Abstract
Identifying pathways by which genetic Alzheimer’s disease (AD) risk factors exert neurocognitive effects in young adults is essential for the effort to develop early interventions to forestall or prevent AD onset. Here, in a brain-imaging cohort of 59 young adults, we investigated effects of a variant within the clusterin (CLU) gene on working memory function and grey matter volume in cortical areas that support working memory. In addition, we investigated the extent to which effects of CLU genotype on working memory were independent of variation in the strongest AD risk factor gene apolipoprotein E (APOE). CLU is among the strongest genetic AD risk factors and, though it appears to share AD pathogenesis-related features with, APOE, it has been far less well studied. CLU genotype was associated with working memory performance in our study cohort. Notably, we found that variation in grey matter volume in a parietal region, previously implicated in maintenance of information for working memory, mediated the effect of CLU on working memory performance. APOE genotype did not affect working memory within our sample, and did not interact with CLU genotype. To our knowledge, this work represents the first evidence of a behavioral effect of CLU genotype in young people. In addition, this work identifies the first gene-brain-cognition mediation effect pathway for the transmission of the effect of an AD risk factor. Relative to conventional pairwise associations in cognitive neurogenetic research, gene-brain-cognition mediation modeling provides a more integrated understanding of how genetic effects transmit from gene to brain to cognitive function.
Keywords: Clusterin, Working Memory, MRI, Gene-Brain-Cognition Mediation Modeling
Alzheimer’s disease (AD) is the most common form of dementia, affecting about 11% of people over 65 years old and 32% of people over 85, creating a pressing public health concern (Thies & Bleiler, 2013). AD etiology has a strong genetic component, with heritability between 60–80% (Gatz et al., 2006). The earliest and most consistent genetic finding in AD genetics is the association between the ε4 variant of the apolipoprotein E (APOE) gene and risk for AD (Corder et al., 1993). However, genome-wide association studies (GWAS) have highlighted other risk genes. In 2009, two independent GWAS reported AD risk associated with a single nucleotide polymorphism (SNP), rs11136000, in the clusterin (CLU) gene, also known as apolipoprotein J (APOJ), which encodes a protein similar to APOE (Harold et al., 2009; Lambert et al., 2009). The CLU-T allele is protective against AD (0.86 odds ratio), whereas the CLU-C allele (defined here as “normal risk”) is associated with relatively greater risk of AD compared to the T allele. CLU has been implicated in AD risk across multiple studies (Carrasquillo et al., 2010; Corneveaux et al., 2010; Jun et al., 2010; Kamboh et al., 2012; Seshadri et al., 2010), and meta-analyses demonstrate it to be the third most significantly associated genetic risk factor associated with AD (Alzgene: http://www.alzgene.org; (Bertram & Tanzi, 2010).
Given the recent association of CLU with Alzheimer’s disease, it is important to investigate whether differences in CLU rs11136000 risk status are associated with differences in brain structure or cognitive function. Key questions remain about the effects of CLU in young healthy people, which may support the development of early biomarkers that present before the onset of AD. Memory-related neurocognitive variables that are compromised in AD pathogenesis are strong candidates for investigation, including working memory, which was the focus of the present investigation.
Effects of CLU rs11136000 on cognition and brain functioning in elderly populations have been examined by some recent studies. In a large sample of individuals in their nineties, homozygous C allele carriers had worse cognitive functioning as measured by a composite score consisting of verbal fluency, forward and backward digit span, and immediate and delayed recall (Mengel-From, Christensen, McGue, & Christiansen, 2011). A postmortem study found the C allele was significantly associated with the presence of late stage AD related senile plaques compared to the T allele (Kok et al., 2011). Another study investigating longitudinal effects of rs11136000 in elderly individuals did not find any difference in memory performance measured by the California Verbal Learning Test in individuals who remained cognitively normal throughout the study. However, that study did find that, for individuals that converted to mild cognitive impairment (MCI) or AD, C allele carriers showed quicker rates of decline than the T allele carriers (Thambisetty et al., 2013).
Few studies have investigated CLU rs11136000 in healthy younger adults to assess whether AD risk-related effects can be detected prior to AD onset. A study investigating white matter integrity using diffusion tensor imaging found that the CLU C allele was associated with lower fractional anisotropy, a measure of white matter integrity. This investigation encompassed multiple areas involved in AD pathology, including the fornix and cingulum (Braskie et al., 2011). An fMRI investigation of functional connectivity during a memory recall task found lower connectivity in C carriers between the prefrontal cortex (PFC) and the hippocampus, but equivalent recall scores (Erk et al., 2011). A study investigating emotional working memory using images of expressive human faces as stimuli did not observe differences in working memory performance, but homozygous C allele carriers had elevated activity in the dorsolateral PFC, hippocampus, and the dorsal posterior cingulate (Lancaster et al., 2011).
Because working memory is susceptible to AD pathology (Baddeley, Logie, Bressi, Sala, & Spinnler, 1986; Belleville, Peretz, & Dominique, 1996; Stopford, Snowden, Thompson, & Neary, 2007; Stopford, Thompson, Neary, Richardson, & Snowden, 2012), and has previously demonstrated effects of early AD risk in young APOE-ε4 carriers (Wishart et al., 2006), it is a priority to clarify whether CLU genotype affects working memory in young people when not confounded by face and emotion processing. AD does not affect the processing of emotional cues in the same way that it affects memory, especially in early and moderate stages of the disease (Bucks & Radford, 2004; Dequeker et al., 2002; Evans-Roberts & Turnbull, 2011; Henry, Rendell, Scicluna, Jackson, & Phillips, 2009). Thus, it is not likely that these processes would show the same effects of early AD risk that may be seen in working memory, and differences in working memory ability might be masked by compensatory strategies based on emotion and face processing when such strategies are available.
Here, we investigated the effect of CLU genotype on working memory, without the influence of emotional processing, using a verbal n-back task. Working memory is underpinned by frontoparietal brain networks (for a review see (Collette, Hogge, Salmon, & Van der Linden, 2006)). PET studies with fluorodeoxyglucose (F18) have found decreased glucose metabolism in the PFC, parietal cortex, or both, associated with worse working memory performance in AD patients (Collette, Salmon, Linden, Degueldre, & Franck, 1997; Desgranges et al., 1998; Kalpouzos et al., 2005). An fMRI study of AD patients found decreased BOLD activity in PFC, which was related to worse working memory performance (Lim et al., 2008). The same parietal and frontal cortical regions that underlie working memory are subject to atrophy during the course of AD (Buckner et al., 2005; Jacobs, Boxtel, Jolles, Verhey, & Uylings, 2012; Whitwell et al., 2007). Lastly, regional grey matter volume in frontoparietal cortex has been shown to positively correlate with working memory performance over the human lifespan (Bartrés-Faz et al., 2009; Colom, Jung, & Haier, 2007).
Given that individuals with AD manifest deficits in working memory and have brain atrophy in brain regions involved in working memory, and that grey matter volumes in these areas predict better working memory across the lifespan, we chose to use voxel-based morphometry (VBM) to investigate CLU effects on grey matter volume in regions associated with working memory. Specifically, we sought to test the hypothesis that differences in grey matter volume in the frontoparietal network involved in n-back performance mediate the effect of CLU genotype on working memory performance in healthy young adults. This approach is notable given the paucity of gene-brain-cognition pathways that have thus far been tested in the developing cognitive neurogenetics literature. We have recently demonstrated the viability of gene-brain-cognition mediation modeling (Green, Kraemer, Deyoung, Fossella, & Gray, 2013), which represents a more cohesive integration of genetic, neural, and behavioral datasets than conventional pairwise associations (Green & Dunbar, 2012; Green et al., 2008, 2013).
Though the literature on CLU in young people is limited, it is possible to formulate hypotheses based on the somewhat more developed APOE literature. APOE and CLU appear to affect similar pathways leading to the development of AD (Green, et al., 2014; Wu, Yu, Li, & Tan, 2012). The apoE and apoJ proteins share a number of important characteristics: they are among very few proteins associated with brain lipoproteins (Elliott, Weickert, & Garner, 2010; Koch et al., 2001); they interact with a shared set of cell surface receptors (Kounnas et al., 1995); they promote neurite outgrowth (Kang et al., 2005; Nathan et al., 1994); and elimination of apoE or apoJ in an AD mouse model caused similar effects on Aβ accumulation (DeMattos et al., 2004). In young adults, APOE-ε4 genotype has been associated with increased neuronal activity in various operations such as memory encoding (Dennis et al., 2010; Filippini et al., 2009). Alterations in brain structure have also been noted, such as decreased structural integrity of white matter tracts in ε4 allele carriers as measured by diffusion tensor imaging (Persson et al., 2006). However, the ε4 allele has also been associated with enhanced cognitive performance in young people, including verbal fluency and decision making, as well as enhanced neural efficiency and better delayed recall during an episodic memory task (Marchant, King, Tabet, & Rusted, 2010; Mondadori et al., 2007). These somewhat counterintuitive findings of AD risk-associated benefits suggest that AD genetic risk factors may have antagonistic pleiotropic effects in some cognitive domains (i.e., beneficial effects during prime reproductive years that may lead to neurocognitive costs later in life; (for example see (Han & Bondi, 2008; Tuminello & Han, 2011)).
Based on this literature, we generated two alternative hypotheses. If the CLU-C allele confers early neurocognitive components of AD-related deficits, we should observe lower working memory performance scores measured by the n-back task. However, if CLU-C has antagonistic pleiotropic effects similar to those observed for APOE-ε4, we should see better working memory performance for the CLU-C allele carriers. In both cases, we hypothesized that differences in performance should be mediated by differences in cortical grey matter volume in regions specific to n-back performance (i.e., a gene-brain-cognition pathway). Though the CLU gene was this study’s primary target of investigation, we also considered APOE genotype to investigate whether any effects associated with CLU were dependent or independent of APOE variation.
Materials and Methods
Participants
Fifty-nine undergraduates and community members participated in the behavioral n-back task and also underwent structural MRI. Participants were right-handed native English speakers with no history of mental illness, serious brain injury, or psychoactive drug use (specifically, medications and illicit drugs; excepting alcohol, tobacco, and caffeine), provided informed consent for participation in behavioral and fMRI sessions. All procedures received IRB approval prior to the study. Four participants were excluded due to indeterminate genotyping results, such that a total of 55 participants were included in our analyses. These participants (29 male) ranged in age from 18–36 years (mean = 22.8, SD = 4.14).
Genotyping
We performed genotyping of the clusterin polymorphism at CLU rs11136000, APOE polymorphism at rs429358 (codon 112), and APOE polymorphism at rs7412 (codon 158) using the TaqMan assay (Applied Biosystems). A reaction volume of 25 μL containing 50 ng DNA, 5 mL MgCl2 and 1X TaqMan Universal PCR Master Mix containing AmpliTaq Gold DNA Polymerase was amplified using 40 cycles of 15s at 95°C and 1 min at 60°C. A total of 0.2μM of each of the sequence-specific probes 5′-6FAM ACCAAAGCCACACCAGCTATCAAAA[T]TCTCTAACGGGCCCTTGCCACTTGA-TAMRA-3′ and 5′-VIC-ACCAAAGCCACACCAGCTATCAAAA[C]TCTCTAACGGGCCCTTGCCACTTGA-TAMRA-3′ was used in the allelic discrimination assay for rs11136000. For the allelic discrimination assay for APOE, sequence specific probes were also used: 5′ VIC-CCGCGATGC CGATGACCTGCAGAAG[C]GCCTGGCAGTGTACCAGGCCGGGGC–TAMRA-3′ and 5′FAM-CCGCGATGCCGATGACCTGCAGAAG[T]GCCTGGCAGT GTACCAGGCCGGGGC-TAMRA-3′ for rs7412 and 5′ VIC-GCTGGGCGCG GACATGGAGGACGTG[C]GCGGCCGCCTGGTGCAGTACCGCGG-TAMRA-3′ and 5′FAM-GCTGGGCGCGGACATGGAGGACGTG[T]GCGGCCGCCTGGT GCAGTACCGCGG-TAMRA-3′ for rs429358. Allele detection and genotype calling were performed using the ABI 7700 and Sequence Detection Software (Applied Biosystems).
n-Back Task
Participants completed a 3-back working memory task in which they were presented with words in a serial fashion on a computer screen (task duration: approximately 7 minutes). Participants were instructed to make a response when a presented word (all stimuli were nouns) was the same word that had been presented three stimuli ago. A total of 192 trials were presented. Each trial was 2 seconds in duration, with 250-millisecond ISI. Participants’ accuracy (percent correct) was the measure of task performance.
Image Acquisition, Processing, and Analysis
MRI images were acquired using a 3-T Allegra System (Siemens, Erlangen, Germany). Whole-brain structural T1-weighted magnetization-prepared rapid gradient-echo (MPRAGE) image was acquired for each subject (FOV = 256 mm; 256 × 256 matrix; 1 × 1 mm in-plane resolution, 1.25 mm thick axial slices, 1 average). All MPRAGE images were processed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) on MATLAB 2010b (http://www.mathworks.com/products/matlab/).
Voxel-Based Morphometry (VBM) was performed using the DARTEL toolbox for SPM (J Ashburner & Friston, 2000; John Ashburner, 2007). All default settings were used except were noted otherwise. Images were aligned into AC/PC orientation and then segmented into grey matter, white matter, and cerebrospinal fluid (CSF). All participants grey and white matter images were then simultaneously registered together to create a study specific template to reduce between-participant variability. The template was then used to normalize all images into the standard MNI space using the DARTEL Normalize to MNI Space program, utilizing the “preserve amount” option to retain the volumetric data of the original images, and the images were smoothed using a Gaussian kernel with 8 mm full-width half maximum (FWHM).
Statistical analysis of the grey matter images was performed using the multiple regression option of SPM. All participants’ grey matter images as a single group were regressed on working memory performance on the 3-back task, measured as percent correct. Age was added as a regressor to control any effects of age on grey matter. A minimum voxel intensity of 0.2 was applied to help omit spurious grey matter findings from the segmentation process. In addition, a mask was made and applied during volumetric analyses from the SPM a priori probabilistic grey matter image, with a minimum threshold of 0.5 to ensure that grey matter was within the top 50% concordance for the probabilistic image. To control for global brain volume differences between participants, each participant’s intracranial brain volume was used to normalize their grey matter volumes. The intracranial brain volumes were calculated by adding the volumes found in the native space grey matter, white matter, and CSF images of each participant. To limit the voxel-wise search to prefrontal and parietal cortical areas that have been consistently observed in fMRI studies using the n-back task, a mask was applied which was created using WFU PickAtlas (version 3.0.3) containing Brodmann areas 7, 9, 40, 44, and 46 (Collette et al., 2006; Maldjian, Laurienti, Kraft, & Burdette, 2003).
Correction for multiple comparisons was performed using the Non-Stationary (NS) Cluster Extent Correction toolbox for SPM with a corrected value of p <0.05 being considered significant (Hayasaka, Phan, Liberzon, Worsley, & Nichols, 2004; Smith & Nichols, 2009). Previous studies using NS Cluster Correction have used uncorrected height thresholds ranging from 0.001 to 0.01. We used a moderately stringent height threshold of 0.005 (Gotman et al., 2005; Meda et al., 2008; Rayhan et al., 2013). Caret (version 5.65) was used for all brain image visualization (Van Essen et al., 2001).
For later use in the mediation analysis (described below), masks were created for areas that showed significant correlation between grey matter volume and working memory performance, which included regions in both the left and right parietal cortex (see results). Using the masks, each subject’s MNI space mean GM volume was extracted from each of these areas separately. As with the VBM analysis, the grey matter volumes were then normalized by the participant’s intracranial brain volumes.
Mediation Analysis
Mediation modeling was done in AMOS 16 (SPSS Inc., Chicago, IL), a structural equation modeling add-on to the SPSS statistical software using the technique described by (Green et al., 2013). Inputs were genotype, grey matter volume, and behavioral performance on the 3-back for all participants. Bias-corrected bootstrapping 95% confidence intervals were calculated over 500 bootstrap samples. Estimation was achieved via scale-free least squares to enable significance measures while accounting for the non-uniform scaling of the genotype variable relative to the working memory and volumetric measures. The unique (unshared) variance of both mediator ROIs was used to calculate individual path coefficients. This ensured that each ROI’s contribution to the mediation was not driven by covariance/colinearity with the other ROI. The independent variable in the mediation model was CLU genotype (T/T, T/C vs. C/C) for the rs11136000 SNP. The dependent variable was 3-back percent correct. We modeled the mediated relationships between CLU genotype and 3-back performance (i.e., the indirect path from CLU genotype to 3-back performance via grey matter volume in our ROIs). The model also included the unmediated relationship (direct path) from CLU genotype to 3-back performance.
Results
Genetic
The genotype distribution for CLU was 9 T/T, 13 C/C, and 33 C/T. Hardy–Weinberg-Equilibrium was checked using a χ2-goodness of fit test (χ2 = 2.34, p = 0.126). The genotype distribution of APOE was 2 ε2/ε2, 6 ε2/ε3, 38 ε3/ε3, 7 ε4/ε3, 2 ε4/ε2 and 0 ε4/ε4. The APOE genotype distribution was also in Hardy-Weinberg-Equilibrium (χ2 = 5.73, permutation derived p = 0.252 using the Genetics plugin, version 1.3.8, for the R statistical computing software, http://cran.r-project.org/package=genetics).
Behavioral
Homozygous CLU-C/C individuals were compared to CLU-T carriers. Previous reports have found no dose-dependent effects for T-carriers (i.e., between C/T and T/T individuals) and have thus grouped them together (Lancaster et al., 2011; Mengel-From et al., 2011), although some studies have indicated that the C allele may have some dose-dependent effects (Braskie et al., 2011; Erk et al., 2011; Thambisetty et al., 2013). In order to determine which grouping to use in relation to 3-back performance, we first verified the 3-back scores were normally distributed (Shapiro-Wilk test, W = 0.979, p = 0.442). We then used parametric methods to directly compare the percent correct between the C/T and T/T groups, and found no significant differences (t = 0.055, p = 0.956, two-tailed; T/T mean = 84.0%, SD = 11.6%; C/T mean = 83.3%, SD = 7.3%). Therefore, for all subsequent analyses involving CLU genotype, the T/T and C/T groups were combined as the reduced-risk group.
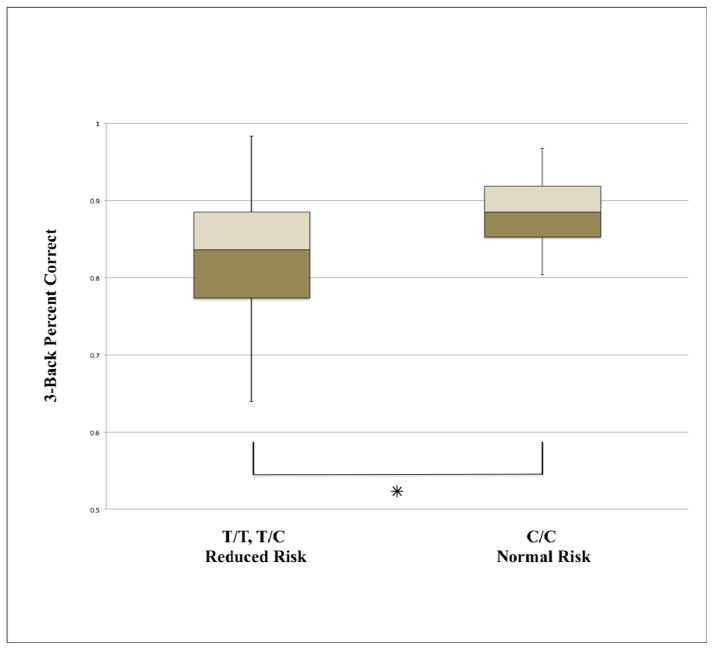
The normal-risk and reduced-risk groups for CLU did not differ with respect to age (t = −0.747, p = 0.459), gender (χ2 = 1.06, p = 0.304) or ethnicity (Fisher’s exact test, p = 0.246). To assess effects of APOE genetic risk, the 9 APOE–ε4 allele carriers were compared to the non-ε4 carriers. The ε4 carriers did not differ from the non-ε4 carriers in age (t = −0.988, p = 0.348), gender (Fisher’s exact test, p = 1.0) or ethnicity (Fisher’s exact test, p = 0.852). Table 1 provides demographic data. We then entered both CLU and APOE into a 2 (CLU genotype group) x 2 (APOE genotype group) ANOVA in order to evaluate their main effects on 3-back performance as well as their interaction. There was only a significant main effect for CLU (F = 4.566, p = 0.037), but not for APOE (F = 0.215, p = 0.645) or the interaction between CLU and APOE (F = 0.389, p = 0.536). Post-hoc analysis of the CLU main effect for the normal risk (C/C) vs. reduced-risk group (T/T, T/C) revealed significantly better 3-back working memory performance among the normal-risk group (Figure 1; t = −2.85, p = 0.007, two-tailed; Cohen’s d = 0.77; C/C mean = 88.3%, SD = 4.4%; T/T, T/C mean = 83.3%, SD = 8.1%). A direct comparison of APOE genotypes indicated no difference between ε4 carriers and ε4 non-carriers (t = −0.181, p = 0.857, two-tailed; Cohen’s d = 0.07; ε4 carriers mean = 84.9%, SD = 7%; non-ε4 carriers mean = 84.4%, SD = 7.8%). Therefore, all subsequent brain-based analyses were carried out for CLU but not for APOE.
Table 1.
Participant demographics by genotype group
| Genotype Group | Group Size | Gender | Ethnicity | Age | APOE–ε4 Status |
|---|---|---|---|---|---|
| CLU T/T & T/C | 42 | 19 F; 23 M | 34 Caucasian | 23 years (SD = 4.14) | 34 non–ε4 |
| 3 African American | Range = 18–36 years | 8 ε4 | |||
| 4 Asian | |||||
| 1 Latino | |||||
| CLU C/C | 13 | 8 F; 5 M | 8 Caucasian | 22.1 years (SD = 4.21) | 12 non–ε4 |
| 1 African American | Range = 18–34 years | 1 ε4 | |||
| 4 Asian | |||||
| 0 Latino |
Figure 1.
A boxplot comparison of working memory performance as measured by percent correct of the 3-back task between the reduced risk T/T, T/C group and the normal risk C/C CLU rs11136000 genotypes. (✳: p = 0.007; C/C mean = 88.3%, SD = 4.4%, range = 80%–97%; C/T, T/T mean = 83.3%, SD = 8.1%, range = 64%–98%).
Voxel-Based Morphometry (VBM)
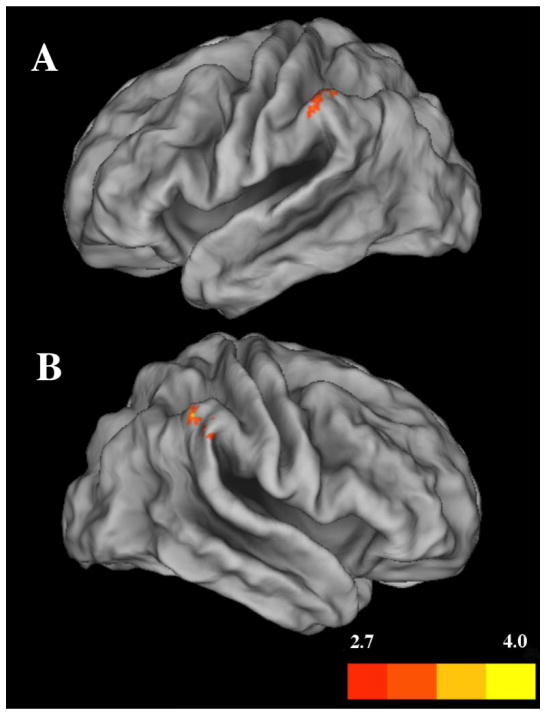
We next investigated whether variation in grey matter volume predicted performance on the 3-back task. We regressed 3-back accuracy on grey matter volume, combining all participants into one group. We confined the regression to frontal and parietal cortical areas that are consistently activated during n-back tasks using functional imaging, including Brodmann areas 7, 9, 40, 44, and 46 (for a review see (Collette et al., 2006)). This analysis identified significant positive associations in both the left and right inferior parietal cortex (Figure 2), contained in Brodmann area 40 within the supramarginal gyrus (p < 0.05 non-stationary cluster corrected; see Table 2). No negative associations were observed
Figure 2.
Voxel-wise positive regression with all participants as one group for 3-back performance, measured as percent correct, with cortical grey matter volume for A) the left supramarginal gyrus and B) the right supramarginal gyrus. Color bar represents t-values; p < 0.05 non-stationary cluster corrected.
Table 2.
Regional grey matter volume associated with 3-back performance
| Anatomical Region | Cluster Size | Corrected Cluster P-Value | Peak MNI Coordinates | Uncorrected T-Value | Peak Voxel P-Value | Mean Voxel Volume by Cluster for CLU Genotype Groups | ||
|---|---|---|---|---|---|---|---|---|
| x | y | z | ||||||
| Right Supramarginal Gyrus | 215 | 0.014 | 51 | −41 | 48 | 3.59 | 0.000 |
T/T, T/C 0.28 (SD = 0.040) range = 0.21–0.37 C/C 0.30 (SD = 0.037) range = 0.26–0.36 |
| 48 | −48 | 50 | 3.29 | 0.001 | ||||
| 62 | −32 | 38 | 2.88 | 0.003 | ||||
|
| ||||||||
| Left Supramarginal Gyrus | 64 | 0.035 | −54 | −38 | 47 | 3.08 | 0.002 |
T/T, T/C 0.26 (SD = 0.052) range = 0.12–0.36 C/C 0.28 (SD = 0.036) range = 0.21–0.36 |
Mediation Analysis
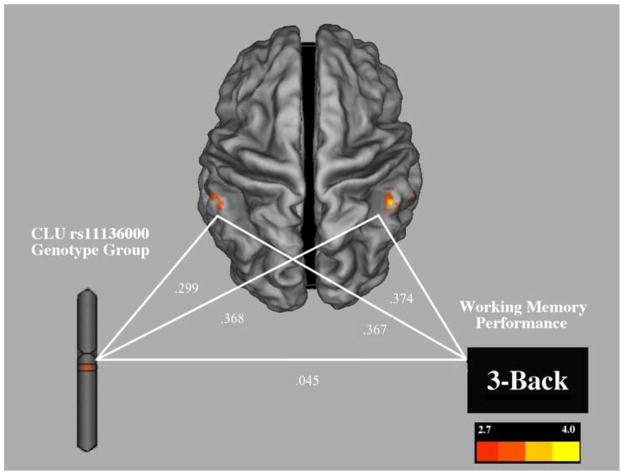
Given that the CLU C/C normal-risk group demonstrated better working memory performance, and that greater grey matter volume in the left and right supramarginal gyri predicted better working memory performance, we next tested a gene-brain-cognition mediation model. We constructed a path-analytic mediation model to test the hypothesis that grey matter volume mediated the effect of CLU genotype on working memory performance. The model demonstrated good fit (Goodness of fit index = 0.948; Root mean square residual = 0.05). Testing the mediation model revealed that grey matter volume in the left and right supramarginal gyri significantly mediated the effect of CLU genotype on 3-back performance, indirect effect = 0.247, p = 0.018, 95% CI = 0.103–0.586 (Figure 3; Table 3).
Figure 3.
Gene-brain-cognition mediation model illustrating grey matter volume in the left and right supramarginal gyri mediating the direct effect between genotype group and 3-back performance, with path coefficients indicated. Color bar represents t-values.
Table 3.
Mediation model parameters
| Parameter | Estimate | 95% Confidence Interval | p |
|---|---|---|---|
| Left Parietal → 3-back | .367 | .191–.557 | .018 |
| Right Parietal → 3-back | .374 | .194–.543 | .016 |
| CLU Risk → Left Parietal | .299 | .066–.507 | .084 |
| CLU Risk → Right Parietal | .368 | .102–.595 | .033 |
| CLU Risk → 3-back | .045 | −.295–.236 | .916 |
Note: The direct effect of CLU Genotype on 3-back performance (i.e., modeled without indirect effect) was β = .28, p = .038, which was reduced by the inclusion of the indirect effect, indicating mediation.
Discussion
This investigation of a poorly understood AD risk factor found that young, healthy individuals homozygous for the CLU-C normal risk allele displayed significantly better working memory performance than carriers of the reduced risk T allele and that this effect was mediated by supramarginal gyrus gray matter volume. In contrast, we did not observe APOE genotype effects on working memory performance. To our knowledge, this is the first report of a behavioral effect for CLU rs11136000 in young people. In addition, this work identified the first effect pathway for the transmission of the effect of an AD risk factor from gene to brain to cognition. Specifically, the data indicate a gene-brain-cognition effect pathway (Green et al., 2013), whereby CLU genotype influences working memory via variation in parietal grey matter volume.
Cognitive neurogenetic research has typically examined individual pairwise relationships (e.g., gene-brain and gene-cognition relationships) rather than using integrated gene-brain-cognition models to test neural mediation of genetic effects on cognition. Mediation models have the potential to be more informative than pairwise relationships, not only because they integrate multiple data types into a single model to identify effect pathways from gene to brain to cognition, but because they can do so in a way that accounts for the complexity of multiple contributing brain regions. Mediation models also provide a useful framework for directional/causal hypotheses, which are generally appropriate for cognitive neurogenetics because effects generally transmit from genes to brain and cognition, rather than vice versa.
The finding that the normal-risk CLU genotype was associated with better working memory performance suggests that the CLU-C allele may have antagonistic pleiotropic effects, at least concerning working memory. This evidence is consistent with the evidence for antagonistic pleiotropy associated with the strongest AD risk factor allele (APOE-ε4) (Tuminello & Han, 2011), and suggests that long term effects of genetically programmed neural advantages in youth may be a general (i.e., not APOE-specific) contributor to AD susceptibility late in life. To our knowledge, this is the first report of an antagonistic pleiotropic effect of the CLU rs11136000 polymorphism.
A prior study in younger adults (Lancaster et al., 2011) did not observe CLU-related differences in working memory, but used a paradigm that differed from ours in both in the modality (faces versus words) and the inclusion of a strong emotional component (emotive facial expressions). The rs11136000 polymorphism has not been investigated in relation to emotional processing, and any such effects could confound working memory effects in a behavioral measure of working memory using emotional facial expressions (for example see (Mather et al., 2006)). As noted above, it is unlikely that AD risk-associated effects on emotional processing parallel effects on working memory since AD pathogenesis affects these types of cognition quite differently (Bucks & Radford, 2004; Dequeker et al., 2002; Evans-Roberts & Turnbull, 2011; Henry et al., 2009). In addition, the study by Lancaster et al. (2011) included an age range extending well into middle age (18–51 years), which is considerably older than our subject cohort. Thus, it is not clear whether a lack of effect in that study may have reflected changing effects of the CLU-C allele across the adult lifespan. If CLU-C exhibits antagonistic pleiotropic effects, as our data suggest, then it is likely that strong performance by younger C allele carriers in the Lancaster et al. (2011) cohort was counterweighted by poorer performance in middle aged C allele carriers, causing an effect to wash out.
Working memory is a complex function that involves multiple regions in parietal and frontal cortices. The specificity of the effects we observed to parietal working memory regions may enable inferences concerning the specific components of working memory that are affected by CLU variation. Working memory research has begun to parse different cognitive contributions of regions within the frontoparietal working memory network. PFC appears to support the selection of items to be stored in working memory as well as the filtering out of irrelevant items that should not be stored (McNab & Klingberg, 2007). In contrast parietal cortex appears to be involved more directly with the storage and maintenance component of working memory, and parietal activity is a determinant of item capacity limit (Todd & Marois, 2004). Thus, our findings suggest that the effect of CLU on working memory may be most directly due to impacts on the storage and maintenance component.
Conclusion
These data provide new evidence for neurocognitive markers of CLU-related AD risk in young people, extending the few prior studies of CLU in young people, which have identified potential early brain-imaging markers (Braskie et al., 2011; Erk et al., 2011; Lancaster et al., 2011). Our findings provide the first indication of a CLU genotype effect on cognitive performance. The observed cognitive effect is particularly noteworthy because it is in an area of cognition, working memory, which is highly susceptible to AD pathogenesis (Baddeley et al., 1986; Belleville et al., 1996; Stopford et al., 2007, 2012). This work utilized a gene-brain-cognition mediation modeling approach to provide the first indication of a pathway by which the effect of an AD risk factor is transmitted from gene to brain to cognition. The data preliminarily support an antagonistic pleiotropy account of CLU genotype in young people, though additional study, including convergence across cognitive tasks, will be important for resolving this issue.
CLU genotype was associated with working memory performance in young people
Parietal grey matter volume mediated the effect of CLU on working memory
First evidence of CLU effects on cognitive function in young people
First gene-brain-cognition effect pathway identified for an Alzheimer’s risk gene
Results indicate support for antagonistic pleiotropy in Alzheimer’s genetic risk
Acknowledgments
This work was supported by NIH R01 AG035379, P01 AG030128 (GWR), grants from Partners in Research and the American Legacy Foundation (AEG), NIH/NINDS 5T32NS041218 (AMD), and NIH/NINDS T32NS041231 (BWS). Samples from the National Cell Repository for Alzheimer’s Disease, which receives government support under a cooperative agreement grant U24 AG21886 awarded by the National Institute on Aging, were used in this study. We thank Rusan Chen for helpful comments on an earlier version of the manuscript.
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J, Friston KJ. Voxel-based morphometry-The methods. NeuroImage. 2000;11(6 Pt 1):805–21. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner John. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Baddeley A, Logie R, Bressi S, Sala S Della, Spinnler H. Dementia and working memory. The Quarterly Journal of Experimental Psychology Section A. 1986;38(4):603–618. doi: 10.1080/14640748608401616. [DOI] [PubMed] [Google Scholar]
- Bartrés-Faz D, Sole-Padullés C, Junqué C, Rami L, Bosch B, Bargalló N, Molinuevo JL. Interactions of cognitive reserve with regional brain anatomy and brain function during a working memory task in healthy elders. Biological Psychology. 2009;80(2):256–259. doi: 10.1016/j.biopsycho.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Belleville S, Peretz I, Dominique M. Examination of the working memory components in normal aging and in dementia of the Alzheimer type. Neuropsychologia. 1996;34(3):195–207. doi: 10.1016/0028-3932(95)00097-6. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Alzheimer disease: New light on an old CLU. Nature reviews Neurology. 2010;6(1):11–3. doi: 10.1038/nrneurol.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Thompson PM. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31(18):6764–70. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, Larossa G, Sachs R, Fotenos AF, Mintun MA. Molecular, structural, and functional characterization of Alzheimer’s disease: Evidence for a relationship between default activity, amyloid, and memory. Neurobiology of Disease. 2005;25(34):7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucks RS, Radford SA. Emotion processing in Alzheimer’s disease. Aging & mental health. 2004;8(3):222–32. doi: 10.1080/13607860410001669750. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter Ta, Ma L, Bisceglio GD, Zou F, Younkin SG. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Archives of neurology. 2010;67(8):961–4. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette F, Hogge M, Salmon E, Van der Linden M. Exploration of the neural substrates of executive functioning by functional neuroimaging. Neuroscience. 2006;139(1):209–221. doi: 10.1016/j.neuroscience.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Collette F, Salmon E, Van Der Linden M, Degueldre C, Franck G. Functional anatomy of verbal and visuospatial span tasks in Alzheimer’s disease. Human Brain Mapping. 1997;5(2):110–118. doi: 10.1002/(SICI)1097-0193(1997)5:2<110::AID-HBM4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Colom R, Jung RE, Haier RJ. General intelligence and memory span: Evidence for a common neuroanatomic framework. Cognitive Neuropsychology. 2007;24(8):867–878. doi: 10.1080/02643290701781557. [DOI] [PubMed] [Google Scholar]
- Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, Pericak-Vance M. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Huentelman MJ. Association of CR1, CLU and PICALM with Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. Human molecular genetics. 2010;19(16):3295–301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Holtzman DM. ApoE and clusterin cooperatively suppress Aβ levels and deposition. Neuron. 2004;41(2):193–202. doi: 10.1016/s0896-6273(03)00850-x. Retrieved from http://www.sciencedirect.com/science/article/pii/S089662730300850X. [DOI] [PubMed] [Google Scholar]
- Dennis Na, Browndyke JN, Stokes J, Need A, Burke JR, Welsh-Bohmer Ka, Cabeza R. Temporal lobe functional activity and connectivity in young adult APOE epsilon4 carriers. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2010;6(4):303–311. doi: 10.1016/j.jalz.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeker J, Boller F, El Massioui F, Degreef H, Busschots AM, Devouche E, Starkstein SE. Processing emotional information in Alzheimer’s disease: Effects on memory performance and neurophysiological correlates. Dementia and Geriatric Cognitive Disorders. 2002;14(2):104–112. doi: 10.1159/000064932. [DOI] [PubMed] [Google Scholar]
- Desgranges B atrice, Baron JC, de La Sayette V, Petit-Taboue MC, Karim B, Landeau B, Eustache F. The neural substrates of memory systems impairment in Alzheimer’s disease - A PET study of resting brain glucose utilization. Brain. 1998;121(Pt 4):611–631. doi: 10.1093/brain/121.4.611. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Weickert CS, Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clinical lipidology. 2010;51(4):555–573. doi: 10.2217/CLP.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, Walter H. Hippocampal function in healthy carriers of the CLU Alzheimer’s disease risk variant. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31(49):18180–4. doi: 10.1523/JNEUROSCI.4960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans-Roberts CEY, Turnbull OH. Remembering relationships: preserved emotion-based learning in Alzheimer’s disease. Experimental aging research. 2011;37(1):1–16. doi: 10.1080/0361073X.2011.536750. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds Ca, Fratiglioni L, Johansson B, Mortimer Ja, Berg S, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Archives of general psychiatry. 2006;63(2):168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Gotman J, Grova C, Bagshaw a, Kobayashi E, Aghakhani Y, Dubeau F. Generalized epileptic discharges show thalamocortical activation and suspension of the default state of the brain. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(42):15236–40. doi: 10.1073/pnas.0504935102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AE, Dunbar KN. Mental Function as Genetic Expression: Emerging Insights From Cognitive Neurogenetics. In: Holyoak KJ, Morrison RG, editors. The Oxford Handbook of Thinking and Reasoning. 1. New York: Oxford University Press; 2012. pp. 90–111. Retrieved from http://books.google.com/books?hl=en&lr=&id=DwZTCoV3cP4C&pgis=1. [Google Scholar]
- Green AE, Gray JR, DeYoung CG, Mhyre TR, Padilla R, DiBattista AM, Rebeck GW. A combined effect of two Alzheimer’s risk genes on medial temporal activity during executive attention in young adults. Neuropsychologia. 2014;56:1–8. doi: 10.1016/j.neuropsychologia.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AE, Kraemer DJM, Deyoung CG, Fossella Ja, Gray JR. A gene-brain-cognition pathway: prefrontal activity mediates the effect of COMT on cognitive control and IQ. Cerebral cortex. 2013;23(3):552–9. doi: 10.1093/cercor/bhs035. [DOI] [PubMed] [Google Scholar]
- Green AE, Munafò MR, DeYoung CG, Fossella JA, Fan J, Gray JR. Using genetic data in cognitive neuroscience: from growing pains to genuine insights. Nature Reviews Neuroscience. 2008;9(9):710–720. doi: 10.1038/nrn2461. [DOI] [PubMed] [Google Scholar]
- Han SD, Bondi MW. Revision of the apolipoprotein E compensatory mechanism recruitment hypothesis. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2008;4(4):251–4. doi: 10.1016/j.jalz.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Williams A. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature genetics. 2009;41(10):1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. NeuroImage. 2004;22(2):676–87. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Henry JD, Rendell PG, Scicluna A, Jackson M, Phillips LH. Emotion experience, expression, and regulation in Alzheimer’s disease. Psychology and Aging. 2009;24(1):252–257. doi: 10.1037/a0014001. [DOI] [PubMed] [Google Scholar]
- Jacobs HIL, Van Boxtel MPJ, Jolles J, Verhey FRJ, Uylings HBM. Parietal cortex matters in Alzheimer’s disease: An overview of structural, functional and metabolic findings. Neuroscience and Biobehavioral Reviews. 2012;36(1):297–309. doi: 10.1016/j.neubiorev.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Schellenberg GD. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Archives of neurology. 2010;67(12):1473–84. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpouzos G, Eustache F, de la Sayette V, Viader F, Chételat G, Desgranges B. Working memory and FDG-PET dissociate early and late onset Alzheimer disease patients. Journal of Neurology. 2005;252(5):548–558. doi: 10.1007/s00415-005-0685-3. [DOI] [PubMed] [Google Scholar]
- Kamboh MI, Minster RL, Demirci FY, Ganguli M, Dekosky ST, Lopez OL, Barmada MM. Association of CLU and PICALM variants with Alzheimer’s disease. Neurobiology of aging. 2012;33(3):518–21. doi: 10.1016/j.neurobiolaging.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Shin Y, Shim Y, Jeong S, Park I, Min B. Clusterin interacts with SCLIP (SCG10-like protein) and promotes neurite outgrowth of PC12 cells. Experimental Cell Research. 2005;309(2):305–315. doi: 10.1016/j.yexcr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Koch S, Donarski N, Goetze K, Kreckel M, Stuerenburg HJ, Buhmann C, Beisiegel U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J Lipid Res. 2001;42(7):1143–1151. Retrieved from http://www.jlr.org/content/42/7/1143.long. [PubMed] [Google Scholar]
- Kok EH, Luoto T, Haikonen S, Goebeler S, Haapasalo H, Karhunen PJ. CLU, CR1 and PICALM genes associate with Alzheimer’s-related senile plaques. Alzheimer’s research & therapy. 2011;3(2):12. doi: 10.1186/alzrt71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounnas MZ, Moir RD, Rebeck GW, Bush AI, Argraves WS, Tanzi RE, Strickland DK. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted β-amyloid precursor protein and mediates its degradation. Cell. 1995;82(2):331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics. 2009;41(10):1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lancaster TM, Baird A, Wolf C, Jackson MC, Johnston SJ, Donev R, Linden DEJ. Neural hyperactivation in carriers of the Alzheimer’s risk variant on the clusterin gene. European neuropsychopharmacology: the journal of the European College of Neuropsychopharmacology. 2011;21(12):880–4. doi: 10.1016/j.euroneuro.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Lim HK, Juh R, Pae CU, Lee BT, Yoo SS, Ryu SH, Lee CU. Altered verbal working memory process in patients with Alzheimer’s disease - An fMRI investigation. Neuropsychobiology. 2008;57(4):181–187. doi: 10.1159/000147471. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage. 2003;19(3):1233–1239. doi: 10.1016/S1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Marchant NL, King SL, Tabet N, Rusted JM. Positive effects of cholinergic stimulation favor young APOE e4 carriers. Neuropsychopharmacology. 2010;35(5):1090–1096. doi: 10.1038/npp.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather M, Mitchell KJ, Raye CL, Novak DL, Greene EJ, Johnson MK. Emotional arousal can impair feature binding in working memory. Journal of cognitive neuroscience. 2006;18(4):614–25. doi: 10.1162/jocn.2006.18.4.614. [DOI] [PubMed] [Google Scholar]
- McNab F, Klingberg T. Prefrontal cortex and basal ganglia control access to working memory. Nature neuroscience. 2007;11(1):103–107. doi: 10.1038/nnXXXX. [DOI] [PubMed] [Google Scholar]
- Meda SA, Gelernter J, Gruen JR, Calhoun VD, Meng H, Cope NA, Pearlson GD. Polymorphism of DCDC2 reveals differences in cortical morphology of healthy individuals-A preliminary voxel based morphometry study. Brain imaging and behavior. 2008;2(1):21–26. doi: 10.1007/s11682-007-9012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel-From J, Christensen K, McGue M, Christiansen L. Genetic variations in the CLU and PICALM genes are associated with cognitive function in the oldest old. Neurobiology of aging. 2011;32(3):554.e7–11. doi: 10.1016/j.neurobiolaging.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondadori CRA, De Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF, Henke K. Better memory and neural efficiency in young apolipoprotein E e4 carriers. Cerebral Cortex. 2007;17(8):1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- Nathan B, Bellosta S, Sanan D, Weisgraber K, Mahley R, Pitas R. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264(5160):850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- Persson J, Lind J, Larsson A, Ingvar M, Cruts M, Van Broeckhoven C, Nyberg L. Altered brain white matter integrity in healthy carriers of the APOE epsilon4 allele: a risk for AD? Neurology. 2006;66(7):1029–33. doi: 10.1212/01.wnl.0000204180.25361.48. [DOI] [PubMed] [Google Scholar]
- Rayhan RU, Stevens BW, Raksit MP, Ripple Ja, Timbol CR, Adewuyi O, Baraniuk JN. Exercise challenge in Gulf War Illness reveals two subgroups with altered brain structure and function. In: Valdes-Sosa PA, editor. PLoS ONE. 6. Vol. 8. 2013. p. e63903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Lambert JC. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA: the journal of the American Medical Association. 2010;303(18):1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- Stopford CL, Snowden JS, Thompson JC, Neary D. Distinct memory profiles in Alzheimer’s disease. Cortex. 2007;43(7):846–857. doi: 10.1016/S0010-9452(08)70684-1. [DOI] [PubMed] [Google Scholar]
- Stopford CL, Thompson JC, Neary D, Richardson AMT, Snowden JS. Working memory, attention, and executive function in Alzheimer’s disease and frontotemporal dementia. CORTEX. 2012;48(4):429–446. doi: 10.1016/j.cortex.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Resnick SM. Alzheimer risk variant CLU and brain function during aging. Biological psychiatry. 2013;73(5):399–405. doi: 10.1016/j.biopsych.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies W, Bleiler L. Alzheimer’s disease facts and figures Alzheimer’s Association. Alzheimer’s & Dementia. 2013;9(2):208–245. doi: 10.1016/j.jalz.2013.02.003. Retrieved from http://www.alz.org/downloads/facts_figures_2013.pdf. [DOI] [PubMed] [Google Scholar]
- Todd JJ, Marois R. Capacity limit of visual short-term memory in human posterior parietal cortex. Nature. 2004;428(6984):751–4. doi: 10.1038/nature02466. [DOI] [PubMed] [Google Scholar]
- Tuminello ER, Han SD. The apolipoprotein e antagonistic pleiotropy hypothesis: review and recommendations. International journal of Alzheimer’s disease. 2011;2011:726197. doi: 10.4061/2011/726197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury Ha, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. Journal of the American Medical Informatics Association: JAMIA. 2001;8(5):443–59. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CRJ. 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain. 2007;130(Pt 7):1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, Rabin LA, Santulli RB, Flashman LA, Guerin SJ, McAllister TW. Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. The American journal of psychiatry. 2006;163(9):1603–10. doi: 10.1176/appi.ajp.163.9.1603. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yu J, Li Y, Tan L. Advances in Clinical Chemistry. 1. Vol. 56. Elsevier Inc; 2012. Clusterin in Alzheimer’s disease; pp. 155–173. [DOI] [PubMed] [Google Scholar]