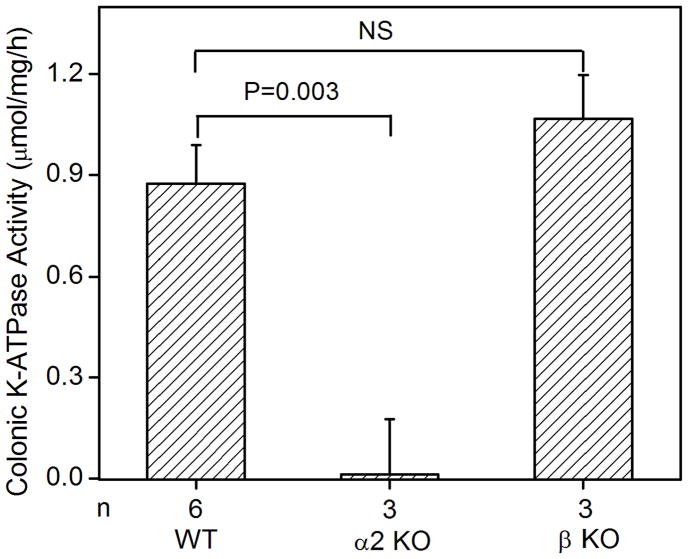
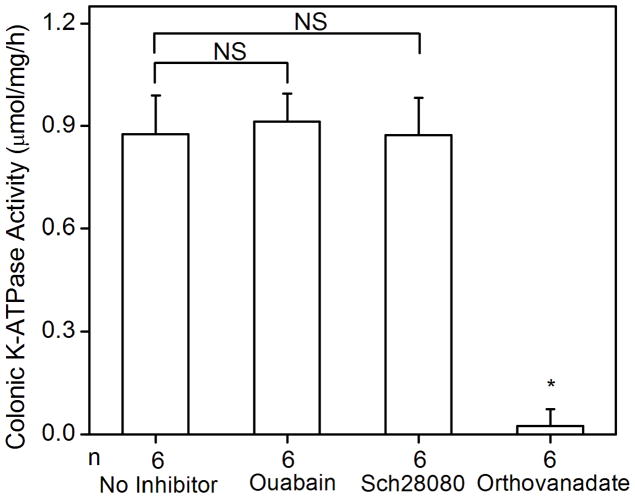
The colonic K-ATPase activity in the membrane vesicles extracted from the distal colon of wild type mice was 0.88 ± 0.11 μmol/mg/h (n = 6). K-ATPase activity of the colonic membrane was absent in the age-matched HKα2 knockout mice (Fig. 2A), which demonstrated that the K-ATPase activity measured in the wild type distal colon was directly attributable to the colonic HKα2 subunit. Since HKβ is able to support the H,K-ATPase activity when it is co-expressed with HKα2 in vitro [15, 19], we used the HKβ knockout mouse as a tool to determine the role of HKβ in the K-ATPase activity in mouse distal colon. We measured the K-ATPase activity from HKβ WT distal colon, which is statistically indistinguishable from the activity of the colonic membranes from C57BL/6 Black 6 HKα2 WT. The K-ATPase activity in the colonic membrane vesicles of HKβ knockout mice was 1.06 ± 0.13 μmol/mg/h (n = 3), which is not significantly different from the activity of the wild type (Fig. 2A). Therefore, HKβ is not essential for the K-ATPase activity in the mouse distal colon.
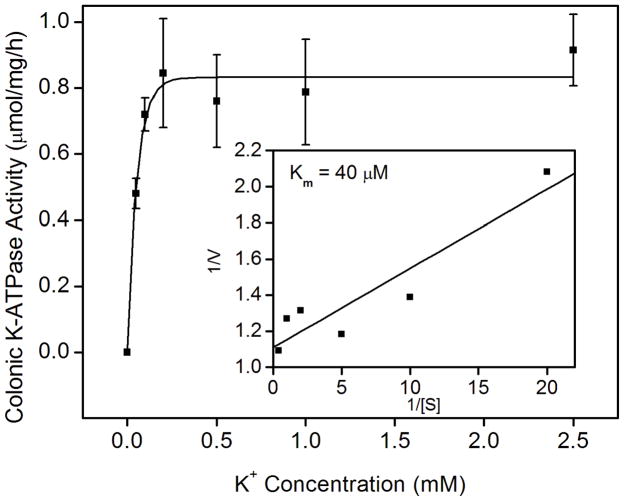
Fig. 2.
K-ATPase activity in the colonic membrane vesicles of mice. (A) Colonic K-ATPase activity was determined in the colonic membrane vesicles of HKα2 wild type (WT), HKα2 knockout (α2 KO), and HKβ knockout (β KO) mice. Values (in micromole ATP hydrolyzed per mg membrane protein per hour) are means ± se from the indicated number of animals (n). Comparisons between groups were performed by Student’s t-test. NS stands for no statistically significant difference between groups. (B) K-dependent ATPase activity was measured in the colonic membrane vesicles extracted from wild type mice. Maximal stimulation of K-ATPase activity was reached at K+ concentration of 200 μM. Inset, Lineweaver-Burk plot of 1/V versus 1/[S] indicates apparent Km of 40 μM for the colonic K-ATPase activity. (C) Colonic K-ATPase activity was measured in the absence of inhibitor or in the presence of 2 mM ouabain, or 100 μM Sch28080, or 100 μM orthovanadate. Values are means ± se from the indicated number of animals (n). * indicates P<0.001, statistically different from the colonic K-ATPase activity in the absence of inhibitor by Student’s t-test.
We further studied the activation and pharmacological properties of the colonic K-ATPase. A K+ concentration of 0.2 mM fully stimulated the colonic K-ATPase activity. The apparent Km for K+ derived from the double-reciprocal plot was 40 μM (Fig. 2B). Sch28080, a specific inhibitor of the gastric H,K-ATPase, did not inhibit the colonic K-ATPase activity. Ouabain, a specific inhibitor of Na,K-ATPase, had no effect on the colonic K-ATPase activity. On the other hand, orthovanadate completely inhibited the K-ATPase activity in distal colon membrane vesicles (Fig. 2C).
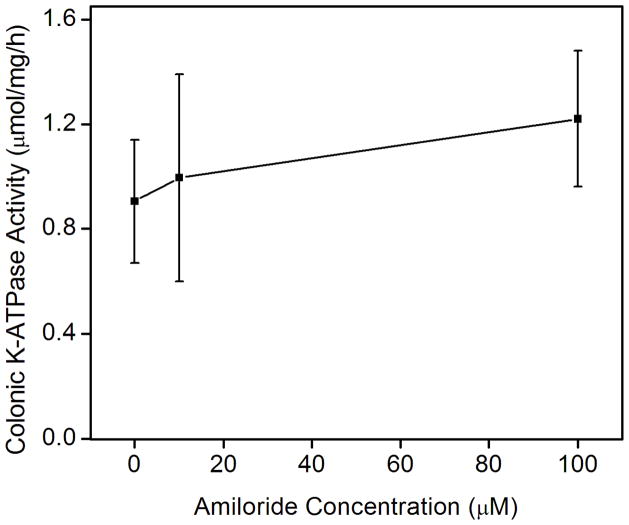
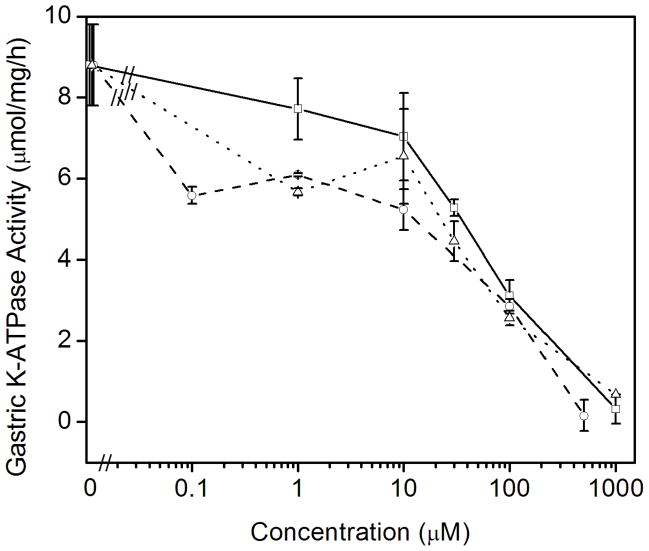
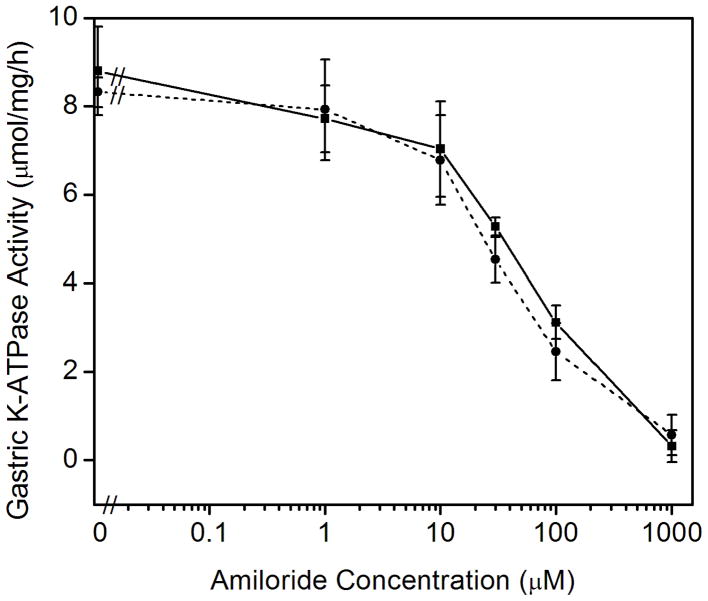
Previous data has shown that P-type ATPases bind amiloride derivatives [30]. Therefore, we tested whether amiloride or the high affinity amiloride analogues that inhibit either the epithelial Na+ channel (ENaC) benzamil or the Na-H exchangers 5-N-ethyl-N-isopropylamiloride (EIPA) could inhibit the colonic and gastric K-ATPase activities. Amiloride is routinely used for the inhibition of the sodium transport and is a potent inhibitor of sodium ion uptake: it inhibits most Na+-transport systems such as ENaC in urinary epithelia at low concentrations (< 1 μM) and the plasma membrane Na/H exchanger at higher concentrations (1–100μM). Benzamil inhibits ENaC more selectively than amiloride whereas EIPA is a more specific inhibitor of Na-H exchangers than amiloride [31]. To our surprise, we found that amiloride specifically inhibited the K-ATPase activity of the gastric, HKα1-containing H,K-ATPase. Amiloride had no effect on the K-ATPase activity of the colonic, HKα2-containing H,K-ATPase (Fig. 3A). In contrast, 100 μM amiloride caused ~65% decrease in gastric K-ATPase activity, and 1.0 mM amiloride completely inhibited the gastric K-ATPase activity. Amiloride derivatives benzamil and EIPA had similar inhibitory effects on the gastric K-ATPase activity (Fig. 3B). Nigericin, a polyether antibiotic and ionophore that permits electroneutral exchange of K+ and H+ across the membrane, was used to insure the ion permeability of the membrane vesicles. The addition of 1 μM nigericin had no effect on the inhibition of gastric K-ATPase activity by amiloride (Fig. 3C).
Fig. 3.
Effects of amiloride and derivatives on the colonic and gastric K-ATPase activity. (A) Colonic K-ATPase activity was measured as a function of the concentration of amiloride. (B) Gastric K-ATPase activity was measured as a function of the concentration of amiloride (□, solid line), or benzamil (○, dashed line), or EIPA (△, dotted line). (C) K-ATPase activity was measured as a function of the concentration of amiloride in the absence (■, solid line) or presence of 1 μM nigericin (●, short dashed line). Values are means ± se of 3 animals.
4. Discussion
By comparing the colonic K-ATPase activity in the native tissue of the wild type and knockout mice of HKα2, we demonstrated that HKα2 is responsible for colonic K-ATPase activity and that the mouse colonic K-ATPase activity was ouabain and Sch28080-insensitive. However, earlier studies obtained from the functional expression systems with HKα2 from different species demonstrate different degrees of ouabain-sensitivity, for example, human nongastric H,K-ATPase is sensitive to ouabain inhibition with Ki of ~13 μM [18] and a similar sensitivity was observed in Bufo marinus [13]. The ouabain sensitivity of the rat colonic H,K-ATPase from different studies is comparable, in the range of 0.4–1 mM, whereas in some studies 1 mM does not inhibit the activity. Sch28080 usually does not inhibit the rat colonic H,K-ATPase. The amino acid identity of colonic H,K-ATPase between species is more divergent than that of gastric H,K-ATPase and Na,K-ATPase, which could contribute to its wide range of sensitivity to ouabain inhibition. The mouse variant is very similar to the rat in amino acid composition and may have similar properties.
Additionally, it is possible that there is a difference between the biochemical properties of membrane vesicle preparations from the native cells and oocytes/insect cell expression systems. We used membrane fractions purified from mouse native tissue and showed that mouse colonic H,K-ATPase is ouabain and Sch28080-insensitive. The colonic pump was initially characterized as a ouabain-insensitive H,K-ATPase in the rabbit descending colon epithelium membranes [7]; a both ouabain- and Sch28080-insensitive Na+-independent K-ATPase activity was identified in the apical membrane of rat distal colon [9]. As well, rat colonic H,K-ATPase extracted from baculovirus expression system exhibited very low affinity for ouabain (IC50 = 4 mM) in the presence of K+, but was sensitive to ouabain inhibition (IC50 = 0.05 mM) in the presence of Na+ [23]. By comparison, in the present study, we examined the differential properties of H,K-ATPase from native membrane vesicles of wild type and knockout animals, and demonstrated that mouse colonic H,K-ATPase is not sensitive to ouabain inhibition in the presence of K+. The native membrane vesicles have a heterogeneous collection of ATPases including H,K-ATPase and Na,K-ATPase, and ouabain is a poor inhibitor of rodent Na,K-ATPase. Thus there are limits for using native membrane vesicles to clearly define the ouabain-sensitivity of mouse colonic H,K-ATPase in the presence of Na+. The ouabain-sensitivity of mouse colonic H,K-ATPase could be determined under different ion conditions using an over-expression system such as the baculovirus expression system, provided that the lipid properties of the expression cell system have a composition similar to the cells that natively express the enzyme.
Recently, our laboratory has found that ouabain inhibits the HKα2 containing H,K-ATPase as determined by studies in isolated perfused tubule in wild type and knockout mice [32]. Dherbecourt et al. also identified that HKα2 is responsible for a ouabain- and Sch28080-sensitive K-ATPase activity in renal collecting duct using wild type and knockout mice fed a potassium-depleted diet [33]. The explanation for the difference in the pharmacological profiles of the K-ATPase activity in mouse distal colon versus the renal collecting duct is presently unclear. Although dietary K-depletion upregulates renal H,K-ATPase, K-depletion has no effect on colonic H,K-ATPase [34–36]. Therefore, the colonic and renal H,K-ATPase have different regulatory mechanisms and likely have different functions.
Kinetic studies showed that the apparent Km for K+ for the gastric H,K-ATPase is 77 μM and is 40 μM for the colonic H,K-ATPase, indicating that colonic and gastric H,K-ATPase have moderate differences in sensitivity to K+ stimulation. However, a lower affinity for K+ with Km around 1mM was reported for rat colonic H,K-ATPase [15, 16, 21–23]. It is worth noting that Km studies of the rat colonic H,K-ATPase were performed in the functional expression systems. In the other hand, we measured the Km for K+ of the colonic H,K-ATPase with membrane vesicles from mouse native tissue, which has the advantage of examining the enzyme where the necessity of ancillary proteins or full maturation of the holoenzyme complex in vivo is not an issue. Similar differences in Km for K+ were reported for the colonic H,K-ATPase from guinea pig as well. When it is expressed in HEK-293 cells, the guinea pig distal colon H,K-ATPase has a Km value of 0.68 mM [19], which is much higher than the 0.055 mM Km for K+ reported for the colonic H,K-ATPase from guinea pig colonic epithelia cells [37]. One explanation for this discrepancy is that the β-subunit used in the functional expression systems (either gastric HKβ or NaKβ) associated with the colonic H,K-α subunit may have conferred different affinity for cations. The colonic H,K-ATPase α-subunit is likely to associate with its endogenous β-subunit in the colon, resulting in higher affinity for K+. Data from other native tissues also showed relatively high affinity to K+, for examples, a value of 0.2 mM was reported by using the microsomes from native tissue [38] and microdissected segments of rabbit nephron [39]. Oligomycin was used in our assay solution to inhibit background ATPase activity. However, Swarts et al. reported that oligomycin inhibits the rat colonic H,K-ATPase expressed in sf9 cells [23]. If the similar oligomycin inhibition of the enzyme activity occurs in the mouse native tissue then the use of oligomycin in the assay might influence the value of the K-ATPase activity measured and K+ affinity determined.
We detected the K-ATPase activity in the colonic membrane vesicles of HKβ knockout mice. Although HKβ and NaKβ1 are equally capable of supporting K-ATPase activity when they are co-expressed with HKα2 in vitro [15, 19], the presence of colonic K-ATPase activity in HKβ knockout mice demonstrates that the HKβ subunit is not necessary for the colonic K-ATPase activity in the mouse distal colon. Our data support the association of HKα2 with a β-subunit other than HKβ subunit. NaKβ1 is the most likely candidate since immunoprecipitation and immunohistochemistry experiments demonstrated that HKα2 assembled with NaKβ1 in rat distal colon, renal medulla, and prostate [40–42]. In fact, coexpression of rat HKα2 and NaKβ1 in HEK293 cells yielded a ouabain- and Sch28080-insensitive, orthovanadate-sensitive K-ATPase activity and rubidium uptake [21].
The loss of the ouabain-resistant and Sch28080-sensitive K-ATPase enzymatic activity in HKα1 and HKβ knockout mice accounts for their achlorhydria phenotype [26, 27]. An amiloride concentration of up to 1 mM is widely used for the inhibition of sodium transport by NHE3, the predominant Na-H exchanger at the apical membrane of the proximal tubule [31]. Our data suggest that in our preparation the inhibition of gastric K-ATPase activity by amiloride was not caused by the change in the membrane potential since amiloride inhibited gastric K-ATPase activity in the presence of nigericin as well. This observation is consistent with the finding of Ganser and Forte that nigericin has no additional stimulatory effect on the K-dependent portion of ATPase activity in bullfrog oxyntic cell microsomes [43]. Our finding that the inhibition of gastric H,K-ATPase activity by amiloride, benzamil, and EIPA with similar dose dependence supports the hypothesis that amiloride inhibits HKα1 by a mechanism that is not dependent on inhibition of ENaC or a Na-H exchanger. Although amiloride inhibits Na+-transport in the mammalian stomach [44], a previous study has shown that amiloride treatment has no effect on collecting duct H,K-ATPase activity during rat distal renal tubular acidosis [45]. Whether this reflects the small abundance of this enzyme under “normal” conditions will require further study. However, a photolabile amiloride derivative – 5-(N-ethyl-[2-methoxy-4-nitrobenzyl]) amiloride (NENMBA) was reported to inhibit purified canine Na,K-ATPase activity through its binding and disruption of the cation transport domain of the α-subunit of the Na,K-ATPase [30]. Given the observations of Swarts and coworkers [46], it is possible that amiloride inhibits the gastric H,K-ATPase by a similar mechanism.
In conclusion, we used genetically engineered HKα2 knockout mice to demonstrate that an orthovanadate-sensitive, ouabain- and Sch28080-resistant K-ATPase activity in the mouse distal colon was encoded by HKα2. Amiloride inhibited the gastric but not the colonic H,K-ATPase. The gastric H,K-ATPase β subunit is not essential for the K-ATPase activity of HKα2 in mouse distal colon. The characterization of the pharmacological properties of the mouse gastric and colonic H,K-ATPases presented here will be important for the planning and interpretation of future studies concerning these enzymes in knockout mouse models.
Acknowledgments
The authors wish to thank Alicia Rudin and Jeannette Lynch for their assistance in the animal studies. This work was supported by the Department of Veterans Affairs and the National Institutes of Health, Grant DK-49750.
Footnotes
General Significance: Characterization of the pharmacological profiles of the H,K-ATPases is useful for understanding the relevant knockout animals and for considering the specificity of the inhibitors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gumz ML, Lynch IJ, Greenlee MM, Cain BD, Wingo CS. The Renal H+, K+ ATPases: Physiology, Regulation, and Structure. Am J Physiol Renal Physiol. 2009 doi: 10.1152/ajprenal.90723.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano S, Morii M, Takeguchi N. Molecular and cellular regulation of the gastric proton pump. Biol Pharm Bull. 2004;27:1–12. doi: 10.1248/bpb.27.1. [DOI] [PubMed] [Google Scholar]
- 3.Lecain E, Robert JC, Thomas A, Tran Ba Huy P. Gastric proton pump is expressed in the inner ear and choroid plexus of the rat. Hear Res. 2000;149:147–154. doi: 10.1016/s0378-5955(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 4.Shull GE, Lingrel JB. Molecular cloning of the rat stomach (H+ + K+)-ATPase. The Journal of Biological Chemistry. 1986;261:16788–16791. [PubMed] [Google Scholar]
- 5.Hersey SJ, Sachs G. Gastric acid secretion. Physiol Rev. 1995;75:155–189. doi: 10.1152/physrev.1995.75.1.155. [DOI] [PubMed] [Google Scholar]
- 6.Jaisser F, Beggah AT. The nongastric H+-K+-ATPases: molecular and functional properties. Am J Physiol. 1999;276:F812–F824. doi: 10.1152/ajprenal.1999.276.6.F812. [DOI] [PubMed] [Google Scholar]
- 7.Gustin MC, Goodman DB. Isolation of brush-border membrane from the rabbit descending colon epithelium. Partial characterization of a unique K+-activated ATPase. The Journal of Biological Chemistry. 1981;256:10651–10656. [PubMed] [Google Scholar]
- 8.Del Castillo JR, Rajendran VM, Binder HJ. Apical membrane localization of ouabain-sensitive K(+)-activated ATPase activities in rat distal colon. Am J Physiol. 1991;261:G1005–G1011. doi: 10.1152/ajpgi.1991.261.6.G1005. [DOI] [PubMed] [Google Scholar]
- 9.Codina J, Pressley TA, DuBose TD., Jr The colonic H+,K+-ATPase functions as a Na+-dependent K+(NH4+)-ATPase in apical membranes from rat distal colon. The Journal of Biological Chemistry. 1999;274:19693–19698. doi: 10.1074/jbc.274.28.19693. [DOI] [PubMed] [Google Scholar]
- 10.Crowson MS, Shull GE. Isolation and characterization of a cDNA encoding the putative distal colon H+,K(+)-ATPase. Similarity of deduced amino acid sequence to gastric H+,K(+)-ATPase and Na+,K(+)-ATPase and mRNA expression in distal colon, kidney, and uterus. The Journal of Biological Chemistry. 1992;267:13740–13748. [PubMed] [Google Scholar]
- 11.Fejes-Toth G, Rusvai E, Longo KA, Naray-Fejes-Toth A. Expression of colonic H-K-ATPase mRNA in cortical collecting duct: regulation by acid/base balance. Am J Physiol. 1995;269:F551–F557. doi: 10.1152/ajprenal.1995.269.4.F551. [DOI] [PubMed] [Google Scholar]
- 12.Grishin AV, Sverdlov VE, Kostina MB, Modyanov NN. Cloning and characterization of the entire cDNA encoded by ATP1AL1--a member of the human Na,K/H,K-ATPase gene family. FEBS Lett. 1994;349:144–150. doi: 10.1016/0014-5793(94)00655-5. [DOI] [PubMed] [Google Scholar]
- 13.Jaisser F, Horisberger JD, Geering K, Rossier BC. Mechanisms of urinary K+ and H+ excretion: primary structure and functional expression of a novel H,K-ATPase. J Cell Biol. 1993;123:1421–1429. doi: 10.1083/jcb.123.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnay M, Crambert G, Kharoubi-Hess S, Geering K, Horisberger JD. Electrogenicity of Na,K- and H,K-ATPase activity and presence of a positively charged amino acid in the fifth transmembrane segment. The Journal of Biological Chemistry. 2003;278:19237–19244. doi: 10.1074/jbc.M300946200. [DOI] [PubMed] [Google Scholar]
- 15.Codina J, Kone BC, Delmas-Mata JT, DuBose TD., Jr Functional expression of the colonic H+,K+-ATPase alpha-subunit. Pharmacologic properties and assembly with X+,K+-ATPase beta-subunits. The Journal of Biological Chemistry. 1996;271:29759–29763. doi: 10.1074/jbc.271.47.29759. [DOI] [PubMed] [Google Scholar]
- 16.Cougnon M, Planelles G, Crowson MS, Shull GE, Rossier BC, Jaisser F. The rat distal colon P-ATPase alpha subunit encodes a ouabain-sensitive H+, K+-ATPase. The Journal of Biological Chemistry. 1996;271:7277–7280. doi: 10.1074/jbc.271.13.7277. [DOI] [PubMed] [Google Scholar]
- 17.Crambert G, Horisberger JD, Modyanov NN, Geering K. Human nongastric H+-K+-ATPase: transport properties of ATP1al1 assembled with different beta-subunits. Am J Physiol. 2002;283:C305–C314. doi: 10.1152/ajpcell.00590.2001. [DOI] [PubMed] [Google Scholar]
- 18.Modyanov NN, Mathews PM, Grishin AV, Beguin P, Beggah AT, Rossier BC, Horisberger JD, Geering K. Human ATP1AL1 gene encodes a ouabain-sensitive H-K-ATPase. Am J Physiol. 1995;269:C992–C997. doi: 10.1152/ajpcell.1995.269.4.C992. [DOI] [PubMed] [Google Scholar]
- 19.Asano S, Hoshina S, Nakaie Y, Watanabe T, Sato M, Suzuki Y, Takeguchi N. Functional expression of putative H+-K+-ATPase from guinea pig distal colon. Am J Physiol. 1998;275:C669–C674. doi: 10.1152/ajpcell.1998.275.3.C669. [DOI] [PubMed] [Google Scholar]
- 20.Grishin AV, Bevensee MO, Modyanov NN, Rajendran V, Boron WF, Caplan MJ. Functional expression of the cDNA encoded by the human ATP1AL1 gene. Am J Physiol. 1996;271:F539–F551. doi: 10.1152/ajprenal.1996.271.3.F539. [DOI] [PubMed] [Google Scholar]
- 21.Sangan P, Thevananther S, Sangan S, Rajendran VM, Binder HJ. Colonic H-K-ATPase alpha- and beta-subunits express ouabain-insensitive H-K-ATPase. Am J Physiol. 2000;278:C182–C189. doi: 10.1152/ajpcell.2000.278.1.C182. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Rajendran VM, Mann AS, Kashgarian M, Binder HJ. Functional expression and segmental localization of rat colonic K-adenosine triphosphatase. J Clin Invest. 1995;96:2002–2008. doi: 10.1172/JCI118247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swarts HGP, Koenderink JB, Willems PHGM, De Pont JJHHM. The non-gastric H,K-ATPase is oligomycin-sensitive and can function as an H+,NH4(+)-ATPase. The Journal of Biological Chemistry. 2005;280:33115–33122. doi: 10.1074/jbc.M504535200. [DOI] [PubMed] [Google Scholar]
- 24.Swarts HGP, Koenderink JB, Willems PHGM, De Pont JJHHM. The human non-gastric H,K-ATPase has a different cation specificity than the rat enzyme. Biochim Biophys Acta. 2007;1768:580–589. doi: 10.1016/j.bbamem.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Meneton P, Schultheis PJ, Greeb J, Nieman ML, Liu LH, Clarke LL, Duffy JJ, Doetschman T, Lorenz JN, Shull GE. Increased sensitivity to K+ deprivation in colonic H,K-ATPase-deficient mice. J Clin Invest. 1998;101:536–542. doi: 10.1172/JCI1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scarff KL, Judd LM, Toh BH, Gleeson PA, Van Driel IR. Gastric H(+),K(+)-adenosine triphosphatase beta subunit is required for normal function, development, and membrane structure of mouse parietal cells. Gastroenterology. 1999;117:605–618. doi: 10.1016/s0016-5085(99)70453-1. [DOI] [PubMed] [Google Scholar]
- 27.Spicer Z, Miller ML, Andringa A, Riddle TM, Duffy JJ, Doetschman T, Shull GE. Stomachs of mice lacking the gastric H,K-ATPase alpha -subunit have achlorhydria, abnormal parietal cells, and ciliated metaplasia. The Journal of Biological Chemistry. 2000;275:21555–21565. doi: 10.1074/jbc.M001558200. [DOI] [PubMed] [Google Scholar]
- 28.Reenstra WW, Forte JG. Isolation of H+,K(+)-ATPase-containing membranes from the gastric oxyntic cell. Methods Enzymol. 1990;192:151–165. doi: 10.1016/0076-6879(90)92068-o. [DOI] [PubMed] [Google Scholar]
- 29.Forbush B., 3rd Assay of Na,K-ATPase in plasma membrane preparations: increasing the permeability of membrane vesicles using sodium dodecyl sulfate buffered with bovine serum albumin. Anal Biochem. 1983;128:159–163. doi: 10.1016/0003-2697(83)90356-1. [DOI] [PubMed] [Google Scholar]
- 30.Ellis-Davies GC, Kleyman TR, Kaplan JH. Photolabile amiloride derivatives as cation site probes of the Na,K-ATPase. The Journal of Biological Chemistry. 1996;271:10353–10358. doi: 10.1074/jbc.271.17.10353. [DOI] [PubMed] [Google Scholar]
- 31.Kleyman TR, Cragoe EJ., Jr Cation transport probes: the amiloride series. Methods Enzymol. 1990;191:739–755. doi: 10.1016/0076-6879(90)91045-8. [DOI] [PubMed] [Google Scholar]
- 32.Lynch IJ, Greenlee MM, Gumz ML, Rudin A, Xia SL, Wingo CS. Heterogeneity of H-K-ATPase-mediated acid secretion along the mouse collecting duct. American Journal of Physiology. 2010;298:F408–415. doi: 10.1152/ajprenal.00333.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dherbecourt O, Cheval L, Bloch-Faure M, Meneton P, Doucet A. Molecular identification of Sch28080-sensitive K-ATPase activities in the mouse kidney. Pflugers Arch. 2006;451:769–775. doi: 10.1007/s00424-005-1508-1. [DOI] [PubMed] [Google Scholar]
- 34.Codina J, Delmas-Mata JT, DuBose TD., Jr Expression of HKalpha2 protein is increased selectively in renal medulla by chronic hypokalemia. Am J Physiol. 1998;275:F433–F440. doi: 10.1152/ajprenal.1998.275.3.F433. [DOI] [PubMed] [Google Scholar]
- 35.Jaisser F, Escoubet B, Coutry N, Eugene E, Bonvalet JP, Farman N. Differential regulation of putative K(+)-ATPase by low-K+ diet and corticosteroids in rat distal colon and kidney. Am J Physiol. 1996;270:C679–C687. doi: 10.1152/ajpcell.1996.270.2.C679. [DOI] [PubMed] [Google Scholar]
- 36.Sangan P, Rajendran VM, Mann AS, Kashgarian M, Binder HJ. Regulation of colonic H-K-ATPase in large intestine and kidney by dietary Na depletion and dietary K depletion. Am J Physiol. 1997;272:C685–C696. doi: 10.1152/ajpcell.1997.272.2.C685. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, Suzuki T, Suzuki Y. Ouabain-sensitive K(+)-ATPase in epithelial cells from guinea pig distal colon. Am J Physiol. 1990;258:G506–G511. doi: 10.1152/ajpgi.1990.258.4.G506. [DOI] [PubMed] [Google Scholar]
- 38.Forte JG, Ganser A, Beesley R, Forte TM. Unique enzymes of purified microsomes from pig fundic mucosa. K+-stimulated adenosine triphosphatase and K+-stimulated pNPPase. Gastroenterology. 1975;69:175–189. [PubMed] [Google Scholar]
- 39.Doucet A, Marsy S. Characterization of K-ATPase activity in distal nephron: stimulation by potassium depletion. Am J Physiol. 1987;253:F418–F423. doi: 10.1152/ajprenal.1987.253.3.F418. [DOI] [PubMed] [Google Scholar]
- 40.Codina J, Delmas-Mata JT, DuBose TD., Jr The alpha-subunit of the colonic H+,K+-ATPase assembles with beta1-Na+,K+-ATPase in kidney and distal colon. The Journal of Biological Chemistry. 1998;273:7894–7899. doi: 10.1074/jbc.273.14.7894. [DOI] [PubMed] [Google Scholar]
- 41.Kraut JA, Hiura J, Shin JM, Smolka A, Sachs G, Scott D. The Na(+)-K(+)-ATPase beta 1 subunit is associated with the HK alpha 2 protein in the rat kidney. Kidney Int. 1998;53:958–962. doi: 10.1111/j.1523-1755.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 42.Pestov NB, Korneenko TV, Radkov R, Zhao H, Shakhparonov MI, Modyanov NN. Identification of the beta-subunit for nongastric H-K-ATPase in rat anterior prostate. Am J Physiol. 2004;286:C1229–C1237. doi: 10.1152/ajpcell.00393.2003. [DOI] [PubMed] [Google Scholar]
- 43.Ganser AL, Forte JG. Ionophoretic stimulation of K+-ATPase of oxyntic cell microsomes. Biochem Biophys Res Commun. 1973;54:690–696. doi: 10.1016/0006-291x(73)91478-2. [DOI] [PubMed] [Google Scholar]
- 44.Machen TE, Silen W, Forte JG. Na+ transport by mammalian stomach. Am J Physiol. 1978;234:E228–E235. doi: 10.1152/ajpendo.1978.234.3.E228. [DOI] [PubMed] [Google Scholar]
- 45.Eiam-Ong S, Dafnis E, Spohn M, Kurtzman NA, Sabatini S. H-K-ATPase in distal renal tubular acidosis: urinary tract obstruction, lithium, and amiloride. Am J Physiol. 1993;265:F875–F880. doi: 10.1152/ajprenal.1993.265.6.F875. [DOI] [PubMed] [Google Scholar]
- 46.Swarts HG, Klaassen CH, Schuurmans Stekhoven FM, De Pont JJ. Sodium acts as a potassium analog on gastric H,K-ATPase. The Journal of Biological Chemistry. 1995;270:7890–7895. doi: 10.1074/jbc.270.14.7890. [DOI] [PubMed] [Google Scholar]