Abstract
Alcoholism is a complex psychiatric disorder that has a multifactorial etiology. Epigenetic mechanisms are uniquely capable of accounting for the multifactorial nature of the disease in that they are highly stable and are affected by environmental factors, including alcohol itself. Chromatin remodeling causes changes in gene expression in specific brain regions contributing to the endophenotypes of alcoholism such as tolerance and dependence. The epigenetic mechanisms that regulate changes in gene expression observed in addictive behaviors respond not only to alcohol exposure, but also to comorbid psychopathology such as the presence of anxiety and stress. This review summarizes recent developments in epigenetic research that may play a role in alcoholism. We propose that pharmacologically manipulating epigenetic targets, as demonstrated in various preclinical models, holds great therapeutic potential in the treatment and prevention of alcoholism.
Keywords: Alcoholism, Anxiety, amygdala, Negative affective state, Histone acetylation, DNA methylation, Synaptic plasticity
Introduction
Alcoholism can be characterized as a chronic relapsing brain disorder. The pathogenesis of alcoholism is driven by both positive and negative affective states, which promote alcohol drinking, most likely due to allostatic adaptations in specific brain circuitry (Koob, 2003; Koob & Kreek, 2007; Pandey, 2004). This leads to a recurrent compulsion to consume alcohol and ultimately to a loss of control over intake, arising due to the development of a negative emotional state during withdrawal. This has been referred to as the “dark side of addiction”, that ultimately results to maintenance of addiction (Koob & Le Moal, 2005; Koob, 2013).
The comorbidity of anxiety and alcohol use disorders (AUD) is an important criterion to consider for the propensity of an individual to become an alcoholic (Kushner, Abrams, & Borchardt, 2000). Individuals with anxiety disorders have the tendency to transition from occasional drinking to alcohol dependence more rapidly than individuals who have no comorbid anxiety (Kushner, Maurer, Menary, & Thuras, 2011). Short-term consumption of alcohol to relieve anxiety escalates to chronic intake and withdrawal, leading to an exacerbation of anxiety symptoms at which point the individual drinks to “self-medicate” to counter the heightened anxiety (Robinson, Sareen, Cox, & Bolton, 2009). Interestingly, when the negative state is augmented by the presence of comorbid stress-related psychiatric disorders, including mood and anxiety disorders, the risk of becoming an alcoholic is increased (Grant et al., 2004; Schuckit & Hesselbrock, 1994).
Both genetic and epigenetic (environment influenced) factors affect alcohol consumption and have been shown to aid in the transition from use to abuse to addiction via neuroadaptations occurring in the brain (Cloninger, 1987; Moonat & Pandey, 2012; Pandey, Ugale, Zhang, Tang, & Prakash, 2008; Starkman, Sakharkar, & Pandey, 2012). Epigenetics can be loosely defined as a means of “stably” propagating a change or “heritability” of a cellular state that is not encoded in the DNA itself (Holliday, 1987). The brain essentially consists of post-differentiated non-dividing cells, and epigenetic regulation of neuronal pathways become critical to the establishment and maintenance of neuronal homeostasis and plasticity in the brain by mediating both transient and stable changes in gene expression. Epigenetic regulation in post-mitotic cells, such as neurons, brings about some debate since it is not propagated through cell divisions and therefore has prompted researchers to classify it into a subfield called “Neuroepigenetics” (Day & Sweatt, 2010). Epigenetic regulatory mechanisms include covalent modifications to histones, transcription factor-mediated energy-dependent chromatin remodeling, DNA modification by methylation at cytosine residues and control of gene expression by non-coding RNAs (Bonasio, Tu & Reinberg, 2010). Neuroplasticity is a fundamental property of the brain that allows it to dynamically adapt following exposure to alcohol and other drugs of abuse. One mechanism by which neuroplasticity to drugs of abuse such as ethanol and cocaine may occur is via epigenetic modifications that regulate gene expression i.e. histone covalent modifications and DNA methylation mechanisms (Moonat & Pandey, 2012; Robison & Nestler, 2011).
In addition to neuroplasticity, ethanol has been shown to affect epigenetic pathways in peripheral tissues such as the gastro-intestinal and biliary systems (Shukla & Lim, 2013). Fetal development may also be under epigenetic regulation (Liu, Balaraman, Wang, Nephew, & Zhou, 2009; Singh, Shiue, Schomberg & Zhou, 2009). Despite four decades of clinical and preclinical research documenting the teratogenicity of ethanol, knowledge of the epigenetic changes induced by prenatal ethanol exposure is only recently emerging. Studies investigating fetal alcohol spectrum disorders (FASD) have shown an important role played by the epigenome in the pathogenesis of FASD (Perkins, Lehmann, Lawrence, & Kelly, 2013; Resendiz, Chen, Ozturk, & Zhou, 2013). Interestingly, early ethanol exposure has been shown to affect epigenetic regulation of genes involved in imprinting (Haycock & Ramsay, 2009), neural and glial development (Liu et al., 2009), cell cycle regulation (Hicks, Middleton, & Miller, 2010) and nervous system growth (Zhou et al., 2011).
This review will focus on two major forms of epigenetic regulation, histone covalent modifications and DNA methylation, and their influence on ethanol-related behaviors and vice-versa in the brain and periphery.
1. Epigenetic Regulation due to Histone Covalent Modifications
Histone covalent modifications that have been identified as functionally important in regulating transcription include acetylation (lysine residues), methylation (arginine and lysine residues), phosphorylation (serine and threonine residues), ubiquitination, sumoylation, ADP-ribosylation (lysine residues) and proline isomerization (Allis et al., 2007; Kouzarides, 2007). Of these, acetylation and phosphorylation of histones H3 and H4 most commonly activate transcription while sumoylation has been implicated in repression. Some modifications need to be interpreted in a context-specific manner, e.g. methylation, which can cause either activation or repression (Lachner, O’Sullivan, & Jenuwein, 2003). Lysine (K) residues are an important substrate for histone modifications since they undergo acetylation, methylation, ubiquitination and sumoylation. Furthermore, different patterns of modifications give rise to varying cellular outcomes. For example, lysine methylation at histone H3 residues K4, K36 and K79 (H3K4, H3K36 and H3K79) causes activation; however, methylation at H3K9, H3K27 and H4K20 causes transcriptional repression (Allis et al., 2007). Histone covalent modifications are dynamically regulated by the actions of deacetylases, serine-threonine phosphatases, lysine demethylases, deiminases and ubiquitin proteases that act to reverse the corresponding modifications. Thus, the epigenetic regulation of gene expression becomes a very complex phenomenon that warrants a greater understanding of how these histone modifications interact with each other. This has been likened to a functional ‘code’ that presumably may lead to specific functional outcomes when specific combinations of modifications are present (Strahl & Allis, 2000). We will first describe histone acetylation and deacetylation mechanisms in the brain and their implications in alcoholism and other brain disorders.
1A. Role of Histone Acetylation and Deacetylation in Transcriptional Regulation
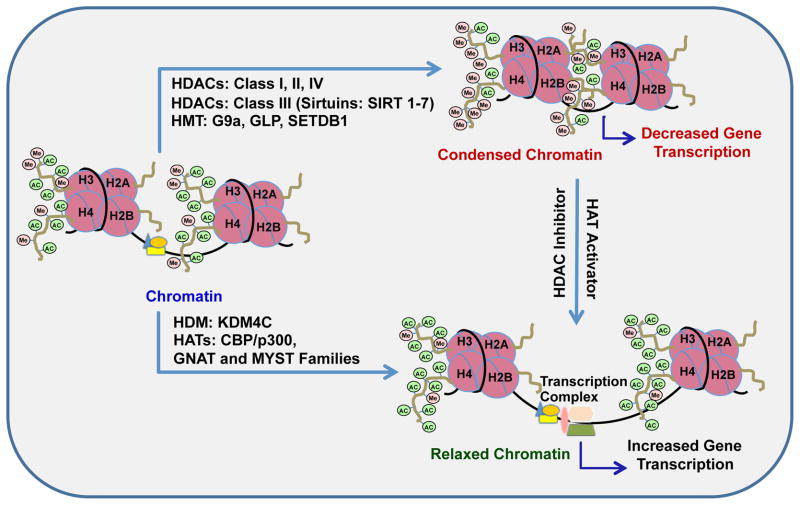
One of the well-characterized histone covalent modifications is lysine acetylation and its opposing mark deacetylation, which are facilitated by two groups of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetylation helps neutralize the basic charge of lysine by directly promoting the unraveling of DNA through minimizing nucleosomal contacts (Bannister & Kouzarides, 2011). HATs are associated with transcriptional initiation and elongation, genome stability and cell cycle-regulated DNA repair (Masumoto, Hawke, Kobayashi, & Verreault, 2005; Wang et al., 2009). Gene activation is enabled through acetylation of specific lysine tails predominantly located on the N- termini of core histones (H2A, H2B, H3 and H4) resulting in an increase in transcription. Numerous lysine residues undergo acetylation including K5, K8, K12, K16 in histone H4 and K9, K14, K18 and K56 in histone H3 (Allis et al., 2007). The three major families of HATs are: the GNAT family (Gcn5-associated N-acetyltransferase) which include Gcn5, PCAF and others; the MYST family (named after its founding members MOZ, Ybf2/Sas3, Sas2, Tip60) and the global co-activators CREB binding protein/p300 (CBP/p300) family (Sterner & Berger, 2000). (Figure 1)
Figure 1.
Schematic representation of the epigenetic mechanisms of acetylation and methylation of lysine (K) residues at histone H3 and H4 tails that mediate the switching between ‘open’ (relaxed) and ‘closed’ (condensed) chromatin structures. Acetylation of histone H3 at K9, K14, K18 and K56 and of H4 at K5, K8, K12 and K16 are believed to relax the chromatin structure making it more accessible to transcription factors and chromatin remodelers, thereby promoting transcription, whereas methylation at these lysine residues condenses the chromatin followed by reduced gene transcription. Histone deacetylases (HDACs) remove acetyl groups and histone methyltransferases (HMTs) attach methyl groups at lysine residues. HDACs are classified into four different classes (class I–IV), of which the class III, sirtuins, are NAD+ dependent and comprise of seven different isoforms (SIRT 1–7). HDACs classes I (HDAC 1, 2, 3, and 8), II (HDAC 4–7, 9, and 10) and IV (HDAC11) are Zn+ dependent enzymes. Histone methyltransferases such as G9a, GLP and SETDB1 can catalyze the methylation of H3-K9 contributing to the restrictive state of the chromatin. Histone demethylases (HDMs) (e.g. KDM family) remove the methyl groups from lysine residues associated with repressive marks (e.g. H3K9) in the tails of histones facilitating the relaxation of chromatin. Histone acetyltransferases (HATs) such as GNAT and MYST families of enzymes and CREB-binding protein (CBP)-p300 complex transfer acetyl groups leading to relaxed chromatin (Allis et al., 2007; Kouzarides, 2007).
CBP/p300 are an important family of HATs that act to remodel chromatin architecture functioning as part of different multimeric complexes as well as directly via binding phosphorylated-CREB (cyclic-AMP-response element binding protein). CREB is a critical neuronal transcription factor that regulates normal physiological function and is especially involved in the regulation of the epigenome during alcoholism (Mayr & Montminy, 2001; Starkman et al., 2012). CREB binds to the canonical cAMP responsive element (CRE) consensus sequence at target gene promoters to activate gene expression (Montminy & Bilezikjian, 1987). Phosphorylation at serine 133 serves to activate CREB (pCREB), which recruits the HAT, CBP (Chrivia et al., 1993; Parker et al., 1996), which in conjunction with another protein, p300, mediates chromatin remodeling (Arany, Newsome, Oldread, Livingston, & Eckner, 1995). CBP and p300 have been implicated as targets with therapeutic potential in a variety of neurodegenerative disorders (Valor, Viosca, Lopez-Atalaya, & Barco, 2013). Landmark studies have shown that CBP is important for memory consolidation using a rodent model of Rubinstein-Taybi syndrome (RTS) (Korzus, Rosenfeld, & Mayford, 2004). Mutations in CBP have been shown to impair long-term potentiation and memory and alter acetylation leading to cognitive and memory deficits (Barrett et al., 2011; Alarcón et al., 2004).
Generally, as discussed above, transcriptional activators and co-activators recruit HATs such as CBP, whereas repressors and co-repressors recruit HDACs. HDACs reverse the activity of HATs and cause a decrease in transcription through removal of acetyl groups from histones. HDACs are grouped into four classes: class I HDACs (1,2,3 and 8), class II HDACs (4,5,6,7 and 9) and class IV HDACs are similar in that they require zinc for activity whereas class III HDACs or sirtuins are structurally unique requiring NAD+ for enzymatic activity (Blander & Guarente, 2004; Haberland, Montgomery, & Olson, 2009). Regulating the amount of histone acetylation by manipulating HDAC activity is an attractive therapeutic option for treating brain disorders. HDAC2 appears to be an important molecule in maintaining normal neuronal cognitive function and regulating hippocampal memory with implications for remote fear memory and treatment of post-traumatic stress disorders (Gräff et al., 2012, 2014). Inhibition of HDAC activity may serve as a useful pharmacological approach in the treatment of learning and memory disorders (Fischer, Sananbenesi, Wang, Dobbin, & Tsai, 2007) as HDAC isoform-specific regulation of learning and memory has been demonstrated in both vertebrate and invertebrate models (Fitzsimons, Schwartz, Given, & Scott, 2013; Fitzsimons & Scott, 2011; Guan et al., 2009; Kim et al., 2012; McQuown et al., 2011; Wang, Zang et al., 2011). The class III HDACs, sirtuins, due to their NAD+ dependency, are believed to be critical in the regulation of biological and circadian rhythms, cell survival, metabolic activity, aging and neurodegeneration (Hirayama et al., 2007; Nakahata et al., 2008; Bishop & Guarente, 2007; Cohen et al., 2004; Kim et al., 2007). Dysregulation of the circadian clock has been implicated in psychiatric disorders and alcohol phenotypes in both vertebrate and invertebrate systems (Mansour et al., 2006; Pohl et al., 2013; Sarkar, 2012). Hence HDAC manipulation thereby affecting circadian pathways, could be an important avenue for therapeutic intervention. Recently, the sirtuin isoform SIRT1 has been shown to modulate CREB function and affect learning and memory pathways via its regulation of microRNA-134 (Gao et al., 2010). This serves to illustrate the complexity of HDAC inhibition and warrant a greater understanding of specific HDAC isoform function. In recent years, inhibition of HDACs has been a promising new avenue for a variety of human brain disorders (Kazantsev & Thompson, 2008). The HDAC inhibitor SAHA (marketed as Vorinostat) has been approved for treatment of cancers (Richon, 2006). Let us now briefly review the histone acetylation and deacetylation mechanisms that play a role in ethanol-mediated phenotypes in both central and peripheral tissues. We will then briefly review these pathways in the context of other drugs of abuse in order to understand the shared epigenetic pathways that lead to addiction.
1B. Alcohol and Histone Acetylation and Deacetylation Mechanisms in the Brain
CREB signaling pathways have been implicated in ethanol and other drugs of abuse phenotypes in both vertebrate and invertebrate models (Levine et al., 2005; Pandey, Roy, & Zhang, 2003; Pandey, 2003, 2004; Wang, Ghezzi, Yin, & Atkinson, 2009). In vertebrate models, CREB signaling in the amygdaloid circuitry plays an important role in regulating ethanol-related behaviors and in mediating the anxiolytic effects of ethanol (Pandey et al., 2003; Pandey, Zhang, Roy, & Xu, 2005; Pandey, 2003, 2004). In the central and medial nucleus of the amygdala, after acute administration of ethanol we observed increased CREB phosphorylation, CBP levels, histone H3 and H4 acetylation, and neuropeptide Y (NPY) expression in addition to producing anxiolytic effects. On the other hand, withdrawal from chronic ethanol exposure produced opposite effects, resulting in a reduction in CREB phosphorylation, CBP and NPY levels corresponding to development of anxiety-like behaviors (Pandey, Ugale, et al., 2008; Pandey, Zhang, et al., 2008) (Figure 2). The neurotrophin BDNF (brain-derived neurotrophic factor), a critical modulator of synaptic plasticity and a downstream target gene of CREB, has also been implicated in the comorbidity of alcoholism and anxiety (Pandey, Zhang, Roy, & Misra, 2006; Poo, 2001). BDNF activity via CREB phosphorylation regulates activity regulated cytoskeleton-associated protein (Arc) and dendritic spine density and we showed that this pathway was positively regulated during acute ethanol exposure (anxiolysis) and negatively regulated during withdrawal from chronic ethanol exposure (anxiety-like behaviors) (Moonat, Sakharkar, Zhang, & Pandey, 2011; Pandey, Zhang, et al., 2008). Ethanol exposure has been shown to decrease CBP expression and histone acetylation in the developing cerebellum and this presumably is responsible for the motor deficits associated with fetal alcohol spectrum disorders (Guo et al., 2011). In summary, these findings clearly implicate the functional importance of CREB signaling pathways in anxiety and alcohol use disorders.
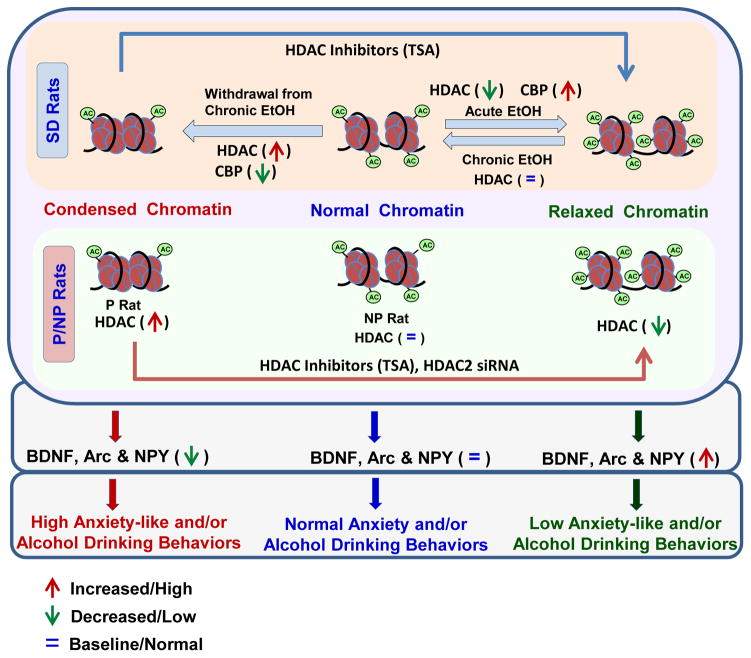
Figure 2.
Schematic representation of the histone acetylation mechanisms in the amygdala of Sprague-Dawley (SD) and alcohol preferring (P) and –non-preferring (NP) rats, that are operative in the modulation of ethanol drinking and anxiety-like behaviors. Acute ethanol exposure of SD rats inhibits the histone deacetylase (HDAC) activity and increases CBP expression, which has intrinsic histone acetyltransferase activity, leading to the hyperacetylation of histones. This relaxes chromatin and increases expression of activity-regulated cytoskeleton-associated protein (Arc), brain-derived neurotrophic factor (BDNF) and neuropeptide Y (NPY) resulting in acute ethanol’s anxiolytic effects. On the contrary, withdrawal from chronic ethanol treatment increases HDAC activity and decreases CBP levels leading to hypoacetylation of histones and a condensed state of chromatin that reduces BDNF, Arc and NPY, which precipitate anxiety-like behaviors. Treatment with trichostatin A (TSA), a pan-HDAC inhibitor, in alcohol-withdrawn SD rats was able to inhibit HDAC activity thereby increasing histone acetylation and expression of Arc, BDNF and NPY and further attenuated the withdrawal-related anxiety-like behaviors. Similarly, innately higher HDAC activity and HDAC2 expression in the amygdala of P rats compared to NP rats is associated with hypoacetylation of histones and low levels of Arc, BDNF and NPY and high levels of anxiety-like and alcohol-drinking behaviors. Inhibition of HDAC activity by treatment with TSA or knockdown of HDAC2 mRNA levels by using HDAC2 siRNA was able to correct the deficits in innate hypoacetylation and expression of Arc, BDNF and NPY thereby attenuating the high anxiety-like behavior and alcohol-preference in P rats (Pandey, Ugale et al., 2008; Moonat et al., 2013; You et al., 2014; Sakharkar, Zhang, et al., 2014).
1C. Role of HDACs in Alcoholism
Understanding the role played by HDACs in alcoholism and other drugs of abuse disorders is critical to understanding the etiology of addictive behaviors and at the same time providing a therapeutic avenue to treat these conditions. In a series of recent studies using preclinical models, Pandey’s laboratory have extensively investigated the HDAC-mediated chromatin remodeling pathways in the amygdala that play a role in the comorbidity of anxiety and alcohol-use disorders using both unselected and selected alcohol-preferring rat models. Inhibition of HDAC using Trichostatin A (TSA) was found to alleviate the symptoms of anxiety and reverse deficits in histone acetylation, NPY expression, and BDNF-Arc pathway in the amygdala upon withdrawal from chronic ethanol in adult male Sprague Dawley rats (Pandey, Ugale, et al., 2008; You, Zhang, Sakharkar, Teppen, & Pandey, 2014) (Figure 2). In addition, TSA treatment was able to normalize deficits in BDNF-Arc signaling and dendritic spine density in the amygdala upon withdrawal from chronic ethanol exposure (You et al., 2014). Furthermore, HDAC inhibition was able to reverse the development of rapid tolerance to the anxiolytic effects of ethanol and increase histone acetylation and NPY levels in the amygdala (Sakharkar, Zhang, Tang, Shi, & Pandey, 2012). The popular genetic model of alcohol consumption, alcohol-preferring (P) and -nonpreferring (NP) have been used to successfully characterize alcohol drinking behaviors and their comorbidity with anxiety-like behaviors (Pandey et al., 2005; Prakash, Zhang, & Pandey, 2008; Zhang et al., 2010). It was shown that innate deficits in the BDNF-Arc pathway, by regulating dendritic spine density, was operative in regulating the anxiety-like and excessive ethanol-drinking behaviors of P rats as compared to NP rats (Moonat et al., 2011) (Figure 2). The epigenetic mechanisms of these deficits were further investigated, and it was found that innate HDAC2 levels were higher in the amygdala of P as compared with NP rats, which corresponded to lower H3K9 acetylation globally and specifically at the promoters of BDNF and Arc genes corresponding to decreased expression of BDNF and Arc. These ultimately resulted in lower dendritic spine density in the amygdala. These alterations were ameliorated by infusion of HDAC2 siRNA into the central nucleus of amygdala (CeA), resulting in anxiolytic effects and reducing voluntary ethanol intake (Moonat, Sakharkar, Zhang, Tang, & Pandey, 2013) (Figure 2). Similarly, TSA treatment reduced the innately anxiety-like behaviors and ethanol intake in P rats, via inhibition of HDAC activity due to a reduction in HDAC2 protein levels in the CeA and medial nucleus of amygdala (MeA). TSA treatment also corrected the deficits in H3K9 acetylation levels globally and specifically at the NPY gene promoter, which correlated with increased NPY expression in the CeA and MeA of P rats (Sakharkar, Zhang, et al., 2014). Taken together, these data suggest that epigenetic modifications involving HDACs (namely HDAC2) may regulate amygdaloid chromatin assembly, and alter the expression of genes implicated in alcohol preference, tolerance and dependence. HDAC2 specific inhibition in the amygdala might be a therapeutic option to reduce the dysphoric symptoms of alcohol-use that are comorbid with anxiety and characterize the negative affective state of alcoholism (Moonat et al., 2013). Similarly, others have also shown that ethanol intake reduced histone H4 acetylation in the nucleus accumbens (NAc) in two rodent models and that HDAC inhibitor treatment (using both TSA and the FDA-approved HDAC inhibitor SAHA) reduced excessive ethanol drinking in mice and ethanol-seeking behaviors in rats (Warnault, Darcq, Levine, Barak, & Ron, 2013). In an ethanol-induced behavioral sensitization model, ethanol was shown to increase H4 acetylation in the NAc, which corresponded to a decrease in HDAC activity in the striatum (Botia, Legastelois, Alaux-Cantin, & Naassila, 2012). This was attenuated by treatment with the HDAC inhibitor, sodium butyrate, and associated with increased BDNF expression in the striatum (Legastelois, Botia, & Naassila, 2013). Opioid signaling pathways in the amygdala have also been identified as an epigenetic target of ethanol. Alcohol dependence has been shown to be associated with abnormal regulation of the Dynorphin/κ-opioid system with increased consumption linked to activation of this system contributing to the negative affective state. This may play a role in the motivational aspects of dependence and drug seeking and compulsive behaviors (Walker & Koob, 2008). Epigenetic regulation of the prodynorphin gene promoter following ethanol administration showed an increase in H3K9 acetylation (activating) and a decrease in H3K27 tri-methylation (repressive) marks in the amygdala of rats (D’Addario et al., 2013).
In summary histone acetylation and deacetylation mechanisms especially in key neuronal circuitry such as the amygdala, are an important regulatory neuro-mechanism that may be involved in the development and maintenance of alcoholism. Thus refining and optimizing HDAC inhibitor therapy provides an attractive option to treat alcoholism and its underlying causes.
1D. Alcohol and Histone Acetylation and Deacetylation Mechanisms in Non-neuronal Tissues
Histone covalent modifications have been implicated in alcohol phenotypes in peripheral tissues as well (Kim & Shukla, 2006). H3K9 acetylation was found to be increased in the liver of rats that had intra-gastric administration of ethanol (Kim & Shukla, 2006). Ethanol was shown to increase histone H3K9 acetylation in a dose and time-dependent manner in primary cultures of hepatocytes, without affecting H3K14 acetylation, demonstrating the specificity of ethanol in regulating covalent modifications to histone proteins. TSA administration was able to mimic ethanol’s effects on hepatocytes (Park, Miller, & Shukla, 2003). However, chronic ethanol treatment did not change global H3K9 acetylation, but did result in an increase in H3K9 acetylation in the promoter and coding regions of the alcohol dehydrogenase 1 gene (Park, Lim, & Shukla, 2012).
The above examples from both central and peripheral tissues reveal that histone acetylation and deacetylation changes, by bringing about dynamic regulation of gene transcription, provide a mechanism by which alcohol may alter the transcriptome, leading to altered neuronal plasticity and predisposition to addictive behaviors and the associated morbidity.
1E. Other Drugs of Abuse and Histone Acetylation and Deacetylation Mechanisms
Addiction mechanisms are conserved at the neuroanatomic and molecular level and so understanding the basis of acetylation and deacetylation mechanisms that regulate other drugs of abuse (e.g. cocaine) phenotypes gives us a clue as to which neuroanatomic regions and underlying molecular pathways are shared mechanisms among these different classes of drugs with addictive potential (Robison & Nestler, 2011). CREB and corticotropin releasing factor interactions have been implicated in stress-induced cocaine reward potentiation (Kreibich et al., 2009). CBP-mediated acetylation has also been shown to regulate cocaine behaviors. For example, mice that are haploinsufficient for CBP show decreased sensitivity to cocaine, due to decreased histone acetylation at the fosB promoter (Levine et al., 2005). Cocaine was shown to induce c-fos expression and increase acetylation at the c-fos promoter, which were reduced in a CBP focal knockout in the NAc that also reduced cocaine sensitivity (Malvaez, Mhillaj, Matheos, Palmery, & Wood, 2011). Acute cocaine was shown to rapidly induce H4 acetylation, but not H3, at the cFos promoter, whereas chronic cocaine administration induced histone H3 acetylation at the BDNF promoter (Kumar et al., 2005). HDAC inhibition through pharmacological approaches using sodium butyrate and TSA and using genetic approaches (HDAC5 knockout), was able to potentiate cocaine’s behavioral effects whereas overexpression of HDAC4 and HDAC5 were able to decrease the behavioral responses to cocaine (Kumar et al., 2005; Renthal et al., 2007). Others have also shown HDAC isoform specific regulation of cocaine behaviors (Host, Dietrich, Carouge, Aunis, & Zwiller, 2011; Malvaez et al., 2013; Schroeder et al., 2008). In the case of the sirtuin class of HDACs, chronic cocaine exposure was shown to induce SIRT1 and SIRT2 and regulate cocaine self-administration and reward. This was shown to occur due to increased H3 acetylation and delta-fosB binding to the SIRT promoters (Renthal et al., 2009). Regulation of HDAC activity has also been implicated in enhancement of morphine-induced locomotor sensitization and conditioned place preference (Sanchis-Segura, Lopez-Atalaya, & Barco, 2009) and in nicotine preference (Pastor, Host, Zwiller, & Bernabeu, 2011). All these studies emphasize the complexity of HDAC inhibition and bring into context the different variables that have to be addressed such as the dosage of the drug, brain region studied, class and dosage of HDAC inhibitor, timing of administration and innate genetic differences in the employed animal models. Examinations of HDAC mediated pathways in ethanol and cocaine studies reveal the importance of understanding the regulation and mode of action of HDACs in order to advance the field of addiction and develop appropriate therapeutic interventions.
1F. Alcohol and Histone Methylation Mechanisms
Histone lysine methylation is an important modification that is associated with chromatin remodeling and gene regulation and has been implicated in drug-induced neuronal plasticity mechanisms, memory formation and cognition (Gupta et al., 2010; Maze et al., 2010, 2011; Schaefer et al., 2009; Shinkai & Tachibana, 2011). Tri-methylation of H3K4 (H3K4me3) is associated with active chromatin and is potentiated by acetylation of H3K9 and 14, providing further evidence of cross-talk between histone modifications and transcriptional regulation (Vermeulen et al., 2007). In a model of contextual fear conditioning, H3K4me3 was shown to be increased in the promoters of BDNF and Zif-268 genes within the CA1 region of the hippocampus. Moreover, infusion of an HDAC inhibitor directly into CA1 caused an increase in the transcriptionally active H3K4me3 and a simultaneous decrease in the transcriptionally repressive H3K9me2 (Gupta et al., 2010). In an invertebrate model of Huntington Disease, lower levels of the H3K4 de-methylase SMCX/Jarid1c have been shown to be neuroprotective, a mechanism that is also conserved in a mouse model (Vashishtha et al., 2013). Contrary to H3K4, methylation at H3K9 is a repressive mark and is involved in alcohol and drugs of abuse phenotypes. Studies reveal a link between G9a (lysine dimethyltransferase), which catalyzes H3K9 methylation, and neurodegeneration in the developing brain that has been exposed to postnatal ethanol, which may have implications for fetal alcohol spectrum disorders (Subbanna et al., 2013). In a chronic ethanol treatment model, both treatment with and withdrawal from ethanol produced an increase in H3K9 acetylation at the NR2B (NMDA receptor 2B) gene promoter and a corresponding decrease in H3K9 methylation (Qiang, Denny, Lieu, Carreon, & Li, 2011). In cultured rat hepatocytes, ethanol decreased the levels of H3K9me2 while concomitantly increasing H3K4me2, with both ultimately contributing to an open chromatin state. This corresponded to increased expression of the alcohol dehydrogenase gene. Furthermore histone H3K9 acetylation was increased in the promoter and coding regions of the gene (Pal-Bhadra et al., 2007; Park et al., 2012). Histone methylation is a dynamic reversible process and is under the control of two opposing groups of enzymes-the lysine methyltransferases and the lysine demethylases (Kouzarides, 2007). Genome-wide association studies have found KDM4C (a lysine demethlyase of the jumonji domain 2 (JMJD2) family) loci associated with alcohol withdrawal (Wang, Liu, Zhang, Wu, & Zeng, 2012). Other drugs of abuse, such as cocaine, resulted in a reduction of G9a and G9a-like protein (GLP) (lysine dimethyltransferases), which corresponded to a decrease in H3K9me2, and an increase in spine density in the NAc and cocaine place preference, whereas overexpression was able to reverse these effects (Maze et al., 2010). In the case of the repressive mark, H3K9me3, repeated cocaine caused a decrease in H3K9me3 binding to non-coding retro-transposon associated inter-genic regions (Maze et al., 2011).
1G. Alcohol and Histone Phosphorylation Mechanisms
Phosphorylation of histones is associated with transcriptional activation, DNA repair and cell-cycle dynamics and is mediated by protein kinases and removed by phosphatases (Banerjee & Chakravarti, 2011; Bannister & Kouzarides, 2011). Histone H3 phosphorylation has been shown to regulate important neuronal pathways. For example, histone H3 phosphorylation at serine 10 has been implicated in a contextual fear-conditioning model in the hippocampus (Chwang, O’Riordan, Levenson, & Sweatt, 2006). The protein kinase ribosomal S6 kinase 2 (RSK2) has been shown to phosphorylate histone H3 and CREB and, in the presence of CBP, mediate a combined acetylation-phosphorylation event that regulates chromatin remodeling (Merienne, Pannetier, Harel-Bellan, & Sassone-Corsi, 2001). The psychostimulant drugs, amphetamines and cocaine, as well as morphine have been found to increase phosphorylation of serine 10 of histone H3 which further results in sequestration of dopamine- and cyclic-AMP regulated phosphoprotein 32 (DARPP-32) in the nucleus which inhibits protein phosphatase-1, thereby promoting phosphorylation. Blocking this phosphorylation event at serine 10 on H3 was able to diminish the behavioral responses to both cocaine and morphine and blocked conditioned place preference for cocaine (Stipanovich et al., 2008). In the periphery, ethanol and its breakdown product, acetaldehyde, activate various MAP kinase-signaling pathways (Aroor & Shukla, 2004). In primary rat hepatocytes, ethanol and acetaldehyde were shown to increase phosphorylation of histone H3 at serine 10 and serine 28 mediated by p38 MAPK in the nucleus (Lee & Shukla, 2007). In the same model it was also shown that ethanol upregulates the activating marks H3K9 acetylation and H3K4 dimethylation and down-regulates H3K9 dimethylation (Pal-Bhadra et al., 2007; Park et al., 2003). This illustrates the complex modulation of histone H3 at different residues by ethanol in the liver, which may give rise to the various pathologic states resulting due to chronic ethanol ingestion contributing to alcoholic liver disease.
Overall, covalent modifications of histone side-chain residues which are under the control of different enzymatic pathways such as acetylation/deacetylation, methylation/demethylation and phosphorylation/dephosphorylation are crucial regulatory mechanisms that change the brain transcriptome upon acute or chronic exposure to alcohol. These contribute to the pathogenesis of alcoholism and the long-term neuroadaptations resulting from and associated with the alcoholic state.
2. Epigenetic Regulation due to DNA Methylation
DNA methylation is a covalent modification to DNA, which occurs on cytosine residues and involves the addition of a methyl group to the C5 position (5-mc) and is catalyzed by DNA methyltransferases (DNMTs) (Bestor, 2000; Klose and Bird 2006). It is a stable mark that is important for diverse cellular processes such as X-chromosome inactivation, transcriptional silencing, allele-specific expression (imprinting) and is fundamentally important for normal embryonic development (Cedar & Bergman, 2012). DNA methylation patterns are stably propagated and studies employing imprinted loci show that DNA methylation patterns are re-established in the developing embryo despite their removal in primordial germ cells (Allegrucci, Thurston, Lucas, & Young, 2005; Li, 2002). Methylation of DNA is thought to mediate transcriptional repression via two ways; directly hindering binding of DNA-binding proteins (Watt & Molloy, 1988) and indirectly through binding of methyl CpG binding proteins and recruitment of HDACs, co-repressors and other heterochromatin associated proteins (Boyes & Bird, 1991; Klose & Bird, 2006; Nan et al., 1998; Ng et al., 1999). In the CNS, DNA methylation is a critical epigenetic regulator for normal brain development, differentiation and maintenance of function. Dysfunction of the pathways that regulate DNA methylation have been implicated in a variety of brain disorders such as abnormal learning and memory (Feng et al., 2010; Miller & Sweatt, 2007), synaptic plasticity (Chen et al., 2003; Levenson et al., 2006), fragile-x syndrome (Sutcliffe et al., 1992), autism spectrum disorder (Zhubi et al., 2014), Rett’s Syndrome, (an X-linked autism spectrum disorder caused by mutations in methyl-CpG binding protein 2, MeCP2) (Amir et al., 1999), schizophrenia and bipolar disorders (Grayson & Guidotti, 2013).
2A. Functions of DNMTs and MBDs
The enzymes that catalyze DNA methylation are known as DNA methyltransferases (DNMT) (Goll & Bestor, 2005). Three active mammalian DNMTs have been identified, DNMT1, 3a and 3b. The first identified and most abundant DNMT, DNMT1, otherwise known as a “maintenance methyltransferase,” can recognize hemi-methylated DNA and perform methylation of the complementary strand. The de novo methyltransferases, DNMT3a and DNMT3b, methylate previously unmethylated cytosines, are essential during development and, in the CNS, have been implicated in synaptic plasticity and learning and memory mechanisms (Feng et al., 2010; Levenson et al., 2006; Miller & Sweatt, 2007; Okano, Bell, Haber, & Li, 1999). Disparately, DNMT3l is a protein structurally related to 3a and 3b, but does not have methyltransferase activity on its own and regulates catalytic activities of DNMT3a and 3b (Hata, Okano, Lei, & Li, 2002). Methyl-CpG binding protein 2 (MeCP2) is a protein that binds methylated CpG dinucleotides via interactions through the methyl-CpG-binding domain (MBD) (Lewis et al., 1992; Meehan, Lewis, McKay, Kleiner, & Bird, 1989). It has been shown to both activate and repress transcription (Chahrour et al., 2008; Jones et al., 1998; Mellen, Ayata, Dewell, Kriaucionis, & Heintz, 2012; Nan et al., 1998) and four families of MBD containing proteins have now been identified including the founding member, MeCP2, and a newly identified MBD-dependent member Kaiso, which recognizes DNA via zinc-finger domains (Klose & Bird, 2006). The role played by MeCP2 in regulating BDNF function and regulating synaptic plasticity has been well characterized (Martinowich et al., 2003; Chen et al., 2003; Zhou et al., 2006). We have shown a deficit in the BDNF system in the CeA and MeA have been associated with anxiety-like and alcohol-drinking behaviors (Moonat et al., 2011; 2013; Pandey et al., 2006). Further studies are needed to understand the functional role of MeCP2 in the regulation of BDNF expression during the comorbidity of anxiety and alcoholism.
2B. DNA Demethylation Pathways in the Brain
The demethylation of DNA is a rapidly emerging field involving a complex interplay of interdependent pathways and mechanisms (Gavin, Chase, & Sharma, 2013; Wu & Zhang, 2014). Activity-dependent DNA demethylation is a dynamic process crucial to neuronal function. The “ten eleven translocation” (TET) enzyme family of proteins converts methylcytosine to hydroxymethylcytosine (hmC), an oxidized form of the enzyme that can be further demethylated (Tahiliani et al., 2009). A pair of studies in 2009 identified the presence of 5-hydroxymethylcytosine (5-hmC) in brain cells (Purkinje cell layer of the cerebellum) and in mouse embryonic stem cells (Kriaucionis & Heintz, 2009; Tahiliani et al., 2009). Genome-wide mapping studies have indicated the presence of 5-hmc in numerous tissues and in great abundance in the brain. Overall 5-hmc is associated with gene bodies, promoters and enhancers most likely implicating its role as a transcriptional activator (Ficz et al., 2011; Guo, Su, Zhong, Ming, & Song, 2011; Mellen et al., 2012; Song et al., 2011; Yu et al., 2012). TET1 has also been implicated as a key molecule in synaptic plasticity and memory mechanisms through regulation of Arc thereby modulating extinction of fear memories (Kaas et al., 2013; Rudenko et al., 2013). Studies have also implicated the Growth arrest and DNA damage (Gadd45) family of proteins to be important for DNA demethylation and suggested a role for this pathway in hippocampal synaptic plasticity associated learning and memory (Barreto et al., 2007; Ma et al., 2009; Sultan, Wang, Tront, Liebermann, & Sweatt, 2012). These studies have brought DNA demethylation into the limelight and more importantly suggest that even a ‘stable’ mark such as cytosine methylation is subject to dynamic regulation. Let us now look at the effects of ethanol on DNA methylation and de-methylation networks and the phenotypic outcomes.
2C. Alcohol and DNA Methylation and Demethylation Mechanisms
DNA methylation/demethylation mechanisms have been implicated in a variety of alcohol phenotypes in both central and peripheral tissues. For example, chronic ethanol treatment of mouse embryonic cortical neurons revealed DNA demethylation at the NMDA receptor (NR2B) gene promoter, which correlated with an upregulation of NR2B expression. However, acute ethanol treatment did not alter the methylation of NR2B gene promoter or NR2B expression in adult mouse cortex or in mouse fetal cortical neurons (Ravindran & Ticku, 2004, 2005). The DNMT inhibitor 5-aza mimicked these chronic ethanol effects independently and also in combination with ethanol (Ravindran & Ticku, 2004, 2009). Using a chronic intermittent ethanol (CIE) exposure model, a decrease in site-specific (proximal to transcription factor binding AP-1 and CRE sites) DNA methylation was observed, which persisted following ethanol withdrawal. Moreover, these changes were associated with CIE-induced NR2B expression (Qiang et al., 2010; Qiang, Denny, & Ticku, 2007). Human post-mortem studies have revealed three single-nucleotide polymorphisms in the prodynorphin gene that are associated with alcohol dependence which overlap with CpG sites and are differentially methylated (Taqi et al., 2011). Prefrontal cortical tissue from post-mortem psychiatric patients display increased levels of DNMT1 and TET1. Furthermore, psychiatric patients with a history of alcohol-abuse showed an even greater induction of TET1, yet normalized DNMT1 levels. These findings suggest that comorbidity of psychosis with alcoholism is a complex phenomenon and consideration needs to be taken in the treatment of individuals with comorbid disorders (Guidotti et al., 2013).
Prenatal alcohol exposure predisposes the offspring to neurofacial deficits and growth retardation resulting in a spectrum of mild to severe deficits, which comprise the Fetal Alcohol Spectrum Disorders (FASD) (Idrus & Thomas, 2011). Dysregulation of methylation pathways is an important causative factor for FASD development. In a mouse model of FASD, alcohol exposure during early neurulation caused global methylation pattern differences (Liu et al., 2009). Alcohol has been shown to inhibit DNA methylation and affect programming of neural stem cells thereby affecting their differentiation and causing disruptions in early embryonic development (Zhou et al., 2011). Prenatal alcohol exposure was shown to increase DNMT1 and MeCP2 expression and reduce hypothalamic pro-opiomelanocortin (POMC) gene expression (Bekdash, Zhang, & Sarkar, 2013).
Ethanol induced hyperhomocysteinemia and its NMDA-agonistic byproducts have been suggested as a possible predictor of ethanol withdrawal seizures (Bleich et al., 2004). Elevated homocysteine levels in the blood of alcoholic patients have been associated with hypermethylation at the promoter of the homocysteine-induced endoplasmic reticulum protein gene, corresponding to lower mRNA expression in the blood cells (Bleich et al., 2006). Elevated homocysteine levels have also been implicated in hypermethylation of the alpha-synuclein gene promoter, which may be responsible for altered mRNA and protein expression and was linked to alcohol craving (Bönsch et al., 2004; 2005; Bönsch, Greifenberg, et al., 2005). Chronic alcohol consumption could be associated with altered methylation patterns of various genes acting via s-adenosylmethionine (SAM) metabolism and altering homocysteine levels. Approaches that can detect altered transcripts and corresponding changes in methylation levels peripheral blood samples such as lymphocytes and mononuclear cells can be used as potential tool for the development of biomarkers (Biermann et al., 2009; Hillemacher et al., 2009; Zhang et al., 2013; Zhao et al., 2013).
2D. Alcohol and DNA Methylation Mechanisms in Non-neuronal Tissues
The liver is an important target for ethanol induced DNA methylation changes that affect normal physiological function. Chronic ethanol consumption leads to abnormal regulation of the methylation pathway by inhibiting methionine adenosyl transferase the enzyme that converts methionine to SAM. This enzymatic inhibition leads to global DNA hypomethylation (French, 2013; Varela-Rey, Woodhoo, Martinez-Chantar, Mato, & Lu, 2013). Methylation patterns can also be affected by altered DNMT activity. For example, alcoholic patients have lower expression levels of DNMT3b (Bonsch et al., 2006) and acetaldehyde, the breakdown product of alcohol, which has been shown to inhibit DNMT activity (Garro, McBeth, Lima, & Lieber, 1991). Ethanol-induced hepatic steatosis (fatty liver) was reduced in DNMT1 hypomorphic mice suggesting a role for DNMTs in the progression of hepatic liver disease (Kutay et al., 2012). The effects of alcohol on methylation patterns have been implicated in advanced disease states such as alcoholic liver disease and cancer and further research may therefore provide promising therapeutic targets for pharmacological intervention (Varela-Rey et al., 2013).
2E. Other Drugs of Abuse and DNA Methylation Mechanisms
Drugs of abuse such as cocaine also regulate DNA methylation pathways. Cocaine exposure causes a complex expression pattern of DNMT3a expression. There was a biphasic response in that early induction of DNMT3a in the nucleus accumbens (NAc), was followed by down-regulation upon both acute and chronic cocaine exposure. During chronic cocaine exposure, a persistent and stable enhancement was observed in the NAc suggesting a role for DNMTs in the homeostatic feedback regulation of cocaine-mediated effects (LaPlant et al., 2010). Striatal MeCP2 was induced by cocaine self-administration and shown to positively regulate BDNF expression by a homeostatic regulation via microRNA-212 (Im, Hollander, Bali, & Kenny, 2010). MeCP2 signaling in the NAc is also involved in the behavioral responses to psychostimulants such as amphetamines (Deng et al., 2010). The induction of DNMT3a and 3b in the NAc by acute cocaine exposure was shown to increase binding of MeCP2 at the protein phosphatase-1 catalytic subunit promoter implicating DNMT involvement in the behavioral sensitization to cocaine. Consequently, use of the DNMT inhibitor zebularine was able to delay the onset of cocaine behavioral sensitization (Anier, Malinovskaja, Aonurm-Helm, Zharkovsky, & Kalda, 2010). Furthermore, infusion of the DNMT inhibitor 5-aza-2-deoxycytidine (5-aza) into the hippocampus or prefrontal cortex was able to attenuate either the acquisition (hippocampus) or expression (prefrontal cortex) of cocaine-induced conditioned place preference behaviors (cocaine reward) in C57BL/6 mice (Han et al., 2010). These examples illustrate the fact that alterations of DNA methylation/demethylation pathways play a role in the phenotypes of drugs of abuse such as cocaine, implicating these pathways in drug addiction mechanisms. The use of DNMT inhibitors enables us to control these pathways pharmacologically and epigenetically modulate target gene expression that control drug phenotypes (Warnault, Darcq, Levine, Barak, & Ron, 2013).
2F. DNMT inhibitors as a Therapy for Drug Addiction
Drugs used in manipulating DNMT activity and affect DNA methylation patterns have shown promise in both in-vitro and in-vivo models to dissect neuronal function. Two DNMT inhibitors, zebularine and 5-aza were used to modulate LTP induction, synaptic plasticity and learning and memory behaviors in the hippocampus (Levenson et al., 2006; Lubin, Roth, & Sweatt, 2008). Zebularine and 5-aza have been used to study cocaine-regulated phenotypes and have been used to delay the onset of cocaine sensitization and attenuate cocaine reward respectively (Anier et al., 2010; Han et al., 2010). In an excessive alcohol-drinking paradigm, 5-aza was able to reduce excess ethanol intake in mice (Warnault et al., 2013). Newer direct DNMT catalytic activity inhibitors, such as the pan-DNMT inhibitor RG108 have been developed and have been successfully used in regulating remote memories (Miller et al., 2010).
Alternate approaches use HDAC inhibition to regulate DNA methylation. It is well established that DNA methylation and histone modifications are interdependent and lead to coordinated patterns of regulation that modulate various phenotypes such as learning and memory, synaptic plasticity, somatic cell reprogramming and cancer biology (Cedar & Bergman, 2009; Miller, Campbell, & Sweatt, 2008). Marking sites with active methylated H3K4 appears to be one of the ways for CpG island-rich promoters in somatic cells to prevent DNA methylation and thus protected from de novo methylation (Weber et al., 2007). These findings are consistent with other studies where increased DNA methylation has been shown to be associated with the absence of H3K4 methylation and the presence of H3K9 methylation (Meissner et al., 2008). In alcoholic post-mortem brain, loci of hypomethylation have been well correlated with increased histone H3K4 trimethylation (Ponomarev, Wang, Zhang, Harris, & Mayfield, 2012). The histone methyltransferase G9a has been shown to exhibit dual functionality through SET domain- mediated H3K9 tri-methylation mediated heterochromatin formation as well as ANK domain- recruitment of DNMT3a and 3b to initiate de novo methylation.
Taken together these processes favor a repressive chromatin formation and inactivate early embryonic genes (Epsztejn-Litman et al., 2008). These studies suggest that instead of genomic context, such as presence of a CpG island, a better predictor of DNA methylation could be histone marks or the epigenomic status of the given loci. In the case of HDAC inhibitors, they are capable of inducing demethylation in the brain (Weaver et al., 2004). The HDAC inhibitor, Valproate has been used in neuropsychiatric disorders and recent findings show that it induces DNA demethylation in adult mouse brain (Dong, Chen, Gavin, Grayson, & Guidotti, 2010). Similarly, TSA and valproic acid have been shown to increase histone acetylation and decrease methylation. Interestingly in the same study 5-aza, only decreased methylation without affecting acetylation of H3 and H4 (Mitchell, Chen, Kundakovic, Costa, & Grayson, 2005). It is interesting to note that the class III histone deacetylase, SIRT1 has been shown to directly deacetylate DNMT1 at specific lysine residues and modulate its activity (Peng et al., 2011).
Manipulating DNMT function in the brain using these approaches enables us to understand the the functioning of these pathways at the molecular level following alcohol exposure. However, DNA methylation/demethylation is a crucial process involved in normal development and physiological function, with dysregulation of these pathways causing a myriad of pathologies. Therefore DNMT inhibition has to be specific and carefully controlled since non-methylation specific effects in pathways other than those intended, can influence several concurrent phenotypes. Moreover as mentioned above HDAC inhibition can also have cross-talk with DNMT function. Caveats such as these stress the need for a refinement of our scientific tools in order to increase understanding of the epigenetic basis and treat a complex disorder such as addiction.
3. Epigenetics of Adolescent Drinking and Implications for Alcohol Phenotypes in Later Life
The neurodevelopmental implications of alcohol drinking during the adolescent period are becoming a cause for great concern warranting a greater understanding of these brain adaptations and how they affect the progression of alcoholism later in life (Guerri & Pascual, 2010; Spear, 2011). Epigenetic regulation of critical neural pathways has been recognized as a major effector in mediating these changes in neuroplasticity. Intermittent ethanol treatment of adolescent Wistar rats has been shown to increase histone H3K9 and H4K12 acetylation in the frontal cortex and NAc with a concomitant decrease in the striatum. However, these alterations in histone acetylation due to intermittent ethanol exposure were not preserved in adult rats further highlighting the vulnerability of the adolescent brain to binge-like alcohol drinking, the most common pattern of drinking among adolescents (Pascual, Boix, Felipo, & Guerri, 2009). In a subsequent study, Pascual et al demonstrated that the effects of ethanol on histone acetylation in the prefrontal cortex at specific gene promoters (cFos, Cdk5, FOSB and BDNF) and their potentiation through HDAC inhibition are associated with ethanol-induced conditioned place aversion and reinstatement (Pascual et al., 2012). Recently, we observed that acute ethanol inhibits HDAC and DNMT activities in the amygdala and only DNMT activity in the bed nucleus of stria terminalis of adolescent rats and produces anxiolytic-like effects (Sakharkar, Tang, et al., 2014). Adolescent rats, unlike adult rats, do not show the development of rapid tolerance to ethanol-induced anxiolysis and inhibition of HDAC in the amygdala (Sakharkar et al., 2012; Sakharkar, Tang, et al., 2014). Because the age of first alcohol exposure is believed to be an important predictor of increased risk of alcohol abuse, these studies necessitate further investigation to identify possible epigenetic marks or mechanisms involved in psychiatric and substance abuse disorders associated with adolescent alcohol exposure (DeWit, Adlaf, Offord, & Ogborne, 2000; Hawkins et al., 1997).
4. Epigenetic Regulation of Glial-Neuronal Interactions and Alcohol
Although much research has focused on neuronal populations in alcohol use, glial cells comprise a vital component in the proper functioning of the nervous system and regulate synaptic plasticity (Dallerac, Chever, & Rouach, 2013; Oliet, Piet, & Poulain, 2001). The role of glia in neuroinflammatory processes that regulate neuronal function is well recognized in the context of drug phenotypes (Miguel-Hidalgo, 2009). Studies have shown that ethanol induces the transcription factor, NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) which induces a host of pro-inflammatory molecules that may be epigenetically regulated (Crews, Zou, & Qin, 2011; Ponomarev et al., 2012). Cocaine exposure has also been shown to induce NF-κB expression in the NAc where it regulates dendritic spines and sensitization (Russo et al., 2009). NF-κB and related transcription factors integrate various signals and regulate, via epigenetic and chromatin-remodeling pathways, corresponding targets to bring about distinct cellular outcomes (Meffert, Chang, Wiltgen, Fanselow, & Baltimore, 2003; Natoli, Saccani, Bosisio, & Marazzi, 2005). Neuroinflammation induced following chronic ethanol use and withdrawal was shown to occur via Toll-like receptor signaling (TLR4) and activation of the NF-kB pathway and glial derived proinflammatory cytokines, which was reduced in TLR4- deficient mice (−/−) (Alfonso-Loeches, Pascual-Lucas, Blanco, Sanchez-Vera, & Guerri, 2010). Interestingly, HAT activity and acetylation of histones H3 and H4 were reduced in wild-type mice undergoing withdrawal after chronic ethanol exposure. However these changes were not observed in the TLR4 (−/−) mice (Pascual, Baliño, Alfonso-Loeches, Aragón, & Guerri, 2011). These findings suggest that TLR4 signaling via alterations of chromatin structure can promote the neuroinflammatory changes observed after chronic ethanol exposure (Alfonso-Loeches et al., 2010; Pandey, 2012; Pascual et al., 2011). It has been recently shown that activation of proinflammatory cytokine signaling by ethanol is mediated by inhibition of HDAC activity. This provides a novel therapeutic option to manage the neuroinflammatory sequelae resulting from ethanol exposure (Zou and Crews, 2014).
Ethanol exposure affects DNA methylation in glia and increases tissue plasminogen activator (tPA) expression, which is involved in the inhibition of neuritogenesis and regulates neuronal plasticity (Zhang, Kusumo, Sakharkar, Pandey, & Guizzetti, 2014). Additionally, amygdaloid tPA has been implicated in regulating stress responses during acute cocaine withdrawal (Zhou, Maiya, Norris, Kreek, & Strickland, 2010). These examples illustrate the importance that glial and neuronal compartments be considered as not discrete entities, but rather as highly inter-connected structures which collaboratively influence neuronal plasticity (Todd, Serrano, Lacaille, & Robitaille, 2006). Dissecting out the epigenetic pathways in glia and neurons involved in chronic drug-induced neuroinflammation and synaptic plasticity will provide increased treatment opportunities to address the neuroinflammatory changes following chronic ethanol exposure (Kovacs, 2012).
5. Genome-Wide Approaches to Understand the Basis of Alcoholism
Post-mortem brain studies have applied next generation sequencing approaches to detect changes in histone modifications underlying alcohol and cocaine use. Investigating human post-mortem brain from cocaine addicts and alcoholic patients, a parallel RNA-Sequencing (RNA-Seq) and ChIP-Sequencing(ChIP-Seq) experiment was performed in which RNA-Seq expression data was correlated with the H3K4me3 mark (Zhou, Yuan, Mash, & Goldman, 2011). The RNA-Seq data revealed 29 genes that regulate important neuronal functions showing coordinated regulation between cocaine and alcohol. For the ChIP-Seq data, around 250 H3K4me3 peaks were shared between the two treatments suggesting a shared epigenetic component between these two drugs of abuse (Zhou, Yuan, et al., 2011). A similar approach has been used in Drosophila to construct a gene network based on genome-wide histone H4 acetylation fold changes that occur at genomic loci following exposure to two drugs, benzyl alcohol and ethanol, utilizing the fact that these drugs produce mutual cross-tolerance. Among the 144 shared candidates discovered, the top gene clusters based on gene ontology belonged to the categories of transcriptional regulation, ion channels and synaptic plasticity genes (Ghezzi et al., 2013). Next-generation sequencing approaches have been used to determine global methylation levels in human post-mortem brains of alcoholics. Alcoholic brain tissue display an overall decrease in methylation, especially at long-terminal repeat containing endogenous retroviruses which corresponded to their activation as well as upregulation of GC rich genes and associated histone H3K4 tri-methylation (Ponomarev et al., 2012). These epigenetic alterations may contribute to the widespread changes in gene expression and in the global transcriptome following chronic ethanol exposure. These methodologies offer a novel way to identify and characterize epigenetic networks that are unique to a single drug and also that are shared following exposure to different drugs sharing similar phenotypes and/or affecting the same neuroanatomical circuits.
The future of next-generation sequencing approaches to unravel the epigenetic pathways that regulate drug phenotypes lies in combining approaches so meaningful networks and pathways emerge. A concerted effort such as the ENCODE (Encyclopedia of DNA Elements) project, but specifically for drug addiction is required for the field to advance (Ecker et al., 2012). This way candidate pathways and hypotheses can be tested based on unbiased approaches.
6. Using Invertebrate Models to Study the Basis of Ethanol Phenotypes
The invertebrate model, Drosophila melanogaster, has been used to identify genetic networks involved in ethanol behaviors whose components have also been identified in pre-clinical animal models to regulate ethanol behaviors (Atkinson 2009). Drosophila neuropeptide F (NPF), which is the homolog of mammalian NPY, mediates sensitivity to ethanol sedation and NPY signaling in rat amygdala has been shown to regulate anxiety and alcohol tolerance (Pandey, Ugale, et al., 2008; Pandey, Zhang, et al., 2008; Sakharkar et al., 2012; Wen, Parrish, Xu, Wu, & Shen, 2005; Zhang et al., 2010). The Drosophila BK-type Ca2+-activated potassium channel is involved in rapid tolerance to ethanol and has also been shown in the invertebrate model, C. elegans and in higher vertebrates to mediate ethanol sensitivity and tolerance (Cowmeadow, Krishnan, & Atkinson, 2005; Cowmeadow et al., 2006; Davies et al., 2003; Liu, Asuncion-Chin, Liu, & Dopico, 2006; Pietrzykowski et al., 2008). Epigenetic regulation of the BK channel gene promoter is complex and exhibits a spatio-temporal pattern of histone H4 acetylation following benzyl alcohol sedation in adult Drosophila brain. Acetylation increases become manifest at four hours after sedation and remain elevated 24 hours after sedation, however at different regions of the gene promoter. This presumably arises due to decreased acetylation at one region in the promoter being compensated by an increase in another region, which in turn may arise due to transcription factor activity or other cis- and/or trans- acting mechanisms (Wang, Krishnan, Ghezzi, Yin, & Atkinson, 2007). These data demonstrate the importance of genomic location in measuring histone modification changes as well as choosing the appropriate time-points. CREB signaling pathways in higher vertebrates regulate chromatin remodeling and mediate anxiety and ethanol-drinking behaviors (Moonat, Starkman, Sakharkar, & Pandey, 2010; Pandey, Ugale, et al., 2008) and have also been shown to affect ethanol tolerance in Drosophila (Wang, Ghezzi, et al., 2009). Drosophila CREB activates expression of the BK-type Ca2+ activated K channel gene (dslo) by binding to the promoter in response to benzyl alcohol sedation leading to a behavioral tolerance to the drug on a subsequent exposure (Wang, Ghezzi, et al., 2009). Furthermore, a mutation in the Drosophila homolog of CBP, nejire (nej), regulates tolerance to benzyl alcohol and ethanol (Ghezzi et al., 2013). Invertebrate models, such as Drosophila, have been invaluable in identifying genetic networks and pathways that regulate disease phenotypes that are conserved between invertebrate and mammalian models due to their forward and reverse genetic approaches (Atkinson, 2009). The identification of conserved epigenetic regulatory networks regulating behaviors related to ethanol exposure in both vertebrates and invertebrates, such as CREB signaling, serve to illustrate the significance of invertebrate genetic models to further our understanding of epigenetic mechanisms of alcoholism.
7. Inheritance of a Drug Phenotype
Can acquired traits be drafted into an “Epigenome” and passed down to future generations? Experience-driven epigenetic changes alter the genome conveying a modified chromatin state that stably persists throughout subsequent mitotic divisions possibly inherited by progeny through the germline. However, the mechanisms of how this happens are unclear. The adverse effects of prenatal ethanol have been shown to be transmitted across generations via DNA methyl marks on the hypothalamic proopiomelanocortin gene through male germline (Govorko, Bekdash, Zhang, & Sarkar, 2012). Additionally, a recent study revealed that fear memories can be passed on across generations via hypomethylation of odorant receptors (Dias & Ressler, 2014). These transgenerational effects have been demonstrated in cocaine addiction also, where cocaine has been shown to promote acetylation of the BDNF gene promoter in male spermatozoa of sires that self-administered cocaine. This resulted in male offspring that showed resistance to cocaine reinforcement and exhibited increased acetylation of histone H3 and BDNF mRNA and protein expression in the medial prefrontal cortex (Vassoler, White, Schmidt, Sadri-Vakili, & Pierce, 2013). Overall, these examples provide evidence that an epigenetic state, which maintains a changed behavior of a system in a stable manner, can be heritable and co-exist with our genome for subsequent generations.
Conclusions
Epigenetic states are initiated by environmental cues, developmental stimuli, cellular events and their sequelae and mediated by various cellular mechanisms including histone modifications, DNA methylation signals, non-coding RNAs and transcription factor networks (Bonasio and Reinberg, 2010). These mechanisms ultimately shape the chromatin architecture and fine-tune the transcriptome so that it can respond even in the absence of the initiating cue. This results in an “epigenome” that co-exists with the genome and is stably maintained in mitotic and post-mitotic cells, such as neurons, in a self-sustaining fashion (Dulac, 2010). A process that is critical for normal cellular physiology and function. Environmental factors, such as ethanol, usually dysregulate neuronal functions via epigenetic modifiers (Moonat et al., 2013) and lead to positive and negative affective states of addiction (Pandey 2004; Koob & Volkow, 2010; Arora et al., 2013). Epigenetic targets therefore hold the promise for improved disease management, especially for a disease such as alcoholism whose endophenotypes are under complex epigenetic regulation. A deeper knowledge of the epigenomics of alcoholism will enable us to identify the checkpoints and the processes that take us down the vicious path of addiction and the interventions necessary to reverse that trajectory (Figure 3).
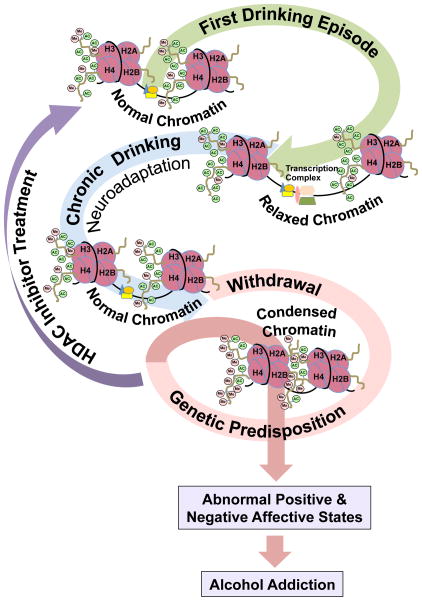
Figure 3.
Hypothetical representation of chromatin remodeling driven by histone acetylation mechanisms in the amygdala that may be operative in the process of alcohol addiction. The first drinking episode (comparable to an acute ethanol treatment in animal models) may cause more acetylation of the histone tails due to HDAC inhibition that results in an ‘open’ or relaxed chromatin structure leading to euphoric effects e.g. anxiolysis. Repeated exposure to alcohol (chronic drinking) may lead to neuroadaptive changes in these epigenetic mechanisms translating into the development of chronic tolerance to these euphoric effects of ethanol. Consequentially, cessation from drinking may precipitate withdrawal-related anxiety due to a rebound phenomenon and underlying epigenetic causes, e.g. hypoacetylation due to an increase in HDAC activity further ‘closing’ or condensing the chromatin structure, which manifests in the negative affective state or the ‘dark side of addiction’ (Koob & Le Moal, 2005). This leads to the development of dysphoric symptoms such as anxiety and depression and subjects tend to self-medicate with ethanol to relieve these dysphoric symptoms, resulting in alcohol abuse and dependence. As seen from preclinical data (Figure 2), this cycle of withdrawal leading to self-medication can be broken with HDAC inhibitors, which by promoting histone acetylation can revert a ‘closed’ chromatin structure to an ‘open’ configuration and can lead to a normal behavioral state (Pandey, Ugale et al., 2008; Arora et al., 2013; Pandey, Ugale, et al., 2008; You et al., 2014). Innately condensed chromatin due to higher HDAC2 expression and deficits in histone acetylation in the amygdala may also be involved in anxiety and alcoholism (Moonat et al., 2013; Sakharkar, Zhang, et al., 2014).
This review briefly summarized different epigenetic pathways that contribute to the phenotypes observed after alcohol use leading to continued use and dependence, which ultimately predisposes to alcoholism. We also provided examples of the epigenetic basis of other brain diseases in order to emphasize the importance of epigenetic regulation in brain disorders. Chronic ethanol changes the epigenome, thereby regulating the transcriptome, which alters neuroplasticity in specific neuroanatomical substrates, and eventually, leading to the phenotypic changes seen in alcoholism. This plasticity leads to the negative dysphoric states, such as anxiety, that are usually comorbid with alcoholism and are alleviated by consumption of ethanol or HDAC inhibition in the amygdala (Pandey, Ugale, et al., 2008; Moonat et al., 2013; Sakharkar, Zhang, et al., 2014). Although the DNA methylation pathways and mechanisms that control this transition are unclear, data is emerging that histone acetylation/de-acetylation pathway proteins that are responsible for covalent modifications to histones, play an important part in the central nucleus of amygdala in the co-morbidity of anxiety and alcohol use disorders. We have shown that the anxiolysis produced by acute ethanol exposure is associated with chromatin de-condensation and repeated ethanol exposure gradually leads to a persistent drinking phenotype and upon withdrawal, causes chromatin condensation and the phenotypes of increased anxiety, which in-turn promote increased consumption. This phenomenon is dependent on HDAC activity and, as long as ethanol is present in the bloodstream, HDAC activity is inhibited. However upon withdrawal, a rebound increase in HDAC activity and chromatin condensation occurs (Figure 3). Administration of HDAC inhibitors at the appropriate time point alleviates the negative dysphoric effects seen upon withdrawal which could prove to be an effective way to reverse the process of alcoholism (Pandey, Ugale, et al., 2008; Moonat et al., 2013; Sakharkar, Zhang, et al., 2014; You et al., 2014). In a similar fashion, positive euphoric effects of alcohol can be modulated by epigenetic changes in the reward circuitry and HDAC inhibitors can reverse HDAC-induced adaptational mechanisms during alcohol withdrawal (Arora et al., 2013). Thus by affecting both the positive and negative states that arise due to alcohol exposure and withdrawal, HDAC inhibition holds great promise as an effective therapeutic option to treat alcoholism and also to further understand the epigenetic alterations that underlie the pathogenesis of alcoholism.
Acknowledgments
The work described here from Dr. Subhash Pandey’s laboratory was supported by National Institute on Alcohol Abuse and Alcoholism Grants AA-016690, AA-019971-NADIA project, and AA-010005, AA-013341 and by the Department of Veterans Affairs (Merit Review Grant; Research Career Scientist award).
References
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J Neurosci. 2010;30(24):8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129(2):137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131(4):633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35(12):2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374(6517):81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Aroor AR, Shukla SD. MAP kinase signaling in diverse effects of ethanol. Life Sci. 2004;74(19):2339–2364. doi: 10.1016/j.lfs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Arora DS, Nimitvilai S, Teppen TL, McElvain MA, Sakharkar AJ, You C, Pandey SC, Brodie MS. Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: reversal by histone deacetylase inhibitors. Neuropsychopharmacology. 2013;38(9):1674–1684. doi: 10.1038/npp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson NS. Tolerance in Drosophila. J Neurogenet. 2009;23(3):293–302. doi: 10.1080/01677060802572937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol. 2011;31(24):4858–4873. doi: 10.1128/MCB.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36(8):1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in beta-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res. 2013;37(7):1133–1142. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Biermann T, Reulbach U, Lenz B, Frieling H, Muschler M, Hillemacher T, Kornhuber J, Bleich S. N-methyl-D-aspartate 2b receptor subtype (NR2B) promoter methylation in patients during alcohol withdrawal. J Neural Transm. 2009;116(5):615–622. doi: 10.1007/s00702-009-0212-2. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8(11):835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bleich S, Degner D, Sperling W, Bonsch D, Thurauf N, Kornhuber J. Homocysteine as a neurotoxin in chronic alcoholism. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28(3):453–464. doi: 10.1016/j.pnpbp.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bonsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30(4):587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16(2):167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Reulbach U, Bayerlein K, Hillemacher T, Kornhuber J, Bleich S. Elevated alpha synuclein mRNA levels are associated with craving in patients with alcoholism. Biological Psychiatry. 2004;56(12):984–986. doi: 10.1016/j.biopsych.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T, Kornhuber J, Bleich S. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin Exp Res. 2005;29(5):763–765. doi: 10.1097/01.alc.0000164360.43907.24. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113(9):1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Botia B, Legastelois R, Alaux-Cantin S, Naassila M. Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice. PLoS One. 2012;7(10):e47527. doi: 10.1371/journal.pone.0047527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Programming of DNA Methylation Patterns. 2012;81(1):97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13(3):322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR. Neurogenetic adaptive mechanisms in alcoholism. Science. 1987;236(4800):410–416. doi: 10.1126/science.2882604. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, Cabo Rd, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29(10):1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Ghezzi A, Al’Hasan YM, Wang YZ, Atkinson NS. Ethanol tolerance caused by slowpoke induction in Drosophila. Alcohol Clin Exp Res. 2006;30(5):745–753. doi: 10.1111/j.1530-0277.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Crews FT, Zou J, Qin L. Induction of innate immune genes in brain create the neurobiology of addiction. Brain Behav Immun. 2011;25(Suppl 1):S4–S12. doi: 10.1016/j.bbi.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Addario C, Caputi FF, Ekstrom TJ, Di Benedetto M, Maccarrone M, Romualdi P, Candeletti S. Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci. 2013;49(2):312–319. doi: 10.1007/s12031-012-9829-y. [DOI] [PubMed] [Google Scholar]
- Dallerac G, Chever O, Rouach N. How do astrocytes shape synaptic transmission? Insights from electrophysiology. Front Cell Neurosci. 2013;7 doi: 10.3389/fncel.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115(6):655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13(11):1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13(9):1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWit DJ, Adlaf EM, Offord DR, Ogborne AC. Age at First Alcohol Use: A Risk Factor for the Development of Alcohol Disorders. Am J Psychiatry. 2000;157(5):745–750. doi: 10.1176/appi.ajp.157.5.745. [DOI] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17(1):89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;5(8):730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465(7299):728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489(7414):52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a are required for the maintenance of DNA methylation and synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447(7141):178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fitzsimons HL, Schwartz S, Given FM, Scott MJ. The Histone Deacetylase HDAC4 regulates long-term memory in Drosophila. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0083903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons HL, Scott MJ. Genetic modulation of Rpd3 expression impairs long-term courtship memory in Drosophila. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0029171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SW. Epigenetic events in liver cancer resulting from alcoholic liver disease. Alcohol Res. 2013;35(1):57–67. doi: 10.35946/arcr.v35.1.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15(3):395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Chase KA, Sharma RP. Active DNA demethylation in post-mitotic neurons: A reason for optimism. Neuropharmacology. 2013;75:233–245. doi: 10.1016/j.neuropharm.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Lew L, Prado FJ, Ong DS, Atkinson NS. Alcohol-induced histone acetylation reveals a gene network involved in alcohol tolerance. PLoS Genet. 2013;9(12) doi: 10.1371/journal.pgen.1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. 2005;74(1):481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry. 2012;72:378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, Holson E, Xu J, Sun J, Neve RL, Mach RH, Haggarty SJ, Tsai LH. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156(1–2):261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJF, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483(7388):222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BF, Stinson FS, Dawson DA, Chou SP, Dufour MC, Compton W, Pickering RP, Kaplan K. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2004;61(8):807–816. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38(1):138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerri C, Pascual M. Mechanisms involved in the neurotoxic, cognitive, and neurobehavioral effects of alcohol consumption during adolescence. Alcohol. 2010;44(1):15–26. doi: 10.1016/j.alcohol.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR. DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcohol Clin Exp Res. 2013;37(3):417–424. doi: 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Crossey EL, Zhang L, Zucca S, George OL, Valenzuela CF, Zhao X. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Li Y, Wang D, Wei C, Yang X, Sui N. Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur J Pharmacol. 2010;642(1–3):93–98. doi: 10.1016/j.ejphar.2010.05.050. [DOI] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129(8):1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hawkins JD, Graham JW, Maguin E, Abbott R, Hill KG, Catalano RF. Exploring the effects of age of alcohol use initiation and psychosocial risk factors on subsequent alcohol misuse. J Stud Alcohol. 1997;58(3):280–290. doi: 10.15288/jsa.1997.58.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol Reprod. 2009;81(4):618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114(6):1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009;43(4):388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Host L, Dietrich JB, Carouge D, Aunis D, Zwiller J. Cocaine self-administration alters the expression of chromatin-remodelling proteins; modulation by histone deacetylase inhibition. J Psychopharmacol. 2011;25(2):222–229. doi: 10.1177/0269881109348173. [DOI] [PubMed] [Google Scholar]
- Idrus NM, Thomas JD. Fetal alcohol spectrum disorders: experimental treatments and strategies for intervention. Alcohol Res Health. 2011;34(1):76–85. [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13(9):1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Jan V, Gert C, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79(6):1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26(13):3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41(2):126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- Kim MS, Akhtar MW, Adachi M, Mahgoub M, Bassel-Duby R, Kavalali ET, Olson EN, Monteggia LM. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32(32):10879–10886. doi: 10.1523/JNEUROSCI.2089-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends in Biochemical Sciences. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164(8):1149–1159. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Alcoholism: allostasis and beyond. Alcohol Clin Exp Res. 2003;27(2):232–243. doi: 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Plasticity of reward neurocircuitry and the ‘dark side’ of drug addiction. Nat Neurosci. 2005;8(11):1442–1444. doi: 10.1038/nn1105-1442. [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Addiction is a Reward Deficit and Stress Surfeit Disorder. Front Psychiatry. 2013;4:72. doi: 10.3389/fpsyt.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kovacs KJ. Microglia and drug-induced plasticity in reward-related neuronal circuits. Front Mol Neurosci. 2012;5:74. doi: 10.3389/fnmol.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich AS, Briand L, Cleck JN, Ecke L, Rice KC, Blendy JA. Stress-induced potentiation of cocaine reward: A role for CRFR1 and CREB. Neuropsychopharmacology. 2009;34(12):2609–2617. doi: 10.1038/npp.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DEH, Truong HT, Russo SJ, LaPlant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Kushner MG, Abrams K, Borchardt C. The relationship between anxiety disorders and alcohol use disorders: a review of major perspectives and findings. Clin Psychol Rev. 2000;20(2):149–171. doi: 10.1016/s0272-7358(99)00027-6. [DOI] [PubMed] [Google Scholar]
- Kushner MG, Maurer E, Menary K, Thuras P. Vulnerability to the rapid (“Telescoped”) development of alcohol dependence in individuals with anxiety disorder. J Stud Alcohol Drugs. 2011;72(6):1019–1027. doi: 10.15288/jsad.2011.72.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay H, Klepper C, Wang B, Hsu SH, Datta J, Yu L, Zhang X, Majumder S, Motiwala T, Khan N, Belury M, McClain C, Jacob S, Ghoshal K. Reduced susceptibility of DNA methyltransferase 1 hypomorphic (Dnmt1N/+) mice to hepatic steatosis upon feeding liquid alcohol diet. PLoS One. 2012;7(8):e41949. doi: 10.1371/journal.pone.0041949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren B, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13(9):1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Shukla SD. Histone H3 phosphorylation at serine 10 and serine 28 is mediated by p38 MAPK in rat hepatocytes exposed to ethanol and acetaldehyde*. Eur J Pharmacol. 2007;573(1–3):29–38. doi: 10.1016/j.ejphar.2007.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legastelois R, Botia B, Naassila M. Blockade of ethanol-induced behavioral sensitization by sodium butyrate: descriptive analysis of gene regulations in the striatum. Alcohol Clin Exp Res. 2013;37(7):1143–1153. doi: 10.1111/acer.12088. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. PNAS. 2005;102(52):19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Liu J, Asuncion-Chin M, Liu P, Dopico AM. CaM kinase II phosphorylation of slo Thr107 regulates activity and ethanol responses of BK channels. Nat Neurosci. 2006;9(1):41–49. doi: 10.1038/nn1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4(7):500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci U S A. 2013;110(7):2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA. CBP in the Nucleus Accumbens Regulates Cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci. 2011;31(47):16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, Kupfer DJ, Monk TH, Devlin B, Nimgaonkar VL. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5(2):150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent bdnf gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436(7048):294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. PNAS. 2011;108(7):3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 Is a Critical Negative Regulator of Long-Term Memory Formation. J Neurosci. 2011;31(2):764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6(10):1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmc enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Pannetier S, Harel-Bellan A, Sassone-Corsi P. Mitogen-Regulated RSK2-CBP Interaction Controls Their Kinase and Acetylase Activities. Mol Cell Biol. 2001;21(20):7089–7096. doi: 10.1128/MCB.21.20.7089-7096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Hidalgo JJ. The role of glial cells in drug abuse. Curr Drug Abuse Rev. 2009;2(1):76–82. doi: 10.2174/1874473710902010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89(4):599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13(6):664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CP, Chen Y, Kundakovic M, Costa E, Grayson DR. Histone deacetylase inhibitors decrease reelin promoter methylation in vitro. J Neurochem. 2005;93(2):483–492. doi: 10.1111/j.1471-4159.2005.03040.x. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328(6126):175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Moonat S, Pandey SC. Stress, epigenetics, and alcoholism. Alcohol Res. 2012;34(4):495–505. doi: 10.35946/arcr.v34.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Pandey SC. The role of amygdaloid brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein and dendritic spines in anxiety and alcoholism. Addict Biol. 2011;16(2):238–250. doi: 10.1111/j.1369-1600.2010.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant Histone Deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biol Psychiatry. 2013;73(8):763–773. doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67(1):73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat Immunol. 2005;6(5):439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23(1):58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases dnmt3a and dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliet SH, Piet R, Poulain DA. Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science. 2001;292(5518):923–926. doi: 10.1126/science.1059162. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at Lys-4 and Lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sci. 2007;81(12):979–987. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. TLR4-MyD88 signalling: a molecular target for alcohol actions. Br J Pharmacol. 2012;165(5):1316–1318. doi: 10.1111/j.1476-5381.2011.01695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. Anxiety and alcohol abuse disorders: a common role for CREB and its target, the neuropeptide Y gene. Trends Pharmacol Sci. 2003;24(9):456–460. doi: 10.1016/S0165-6147(03)00226-8. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H. The decreased phosphorylation of cyclic adenosine monophosphate (cAMP) response element binding (CREB) protein in the central amygdala acts as a molecular substrate for anxiety related to ethanol withdrawal in rats. Alcohol Clin Exp Res. 2003;27(3):396–409. doi: 10.1097/01.ALC.0000056616.81971.49. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Xu T. Deficits in amygdaloid cAMP-responsive element-binding protein signaling play a role in genetic predisposition to anxiety and alcoholism. J Clin Invest. 2005;115(10):2762–2773. doi: 10.1172/JCI24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. The gene transcription factor cyclic AMP-responsive element binding protein: role in positive and negative affective states of alcohol addiction. Pharmacology & Therapeutics. 2004;104(1):47–58. doi: 10.1016/j.pharmthera.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Misra K. Central and medial amygdaloid brain-derived neurotrophic factor signaling plays a critical role in alcohol-drinking and anxiety-like behaviors. J Neurosci. 2006;26(32):8320–8331. doi: 10.1523/JNEUROSCI.4988-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene Arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008;28(10):2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PH, Lim RW, Shukla SD. Gene-selective histone H3 acetylation in the absence of increase in global histone acetylation in liver of rats chronically fed alcohol. Alcohol Alcohol. 2012;47(3):233–239. doi: 10.1093/alcalc/ags004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys Res Commun. 2003;306(2):501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16(2):694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Balino P, Alfonso-Loeches S, Aragon CM, Guerri C. Impact of TLR4 on behavioral and cognitive dysfunctions associated with alcohol-induced neuroinflammatory damage. Brain Behav Immun. 2011;25(Suppl 1):S80–91. doi: 10.1016/j.bbi.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Pascual M, Boix J, Felipo V, Guerri C. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem. 2009;108(4):920–931. doi: 10.1111/j.1471-4159.2008.05835.x. [DOI] [PubMed] [Google Scholar]
- Pascual M, Do Couto BR, Alfonso-Loeches S, Aguilar MA, Rodriguez-Arias M, Guerri C. Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology. 2012;62(7):2309–2319. doi: 10.1016/j.neuropharm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Pastor V, Host L, Zwiller J, Bernabeu R. Histone deacetylase inhibition decreases preference without affecting aversion for nicotine. J Neurochem. 2011;116:636–645. doi: 10.1111/j.1471-4159.2010.07149.x. [DOI] [PubMed] [Google Scholar]
- Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31(23):4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: Impact on the epigenome. Int J Dev Neurosci. 2013;31(6):391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Post-transcriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59(2):274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl JB, Ghezzi A, Lew LK, Robles RB, Cormack L, Atkinson NS. Circadian genes differentially affect tolerance to ethanol in Drosophila. Alcohol Clin Exp Res. 2013;37(11):1862–1871. doi: 10.1111/acer.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene co-expression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2(1):24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Prakash A, Zhang H, Pandey SC. Innate differences in the expression of brain-derived neurotrophic factor in the regions within the extended amygdala between alcohol preferring and nonpreferring rats. Alcohol Clin Exp Res. 2008;32(6):909–920. doi: 10.1111/j.1530-0277.2008.00650.x. [DOI] [PubMed] [Google Scholar]
- Qiang M, Denny A, Chen J, Ticku MK, Yan B, Henderson G. The site specific demethylation in the 5′-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS One. 2010;5(1):e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics. 2011;6(9):1095–1104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny AD, Ticku MK. Chronic intermittent ethanol treatment selectively alters N-methyl-D-aspartate receptor subunit surface expression in cultured cortical neurons. Mol Pharmacol. 2007;72(1):95–102. doi: 10.1124/mol.106.033043. [DOI] [PubMed] [Google Scholar]
- Ravindran CR, Ticku MK. Changes in methylation pattern of NMDA receptor NR2B gene in cortical neurons after chronic ethanol treatment in mice. Brain Res Mol Brain Res. 2004;121(1–2):19–27. doi: 10.1016/j.molbrainres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Ravindran CR, Ticku MK. Effect of 5-azacytidine on the methylation aspects of NMDA receptor NR2B gene in the cultured cortical neurons of mice. Neurochem Res. 2009;34(2):342–350. doi: 10.1007/s11064-008-9783-9. [DOI] [PubMed] [Google Scholar]
- Ravindran CR, Ticku MK. Role of CpG islands in the up-regulation of NMDA receptor NR2B gene expression following chronic ethanol treatment of cultured cortical neurons of mice. Neurochemistry International. 2005;46(4):313–327. doi: 10.1016/j.neuint.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome wide analysis of chromatin regulation by cocaine reveals a novel role for sirtuins. Neuron. 2009;62(3):335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Resendiz M, Chen Y, Ozturk NC, Zhou FC. Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics. 2013;5(1):73–86. doi: 10.2217/epi.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM. Cancer biology: mechanism of antitumour action of vorinostat (suberoylanilide hydroxamic acid), a novel histone deacetylase inhibitor. Br J Cancer. 2006;95(S1):S2–S6. [Google Scholar]
- Robinson J, Sareen J, Cox BJ, Bolton J. Self-medication of anxiety disorders with alcohol and drugs: Results from a nationally representative sample. J Anxiety Disord. 2009;23(1):38–45. doi: 10.1016/j.janxdis.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12(11):623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79(6):1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum SG, Green TA, Robison B, Lesselyong A, Perrotti LI, Bolanos CA, Kumar A, Clark MS, Neumaier JF, Neve RL, Bhakar AL, Barker PA, Nestler EJ. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29(11):3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Tang L, Zhang H, Chen Y, Grayson DR, Pandey SC. Effects of acute ethanol exposure on anxiety measures and epigenetic modifiers in the extended amygdala of adolescent rats. Int J Neuropsychopharmacol. 2014 Jun;26:1–11. doi: 10.1017/S1461145714001047. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Baxstrom K, Shi G, Moonat S, Pandey SC. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol. 2014;17(8):1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36(1):61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34(13):2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- Spear LP. Adolescent neurobehavioral characteristics, alcohol sensitivities, and intake: setting the stage for alcohol use disorders? Child Development Perspectives. 2011;5(4):231–238. doi: 10.1111/j.1750-8606.2011.00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar DK. Circadian genes, the stress axis, and alcoholism. Alcohol Res. 2012;34(3):362–366. doi: 10.35946/arcr.v34.3.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64(5):678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder FA, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, Akbarian S. Drug-induced activation of dopamine D1 receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology. 2008;33(12):2981–2992. doi: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Hesselbrock V. Alcohol dependence and anxiety disorders: what is the relationship? Am J Psychiatry. 1994;151(12):1723–1734. doi: 10.1176/ajp.151.12.1723. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25(8):781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Lim RW. Epigenetic effects of ethanol on the liver and gastrointestinal system. Alcohol Res. 2013;35(1):47–55. doi: 10.35946/arcr.v35.1.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Shiue K, Schomberg D, Zhou FC. Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplantation. 2009;18(10):1197–1211. doi: 10.3727/096368909X12483162197204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkman BG, Sakharkar AJ, Pandey SC. Epigenetics-beyond the genome in alcoholism. Alcohol Res. 2012;34(3):293–305. doi: 10.35946/arcr.v34.3.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64(2):435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, Bertran-Gonzalez J, Brami-Cherrier K, Enslen H, Corbille AG, Filhol O, Nairn AC, Greengard P, Herve D, Girault JA. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453(7197):879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Subbanna S, Shivakumar M, Umapathy NS, Saito M, Mohan PS, Kumar A, Nixon RA, Verin AD, Psychoyos D, Basavarajappa BS. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiology of Disease. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32(48):17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, Harper C, Alkass K, Druid H, Wentzel P, Nyberg F, Yakovleva T, Bakalkin G. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict Biol. 2011;16(3):499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd KJ, Serrano A, Lacaille JC, Robitaille R. Glial cells in synaptic plasticity. J Physiol Paris. 2006;99(2–3):75–83. doi: 10.1016/j.jphysparis.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Valor LM, Viosca J, Lopez-Atalaya JP, Barco A. Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr Pharm Des. 2013;19(28):5051–5064. doi: 10.2174/13816128113199990382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Res. 2013;35(1):25–35. doi: 10.35946/arcr.v35.1.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, Song W, Lau A, Labadorf A, Vogel-Ciernia A, Troncosco J, Ross CA, Bates GP, Krainc D, Sadri-Vakili G, Finkbeiner S, Marsh JL, Housman DE, Fraenkel E, Thompson LM. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci U S A. 2013;110(32):E3027–36. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16(1):42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Walker BM, Koob GF. Pharmacological evidence for a motivational role of kappa-opioid systems in ethanol dependence. Neuropsychopharmacology. 2008;33(3):643–652. doi: 10.1038/sj.npp.1301438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Wu LY, Zeng M. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J Neural Transm. 2012;119(4):425–433. doi: 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- Wang WH, Cheng LC, Pan FY, Xue B, Wang DY, Chen Z, Li CJ. Intracellular trafficking of histone deacetylase 4 regulates long-term memory formation. Anat Rec (Hoboken) 2011;294(6):1025–1034. doi: 10.1002/ar.21389. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghezzi A, Yin JCP, Atkinson NS. CREB regulation of BK channel gene expression underlies rapid drug tolerance. Genes Brain Behav. 2009;8(4):369–376. doi: 10.1111/j.1601-183X.2009.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krishnan HR, Ghezzi A, Yin JCP, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5(10) doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling--a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102(6):2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156(1–2):45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Zhang H, Sakharkar AJ, Teppen T, Pandey SC. Reversal of deficits in dendritic spines, BDNF and Arc expression in the amygdala during alcohol dependence by HDAC inhibitor treatment. Int J Neuropsychopharmacol. 2014;17(2):313–322. doi: 10.1017/S1461145713001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sakharkar AJ, Shi G, Ugale R, Prakash A, Pandey SC. Neuropeptide Y signaling in the central nucleus of amygdala regulates alcohol-drinking and anxiety-like behaviors of alcohol-preferring rats. Alcohol Clin Exp Res. 2010;34(3):451–461. doi: 10.1111/j.1530-0277.2009.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, Hao W, Zhang XY. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18(2):392–403. doi: 10.1111/adb.12037. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kusumo H, Sakharkar AJ, Pandey SC, Guizzetti M. Regulation of DNA methylation by ethanol induces tissue plasminogen activator expression in astrocytes. J Neurochem. 2014;128(3):344–349. doi: 10.1111/jnc.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Zhang R, Li W, Liao Y, Tang J, Miao Q, Hao W. Genome-wide DNA methylation patterns in discordant sib pairs with alcohol dependence. Asia Pac Psychiatry. 2013;5(1):39–50. doi: 10.1111/appy.12010. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh R, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011;35(4):735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Maiya R, Norris EH, Kreek MJ, Strickland S. Involvement of tissue plasminogen activator in stress responsivity during acute cocaine withdrawal in mice. Stress. 2010;13(6):481–490. doi: 10.3109/10253891003786415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. PNAS. 2011;108(16):6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JY, Crews FT. Release of neuronal HMGB1 by Ethanol through decreased HDAC activity activates brain neuroimmune signaling. PLoS One. 2014;9(2):e87915. doi: 10.1371/journal.pone.0087915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]