Abstract
While Notch signaling plays a critical role in the regulation of cartilage formation, its downstream targets are unknown. To address this we performed gain and losses of function experiments and demonstrate that Notch inhibition of chondrogenesis acts via up-regulation of the transcription factor Twist1. Upon Notch activation, murine limb bud mesenchymal progenitor cells in micromass culture displayed an inhibition of chondrogenesis. Twist1 was found to be exclusively expressed in mesenchymal progenitor cells at the onset stage of chondrogenesis during Notch activation. Inhibition of Notch signaling in these cells significantly reduced protein expression of Twist1. Furthermore, the inhibition effect of NICD1 on MPC chondrogenesis was markedly reduced by knocking down of Twist1. Constitutively active Notch signaling significantly enhanced Twist1 promoter activity; whereas mutation studies indicated that a putative NICD/RBPjK binding element in the promoter region is required for the Notch-responsiveness of the Twist1 promoter. Finally, chromatin immunoprecipitation assays further confirmed that the Notch intracellular domain influences Twist1 by directly binding to the Twist1 promoter. These data provide a novel insight into understanding the molecular mechanisms behind Notch inhibition of the onset of chondrogenesis.
Keywords: Notch signaling, Twist1, Chondrogenesis, Mesenchymal progenitor cell
1. Introduction
Osteoarthritis (OA), the most common form of joint disease, is characterized by the destruction of articular cartilage brought on by increased degradation of the extracellular matrix (ECM) and accelerated chondrocyte differentiation and cell death (el Gelse et al., 2012). Restoration of articular cartilage by enhancing mesenchymal progenitor cells (MPCs) in vivo chondrogenesis may have therapeutic potential in both rheumatoid arthritis (RA) and OA.
Notch signaling is essential for many developmental processes, including skeletogenesis. In mammals, Notch signaling is initiated when the ligands Jagged1, or 2 or Delta-like 1, 3, or 4 bind to the single-pass transmembrane cell surface Notch receptors (Notch1–4) of neighboring cells. This interaction induces cleavage of the Notch receptor, via the gamma-secretase complex. Once cleaved, the Notch intracellular domain (NICD) is released into the cytoplasm activating canonical and non-canonical Notch signaling mechanisms (Blaumueller et al., 1997). During canonical Notch signaling the NICD translocates to the nucleus and binds the transcriptional repressor, RBPjκ, converting it into an activator. Coactivators such as Mastermind-like (MAML) proteins are recruited forming a complex that induces the expression of downstream target genes. These genes include specific members of the Hes/Hey family of basic helix-loop-helix transcription factors: Hes1, Hes5, Hes7, Hey1, Hey2, and HeyL (Hsieh et al., 1999; Kopan and Ilagan, 2009; Steidl et al., 2000; Wu et al., 2000). Recently, Hilton et al. demonstrated that “upstream” components of the Notch pathway (PS1/PS2 and N1/N2) were critical in regulating osteoblastic differentiation of bone marrow-derived MPCs in mice (Hilton et al., 2008). In addition, our previous studies demonstrated that Notch signaling suppresses limb bud-derived MPC differentiation toward the chondrocyte, osteoblast, and adipocyte fates, while promoting MPC proliferation during skeletal development and homeostasis (Dong et al., 2010; Kohn et al., 2012). However, the mechanisms underlying the regulation of early chondrogenesis via the Notch/RBPjκ-pathway are still unknown.
Similar to Notch signaling, Twist1, a basic helix-loop-helix transcription factor, also plays important roles in major developmental events such as gastrulation, neural crest migration, and bone formation (Thisse et al., 1987). In mammals, Twist1 is developmentally expressed in mesoderm-derived embryonic tissues and postnatally in adult mesoderm-derived mesenchymal stem cells, where it functions as a major regulator of mesenchymal cell differentiation (Fuchtbauer, 1995). Consistent with its critical role in mesoderm development, mutations that inactivate the Twist1 gene elicit the autosomal dominant inherited disorder Saethre–Chotzen syndrome. This disorder is characterized by a broad range of congenital anomalies, including short stature, craniosynotstoses, high forehead, ptosis, small ears, and maxillary hypoplasia (Ghouzzi et al., 1997; Reardon and Winter, 1994). Although Twist1 has previously been shown to function as an inhibitor and target gene of Wnt signaling during chondrogenesis and chondrocyte differentiation (Dong et al., 2007), it has yet to be shown whether Twist1 plays a role during Notch signaling mediated chondrogenesis.
To further elucidate Notch signaling and its downstream targets that control MPC differentiation and chondrogenesis, we studied MPC chondrogenic differentiation using micromass cultured limb-bud cells. In this model, we found that Notch activation represses early chondrogenesis and chondrogenic gene expression. More importantly, the transcriptional factor, Twist1, was identified as an important target of canonical Notch signaling that contributes to the repression of chondrogenesis.
2. Materials and methods
2.1. Isolation and culture of limb bud MPCs
Embryos were harvested from CD1 pregnant mice at stage embryonic day 11.5 (E11.5) with approval from the animal medical ethics committees of China Medical University. Briefly, pregnant mice were sacrificed by CO2 followed by cervical dislocation. After removal of the uterus, embryos were isolated using a dissecting microscope (Olympus SZX12) and rinsed with sterile ice-cold phosphate buffered saline (PBS). Limb bud-derived MPCs were further isolated as previously described (Wang et al., 2005). Briefly, forelimbs were digested with 1 U/ml dispase for 3–4 h at 37 °C with continuous rotation at 70 rpm using a reciprocal shaking bath. Cells were then filtered through a 40-µm strainer before being re-suspended in 40% DMEM/60% F12 media supplemented with 10% fetal bovine serum (FBS) and antibiotics (Life Technologies, Grand Island, NY, USA). Cells were seeded in micromass at a high-density of 1.0 × 105 cells per 10 µl of media in 12-well plates. The cells were maintained in culture for 2 h before being treated with 10 µM DAPT daily or infected with lentivirus particles. Samples were collected as described at various time points (6 h, 1, 2, 3, 5, and 7 days).
2.2. Construction of the mouse Twist1 promoter-driven luciferase reporter constructs
The full-length mouse Twist1 promoter was amplified from the genomic Twist1 plasmid to contain the promoter region up to nucleotides −2.0 kb using forward primer 5′-TGCCACGTTTTGTTTCCAAG-3′ and reverse primer 5′-AGGTGTCTGAGAGTTGGGC-3′. The PCR product was digested with NheI and HindIII and subsequently cloned into the pGL-3 basic firefly luciferase reporter vector (Promega), and positive clone was designated Twist-luc. To generate the Twist −1432/−1426 mutant, forward primer 5′-AAGGTCCTCAGCTTG TTAACAGGGCAGGAGTTCGAGACGC-3′ and reverse primer 5′-GCGTCTCGAACTCCTGCCCTGTTAACAAGCTGAGGACCTT-3′ were used. Site-directed mutagenesis was done using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s recommendations as previously described (Dong et al., 2006). All constructs were subjected to DNA sequencing to confirm no PCR-generated artifacts.
2.3. Lentivirus-mediated cell infection
cDNAs encoding mouse Notch1-NICD was cloned into the EF.v-CMV lentiviral vector and sequence verified. Lentivirus production and concentration was performed as previously described (Yu et al., 2003). Briefly, VSV.G-pseudotyped recombinant lentiviruses were produced by transient transfection of the transducing vector into 293T cells, along with two packaging vectors: pMD.G, a VSV.G envelope-expressing plasmid, and pCMVΔR8.91 (Invitrogen, Carlsbad, CA, USA), containing the HIV-1 gag/pol, tat and rev genes (1.5 µg:2.0 µg:0.5 µg ratio of these three vectors). Viral supernatants were collected at 24, 48 and 72 h after transfection, and concentrated using filtration columns (Centricon Plus-20, molecular weight cutoff = 100 kDa; Millipore, Bedford, MA, USA). For lentiviral infection, 10 µl high-density (10,000 cell/µl) MPCs were seeded in 12-well plates and incubated for 2 h at 37 °C prior to be added with NICD1 lentivirus condition medium in the presence of 8 µg/ml polybrene. Twist1 shRNA lentiviral particles were obtained from Sigma-Aldrich (SHCLNV-NM_011658). GFP-lentivirus was used in this experiment as a control. The infected micromass cultures were harvested at various time points (1, 2, 3, 5, and 7 days).
2.4. Chondrocyte nodule detection
Alcian blue staining was used to detect chondrocyte nodule formation after 3, 5, and 7 days of culture. Cells in micromass culture were rinsed with PBS and fixed in 10% formaldehyde in PBS for 20 min. Cultures were washed with water three times and stained in 1% Alcian blue in 3% glacial acetic acid for 2 h. Cultures were destained in 70% ethanol two times and stored in water for image capture.
2.5. Immunofluorescence analysis
For immunofluorescence, limb bud cells were plated at 1000 cells/cm2 on coverslips and grown for 6 h after transfection with 3XFlag NICD1 expression plasmid (Addgene, Cambridge MA, USA). PUC19 empty vector was used as control plasmids. Cells were then fixed in 4% paraformaldehyde in PBS for 20 min at room temperature and permeabilized with 0.3% Triton X-100 in PBS for 30 min. Cells were washed in PBS and incubated with 0.5% bovine serum albumin (BSA) dissolved in PBS at room temperature for 20 min. Cultures were then incubated for 2 h at room temperature with the following primary antibodies: rabbit anti-Twist1 (sc-15393, Santa Cruz Biotechnology, CA, USA) diluted 1:50 in PBS. After washing with PBS, cells were incubated with FITC conjugated anti-rabbit IgG for 1 h at room temperature. Reaction controls were performed using a non-immune rabbit immunoglobulin IgG, or by excluding the primary antibody. Cover slips were mounted on slides with PBS/glycerol (1:1). Slides were imaged using fluorescent microscopy.
2.6. Transfection and luciferase assay
The MPC cells were prepared in 12-well plates in triplicate and co-transfected with Twist1-luc reporter plasmids (500 ng/well) and 3XFlag NICD1 expression plasmid by utilizing Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). After 48-h incubation, the cells were washed twice with ice-cold PBS and harvested with a reporter lysis buffer (Promega, Madison, WI). The luciferase activity was analyzed using the dual-luciferase assay system(Promega, Madison, WI) as described previously (Dong et al., 2006) and pRL-SV40 wildtype renilla was used as internal control.
2.7. Chromatin immunoprecipitation
Chromatin immunoprecipitation (Guo et al.) analysis was performed by utilizing a kit purchased from Upstate Biotechnology (Charlottesville, VA) according to the manufacturer’s protocol. The primer sequences for a Twist1 promoter are as follows: forward: 5′-ACCCAGAAAGGGGCACTTG-3′, reverse: 5′-GGCAATTCTGGGTCC GGA-3′. The 180-bp PCR product was resolved on a 2% agarose gel and visualized using GelRed™ Nucleic Acid Gel Staining solution (Biotium, Hayward, CA) and UV illumination.
2.8. Western blot analysis
Forty micrograms of total protein from different samples was used for Western blot analysis. The samples were separated by 10–15% SDS–PAGE gel. These proteins were then transferred to polyvinylidene difluoride (PVDF, Millipore) membranes. After blocking with 5% non-fat milk, the PVDF membranes were incubated with primary antibodies in a Tris-buffered saline (TBS) buffer overnight at 4 °C. These primary antibodies were anti-Twist1 or anti-NICD1. On the following day, PVDF membranes were incubated with appropriate secondary antibodies for 2 h at room temperature. After the membranes had been soaked in an enhanced chemiluminescence reagent (Thermo Scientific) for 5 min, the blots were visualized using X-ray film. β-Actin antibody was used as a loading control. Quantitative band-intensity analysis of Western blots was performed using ImageJ software and normalized to β-actin.
2.9. Immunohistochemistry
For immunohistochemisty, E11.5 limb bud samples were harvested and fixed in 4% paraformaldehyde, dehydrated and embedded in paraffin. Sections (5 µm) were de-paraffinized and rehydrated. Antigen retrieval was performed in 0.01 M sodium citrate buffer in a pressure chamber followed by quenching of endogenous peroxidase activity in 3% H2O2 for 20 min. Blocking of nonspecific antibody binding sites was performed with a 1:20 dilution of normal goat serum for 20 min. Sections were then incubated overnight with a 1:200 dilution of anti-NICD1 antibody, a 1:200 dilution of anti-Twist1 antibody. After incubation with biotinylated secondary antibody for 30 min, antigen was detected following application of horseradish-peroxidase-conjugated streptavidin and a 5-minute incubation with the Romulin AEC Chromogen Kit (Biocare Medical, Concord, CA, USA).
2.10. Real-time PCR analysis
Total RNA was isolated from micromass culture using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). One microgram of RNA was subjected to reverse transcription using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The cDNA was then amplified via real-time PCR using an ABI 7500 Real-time PCR System (Life Technologies, Grand Island, NY, USA) and SYBR® Green Real time PCR Supermix (Bio-Rad, Hercules, CA, USA). The primers used for real-time PCR are listed in Table 1, and β-actin was used as the housekeeping gene. Quantification of the relative expression levels of these target genes was achieved by normalizing to β-actin using the ΔΔCt method.
Table 1.
Mouse gene primers used for real-time RT-PCR experiments.
| Forward primer | Reverse primer | Accession number | |
|---|---|---|---|
| Twist1 | CAGCGGGTCATGGCTAAC | GCAGGACCTGGTACAGGAAG | NM011658 |
| Col2a1 | ACTGGTAAGTGGGGCAAGAC | CCACACCAAATTCCTGTTCA | BC052326 |
| Sox9 | AGGAAGCTGGCAGACCAGTA | CGTTCTTCACCGACT TCCTC | AF421878 |
| Aggrecan1 | CCATGACAACTCACTGAGCG | TTGGAAACACGGCTCCACTT | L07049 |
| β-Actin | AGATGTGGATCAGCAAGCAG | GCGCAAGTTAGGTTTTGTCA | NM007393 |
| Hes1 | TTCCTCCTCCCCGGTGGCTG | TGCCCTTCGCCTCTTCTCCA | NM008235 |
2.11. Statistical analysis
All experiments were repeated at least three times independently. All data were presented as mean ± s.d. Statistical significance among the groups was assessed with one-way ANOVA. The level of significance was P < 0.05.
3. Results
3.1. Chondrogenic differentiation of limb bud MPCs is inhibited by Notch activation
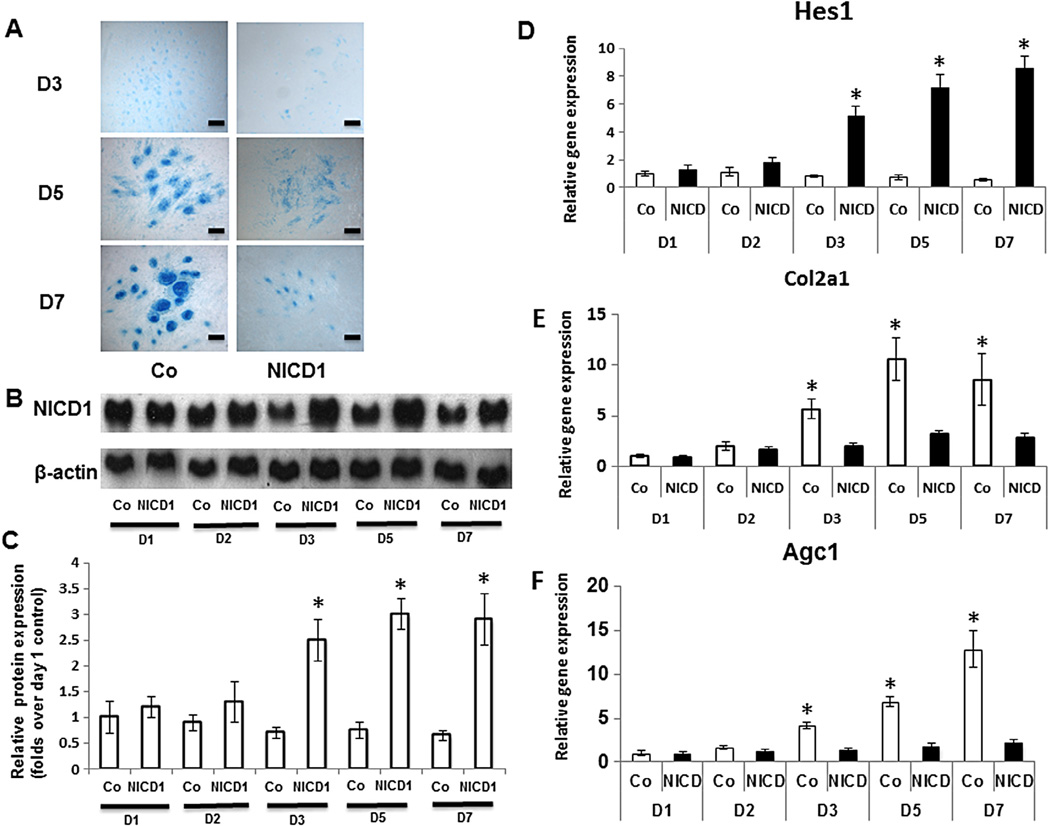
To analyze the effects of Notch activation on MPC chondrogenic differentiation, high-density limb bud cell micromass cultures were employed in this study. Under these conditions, the initial appearance of cartilage matrix and chondrocytes occurred on day 3 of culture and progressed over the next 4 days as illustrated by enhanced number and size of chondrogenic nodules (Fig. 1A). By days 5 and 7 in control cells, a strong Alcian blue staining of cartilage matrix and mature chondrocytes was observed inside the chondrogenic nodules (Fig. 1A). Meanwhile, Notch activation via overexpression of NICD1 in MPCs was confirmed by a dramatically enhanced NICD protein level in western blots at 3, 5 and 7 days post-lentivirus infection (Fig. 1B), but not at days 1 and 2, since lentiviral mediated protein expression needs more than 2 days to reach steady state in regular culture cells. Quantification of NICD1 protein level in Western blots was performed using ImageJ software (Fig. 1C). As we expected, Notch signaling target gene Hes1expression was also significantly enhanced at 3, 5 and 7 days post-NICD1/lentivirus infection, further indicates Notch activation in MPCs (Fig. 1D).
Fig. 1.
Notch signaling represses chondrogenesis in limb bud cell micromass culture. Limb bud cells were cultured in micromass and infected with NICD1 lentivirus before being harvested for Alcian blue staining, western blot and RT-PCR analysis. (A) Overexpression of NICD1 resulted in decreased cartilage nodule formation at days 3, 5, and 7, compared to GFP-lentirvirus infected controls (Co). Scale bars, 100 µm. (B) Western Blot showed a significant increase of NICD protein expression at days 3, 5 and 7 following NICD1 lentiviral infection. (C) Fold change in protein level of NICD1 in Western blots was determined by measuring band intensity. (D) Real time PCR data showed a significant increase of Notch target gene Hes1 expression at days 3, 5 and 7 following NICD1 lentiviral infection compared to GFP-lentirvirus infected controls (Co). (E) In control cells infected with GFP-lentivirus, Col2α1 expression was increased from day 2 to day 5, with maximal increase at day 5. (F) Agc1 expression was increased at days 3, 5 and 7, with maximal increase at day 7. Both Col2α1 and Agc1 expression were significantly down regulated by overexpression of NICD1 at days 3, 5 and 7. Data are means ± s.d. of three independent experiments performed in duplicate and the control gene expression level at day 3 was set at 1. (*, P < 0.05 compared with control at same time point.)
Consistent with our previous in vivo genetic mouse study (Dong et al., 2010), constitutive Notch activation in MPCs significantly inhibited formation of chondrogenic nodules at days 3, 5 and 7 (Fig. 1A). To understand how activation of Notch signaling to inhibit chondrogenesis, expression levels of type II collagen (Col2a1) and Aggrecan1 (Agc1), markers of MPC chondrogenic differentiation, were assayed by real-time PCR (Fig. 1E, F). As expected, Col2a1 and Agc1 expression in control cells were positively correlated with time in culture, indicative of progressive formation of the cartilage matrix. In contrast to control cells, mRNA expression of both Col2a1 and Agc1 in NICD1-expressing cells was down-regulated at time points 3–7 days, in which NICD1 protein expression was highly expressed, suggest an inhibition of cartilage matrix synthesis at that stage.
3.2. Twist1 is induced by Notch activation in MPC micromass culture
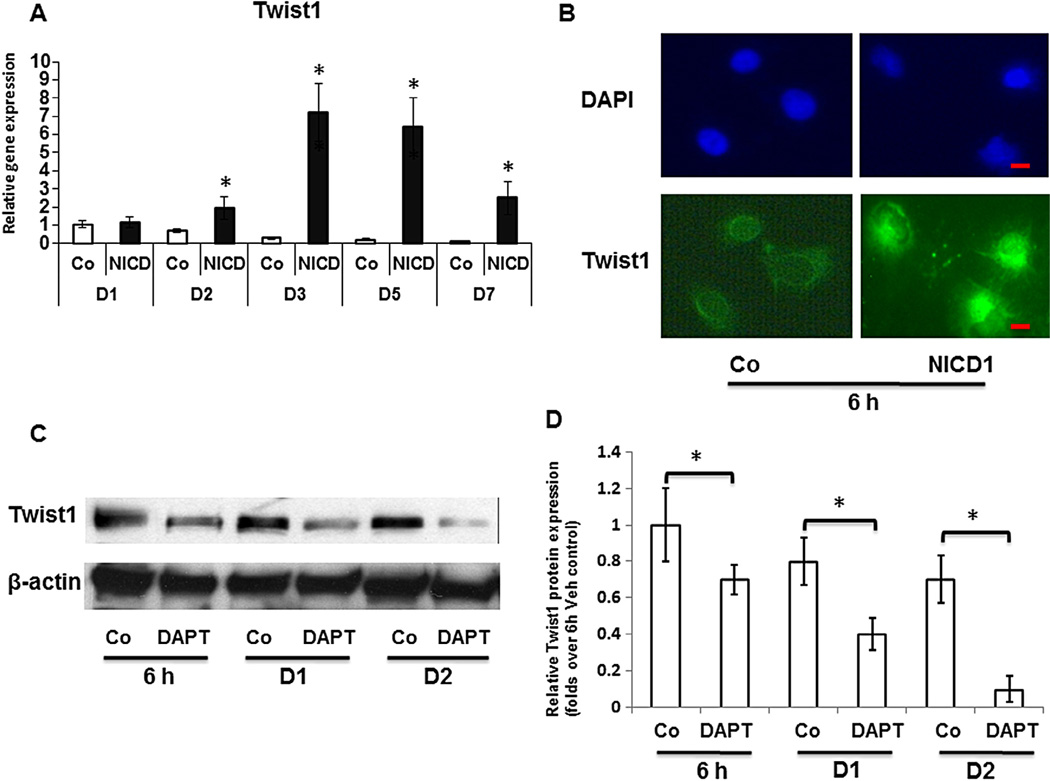
To further elucidate the potential downstream effectors of Notch-induced inhibition of chondrogenesis during limb bud MPC chondrogenic differentiation, the mRNA expression of Twist1, one of the important regulators of stem cell differentiation, was quantified using RT-PCR. As shown in Fig. 2A, Notch activation initially elicited a slightly increase in Twist1 expression after viral infection for 2 days, and a 7-fold increase was shown at day 3 after lentiviral infection, when lentiviral induced NICD1 expression was at steady level. This is indicative of a direct response of Twist1 to NICD1. To further study if there is a quick response of Twist1 to NICD1, we performed transient transfection experiments using NICD1 overexpression plasmid. Immunofluorescence using Twist1 antibody in limb bud cells showed that most of the endogenous Twist1 localized to the cytoplasm in controls, while enhanced Twist1 labeling in both peri-nuclear and nuclear areas was observed as early as 6 h after over-expression of NICD1 (Fig. 2B).
Fig. 2.
Up-regulation of Twist1 expression by Notch signaling in limb bud mesenchymal progenitor cells. (A) Real time PCR analysis reveals that overexpression of NICD1 increased Twist1 RNA levels in the limb bud cells at time points from 2 to 7 days, with a maximal increase at day 3 when compared to control cells (Co). (B) Immunofluorescence assays show that in control cells endogenous Twist1 is localized to the cytoplasm. In contrast, there is a marked increase in peri-nuclear and nuclear labeling of Twist1 in limb bud cells after transit transfection with 3XFLAG NICD1 expressing plasmids for 6 h. Cells were counterstained with DAPI (blue). Scale bars, 5 µm. (C)Western blot shows Twist1 protein levels were visibly reduced by DAPT (10 µM, daily) treatment in MPC micromass culture from 6 h to 48 h. DMSO treated MPC was used as control (Co) and β-actin was used as a loading control. (D) Quantification of Twist1 protein level in Western blots was determined by measuring band intensity. Data are means ± s.d. of three independent experiments. (*, P < 0.05 compared with control at same time point; #, P < 0.05 compared with control at 6 h.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To test whether Notch activation is required for this quick Twist1 expression in MPCs, the effect of DAPT, an inhibitor of Notch signaling, on Twist1 expression was examined during early stage MPC chondrogenesis. Western blots showed the protein levels of Twist1 in control cells were highly expressed from 6 h to 2 days in micromass culture. In contrast, DAPT treatment significantly repressed Twist1 expression at all time points when compared with vehicle treated controls (Fig. 2C). Quantification of Twist1 protein level in Western blots was performed using ImageJ software (Fig. 2D).
3.3. Twist1 is required for Notch-mediated inhibition of chondrogenesis in MPC micromass culture
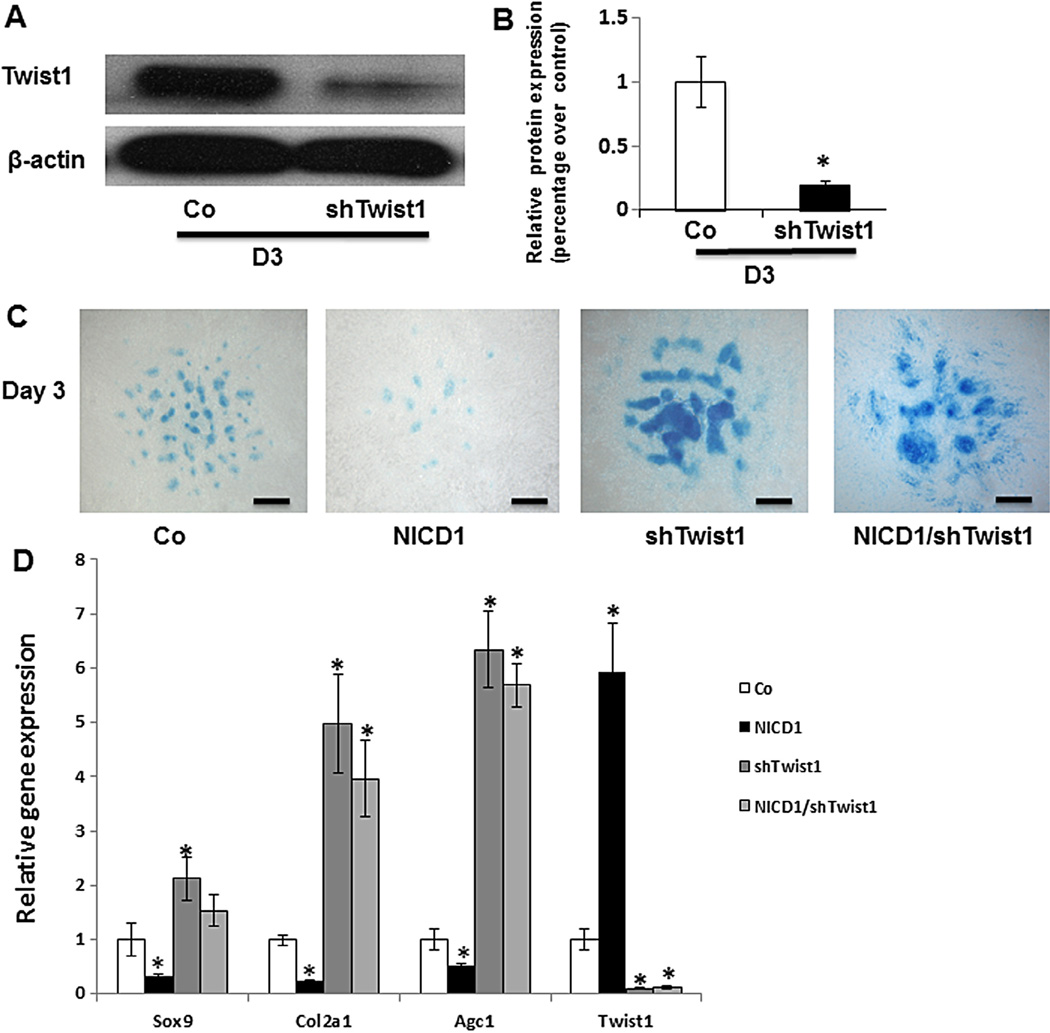
To further explore the specific contribution of Twist1 to Notch-inhibited chondrogenic differentiation in limb bud MPCs, changes in MPC chondrogenic differentiation were measured following knocking down of Twist1 using a lentivirus shRNA infection method. We first checked that shTwist1 was efficient in decreasing Twist protein expression in mesenchymal cells. As shown in Fig. 3A, shTwist1 infection significantly reduced Twist1 expression in MPCs at day 3 post-infection with shTwist1 lentivirus when compared to control cells infected with non-target lentivirus. Quantitative analysis of the blot showed that shTwist1 reduced Twist1 protein expression by about 80% (Fig. 3B), validating the efficiency of Twist silencing in these cells. To analyze the effect of loss of Twist1 on chondrogenic nodule formation in limb bud cell micromass culture, Alcian blue staining was performed at day 3. As shown in Fig. 3C, more chondrogenic nodules were formed in Twist1 knocking down cells when compared to control cells. More importantly, at this time point, when cells were co-infected with both constitutive NICD1 and shRNA of Twist1 lentivirus, chondrogenic nodules were also significantly increased when compared to NICD1 lentivirus alone cells and no significant difference was observed when compared to shTwist1 lentivirus alone cells. Consistent with our Alcian blue staining, real-time PCR data further showed a significant increase in the expression of the MPC chondrogenic marker genes Sox9, Col2a1 and Agc1 in shTwist1 infected cells with or without NICD1 co-infection when compared to control cells (Fig. 3D). Finally, Twist1 RNA level was significantly increased in NICD1 overexpressing cells and reduced by 90% in both shRNA-Twist1 infected cells when compared to control cells. These data suggest that Twist1 is specifically required for Notch-inhibited chondrogenesis.
Fig. 3.
Twist1 is required for Notch activation-mediated inhibition of MPC chondrogenesis. Limb bud cells in micromass were infected with NICD1 or/and shTwist1 lentivirus for 3 days before being harvested for Western blot, Alcian blue staining and RT-PCR analysis. (A) Twist1 protein levels were significantly reduced by Twist1 shRNA lentiviral infection. (B) Quantification of Twist1 protein expression in Western blots showed 80% inhibition of Twist1 expression when compared to control cells (Co). (C) A significant increase in chondrocyte nodule formation was observed in both shTwist1 infected cells with or without co-infection with NICD1, and a drastic reduction in chondrocyte nodule formation was seen in NICD1-expressing cells. Scale bars, 100 µm. (D) Sox9, type II collagen, and Agc1 gene expression were significantly increased in Twist1 knocking down MPCs with or without overexpressing NICD1. Twist1 expression was significantly increased in NICD1 overexpressing cells and decreased in both Twist1 knocking down MPCs with or without overexpressing NICD1. Data are means ± s.d. of three independent experiments performed in duplicate and the GFP-virus control (Co) gene expression level was set at 1. (*, P < 0.05 compared with control.)
3.4. Twist1 expression is directly regulated by Notch signaling
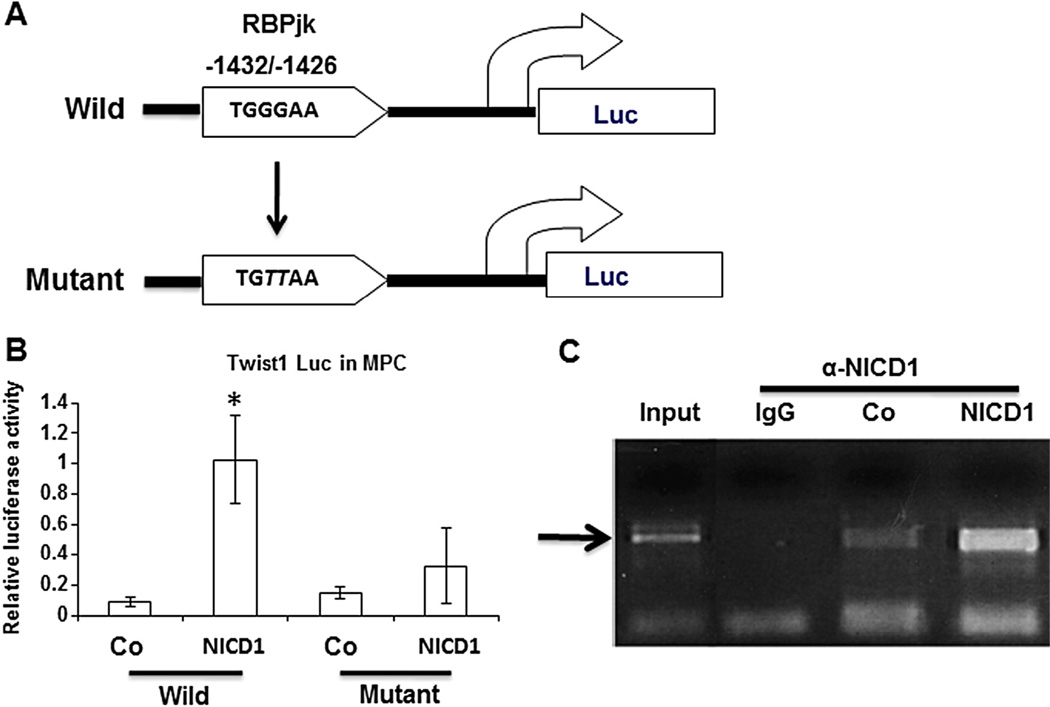
Since Twist1 plays an important role in Notch signaling-mediated MPC chondrogenic differentiation, Twist1 was then investigated as a potential direct downstream target of Notch signaling in MPCs. Co-immunoprecipitation experiments were performed using anti-NICD1 or anti-Twist1 to detect the interaction between NICD1 and Twist1 in MPCs. To test whether Twist1 could be activated by NICD1 at the transcriptional level, a luciferase reporter driven by a 2kb mouse Twist1 promoter fragment containing one potential RBPJk-binding sequence was generated (Fig. 4A) and co-transfected in MPCs with NICD1 expression plasmid. Transient transfection assays showed a 10-fold increase in Twist1 promoter activity upon overexpression of NICD1 (Fig. 4B). To determine if this putative NICD/RBPJk binding site is required for up-regulation of Twist1 promoter activity, point mutations were generated in the RBPJk binding site of the Twist1 promoter (Fig. 4A). Fig. 4B clearly showed that mutation of this putative RBPJK binding site (−1432/−1426) eliminated the luciferase activity of the Twist1 promoter construct induced by NICD1.
Fig. 4.
The RBPJK binding site in Twist1 promoter is required for molecular interaction with NICD1. (A) Top: Bioinformatic analyses uncovered one evolutionarily conserved NICD/RBPJk binding site (TGGGAA) 1426 bps from the 5′ end of the Twist1 transcription start site. Bottom: RBPJK binding site-mutated promoter construct. Nucleotides that mutate from established consensus sequences are highlighted in italic (GG→TT). (B) Luciferase assays showed that NICD1 upregulated gene expression from the wild type promoter, and this transcriptional upregulation is significantly reduced when 3XFLAG NICD1 is co-transfected with the RBPJK binding site mutant Twist1 promoter fragment. Data are means ± s.d. of three independent experiments performed in duplicate and all the results were normalized to internal control (*, P < 0.05 compared with wild control). (C) Chromatin immunoprecipitation was performed on immunocomplex subjected to antibody against NICD1 using chromatin isolated from lentiviral-infected MPCs. PCR results using primers targeting RBPJK binding site showed a specific fragment was detected between the RBPJK response element and Twist1 in control (Co) MPCs, and this fragment formation was enhanced by overexpression of NICD1 in MPCs. The input DNA was used as a positive control for the PCR and IgG as a negative control.
To further test if NICD directly binds to the putative RBPJK binding site of Twist1 promoter, specific NICD/RBPJK binding activity on the Twist1 promoter was quantified using chromatin immunoprecipitation assay (Guo et al., 2010). Soluble chromatin from MPC cells infected with or without NICD1 lentivirus was extracted and anti-NICD1 antibody immunoprecipitated chromatin was analyzed by semi-quantitative PCR using primers flanking the RBPJk binding site in Twist1 promoter. In wild type cells without overexpression of NICD1, endogenous NICD1 was readily detected on the Twist1 promoter but not in lgG control (Fig. 4C), indicating sequence-specific association of NICD1 with the Twist1 promoter in MPC cells. Importantly, overexpression of NICD1 significantly enhanced occupancy of the Twist1 promoter by NICD1.
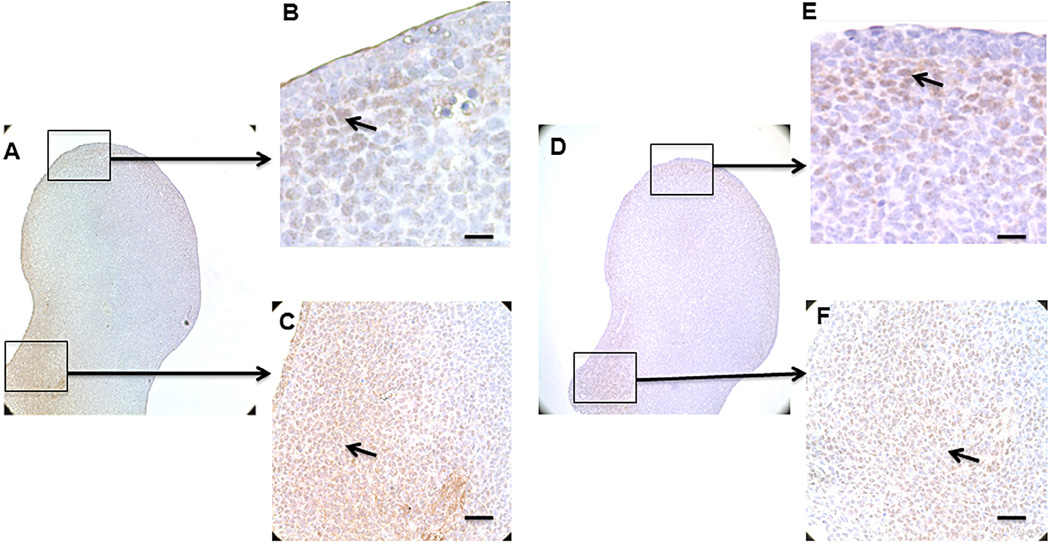
3.5. Twist1 and NICD1 limb bud expression domains overlap
Sine we have established that Twist1 and NICD1 can interact in vitro, we next sought for the in vivo evidence to support this conclusion during embryonic development. We chose to examine the expression domain of both genes in developing vertebrate limb bud (Fig. 5A–F), as our in vitro studies indicated that both genes are expressed in the MPCs isolated from E11.5 limb. The results show that NICD1 is expressed broadly in the distal anterior peripheral mesenchyme (Fig. 5B), as well as posterior mesenchyme (Fig. 5C). Similar to NICD1, the major Twist1 expression domain is also restricted to the posterior proximal (Fig. 5F) and the region of the distal mesenchyme underlying the apical ectodermal ridge (AER) (Fig. 5E). These results demonstrate that Twist1 and NICD1 are coexpressed within distinct regions of the developing vertebrate limb, support the idea that these two factors may functionally interact during in vivo MPC chondrogenic differentiation.
Fig. 5.
Coexpression of Twist1 and NICD1 in the developing limb. Immunochemistry of the E11.5 mouse limb bud. (A) Endogenous NICD1 expression in normal limb bud. (B) Positive labeling for NICD1 is pointed out by black arrow in the distal anterior peripheral mesenchyme. (C) A large population of cells in posterior limb bud was stained by NICD1. (D) Endogenous Twist1 expression in normal limb bud. (E) Positive labeling for Twist1 is restricted to the region of distal mesenchyme underlying the AER. (F) Cells in posterior limb bud were stained positive for Twist1. Scale bars, 50 µm.
4. Discussion
Chondrogenesis is one of the earliest steps of bone development and begins as MPCs aggregate to form the earliest outline of nascent bones. These cell condensations then differentiate into chondrocytes thereby forming the template of the endochondral skeleton (Shimizu et al., 2007). Notch signaling plays a vital role during MPC condensation and chondrocyte differentiation. The effects of Notch signaling on MPCs are complex, as demonstrated by results showing both stimulation and inhibition of differentiation. In assays of osteogenesis using human bone marrow-derived mesenchyme, the application of Jagged-1 (JAG1), the ligand of JAG1 protein in the Notch signaling pathway, is sufficient to induce mesenchymal cell osteoblast differentiation and requires concomitant PKCδ signaling (Zhu et al., 2013). In contrast, our previous study showed ablation of Notch signaling using DAPT, a Notch signaling inhibitor, stimulated MPC chondrogenic, ostegenic and adipogenic differentiation. In addition, transgenic mice expressing constitutive NICD exhibited a skeletal dysplasia with diminished osteochondrogenic differentiation (Dong et al., 2010; Zanotti et al., 2008). These findings suggest that the effects of Notch signaling on cell differentiation are dependent upon the cellular context and stage of differentiation. Furthermore, Notch targets may also differ depending on levels of Notch activation indicates that understanding the effects of Notch signaling on MPC differentiation will require greater knowledge of factors that modify Notch pathways and the specific targets of Notch signaling that regulate MPC differentiation. Since the limb bud cell micromass culture system has been widely used to mimic condensation of the mesenchyme and the formation, maturation of the cartilage anlagen during skeletogenesis (Wang et al., 2013), in this study, we utilized this culture system to study the mechanism of MPC chondrogenic differentiation mediated by Notch signaling.
Consistent with our previous findings from Prx-cre/Rosa26-NICD transgenic mice (Dong et al., 2010), activation of Notch signaling by overexpression of intracellular domain NICD1 in MPC micromass significantly inhibited chondrogenic nodule formation and chondrogenic marker gene expression. These changes are indicative of the inhibitory effect of Notch signaling on mesenchyme chondrogenic differentiation. However, they do not identify the molecular mechanism by which Notch signaling inhibits early chondrogenesis.
The regulation of MPC chondrogenic differentiation and subsequent cartilage formation involves complex signaling pathways including BMPs, TGF-β, Wnt/β-catenin, and Hedgehog, which in turn activate major downstream transcription factors such as Sox9, Runx2, and others (Akiyama et al., 2002; Soung do et al., 2012; Wang et al., 2005). Twist1 has been linked to the regulation of MPC proliferation and differentiation. This is evidenced by a Twist1-induced shift in epithelial cells to a more mesenchymal nature, with increased expression of stem cell markers and acquisition of stem cell properties (Yang et al., 2004). Twist1 also interacts with Runx2 as a target gene of Wnt signaling to regulate chondrocyte differentiation in vitro (Dong et al., 2007). Although Wnt/β-catenin directly activated Twist1 expression in skull progenitors, but conditional Twist1 deletion only partially phenocopied the absence of β-catenin, and Twist1 deletion also partially restored bone formation in the presence of constitutive β-catenin activation suggest other upstream factors may also involved in Twist1 regulation (Goodnough et al., 2012). In this study, we found that Twist1 highly expressed in MPCs, as well as NICD1, suggests that Twist1 may be required for NICD1 mediated maintainess of MPC stem cell-like phenotype. Since Notch signaling up-regulated expression of Twist1 in MPCs as early as 6 h, we speculate that it may function as an upstream mediator of the effect of Twist1 regulation of MPC differentiation. In agreement with this, knocking down of Twist1 in NICD1 overexpressing MPCs showed a loss of inhibitory effect of NICD1 on MPC chondrogenic differentiation, further indicating Notch activation-inhibited MPC chondrogenesis is largely dependent on an underlying activity of Twist1.
To fully elucidate the processes that control Twist1 expression and its interactions with Notch signaling, several biochemistry experiments were performed in this study. A NICD/RBPJk binding site was identified in the Twist1 promoter region and the DNA/protein binding affinity was confirmed by single-point mutation experiment and ChIP assay. In vivo study of expression pattern of NICD1 and Twist1 in limb bud further support our in vitro finding that Notch signaling directly regulates Twist1 expression at both the transcriptional and post-transcriptional levels during MPC chondrogenic differentiation. Although NICD1 and Twist1 are expressed in the limb in overlapping domains, and Twist1 acts as downstream factor of NICD to mediate MPC chondrogenic differentiation, genetic regulations between NICD1 and Twist1 still need to be further studied by intercrossing NICD1 gain-of-function mice and Twist1 loss-of-function mice.
When the individual findings are considered together, this study presents a potential novel mechanism whereby Notch may induce Twist1 to inhibit MPC chondrogenic differentiation via direct Notch-NICD/RBPjk binding to its promoter region. These findings provide additional support for the potential treatment of cartilage defects and osteoarthritis by local modification of Notch signaling in MPCs.
Acknowledgments
This work was supported in part by the following grants: Natural Science Foundation of Liaoning Province (2013021051 to Y.T.), the start-up funds from the Department of Orthopaedic Surgery at LSU Health Sciences Center to Y.D., the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R21AR063803 to Y.D., the National Basic Research Program of China (973 Program grant 2010CB530400), the National Natural Science Foundation of major international cooperation projects (81220108027), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, IRT1270). We also thank Dr. Matthew Hilton and Dr. Edward Schwarz for manuscript review and comments.
Footnotes
Author contributions
Y.T., Y.D., Y.W. obtained the funding; Y.T., Y.D., M.C., Y.W., and X.S. designed all experiments; Y.T., Y.X., and Q. F. performed all the experiments and data collection. Y.T., Y.D. drafted the manuscript. J.M. edited the manuscript.
References
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YF, Soung do Y, Schwarz EM, O’Keefe RJ, Drissi H. Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J. Cell. Physiol. 2006;208:77–86. doi: 10.1002/jcp.20656. [DOI] [PubMed] [Google Scholar]
- Dong YF, Soung do Y, Chang Y, Enomoto-Iwamoto M, Paris M, O’Keefe RJ, et al. Transforming growth factor-beta and Wnt signals regulate chondrocyte differentiation through Twist1 in a stage-specific manner. Mol. Endocrinol. 2007;21:2805–2820. doi: 10.1210/me.2007-0199. [DOI] [PubMed] [Google Scholar]
- el Gelse K, Ekici AB, Cipa F, Swoboda B, Carl HD, Olk A, et al. Molecular differentiation between osteophytic and articular cartilage – clues for a transient and permanent chondrocyte phenotype. Osteoarthritis Cartilage. 2012;20:162–171. doi: 10.1016/j.joca.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev. Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, et al. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat. Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Goodnough LH, Chang AT, Treloar C, Yang J, Scacheri PC, Atit RP. Twist1 mediates repression of chondrogenesis by β-catenin to promote cranial bone progenitor specification. Development. 2012;139:4428–4438. doi: 10.1242/dev.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liu M, Yang D, Bouxsein ML, Saito H, Galvin RJ, et al. Suppression of Wnt signaling by Dkk1 attenuates PTH-mediated stromal cell response and new bone formation. Cell Metab. 2010;11:161–171. doi: 10.1016/j.cmet.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. U.S.A. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, et al. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Winter RM. Saethre-Chotzen syndrome. J. Med. Genet. 1994;31:393–396. doi: 10.1136/jmg.31.5.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Yokoyama S, Asahara H. Growth and differentiation of the developing limb bud from the perspective of chondrogenesis. Dev. Growth Differ. 2007;49:449–454. doi: 10.1111/j.1440-169X.2007.00945.x. [DOI] [PubMed] [Google Scholar]
- Soung do Y, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, et al. Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J. Bone Miner. Res. 2012;27:1585–1597. doi: 10.1002/jbmr.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl C, Leimeister C, Klamt B, Maier M, Nanda I, Dixon M, et al. Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics. 2000;66:195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Holz JD, Rutkowski T, Wang Y, Zhu Z, et al. Runx1 is critical for PTH-induced onset of mesenchymal progenitor cell chondrogenic differentiation. PLoS ONE. 2013;8:e74255. doi: 10.1371/journal.pone.0074255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Belflower RM, Dong YF, Schwarz EM, O’Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J. Bone Miner. Res. 2005;20:1624–1636. doi: 10.1359/JBMR.050516. [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yu X, Zhan X, D’Costa J, Tanavde VM, Ye Z, Peng T, et al. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol. Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149:3890–3899. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Sweetwyne MT, Hankenson KD. PKCdelta is required for Jagged-1 induction of human mesenchymal stem cell osteogenic differentiation. Stem Cells. 2013;31:1181–1192. doi: 10.1002/stem.1353. [DOI] [PubMed] [Google Scholar]