Abstract
Although each T lymphocyte expresses a T-cell receptor (TCR) that recognizes cognate antigen and controls T-cell activation, different T cells bearing the same TCR can be functionally distinct. Each TCR is a heterodimer, and both α- and β-chains contribute to determining TCR antigen specificity. Here we present a methodology enabling integration of information about TCR specificity with information about T cell function. This method involves sequencing of TCRα and TCRβ genes, and amplifying functional genes characteristic of different T cell subsets, in single T cells. Because this approach retains information about individual TCRα-TCRβ pairs, TCRs of interest can be expressed and used in functional studies, for antigen discovery, or in therapeutic applications. We apply this approach to study the clonal ancestry and differentiation of T lymphocytes infiltrating a human colorectal carcinoma.
Single-cell analysis can reveal important functional insights that are masked in bulk analysis of cell populations1–3. Recent technological advances have improved our ability to query expression of multiple genes in single cells simultaneously, thereby helping to resolve the complexity inherent in heterogeneous populations of cells including T lymphocytes. These technologies include time-of-flight mass cytometry (CyTOF), RNA sequencing (RNA-seq) and quantitative RT-PCR4–7. However, these technologies have not thus far been applied in a high-throughput manner to include the most distinctive genes a T cell expresses: the genes that encode the TCR.
The TCR, which determines which complexes of antigenic peptide–major histocompatibility complex (MHC) the T cell responds to, plays a major role in controlling the selection, function and activation of T cells8. Because the TCR expressed in each T cell is composed of α- and β-chain genes that are derived by somatic V(D)J recombination, the TCR repertoire in any given individual is tremendously diverse9. Therefore, the TCR also serves as a unique identifier of a T-cell's ancestry, because it is likely that any two T cells expressing the same TCRαβ pair arose from a common T-cell clone.
There is great potential synergy in pairing TCR sequences (which can reveal information about T-cell ancestry and antigen specificity) with information about expression of genes characteristic of particular T-cell functions. Integrating these two types of information can allow one to comprehensively describe a given T cell. For example, it is becoming clear that T cells responding to different antigens can have very different phenotypic and functional properties, even if these antigens are derived from the same pathogen10. The ability to link T-cell function and TCR specificity will enable one to determine which functional subsets of T cells have undergone clonal expansion and which clones exhibit plasticity, ultimately give rise to progeny that express the same TCRαβ heterodimers, but exhibit diverse functional phenotypes. It will also allow identification of TCRαβ heterodimers expressed in individual T cells of interest without expansion of the T-cell population in vitro, which can result in loss of functional integrity. These heterodimers can be invaluable in studies designed to discover antigens11 or in therapeutic applications12.
Here we present an approach enabling the simultaneous sequencing of TCRα and TCRβ genes and amplification of transcripts of functional interest in single T cells. This approach enables both TCR sequencing and extensive phenotypic analysis in single T cells, linking TCR specificity with information about T-cell function.
Results
Strategy
We and others have successfully sequenced TCR genes from single, sorted T cells using a nested PCR approach followed by Sanger sequencing13–15. Here we devise a strategy enabling simultaneous sequencing of rearranged TCR genes and multiple functional genes in single, sorted T cells through deep sequencing. In addition to enabling the analysis of multiple functional genes in parallel with TCR sequencing, this approach has several advantages over previous TCR sequencing methods that utilize Sanger sequencing13–15. First, it is efficient (5,000-10,000 cells can be sequenced in one sequencing run) and less labor intensive as individual PCR products do not need to be purified and sequenced separately. Second, it is also very accurate as consensus sequences are determined from a high number of independent sequencing reads (often exceeding 1,000) per TCR gene, essentially eliminating the effect of sequencing error. Third, it is well-established that individual T cells can express two TCRα genes16,17. Our approach uniquely enables sequencing of multiple TCRα genes from most single T cells and determination of which of these are functional.
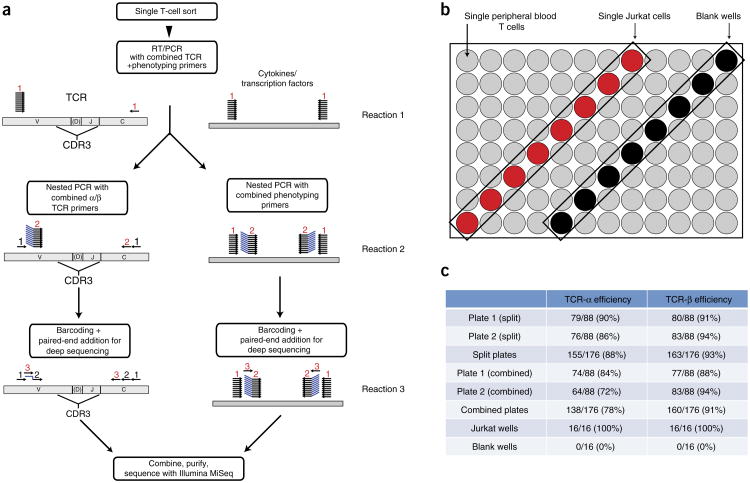
In our method, single T cells are sorted into 96-well PCR plates (Fig. 1a). An RT-PCR reaction is done using 76 TCR primers and 34 phenotyping primers (Supplementary Fig. 1 and Supplementary Tables 1–3). The products are then used in a second PCR reaction—either one that uses nested primers for TCR genes or one that uses nested primers for phenotypic markers, including cytokines and transcription factors. A third reaction is then performed that incorporates individual barcodes into each well (Supplementary Fig. 2)18. The products are combined, purified and sequenced using the Illumina MiSeq platform. The resulting paired-end sequencing reads are assembled and deconvoluted using barcode identifiers at both ends of each sequence by a custom software pipeline to separate reads from every well in every plate (Supplementary Note). The resulting sequences are then analyzed using a program called VDJFasta19, which we have adapted to resolve barcodes and analyze sequences with a customized gene-segment database that includes relevant transcription factors and cytokine genes. The population of annotated sequences above background levels in each well is then measured (see Online Methods for details on data processing). For TCR sequences, the CDR3 nucleotide sequences are then extracted and translated. For phenotypic parameters, the presence or absence of a transcript in a particular well is scored.
Figure 1.
Strategy for single-cell TCR sequencing and phenotyping, and determination of TCR-sequencing efficiency. (a) Strategy for simultaneous TCR-sequence determination and phenotyping of single, sorted T cells. Single T cells were sorted into 96-well plates. The initial RT-PCR reaction (reaction 1) uses 76 TCR primers and 34 phenotyping primers. An aliquot of the product of reaction 1 is used for two separate second nested PCR reactions (reaction 2), one for TCR sequencing and one for phenotyping. Using an aliquot of reaction 2 product as a template, a third PCR reaction is performed that incorporates individual barcodes into each well and enables subsequent sequencing using the Illumina MiSeq platform. For TCR sequencing, the third reaction can be split into separate TCRα and TCRβ reactions (for optimal efficiency), or the two TCR chains can be included in a single reaction. The products of reaction 3 are then combined and sequenced using the Illumina MiSeq platform. (b,c) Accuracy and efficiency of TCR sequencing using this method. (b) Strategy used to validate TCR sequencing. Into each 96-well test plate, individual human peripheral blood T cells were sorted into 80 wells (gray). Single Jurkat T cells were sorted into eight other wells (red), and the remaining eight wells (black) were left empty (blank). For sequencing of these test plates, reaction 3 was initially performed separately for TCRα and TCRβ (split). It was also repeated with TCRα and TCRβ amplified together in the same reaction (combined). (c) Efficiency of TCRα and TCRβ sequencing in split or combined formats. Plate 1 contained 80 single CD45RA+CD4+TCRαβ+ T cells, and plate 2 contained 80 single CD4+ or CD8+ TCRαβ+ T cells sorted from periphera blood of the same healthy human donor. Identical Jurkat sequences were obtained from all Jurkat wells. No sequences were obtained from any empty wells.
TCR sequencing validation
To validate our TCR sequencing methodology, 80 single CD45RA+CD4+TCRαβ+ T cells were sorted from peripheral blood of a healthy human donor into one 96-well plate, and 80 single CD4+ or CD8+TCRαβ+ T cells were sorted from the same sample into a second plate. CD45RA marks naive CD4+ T cells that are not expected to have undergone much clonal expansion20. The Jurkat human T-leukemic cell line was used as a positive control21. Into both plates, individual Jurkat T cells were sorted into eight wells, and eight wells were left blank (Fig. 1b). These plates were initially subjected to the first reaction containing 74 TCR variable (V)-region primers, 2 constant (C)-region primers and 34 phenotyping primers. Phenotyping primers were included in the first reaction to demonstrate that the inclusion of these primers does not interfere with TCR sequencing. The subsequent nested PCR and barcoding reactions were then performed according to the protocol shown in Figure 1a; reaction products were subsequently sequenced and analyzed.
Out of 160 wells into which single, peripheral blood αβ T cells were randomly sorted, productive TCRβ sequences were successfully obtained in 147 wells (92%), and at least one productive TCRα sequence was found in 139 wells (87%) wells (Fig. 1c and Supplementary Table 4). Productive TCR genes have been joined in the proper reading frame by V(D)J recombination without premature stop codons, enabling expression of a complete TCRα or β chain. Paired, productive TCRαβ sequences were found in 131 (82%) wells. Identical Jurkat TCRαβ sequences were found in 16/16 wells into which Jurkat cells were sorted but in no other wells on the plates (Fig. 1c and Supplementary Table 4). There were no sequences above background found in the wells into which no cell was sorted (see Supplementary Fig. 3 for details regarding background). The absence of sequences from wells with no cells and the presence of Jurkat sequences only in the 16 designated Jurkat wells indicates that cross-contamination of wells was minimal. Optimal TCR-sequencing efficiency was obtained when the third PCR reaction (barcoding) was done in two separate plates—one for TCRα and one for TCRβ; there was a only a marginal loss of efficiency when these reactions were done together in one plate (Fig. 1c).
T cells often express two recombined TCRα genes16,17. Sanger sequencing cannot be performed on heterogeneous products, therefore, methods that rely on Sanger sequencing cannot easily identify multiple TCRα chains from a single cell15. Furthermore, the presence of multiple TCRα chains can hinder the efficiency and accuracy of sequencing in methods based on Sanger sequencing. Because our strategy employs deep sequencing, wherein each template is amplified and sequenced independently, we can readily derive multiple TCRα sequences from individual cells. On average (assuming 20 96-well plates on a single sequencing run), we obtain ∼5,000 total TCRα or TCRβ sequences with the same set of barcodes, specifying they are derived from the same well. To distinguish between TCR sequences that differ owing to a sequencing and/or PCR error and those that are probably derived from different TCR genes, our software determines a cutoff value in similarity based upon the assumed rate of sequencing and/or PCR error22. All sequences exceeding this value (e.g., they are very similar to one another) are assumed to derive from the same TCR gene and a consensus sequence is determined. We can therefore accurately identify multiple TCR gene sequences among a heterogeneous group of sequences tagged with the same barcode.
We detected multiple TCRα chains in 80/139 (58%) of wells containing at least one productive TCRα chain sequence (Supplementary Table 4). In contrast, we did not detect multiple TCRβ chains or multiple nonproductive TCRα chains in any wells. This indicates that cross-contamination of wells or the erroneous sorting of two cells into wells is minimal. With the exception of designated Jurkat wells, there were no repeated TCRs present in the first plate containing 80 naive CD45RA+CD4+ T cells. This is consistent with the expectation that naive T cells have undergone minimal clonal expansion and therefore it is unlikely that two identical clones would be in the population of 80 cells sorted into a single plate. In our second plate, which contained 80 total (naive and non-naive) TCRαβ+ T cells, we detected four repeated TCR sequences in 11 different wells (Supplementary Table 5). All these repeated T cells were scattered across the plates and not within close proximity to each other, suggesting that the repetition did not arise as a result of cross-well contamination. For one TCRβ sequence that was repeated across four wells (CAWTLGGNEQFF), each well contained sequences of the same two productive TCRα genes. For a TCRβ (CASSYGDPGGLDGELFF) that was repeated across three wells, the same productive TCRα gene was detected in all three wells. Additionally, within two of these three wells, an identical nonproductive TCRα gene was detected. These findings confirm that detection of two TCRα rearrangements in a particular cell is repeatable and reliable, and not likely to be the result of contamination or error.
We detected two productive TCRα genes in 19/139 (14%) wells containing at least one productive TCRα chain (Supplementary Table 4). This reinforces the importance of single-molecule sequencing methods like ours to determine true TCRαβ heterodimers. Methods that can detect only one TCRα gene per cell may falsely identify a TCRαβ heterodimer because in cells expressing two productive TCRα genes, only one TCRα gene product is thought to be expressed at the T-cell surface16. Further, in cases where only one TCRα chain is detected in a particular cell, there is a possibility that another productive TCRα chain is present but not detected. This possibility is unlikely with our methodology given its efficiency and the fact that we detected all variable (V)-regions in our TCRα data even in the presence of other TCRα chains within the same T cell (Supplementary Fig. 4). However, due to this possibility, all TCRs derived through this method that are reconstituted for use in functional studies should be validated.
Phenotyping validation
In addition to TCR sequencing, this method enables one to simultaneously query multiple phenotypic parameters from single T cells. In our phenotyping panel, we included multiple cytokines and transcription factors that influence T-cell function and define certain T-cell subsets (Supplementary Table 2). Our method is uniquely enabling in this regard, as other methods for measuring expression of cytokines and transcription factors (e.g., flow cytometry) generally require cellular fixation, which compromises the integrity of nucleic acids and makes it difficult to perform TCR sequencing. Furthermore, cellular fixation-based methods for detecting transcription factors are arduous and unreliable, even compared to methods for detecting intracellular cytokine expression23.
For example, the functional diversity of CD4+ T cells is dependent upon expression of various transcription factors24. Some of these transcription factors are termed “master regulators” and their expression has been used to specify particular T-cell lineages. We included T-bet, GATA3, RoRγT (RAR-related orphan receptor gamma T, which is encoded by RORC), BCL-6 and FOXP3 (Forkhead box P3), which have been used to specify helper T type 1 (Th1) cells, Th2, Th17, follicular helper (TfH) and regulatory T (Treg) cells, respectively25,26, in our phenotyping analysis. We also included the runt-related transcription factors Runx1 and Runx3, which influence T-cell differentiation27. Lastly, we also included both pro-inflammatory and inhibitory cytokines that mediate T-cell effector function and define the various T-cell subsets; these include interferon (IFN)-γ (Th1), interleukin (IL)-13 (Th2), IL-17 (Th17), IL-10 and TGFβ (Treg).
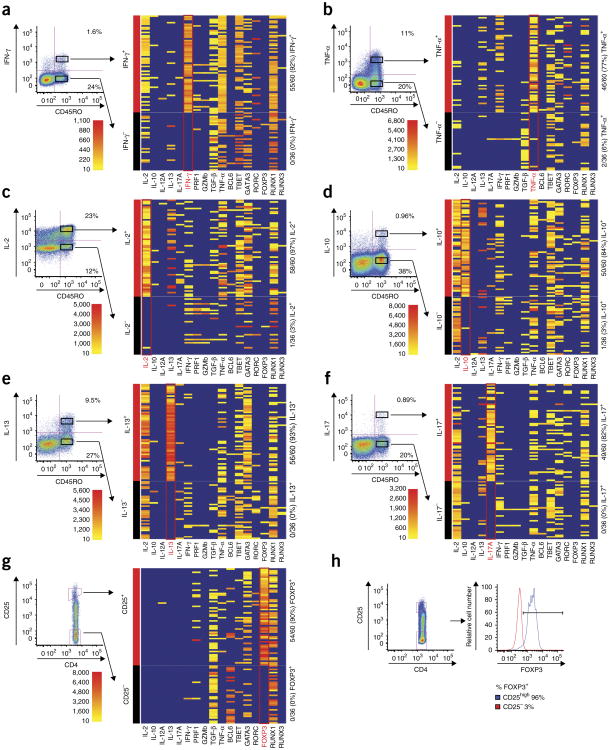
To validate this part of our methodology, we used flow cytometry-based cytokine capture assays (Miltenyi), which enable the determination of cytokine expression without the need for cell fixation28. We tested expression of the following cytokines for which cytokine secretion assays are commercially available: TNF-α, IFN-γ, IL-2, IL-10, IL-13 and IL-17. Into each plate we sorted 60 single CD4+CD45RO+ memory phenotype T cells from healthy human peripheral blood that were positive for protein expression of a particular cytokine and 36 single CD4+CD45RO+ T cells that were negative for cytokine protein expression (Fig. 2 and Supplementary Table 6). These plates were initially amplified with the first reaction containing 74 TCR V-region primers, 2 C-region primers and 34 phenotyping primers. TCR primers were included in the first reaction to demonstrate that their presence does not interfere with subsequent phenotyping reactions. Nested PCR, barcoding and sequencing analysis was performed for phenotypic parameters. We detected transcripts in single, cytokine-positive T cells with 77–97% sensitivity (Fig. 2 and Supplementary Table 7). Our false-positive rate was very low; the specificity of our assay was 94–100% when compared to the relevant cytokine capture assays (Fig. 2 and Supplementary Table 6).
Figure 2.
Accuracy of phenotypic analysis. (a-f) Peripheral blood T cells were stimulated for 3 h with PMA and ionomycin, and expression of the ndicated cytokines was analyzed by cytokine secretion assays that do not require cell fixation: IFN-γ(a), TNF-α(b), IL-2 (c), IL-10 (d), IL-13 (e), IL-17 (f). Sixty single CD45RO+CD4+ T cells that were clearly positive for the indicated cytokine protein and 36 single CD45RO+CD4+ T cells that were clearly negative for the indicated cytokine protein were sorted, and expression of the same cytokine genes were measured by the method depicted in Figure 1a. Seventeen independent phenotypic parameters were assayed in single, sorted cells, and the phenotypic parameter on which cells were sorted is indicated in red. Heatmaps indicate read count of each parameter (x axis) within each particular well (y axis). Scale indicates number of reads obtained from a given well for the indicated parameter. Wells indicated in blue did not display any reads that reached threshold. (g) Unstimulated CD4+ T cells were sorted based upon CD25 expression to validate phenotypic analysis for FOXP3. Sixty single CD4+ T cells with high CD25 expression and 36 single CD4+ T cells that were negative for CD25 expression by flow cytometry were sorted and assayed as in a-f. (h) Expression of CD25 and FOXP3 protein was measured by flow cytometry. Cells from the same donor were fixed and stained with anti-CD25 and anti-FOXP3 antibodies. Histograms on right depict FOXP3 expression in gated CD25high and CD25− populations.
For some of the cytokines genes we validated, there does appear to be a low false-positive rate compared to cytokine secretion assays. Because these wells clearly exceed background levels (Methods and Supplementary Fig. 3), this suggests that these rare cells do indeed express the particular mRNA, although its protein product is not detected. This is not surprising given that cytokine genes are subject to particularly tight regulation, including translational repression that might prevent protein expression even in the presence of mRNA29.
We also readily detected expression of all the transcription factors in our panel in single T cells. For most of these transcription factors, there are no available surface markers that reliably predict expression. An exception is FOXP3, whose expression correlates well with high expression of the surface marker CD25 in CD4+ T cells30. To validate our methodology for FOXP3 expression, we sorted 60 single CD25highCD4+ T cells and 36 single CD25−CD4+ T cells into a single plate. We detected FOXP3 in 54/60 (90%) of CD25high cells and 0/36 (0%) of CD25− cells (Fig. 2g). We also fixed and stained T cells from the same donor with both CD25 and FOXP3 to confirm the correlation between the high expression of CD25 and FOXP3 (Fig. 2h).
We could detect as little as one molecule of template in a given cell, although sensitivity improves with increased template abundance (Supplementary Fig. 5). Whereas frequency of detection improves with template abundance, read number of a given transcript in the cells called as positive does not (Supplementary Fig. 5). This demonstrates that our methodology is binary, in that it indicates presence or absence of a given transcript. Therefore, read number per well should not be considered a quantitative indicator of the abundance of that transcript.
It is very possible that a particular mRNA might be expressed but not detected in a particular cell, especially at lower copy number (Supplementary Fig. 5). Therefore, we expect that false negatives will occur with this method. However, our data show that false positives do not occur at a significant rate (Supplementary Fig. 3). Thus, for practical purposes, the positive predictive value of our assay exceeds its negative predictive value for any given parameter. One should consider this when analyzing data using this methodology.
Despite the many factors that might contribute to discordance between mRNA and protein detection, our data correlate remarkably well with data from cytokine capture assays and with CD25 protein expression in the case of FOXP3 (Fig. 2 and Supplementary Table 6). However, we considered either the cytokine secretion assays or CD25 staining as the gold standard and did not take into account the possibility of true discordance between mRNA and protein expression. Clearly, mRNA expression does not always correlate with protein expression as many genes are subject to post-transcriptional regulation. Cytokine gene expression is subject to particularly complex regulation, including mechanisms affecting translation and/or mRNA stability29. Because there is likely discordance between mRNA and protein expression within cells, the data we show on sensitivity and specificity should be used only as a guide (Supplementary Table 7).
Our strategy can also be customized or expanded. For example, the phenotyping panel can be customized to include additional genes of functional interest. In addition, because we can obtain the sequence of any given parameter, we can also design assays to measure genetic polymorphism, somatic mutation or splice variation of genes in single cells. Of course, because it is difficult to predict the cumulative affect of additional primers in a multiplexed PCR reaction, addition of parameters would likely require additional optimization and validation. But because the presence of additional transcripts does not affect the sensitivity of detection of a given transcript in our current panel of 17 different phenotypic parameters, (Supplementary Fig. 5), substantial expansion of this panel may be possible even with current sequencing technology.
Analysis of tumor-infiltrating lymphocytes
To demonstrate one application of this method, we analyzed tumor-infiltrating lymphocytes (TILs) from a human colorectal cancer sample. Therapies designed to incite anti-tumor T-cell responses have recently shown great promise in the treatment of human cancer31,32, and in colorectal cancer, the presence of TILs correlates strongly with a positive prognosis33,34. To date, however, phenotypic characteristics and TCR sequences of TILs have generally been studied at the bulk population rather than at the single-cell level33–36. Thus, there is some controversy over their function and clinical significance in different tumors37 and no consensus view on their specificity or functional properties.
We applied our methodology to 736 sorted, single CD4+ TILs from one patient volunteer who underwent a colectomy for stage T3N1 rectal adenocarcinoma. For comparison, we also analyzed 372 CD4+ T cells derived from resected adjacent colon tissue from the same patient, as well as peripheral blood T cells from a different healthy donor. TCRβ sequences were successfully obtained from 597 of the 736 CD4+ T cells (81%), and we were able to assign productive, paired TCRαβ sequences to 503 of these (68% of total). In this particular tumor, we detected marked T-cell clonal expansion; the most frequent TCRβ was detected in 52/597 cells, and ten TCRβ sequences were detected in at least 8 cells (Supplementary Table 8). Out of 299 unique TCRβ sequences, the ten most frequent sequences accounted for 215/597 (36%) of the cells where sequences were recovered; 236 sequences (40%) were detected in only one cell (Supplementary Table 8).
Among the 372 CD4+ T lymphocytes derived from resected adjacent colon tissue from the same patient, we obtained TCRβ sequences from 309 cells (83%), and we were able to assign productive, paired TCRαβ sequences to 217 of these (58% of total). In contrast to the TCR repertoire from intratumoral T cells, clonal expansion was minimal, with only four TCR clones detected twice within the population (Supplementary Table 9). Also, not a single TCRαβ heterodimer sequence was shared between T cells in the tumor and adjacent colon tissue. This suggests that expanded T-cell clones present within tumors may be reacting to tumor antigens.
Next we searched for homology between TCR sequences in the intratumoral T-cell population to determine whether T-cell expansion was due to antigen-specific responses. The most highly expanded TCRβ (CASSLASMG VGELFF) sequence within our sample set varied by only two amino acids from another expanded TCRβ (CASSSASGGVGELFF) sequence. These TCRβ sequences comprised, respectively, 52 and 8 of 597 total T cells. These two expanded clones also used the same TCRα chain (CAYRPNYGGATNKLIF), although the TCRα chains used different nucleotide sequences in the two clones and were not present elsewhere within the sample set, indicating that this finding was not a result of cross-contamination (Supplementary Fig. 6 and Supplementary Table 8). Each T-cell clone also expressed a different nonproductive TCRα chain. This finding further confirms that the common TCRα chain was indeed the TCRα chain that was used by these T-cell clones, because both clones expressed only one productive TCRα chain gene. Junctional diversity describes the process of random nucleotide addition (N-nucleotide addition) or subtraction at the junctions of V(D)J rearrangements, which markedly add to TCR diversity9. In both T-cell clones, common sequence motifs within TCRα and TCRβ genes were generated as a result of significant N-nucleotide addition, indicating that these sequences would not be very common by chance (Supplementary Fig. 6). These findings strongly suggest that these two T-cell clones, which comprised over 10% (60/597) of the total CD4+ T cells we analyzed from this tumor, have been selected and activated by the same peptide-MHC ligand.
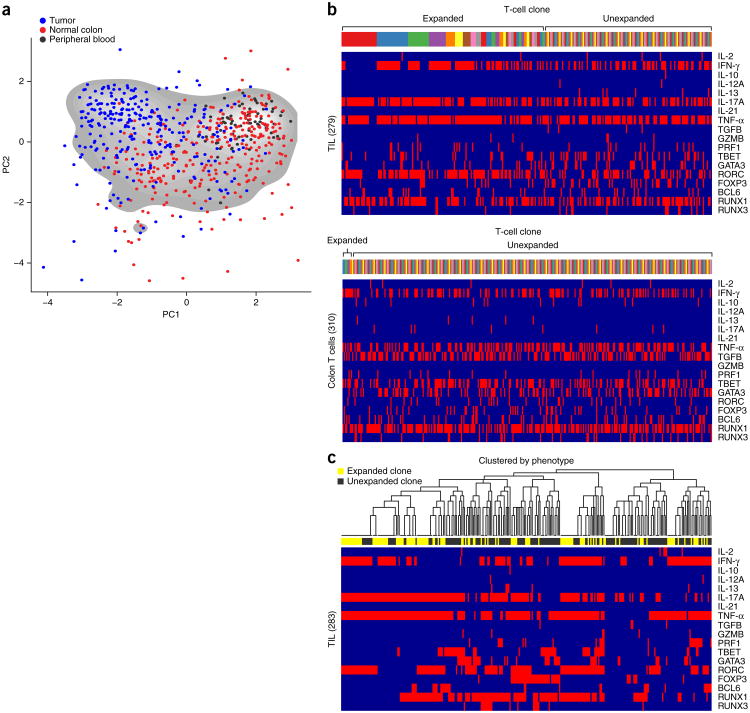
In addition to TCR sequencing, we phenotyped these cells using the 17-parameter panel described above. We stimulated half of the T cells for 3 h with PMA (phorbol 12-myristate 13-acetate) and ionomycin. Consistent with previous findings38–41, the stimulated CD4+ TILs displayed a phenotype distinct from stimulated CD4+ T cells obtained from adjacent colon or peripheral blood (Fig. 3a and Supplementary Tables 10 and 11). A higher percentage of the stimulated TIL cells expressed RORC (146/279, 52%), IL-17 (184/279, 66%), TNF-α (217/279, 78%) and IFN-γ (148/279, 74%; Fig. 3b). To visualize the data, we used principal component analysis, which concentrates the most important sources of variation in larger data sets2. This allows us to readily visualize the phenotypic diversity of CD4+ T cells (Fig. 3a and Supplementary Fig. 7). Although there is substantial overlap between the phenotypes of CD4+ T cells derived from tumor, colon and blood, these three populations of cells cluster discretely (Fig. 3a). Such phenotypic diversity is not as apparent in the absence of stimulation (Supplementary Fig. 8).
Figure 3.
TCR sequencing and phenotypic analysis of single human TILs. (a) T cells were sorted and analyzed using the procedure from Figure 1a. PCA to depict phenotypic diversity of PMA- plus ionomycin-stimulated CD4+ T cells from tumor (blue) and adjacent colon (red) of a single patient, and from peripheral blood of another healthy donor (black). PCA parameter loadings are shown in Supplementary Figure 7. Each dot represents a single T cell. (b) Top two panels: 17-parameter (parameters listed on x axis) phenotypic analysis of stimulated CD4+ T cells from tumor (top) and colon (bottom) of a single patient. Individual T cells are grouped by TCR sequence; each color on the bar above the heat maps represents a distinct TCR sequence. (c) Hierarchical clustering of different cells by phenotype, with expanded (yellow) and unexpanded (black) T-cell clones (read out by TCR sequence) indicated in the horizontal bar above the heat map.
Although CD4+ TILs were largely distinguished from the other populations by their co-expression of IL-17, RORC, TNFα and IFN-γ, there was also notable heterogeneity within each T-cell population (Fig. 3b,c). Also, individual cells frequently co-expressed multiple, different, master-regulator transcription factors, showing that the categorization of CD4+ T cells into specific subsets is not always straightforward (Fig. 3b,c).
A major advantage of our methodology is that it enables us to compare the phenotypic and functional range of T cells that can arise from a single TCR clone. For instance, we observed that compared to unexpanded T cells, a significantly higher percentage of highly expanded (≥10) T-cell clones expressed IL-17 (70/80 versus 65/126, P < 0.005) or RORC (50/80 versus 43/126, P < 0.005). Conversely, FOXP3 was less likely to be expressed in highly expanded versus unexpanded cells (5/80 versus 32/126, P < 0.005, Fig. 3b). When clustering analysis is applied, certain phenotypic clusters are preferentially occupied by unexpanded versus expanded cells or vice versa (Fig. 3c).
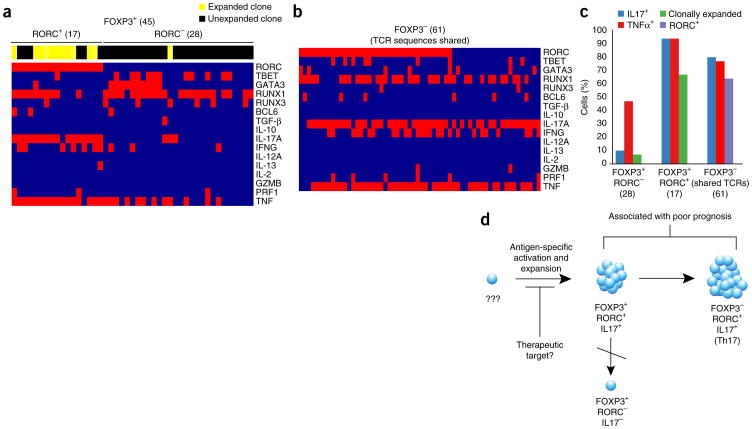
We also looked more closely at the FOXP3+ TILs (Fig. 4). The function of Treg cells in cancer has been the subject of much debate and FOXP3+ T-cell infiltration in tumors has been correlated with both favorable and poor prognoses38–41. Within this particular tumor, we found two distinct subsets of FOXP3+CD4+ T cells, differentiated by their expression of RORC (Fig. 4a). Within FOXP3+RORC+ cells, the overwhelming majority of cells expressed IL-17 (16/17, 94%), whereas IL-17 expression was rare within FOXP3+RORC− cells (3/28, 11%) (Fig. 4a). These two subsets also varied greatly with respect to the degree of clonal expansion. The FOXP3+RORC+ population consisted largely of clones that were expanded within our data set (12/17, 71%), whereas clonal expansion was rare in the FOXP3+RORC− population (1/28, 4%). Incidentally, the only FOXP3+RORC− T cell that was clonally expanded did express IL-17.
Figure 4.
TCR sequencing and phenotypic analysis of FOXP3+ TILs. (a) Clonal expansion (indicated in horizontal bar above heatmaps) and phenotype (parameters indicated on right column) of FOXP3+RORC− T cells and FOXP3+RORC+ T cells. (b) Phenotypic analysis of FOXP3− T cells that share TCR sequences with FOXP3+ populations (total numbers of cells analyzed are indicated in parentheses). (c) Frequency of cells within each indicated population of T cells (x axis) expressing phenotypic markers indicated (right). (d) Model suggested by analysis of TILs. A single T cell is activated by antigen and expands to give rise to FOXP3+RORC+ IL-17–producing T cells, which eventually lose expression of FOXP3 to give rise to FOXP3−RORC+ IL-17–producing T cells. FOXP3+RORC− T cells do not show evidence of clonal expansion, and are thus unlikely to arise from FOXP3+RORC+ cells that lose expression of RORC.
FOXP3+RORC+ IL-17-expressing T cells, described in both human colorectal cancer and in mouse models of polyposis, have been shown to have potent T-suppressive activity while being pro-inflammatory in their expression of IL-17 (refs. 39,40). Although the consequences of FOXP3+ T cell infiltration into tumors are unclear, the presence of IL-17 has been associated with tumorigenesis and poor prognosis41–43. Based on this, IL-17 has been proposed as a therapeutic target. Both FOXP3+RORC+ T cells and FOXP3-RORC+ Th17-phenotype T cells may produce IL-17 within tumors, however, the relationship between these two populations of T cells is unclear. It has been proposed that they are unrelated given the discordance between their numbers within tumors42.
To address this question, we searched for T cells that shared TCRαβ sequences with FOXP3+RORC+ T cells within our data set. We found 61 instances of FOXP3− T cells sharing TCR sequences with FOXP3+RORC+ T cells (Fig. 4b). The majority of these FOXP3− T cells also expressed IL-17 (49/61, 80%) and/or RORC (39/61, 64%). These findings indicate that these two populations of IL-17-expressing T cells share a common ancestry and are consistent with the idea that FOXP3+RORC+ T cells within tumors lose FOXP3 expression to become Th17 cells44,45. The relationship between FOXP3+RORC− T cells and FOXP3+RORC+ T cells is not as clear. We cannot say whether the FOXP3+RORC+ T cells we observed originated as FOXP3+RORC− T cells which underwent clonal expansion. However, we did not detect TCR sequences shared between these two populations, and this suggests that FOXP3+RORC− T cells did not originate from clonally expanded FOXP3+RORC+ T cells.
Interestingly, both of the expanded TIL clones that express highly similar TCRs (Supplementary Fig. 6) contained cells expressing IL-17 and RORC. Among the 27 stimulated cells in the first TCRβ clone (CASSLASMGVGELFF), 24 expressed IL-17 and 16 expressed RORC. One cell co-expressed both FOXP3 and RORC. For the second TCRβ clone (CASSSASGGVGELFF), only two of eight sequences were present in the stimulated sample. Both of these T cells expressed IL-17 and one expressed RORC. Taken together, our data show clear heterogeneity between FOXP3+ T cells within tumors, which might help explain the why the data regarding the function of Treg cells in tumors have been controversial.
Discussion
The approach we describe here enables highly efficient TCR determination and multiparametric phenotypic analysis in single T cells. It requires no proprietary reagents or materials, and can be performed in any standardly equipped laboratory with access to flow cytometry and deep sequencing. To our knowledge, we describe the highest reported efficiency in sequencing TCRs from single T cells. Furthermore, our method is uniquely suited to identifying multiple TCRα chains from single T cells.
We demonstrate the utility of this technology by analyzing TILs, and show how TCR sequences can add an invaluable dimension to multiparametric phenotypic analysis by marking the ancestry of particular T cells, especially when the antigen is not known. For example, we show that FOXP3+RORC+ T cells and FOXP3−RORC+ Th17-phenotype cells can share a common ancestry (indicated by identical TCR sequence), indicating that the activation of a single T cell can lead to subsequent clonal expansion, loss of FOXP3 expression and differentiation to Th17-phenotype cells (Fig. 4d). Furthermore, we show an example of two expanded T-cell clones with highly homologous TCR sequences; among the two clonal populations are members expressing IL-17 and FOXP3. Because these two highly expanded T-cell clones appear to be responding to the same peptide-MHC, antigen-specificity is likely important to the selection of these T cells (Fig. 3d). More work is needed to understand the signals and antigens that lead to activation, loss of FOXP3 expression and clonal expansion of these IL-17–producing T cells within tumors. Also, TILs from colorectal cancer have been shown to be heterogeneous with respect to IL-17 secretion, so these results need to be validated on additional tumor samples40. But given the association of IL-17 with tumorigenesis and poor clinical outcomes, the event(s) responsible for initially activating these cells might represent an attractive therapeutic target.
This technology is also complementary to recently developed methods to determine ligands for TCRs using random peptide-MHC libraries11. These complementary approaches can enable the identification of potentially important disease-associated TCRs and subsequent discovery of their antigens. Many human diseases including cancer, autoimmune disease and infectious diseases are characterized by T-cell activation and proliferation; what antigens are driving these T-cell responses may not be known. Identification of these antigens could be invaluable in understanding the origin of both beneficial and potentially detrimental T-cell responses and provide targets for therapeutic intervention.
Online Methods
Single-cell sorting and flow cytometry
All fluorescence-activated cell sorting (FACS) experiments were performed on ARIA II instruments (Becton Dickinson) in the Stanford Shared FACS Facility. Cytokine capture assays (Miltenyi Biotec) were performed per manufacturer's instructions on freshly isolated human peripheral blood mononuclear cells (PBMCs). PBMCs were collected from a healthy donor who gave informed consent. The Jurkat T-cell leukemia cell line (Clone E6-1) was obtained from ATCC The following antibody clones were used for flow cytometry: anti-CD3 (SK7, BioLegend), anti-CD4 (RPA-T4, BioLegend), anti-CD8 (OKT8, eBiosciences), anti-αβTCR (IP26, BioLegend), anti-CD25 (2A3, Becton Dickinson) and anti-FOXP3 (PCH101, eBiosciences). Dead cells were excluded using a LIVE/DEAD Fixable Dead Cell Stain kit (Invitrogen).
TIL preparation
The Stanford University Institutional Review Board approved all protocols for collection of human tissue and blood. Tissue was collected with informed consent from a patient undergoing colon resection for colon cancer at Stanford University Hospital after initially being processed by the Department of Pathology. Tumor tissue was cut into small pieces and incubated in 10 mM EDTA (EDTA) in PBS for 30 min. Cells in suspension were passed through a 10-μM nylon cell strainer (Becton Dickinson). Tissue was then incubated in RPMI with 5% FCS containing 0.5 mg/ml of Type 4 collagenase for 30 min (Worthington Biochemical). Tissue was periodically disrupted during incubation by passing through a syringe topped with a blunt-ended 16-gauge needle. Lymphocytes were enriched through Percoll (GE Healthcare) gradient centrifugation. Cells were frozen in complete RPMI containing 10% DMSO (dimethylsulfoxide) and 40% FCS (FCS) for later use. Prior to use, cryopreserved lymphocytes were thawed and washed with complete RPMI before overnight recovery at 37°C. Cells were transferred to tubes, washed and resuspended in cytometry buffer (PBS + 0.05% sodium azide + 2 mM EDTA + 2% FCS) for staining. For stimulation, cells were cultured for 3 h at approximately 15 × 106/ml in complete RPMI (10% FCS) and 150 ng/ml PMA + 1 μM ionomycin. At the end of the 3 h stimulation, cells were pipetted vigorously to remove adherent cells from the plate and transferred to tubes, washed and resuspended in cytometry buffer (PBS + 0.05% sodium azide + 2 mM EDTA + 2% FCS).
TCR sequencing and phenotyping
Single-cell sorting was performed using an ARIA II cell sorter (Becton Dickinson). TCR sequence and gene expression analysis from single cells were obtained by a series of three nested PCR reactions as described13. Cells were sorted directly into RT-PCR buffer. For the first reaction, reverse transcription and preamplification were performed with a One-Step RT-PCR kit (Qiagen) using multiplex PCR with multiple Vα and Vβ region primers, Cα and Cβ region primers, and phenotyping primers in a 20-μl reaction (Supplementary Tables 1 and 2). For the PCR reaction #1, the final concentration of each TCR V-region primer is 0.06 μM, each C-region primer is 0.3 μM, each phenotyping primer is 0.1 μM. A 25-cycle first RT-PCR reaction was done per manufacturer's instructions using the following cycling conditions: 50°C 30 min; 95°C 15 min; 94°C 30 s, 62°C 1 min, 72°C 1 min × 25 cycles; 72°C 5 min; 4°C. Next, a 1-μl aliquot of the first reaction was used as a template for second 20-μl PCR using HotStarTaq DNA polymerase (Qiagen) for either TCR sequencing or phenotyping. The following cycling conditions were: 95°C 15 min; 94°C 30 s, 64°C 1 min, 72°C 1 min × 25 cycles (TCR) or 35 cycles (phenotyping); 72°C 5 min; 4°C. For the TCR sequencing reaction, multiple internally nested TCRVα, TCRVβ, TCRCα and Cβ primers (Supplementary Table 1) were used (V primers 0.6 μM, C primers 0.3 μM). For the phenotyping reaction, multiple internally nested phenotyping primers (Supplementary Table 2) were used (0.1 μM). The second set of TCR V-region primers and 5′ phenotyping primers contained a common 23-base sequence at the 5′ end to enable further amplification (during the third reaction) with a common 23-base primer. The second set of 3′ phenotyping primers contained a common 24-base sequence to enable further amplification (during the third reaction). 1-μl aliquot of the second PCR product was used as a template for the third 20-μl PCR reaction, which incorporates barcodes and enables sequencing on the Illumina MiSeq platform. For the third and final PCR reaction for TCR sequencing, amplification was performed with HotStarTaq DNA polymerase for 36 cycles using a 5′ barcoding primer (0.05 μM) containing the common 23-base sequence and a 3′ barcoding primer (0.05 μM) containing sequence of a third internally nested Cα and/or Cβ primer, and Illumina Paired-End primers (0.5 μM each, Supplementary Table 3). For tumor-infiltrating and colonic T-cell analysis, the final barcoding PCR reactions for TCR alpha and TCR beta were combined. When the third reaction was performed together, the 3′ Cα barcoding primer was used in threefold excess to the 3′ Cβ barcoding primer (0.15 μM and 0.5 μM, respectively). In addition to the common 23-base sequence at the 3′ end (that enables amplification of products from the second reaction) and a common 23-base sequence at the 5′ end (that enables amplification with Illumina Paired-End primers), each 5′ barcoding primer contains a unique 5-base barcode that specifies plate and a unique 5-base barcode that specifies row within the plate (Supplementary Fig. 1). These 5′ barcoding primers were added with a multichannel pipette to each of 12 wells within a particular row within a particular plate. In addition to the internally nested TCR C-region sequence and a common 23-base sequence at the 3′ end (that enables amplification with Illumina Paired-End primers), each 3′ barcoding primer contains a unique 5-nucleotide barcode that specifies column. These 3′ barcoding primers were added with a multichannel pipette to each of eight wells within a column within all plates. The third reaction for phenotyping was performed in a similar manner to the TCR sequencing, except that the 3′ primer contains the common 24-base sequence contained in all 3′ primers from the second reaction rather than the internally nested TCR C-region primer. The same 5′ barcoding primers were used for the third phenotyping reaction as the TCR sequencing reaction. After the third and final PCR reaction, each PCR product should have a unique set of barcodes incorporated that specifies plate, row and column and have Illumina Paired-End sequences that enable sequencing on the Illumina MiSeq platform. The PCR products were combined at equal proportion by volume, run on a 1.2% agarose gel, and a band around 350 to 380 bp was excised and gel purified using a Qiaquick gel extraction kit (Qiagen). This purified product was then sequenced.
PCR primer design
All primer sequences are provided in Supplementary Figure 1 and Supplementary Tables 1-3. All primers were designed to have a Tm of 70–72°C (Tm = 4×[GC]+2[AT]). For TCR primers, base degeneracy was incorporated into the primers when necessary to account for TCR polymorphism and ensure amplification of all known functional Vα, Vβ, Cα and Cβ regions identified in the IMGT database (http://www.imgt.org/). V-region primers were designed to be at least 50 bases from the distal end to ensure inclusion of the entire CDR3 region. All TCR and phenotyping primers for the second reaction contain the common sequence CCAGGGTTTTCCCAGTCACGAC at the 5′ end, which enables amplification with barcoding primers during the third reaction. All phenotyping primers for the second reaction contain the common sequence AGCGGATAACAATTTCACACAGGA at the 5′ end, which enables amplification with barcoding primers during the third reaction. After all reactions are performed, TCR primers amplify a segment of the TCR of approximately 250 bp. The final product for sequencing is approximately 380 bp. Phenotyping PCR primers were designed to span introns and amplify all major variants of the genes present in the NCBI database (http://www.ncbi.nlm.nih.gov). After the second reaction is performed, phenotyping primers amplify a gene segment of approximately 200 bp, and the final sequencing product is approximately 350 bp.
Sequencing data analysis
Raw sequencing data were processed and demultiplexed using a custom software pipeline to separate reads from every well in every plate according to specified barcodes. All paired ends are assembled by finding a consensus of at least 100 bases in the middle of the read. The resulting paired-end reads are then assigned to wells according to barcode. Primer dimers are filtered out by establishing minimum length of 100 bases for each amplicon. See Supplementary Note for further details on software. For example, in a recent sequencing run consisting of 2,164 cells, we obtained 2.01 × 107 raw reads, 1.95 × 107 pass-filtered reads (Illumina.com), forward/reverse consensus sequences were obtained and barcodes assigned to 1.66 × 107 reads, with 1.60 × 107 reads above 100 bases. The average read number per well was 7,382 ± 5,366. A consensus sequence is obtained for each TCR gene. Because multiple TCR genes might be present in a given well, our software establishes a cutoff of >95% sequence identity within a given well. All sequences exceeding 95% sequence identity are assumed to derive from the same TCR gene and a consensus sequence is determined. The 95% cutoff conservatively ensures all sequences derived from the same transcript would be properly assigned, even given a PCR rate of 1/9,000 bases, and sequencing error rate up to 0.4%22. TCR V, D and J segments were assigned by VDJFasta19. For phenotyping transcripts, the number of reads containing a 95% match to the customized database of transcription factor and cytokine genes are scored.
Single-well depth and dominance cutoff parameter validation
For both TCR and phenotypic parameters, there is a low background of unrelated sequences (Supplementary Fig. 3). We quantified potential background through high-depth sequencing and set thresholds accordingly. To quantify this background, sequencing was done at high depth. Single, PMA and ionomycin-stimulated T cells were sorted into two plates and processed for TCR and phenotypic analysis. Into these two plates, no cells were sorted into 16 wells, scattered through all columns and rows. Eight of these wells were processed normally with all reagents added. Eight of these wells were left completely blank throughout the analysis with no reagents added. These two plates (as opposed to the usual 20 to 25 plates) were run on a single sequencing run to give a sequencing depth more than tenfold higher than usual.
There was no significant difference in TCR background reads in negative-control wells without sorted T cells, regardless of whether wells were processed with reagents (Supplementary Fig. 3a,b). These data indicate that background is primarily due to error in PCR, sequencing or oligonucleotide synthesis within the barcodes and not due to cross-contamination.
For TCR reads within the two test plates, we validated cutoff criteria by simulated subsampling (Supplementary Fig. 3a,b). Plates were sequenced to an average depth of >45,000 reads per well, and subsampled to depths ranging from 100 to 45,000 average reads per well. By quantifying background signal (negative-control wells), we provide justification for thresholds set in the analysis. For TCR analysis, we establish a threshold normalized depth (based on average number of reads per well in the plate) of 10%. Using normalized depth independently, there is a clear separation between wells containing cells and background signal in negative-control wells at all depths down to 100 reads/well. For TCR analysis, establishing thresholds for clone dominance within the well further excludes the majority of negative-control wells and wells potentially containing more than one cell. For beta chains, a domain dominance cutoff is set at >85%. Domain dominance is determined based on 100% identity in sequence. Thus, this threshold of 85% is considerably lower than 100% because it accounts for the presence of PCR mutation or sequencing error. Because multiple TCRα chains can exist within a given cell, the threshold for domain dominance is more permissive and set to 10%.
For phenotypic parameters, unlike for TCR genes, not all cells express a given parameter. Therefore, background is expected to depend upon number of cells expressing a given parameter as well as read depth. To investigate background for phenotypic parameters, we performed analysis on two plates; we sorted PMA plus ionomycin-stimulated IL-17+ single T cells into 40 wells, PMA plus ionomycin-stimulated IL-17− single T cells into 40 wells, and left 16 wells as negative-control wells. Eight of these negative-control wells were processed normally with all reagents added. Eight of these wells were left completely blank throughout analysis with no reagents added. IL-17+ and IL-17−T cells were sorted because this population gives a variable range of cells expressing all phenotypic parameters within the plate. We assessed background levels of each phenotypic parameter signal in negative-control wells. As was the case with TCR, there was no marked difference in background between negative wells processed with (0.54 background reads/well) or without reagents (0.72 background reads/well), suggesting that background is primarily due to error in PCR, sequencing or oligonucleotide synthesis within the barcodes and not due to cross-contamination. Background was directly proportional to number of reads for each particular parameter on a plate and number of cells expressing a given parameter (Supplementary Fig. 3c). The ratio of reads/negative-control well versus total reads/well for each phenotypic parameter in a given plate is approximately 1.23 × 10−3. This ratio is constant, independent of the frequency of cells expressing a given parameter.
We then performed high-depth analysis on one plate containing 80 wells with single T cells and 16 negative-control wells to further investigate background per well. The plate was sequenced to an average depth of >45,000 reads per well, and subsampled to depths ranging from 100 to 45,000 average reads per well. We individually assessed the two phenotypic parameters with the highest level of background on this plate, RUNX1 and GATA3. For RUNX1 and GATA3, respectively, the ratio of reads/negative-control well versus total reads/well was 1.30 × 10−3 and 1.71 × 10−3 consistent with levels established in the analysis of the prior plate (Supplementary Fig. 3c). This indicates that relative background does not vary significantly, even at high-read depth. We assessed RUNX1 and GATA3 signal in 80 wells containing T cells and 16 negative-control wells (Supplementary Fig. 3d). Setting a threshold to 1 s.d. below the mean of log read counts per well (in all wells within a sequencing run expressing a given parameter) provides a scale-free means of conservatively excluding all background signals for phenotypic parameters. The accuracy of this threshold does not vary as a function of frequency of cells expressing the parameter, as only wells expressing a given parameter are included.
Sensitivity of detection
We further investigated the sensitivity of our method for detection of a particular transcript. A synthetic double-stranded (ds)DNA was constructed that contains binding sites for our IL-17 primers (Supplementary Fig. 5a). The construct is identical to the exogenous IL-17 amplicon except 15 nucleotides of endogenous IL-17 sequence is replaced with a 15-nucleotide random molecular barcode giving a theoretical diversity of >109 (415). This molecular barcode was incorporated to enable tracking of individual molecules during the analysis and further validate quantification (we should not be able to detect more molecular barcodes in a given well than molecules added if quantification was accurate). This synthetic construct was made by PCR using a 124-base 5′ primer incorporating the primer sequences and the molecular barcode (5′ GCG TAA TAC GAC TCA CTA TAG GGA GAC AGA CAA GAA CTT CCC CCG GAC TGT GAT GGT CAA CCT GAA CAT CCA TAA CCG GAA CAT NNN NNN NNN NNN NNN CAA AAG GTC CTC AGA TTA CTA CAA C). To ensure that unique barcodes were not amplified, the template was first amplified by 60-cycle reaction using only the 5′ primer, and then 1 cycle was performed after addition of the 3′ primer. The PCR product was purified and quantified. The product was quantified by Nanodrop 2000 (Thermo Scientific) and Bioanalyzer 2100 (Agilent). Based upon these calculations, serial dilutions were performed and quantities were further verified by performing 50-cycle PCRs using primers within the template sequence. We spiked this synthetic construct into wells at different serial dilutions indicated and performed reactions and analysis on two plates. These two plates were processed identically, except a single, stimulated T cell was added to one of the plates. Into both plates, eight negative-control wells were processed without spiked template or cells.
We demonstrate that the method can detect as little as one molecule of dsDNA template (equivalent to two molecules of mRNA) (Supplementary Fig. 5b). Sensitivity improves with increased copy number and 100% sensitivity is achieved when 8 molecules of dsDNA (equivalent of 16 molecules of mRNA) are spiked into the initial reaction (Supplementary Fig. 5b). Although sensitivity does improve with increased copy number, read count per well does not significantly change (Supplementary Fig. 5c). This indicates that our readout is binary and read depth will not significantly affect sensitivity; in other words, sequencing at a higher depth will not improve identification of low-abundance transcripts in cells. Furthermore, the sensitivity of detection for one particular phenotypic parameter is not affected by the presence of other transcripts, as the sensitivity of detection for this template does not differ when stimulated T cells are added to the reaction and other amplified transcripts are present (Supplementary Fig. 5c). The number of molecular barcodes detected in a well is directly proportional to number of molecules added (Supplementary Fig. 5d). No molecular barcodes were repeated in different wells in our data set after accounting for background and the presence of PCR or sequencing error (Supplementary Fig. 5d).
Mean read counts per well for each phenotypic parameter did not vary markedly for phenotypic parameters present in at least 50 cells in our tumor and colon T-cell data set, which were sequenced to similar read depth (Supplementary Fig. 5e).
Single-cell sequencing accuracy
PCR error occurs at a rate of 1/9,000 bases and sequencing error has been reported to occur at a rate up to 0.4%22. Our method relies on generation of a consensus sequence from 10–10,000 reads, thus establishing single-cell transcript coverage far superior to that provided by genomic sequencing, mitigating the role of PCR error and largely eliminating sequencing error. To determine the accuracy of sequencing, we looked at the incidence of error within phenotyping transcripts that are entirely germline encoded, unlike TCR genes. When consensus sequence was obtained for all phenotyping transcripts within individual wells, the sequences were always identical. This indicates that despite sequencing or PCR error, the consensus sequence derived from a given well from >10 reads is 100% accurate within our data set.
Statistical analysis
To determine sensitivity (SN), specificity (SP), positive predictive value (PPV) and negative predictive value (NPV) (Supplementary Table 7), the following formulas were used: SN = TP/(TP + FN), SP = TN/(FP + TN), PPV = SN × PRV/[(SN × PRV + (1 − SP) × (1 − PRV)], NPV = SP × (1 − PRV)/[(1 − SN) × PRV + SP × (1 − PRV)]. TP = True Positives, TN = True Negative, FP = False Positives, FN = False Negatives. PRV = Prevalence, determined by percentage of cells analyzed that were positive for the given parameter by flow cytometry. Chi-squared test was used to determine the statistical significance of skewing of phenotypic parameters within expanded versus unexpanded T-cell clones.
Supplementary Material
Acknowledgments
We thank members of the Davis laboratory and the Y.-H. Chien laboratory for helpful discussions. We thank E. Newell for critical reading of the manuscript and helpful suggestions. We thank C. Bolen for assistance with data analysis. We thank X. Ji for deep sequencing. Tissue was obtained through the Stanford University Tissue Bank. The Stanford Shared FACS Facility provided use of equipment and the Stanford Functional Genomics Facility provided deep sequencing services. A.H. is supported by a grant from the Simons Foundation. L.H. is supported by a fellowship from the German Research Foundation (D.F.G.). M.M.D. is funded by the US National Institutes of Health and is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. GenBank: KM004027–KM004996
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Author Contributions: A.H. conceived the project and experiments, designed methodology and primers, performed experiments, analyzed data, assisted in the optimization of the software pipeline, generated figures and wrote the manuscript. J.G. designed the software, analyzed data and generated figures. L.H. designed and performed experiments, analyzed data and generated figures. M.M.D. conceived the project and experiments and wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Wills QF, et al. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 2.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 4.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler's guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newell EW, Davis MM. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat Biotechnol. 2014;32:149–157. doi: 10.1038/nbt.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krogsgaard M, Davis MM. How T cells ‘see’ antigen. Nat Immunol. 2005;6:239–245. doi: 10.1038/ni1173. [DOI] [PubMed] [Google Scholar]
- 9.Murphy K, Travers P, Walport M, Janeway C. Janeway's Immunobiology. 8th. Garland Science; 2012. [Google Scholar]
- 10.Newell EW, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birnbaum ME, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinrichs CS, Restifo NP. Reassessing target antigens for adoptive T-cell therapy. Nat Biotechnol. 2013;31:999–1008. doi: 10.1038/nbt.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han A, et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc Natl Acad Sci USA. 2013;110:13073–13078. doi: 10.1073/pnas.1311861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SM, et al. Analysis of the paired TCR alpha- and beta-chains of single human T cells. PLoS ONE. 2012;7:e37338. doi: 10.1371/journal.pone.0037338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dash P, et al. Paired analysis of TCRalpha and TCRbeta chains at the single-cell level in mice. J Clin Invest. 2011;121:288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gascoigne NR, Alam SM. Allelic exclusion of the T cell receptor alpha-chain: developmental regulation of a post-translational event. Semin Immunol. 1999;11:337–347. doi: 10.1006/smim.1999.0190. [DOI] [PubMed] [Google Scholar]
- 17.Malissen M, et al. Regulation of TCR alpha and beta gene allelic exclusion during T-cell development. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 18.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245–248. doi: 10.1038/84701. [DOI] [PubMed] [Google Scholar]
- 21.Yanagi Y, Chan A, Chin B, Minden M, Mak TW. Analysis of cDNA clones specific for human T cells and the alpha and beta chains of the T-cell receptor heterodimer from a human T-cell line. Proc Natl Acad Sci USA. 1985;82:3430–3434. doi: 10.1073/pnas.82.10.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura K, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law JP, et al. The importance of Foxp3 antibody and fixation/permeabilization buffer combinations in identifying CD4+CD25+Foxp3+ regulatory T cells. Cytometry A. 2009;75:1040–1050. doi: 10.1002/cyto.a.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vahedi G, Kanno Y, Sartorelli V, O'Shea JJ. Transcription factors and CD4 T cells seeking identity: masters, minions, setters and spikers. Immunology. 2013;139:294–298. doi: 10.1111/imm.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 27.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assenmacher M, Lohning M, Radbruch A. Detection and isolation of cytokine secreting cells using the cytometric cytokine secretion assay. Curr Protoc Immunol. 2002;4627:6. doi: 10.1002/0471142735.im0627s46. [DOI] [PubMed] [Google Scholar]
- 29.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 30.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 31.Ribas A. Tumor immunotherapy directed at PD-1. N Engl J Med. 2012;366:2517–2519. doi: 10.1056/NEJMe1205943. [DOI] [PubMed] [Google Scholar]
- 32.Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–1198. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 33.Pagès F, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 34.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 35.Gerlinger M, et al. Ultra-deep T-cell receptor sequencing reveals the complexity and intratumour heterogeneity of T-cell clones in renal cell carcinomas. J Pathol. 2013;231:424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer immunol immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasada T, Suekane S. Variation of tumor-infiltrating lymphocytes in human cancers: controversy on clinical significance. Immunotherapy. 2011;3:1235–1251. doi: 10.2217/imt.11.106. [DOI] [PubMed] [Google Scholar]
- 38.deLeeuw RJ, Kost SE, Kakal JA, Nelson BH. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18:3022–3029. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 39.Scurr M, Gallimore A, Godkin A. T cell subsets and colorectal cancer: discerning the good from the bad. Cell Immunol. 2012;279:21–24. doi: 10.1016/j.cellimm.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 41.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer immunol immunother. 2011;60:909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blatner NR, et al. Expression of RORgammat marks a pathogenic regulatory T cell subset in human colon cancer. Sci Transl Med. 2012;4:164ra159. doi: 10.1126/scitranslmed.3004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gounaris E, et al. T-regulatory cells shift from a protective anti-inflammatory to a cancer-promoting proinflammatory phenotype in polyposis. Cancer Res. 2009;69:5490–5497. doi: 10.1158/0008-5472.CAN-09-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.