Abstract
Purpose
The DOCUMENT multicenter trial in the United States validated the performance of an epigenetic test as an independent predictor of prostate cancer risk to guide decision making for repeat biopsy. Confirming an increased negative predictive value could help avoid unnecessary repeat biopsies.
Materials and Methods
We evaluated the archived, cancer negative prostate biopsy core tissue samples of 350 subjects from a total of 5 urological centers in the United States. All subjects underwent repeat biopsy within 24 months with a negative (controls) or positive (cases) histopathological result. Centralized blinded pathology evaluation of the 2 biopsy series was performed in all available subjects from each site. Biopsies were epigenetically profiled for GSTP1, APC and RASSF1 relative to the ACTB reference gene using quantitative methylation specific polymerase chain reaction. Predetermined analytical marker cutoffs were used to determine assay performance. Multivariate logistic regression was used to evaluate all risk factors.
Results
The epigenetic assay resulted in a negative predictive value of 88% (95% CI 85–91). In multivariate models correcting for age, prostate specific antigen, digital rectal examination, first biopsy histopathological characteristics and race the test proved to be the most significant independent predictor of patient outcome (OR 2.69, 95% CI 1.60–4.51).
Conclusions
The DOCUMENT study validated that the epigenetic assay was a significant, independent predictor of prostate cancer detection in a repeat biopsy collected an average of 13 months after an initial negative result. Due to its 88% negative predictive value adding this epigenetic assay to other known risk factors may help decrease unnecessary repeat prostate biopsies.
Keywords: prostate, prostatic neoplasms, epigenomics, methylation, biopsy
Approximately 1 million prostate biopsies are performed yearly in the U.S. alone,1 of which only approximately 25% are positive for carcinoma.2 Pinsky et al reported that about 40% of patients with a negative biopsy undergo a second biopsy, which results in a PCa diagnosis in approximately an additional 15%.3 Also, 40% of the men with a negative repeat biopsy undergo a third biopsy and another 20% undergo a fourth biopsy while still detecting PCa in more than 5% of these patients.3–6
DNA hypermethylation of key genes is ideally suited for early disease detection and, therefore, it could be used to decrease the need for subsequent biopsy. Cancer associated epigenetic changes are biologically relevant, occurring commonly and early during the oncogenic process.7 Some markers show a field effect with epigenetic aberrations a distance from the actual tumor in line with the early observation of DNA methylation changes during oncogenesis. 8 While currently many patients undergo repeat biopsy due to suspicion of missed PCa, those with a low risk epigenetic signature may forego a likely unnecessary repeat biopsy with its associated potential complications.9
As a specific biomarker for PCa, GSTP1 methylation was reported in 1994 with many subsequent validation studies.10 A recent meta-analysis concluded that this gene is methylated in up to 90% of PCa cases.11 Additionally, APC and RASSF1 are important field effect markers that increase the diagnostic sensitivity of the assay.8,12 All 3 genes have prominent tumor suppressive roles in key cancer related pathways, ie GSTP1 acts as a detoxifying agent to prevent genomic damage by carcinogens, APC is a Wnt antagonist in this oncogenic signaling pathway and RASSF1 acts as a negative effector with a pro-apoptotic function in the Ras signaling pathway.13–15
In the previously reported MATLOC study a multiplex epigenetic assay profiling APC, GSTP1 and RASSF1 demonstrated 90% NPV.16 The DOCUMENT study was designed to validate the performance of an epigenetic assay using predetermined marker cutoff values.16 The DOCUMENT study evaluates the same epigenetic assay in a U.S. population of patients with a negative biopsy to identify those at low risk for harboring cancer missed through biopsy sampling error and who could forego an unnecessary repeat biopsy.17
MATERIALS AND METHODS
Patients were selected from 5 geographically dispersed medical centers and grouped into men with 2 consecutive negative biopsies (controls) and those with a negative biopsy followed by a positive biopsy (cases) within 24 months. Each center received institutional review board approval, exemption or waiver to use archived clinical samples for research purposes. A total of 350 patients from Cleveland Clinic, Eastern Virginia Medical School, LHMC, JHU and UCLA were enrolled in this multicenter trial. Two controls were enrolled per case, increasing the cancer prevalence over the estimate of approximately 20% in a general population, as determined by recent studies of PCa screening using 10 to 12-core biopsies,4,5 to allow for more accurate determination of epigenetic assay sensitivity. For direct comparison with the MATLOC cohort the prevalence was set at 18% in all NPV calculations using assay sensitivity and specificity according to the formula, (specificity × (1 – prevalence))/((specificity × (1 – prevalence)) + ((1 –sensitivity) × prevalence)).16 Cohort size was calculated using estimated MATLOC sensitivity and specificity to achieve at least 80% power to demonstrate a NPV higher than 86%, ie the lower boundary of the 95% CI of the MATLOC study.18
The sites identified eligible subjects in a retrograde, consecutive manner from patients biopsied at the clinics starting from the study start date (2011). Study eligibility criteria included a minimum of 8 cores per biopsy collected no earlier than 2007. Excluded from study were initial biopsies with atypia suspicious for cancer, ie atypical small acinar proliferation identified by pathologists at the sites, since this would have triggered repeat biopsy based on histopathology alone.19,20 Blinded central pathology review was performed for all available tissue sections at JHU by one of us (JIE). Atypia (atypical small acinar proliferation) detected after central pathology review and HGPIN were accepted and served as separate categories or risk factors.
We requested 40 μm of each archived, formalin fixed, paraffin embedded tissue core block from the initial biopsy. However, in the event of limited excess tissue a minimum of 20 μm from formalin fixed, paraffin embedded tissue cores were epigenetically profiled based on the 3 genes GSTP1, APC and RASSF1 as previously described.21 After completion of all analyses with the epigenetic assay clinical and patient characteristics were verified and updated with data from subsequent biopsies as necessary.
Epigenetic analysis was performed in random, blinded fashion using a multiplex methylation specific polymerase chain reaction (MDxHealth, Irvine, California). The methylation ratio of all 3 genes was determined relative to that of the ACTB reference gene. Predetermined analytical gene cutoff values for determining methylation status were identical to those used in the independent MATLOC study.16 A positive methylation result for at least 1 gene in at least 1 tissue core produced a positive assay result. In this validation study and because DNA quantity and quality could have been affected by sample age, a minimum of 6 analyzable biopsy cores were required to define a patient methylation as negative and assure accurate results. Since patient outcome is based on the detection of aberrant DNA methylation in at least 1 biopsy, patients with fewer than 6 cores, of which at least 1 was positive, could be included in analysis.
All statistical analysis was done in R (http://www.r-project.org/). An exact binomial test or the chi-square test was used to compare a proportion to a reference value or 2 proportions, respectively, while the Fisher exact test was applied when more than 2 categories were present. Two populations, particularly for ordinal data, were compared with the Wilcoxon (Mann-Whitney) test except for time between biopsies and age, for which the t-test was applied. For comparisons between clinical sites the Kruskal-Wallis test was used for continuous variables and the chi-square test was used for categorical variables.
RESULTS
Patient Characteristics
A total of 350 patients were enrolled in the study, of whom 30 (9%) were excluded because of non-eligibility (2), insufficient DNA (1), insufficient biopsy cores (23) or detection of adenocarcinoma in the first biopsy based on central pathology review (4). Table 1 lists clinical and demographic characteristics of the remaining 320 eligible patients. PSA categories and DRE results showed a significant difference among the sites, chiefly attributable to a high proportion of patients (47% vs 15% at all other sites) with PSA less than 4 ng/ml and a low proportion (21% vs 77%) with a normal DRE at LHMC. The difference in central pathology results between the sites had 2 causes, ie mainly the high percent (81% vs 52%) of samples with benign histology from UCLA and the low incidence of atypia from JHU (1% vs 14%). Eastern Virginia Medical School and UCLA were typed by a lower number of analyzable cores (8.8 vs 10.7). The time between the 2 biopsies was relatively longer at JHU and shorter at LHMC (15.8 vs 9.6 months).
Table 1.
DOCUMENT cohort clinical and demographic characteristics
| Cases | Controls | Overall | p Value | Site Associated p Value | |
|---|---|---|---|---|---|
| No. Ca prevalence (%): | 92 | 228 | 320 (29) | – | |
| Cleveland Clinic | 21 | 49 | 70 (30) | 0.7331 | |
| Eastern Virginia Medical School | 15 | 35 | 50 (30) | ||
| LHMC | 16 | 51 | 67 (24) | ||
| JHU | 24 | 47 | 71 (34) | ||
| UCLA | 16 | 46 | 62 (26) | ||
| Mean/median age | 63.5/64.0 | 61.5/62.0 | 62.1/62 | 0.0478 | 0.1086 |
| No. ethnic background (%): | – | ||||
| Black | 13 | 29 | 42 (13) | ||
| Asian | 4 | 9 | 13 (4) | ||
| White | 70 | 162 | 232 (73) | ||
| Unknown + other | 5 | 28 | 33 (10) | ||
| PSA (ng/ml): | |||||
| Mean/median | 6.4/5.4 | 6.0/5.2 | 6.1/5.3 | 0.4775 | <0.0001 |
| No. less than 4 PSA (%) | 17 (20) | 50 (24) | 67 (23) | 0.7795 | <0.0001 |
| No. 4 or greater-less than 10 (%) | 57 (69) | 142 (67) | 198 (68) | – | |
| No. 10 or greater (%) | 9 (11) | 19 (9) | 28 (10) | – | |
| No. not available (%) | 9 | 17 | 26 | – | |
| No. DRE (%): | |||||
| Normal | 45 (68) | 92 (57) | 137 (60) | 0.15 | <0.0001 |
| Abnormal (nodule) | 18 (27) | 51 (31) | 69 (30) | – | |
| Abnormal (suspicious) | 3 (5) | 19 (12) | 22 (10) | – | |
| Not available | 26 | 66 | 92 | – | |
| No. 1st biopsy central pathology (%): | |||||
| No abnormality | 45 (49) | 140 (61) | 185 (58) | 0.0263 | <0.0001 |
| Atypia | 17 (18) | 20 (9) | 37 (12) | – | |
| HGPIN | 30 (33) | 68 (30) | 97 (31) | – | |
| Mean/median No. analyzable cores | 9.7/10.0 | 10.2/10.0 | 10.1/10.0 | 0.1911 | <0.0001 |
| Mean/median mos between biopsies | 12.6/12.1 | 12.6/11.9 | 12.6/11.9 | 0.4483 | <0.0001 |
| No. α-reductase inhibitor therapy (%) | 5 (7) | 11 (7) | 16 (7) | 0.7827 | 0.0005 |
| Mean/median methylation: | – | ||||
| GSTP1 | 12.8/0.0 | 1.5/0.0 | 4.8/0.0 | <0.0001 | |
| APC | 86.8/43.4 | 44.5/15.9 | 56.7/21.2 | 0.0002 | |
| RASSF1 | 219.9/73.2 | 81.0/54.4 | 120.9/56.4 | 0.0827 | |
| No. GS (%): | |||||
| 6 | 50 (57) | – | – | – | 0.5177 |
| 7 | 30 (34) | – | – | – | – |
| 8 | 5 (6) | – | – | – | – |
| 9 | 3 (3) | – | – | – | – |
| Not available | 4 | – | – | – | – |
| Pos cores: | |||||
| Mean/median No. | 2.1/1 | – | – | – | 0.6681 |
| No. 1 (%) | 41 (51) | – | – | – | 0.6536 |
| No. 2 (%) | 18 (22) | – | – | – | – |
| No. greater than 2 (%) | 22 (27) | – | – | – | – |
| Not available | 11 | – | – | – | – |
Epigenetic Assay Validation
All key clinical parameters (sensitivity, specificity and NPV) were within the 95% CI determined in the MATLOC study (table 2). In this PSA screened population sensitivity was somewhat lower while specificity remained consistent at 64%. Notably the main characteristic of clinical importance (88% NPV) did not statistically differ between the MATLOC and DOCUMENT studies (p = 0.6707). In addition, the 88% NPV was significantly higher than the baseline NPV of 82% (p = 0.0227). That is, there was an 18% false-negative rate based on the adjusted cancer prevalence in repeat biopsies, indicating that the NPV of the epigenetic assay was an improvement over histopathological results alone.16
Table 2.
Epigenetic test performance characteristics
| Sensitivity | Specificity | NPV* | |
|---|---|---|---|
| DOCUMENT: | |||
| No. pts/total No. | 57/92 | 145/228 | 167/189 |
| % (95% CI) | 62 (51–72) | 64 (57–70) | 88 (85–91) |
| % MATLOC (95% CI) | 68 (55–78) | 64 (55–73) | 90 (86–94) |
| p Value | 0.5068 | 1.0000 | 0.6707 |
Patient numbers adjusted to 18% MATLOC study prevalence.
Notably while the number of black patients was too small to accurately determine the performance characteristics in this specific subgroup, sensitivity was 77% (95% CI 46–95), specificity was 66% (95% CI 46–82) and NPV was 93% (85% CI 82–97).
Marker Contribution
All 3 markers had a balanced contribution, accounting for approximately a third of positive cases (table 3). While APC was the most sensitive marker to identify cancer positive cases, GSTP1 showed the best sensitivity vs specificity tradeoff since it was responsible for a third of assay sensitivity but only 17% of false-positive results. The remaining non-specificity was equally divided between APC and RASSF1 in line with the role of these genes in the field effect. Interestingly 72% of correctly identified controls had a completely benign histology compared to only 43% of controls with a positive epigenetic assay result (p <0.0001).
Table 3.
Relative marker contribution
| No. Cases (% pos) | % Pos Controls | |
|---|---|---|
| Total | 92 | – |
| Total pos | 57 (100) | – |
| 1 Marker pos: | 32 (56) | – |
| APC | 11 (19) | |
| GSTP1 | 10 (18) | |
| RASSF1 | 11 (19) | |
| 2 Markers pos: | 13 (23) | – |
| APC + GSTP1 | 7 (12) | |
| APC + RASSF1 | 5 (9) | |
| GSTP1 + RASSF1 | 1 (2) | |
| 3 Markers pos | 12 (21) | – |
| Total marker contribution: | ||
| GSTP1 | 18 (32) | 17 |
| APC | 21 (37) | 41 |
| RASSF1 | 18 (32) | 42 |
Epigenetic Assay Performance Characteristics
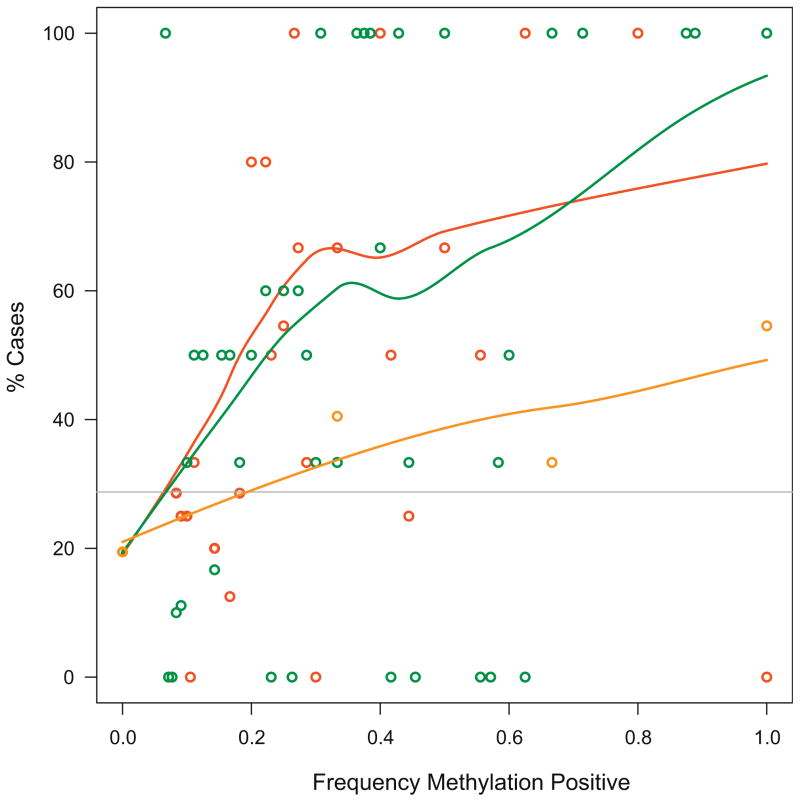
No difference was observed in GS distributions in repeat biopsies of patients with a positive vs a negative epigenetic assay (p = 0.9426). Thus, the GS frequency of each group was comparable to that of the entire cohort (table 1). The epigenetic test performed equally well for low GS 6 and high GS 7 or greater (p = 0.6597). However, only 9% of patients had a GS of 8 or greater. Finally, the likelihood of finding cancer in repeat biopsies following histopathologically negative initial biopsies increased with the number of methylation positive cores, total methylation events or total number of distinct methylated genes (fig. 1).
Figure 1.
Case prevalence trends as function of methylation characteristics. Detection of at least 1 methylated gene in at least 1 biopsy core resulted in significant detection of men with unsampled PCa. Using LOESS fit several trends indicated that likelihood of harboring unsampled cancer increased with increase in observed methylation relative to total amount of analyzable biopsy cores whether expressed as distinct genes (orange curve and circles), methylation positive biopsy cores (red curve and circles) or total number of epigenetic aberrations (green curve or circles). Of overall study population 29% were cases (horizontal line).
DNA Methylation as Significant Independent Predictor of Cancer
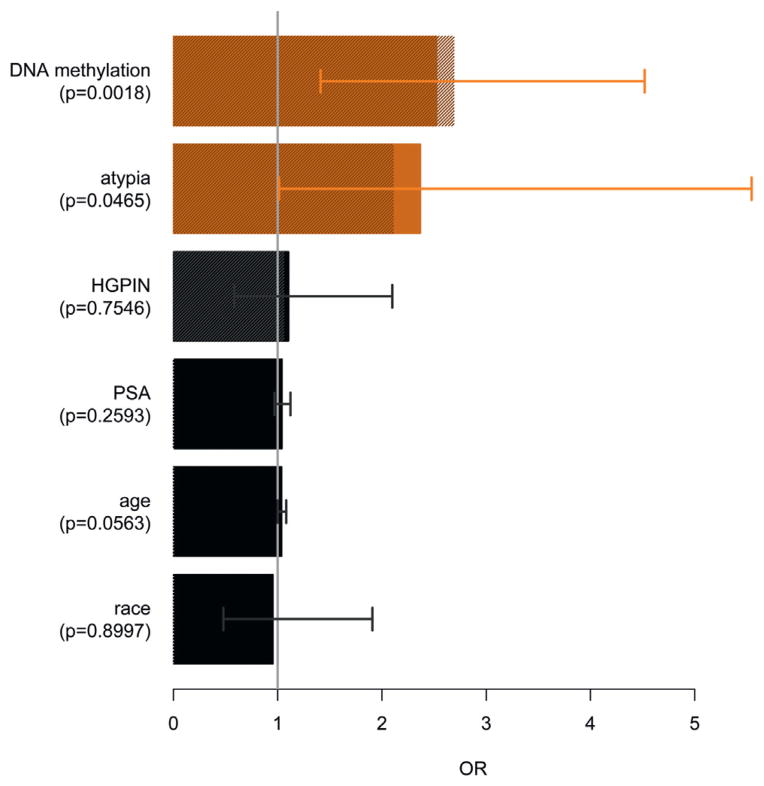
Univariate analysis indicated that DRE, in particular suspicious findings, unexpectedly showed an inverse trend and, therefore, it was excluded from subsequent multivariate analysis (table 4). A multivariate logistic regression model was constructed using DNA methylation, age, PSA, histopathology and race as predictors of cancer in a repeat biopsy. DNA methylation and atypia in the first biopsy were identified as good predictors (fig. 2). Patient age was borderline significant and the associated OR was low. Finally, a model containing only the best performing predictors indicated an important role for DNA methylation and to a lesser extent for atypia but not for HGPIN.
Table 4.
Univariate, multivariate and final models of risk factors with diagnostic potential
| Univariate
|
Multivariate
|
Final
|
||||
|---|---|---|---|---|---|---|
| OR | p Value | OR | p Value | OR | p Value | |
| DNA methylation | 2.85 | <0.0001 | 2.53 | 0.0018 | 2.69 | 0.0002 |
| Histology: | ||||||
| Atypia | 2.64 | 0.0089 | 2.37 | 0.0465 | 2.11 | 0.0531 |
| HGPIN | 1.37 | 0.2550 | 1.11 | 0.7546 | 1.06 | 0.8434 |
| Age | 1.04 | 0.0319 | 1.04 | 0.0563 | – | – |
| PSA | 1.03 | 0.3140 | 1.04 | 0.2593 | – | – |
| Race | 1.10 | 0.7740 | 0.96 | 0.8997 | – | – |
| DRE: | – | – | – | – | ||
| Nodule | 0.72 | 0.3213 | ||||
| Suspicious | 0.32 | 0.0807 | ||||
Figure 2.
Multivariate analysis of potential predictors of cancer on repeat biopsy. OR and 95% CI were determined by logistic regression model. Final model OR (hatched bars) was obtained using only diagnostically important and significant (orange bars) parameters, ie DNA methylation and histopathology of biopsy 1 (atypia and HGPIN). Contribution of atypia in final model was further evaluated by determining ROC AUC. Model with only epigenetic assay had AUC of 0.628. When epigenetic assay was modeled simultaneously with atypia but not with HGPIN, AUC showed minor increase to 0.646.
DISCUSSION
Sampling errors associated with prostatic biopsy raise fear that cancer was not sampled in men at high risk, leading to a high rate of followup procedures that merely confirm the absence of disease. With a NPV of 88% (95% CI 85–91) this multicenter validation study confirms the NPV of the previous MATLOC study16 and indicates that this epigenetic assay could be used to detect or help rule out cancer after an initial histopathologically negative biopsy.
Relative to other molecular assays the use of residual tissue of all available biopsy cores assures adequate sensitivity and may provide additional insight into tumor location.22 Repeat biopsies were performed an average of 1 year after initial biopsy, ie the period between biopsies was shorter than the maximum allowed time of 24 months. To address this shorter time to rebiopsy patient status updates were requested from all participating centers if available.
While assay specificity remained stable, indicating no difference in background methylation levels in this U.S. based cohort vs a European cohort, sensitivity was slightly lower but not significantly so. The predetermined marker cutoffs proved to be robust based on the validated clinical performance. This suggests that the validated gene cutoffs are not overfitted to a particular population and are broadly applicable.
The slightly decreased sensitivity was most likely associated with the higher PSA screening prevalence in the DOCUMENT cohort. Table 1 shows that study patients typically had only 1 cancer positive core compared to 2 in the MATLOC study, indicating lower disease volume. The subgroup of 41 cases (51%) with only 1 cancer positive core in DOCUMENT was larger but not significantly larger than the 37% in MATLOC (p = 0.1458). In contrast, disease grade did not differ in the 2 cohorts and there was no detectable difference in GS (p = 0.4753). However, the lack of a singular pathology review for both populations could have confounded such differences.
Another argument contributing to lowered sensitivity due to a PSA screening effect in DOCUMENT is that the time between biopsies significantly differed from MATLOC cases (p = 0.0002). With a median time to repeat biopsy of 6.5 months in cases the European cohort typically underwent repeat biopsy more than 5 months earlier than the current U.S. population. This could indicate that PSA screening results in biopsying menwith earlier stage, less extensive disease, as reported in large screening studies.23,24 While such disease might be harder to detect on biopsy, negative initial histopathological results combined with a lack of other evidence or symptoms indicating unsampled cancer could then result in a lesser sense of urgency for repeat biopsy. In addition, PSA in all patients at initial biopsy was significantly lower in the U.S. population (p = 0.0051), further strengthening this hypothesis.
Multivariate logistic regression revealed that DNA methylation of GSTP1, APC and/or RASSF1 in initial, histopathologically cancer negative biopsies serves as a significant independent predictor of the presence of unsampled PCa. While the presence of atypia was borderline significant as a predictor, it has 2 major drawbacks. The observation of atypia not including HGPIN is an uncommon event that also has high interobserver variability. Based on the centralized pathology review 17 cases and 20 controls harbored atypia in this cohort, resulting in 18% sensitivity and 91% specificity. Thus, while the presence of atypia is a useful diagnostic parameter, the MATLOC and DOCUMENT studies demonstrate that assessing the methylation status of APC, GSTP1 and RASSF1 would detect more missed cancer cases regardless of histopathological features, resulting in broader, more objective applicability. However, the 2 factors are partially complementary as indicated by the final regression model with a slightly higher positive predictive value (p = 0.6992), ie 32% when corrected for the 18% disease prevalence attributable to the presence of atypia. Notably assessing the predictive value of atypia in the DOCUMENT cohort might have been biased since the cohort included only patients with atypia identified during central pathology review.
Historically DNA methylation of certain genes has been linked to age.25 In addition, this series and another study26 propose a link among all 3 factors, ie DNA methylation, age and cancer, resulting in higher vulnerability for cancer at an older age. Cancer-free controls in this cohort were significantly younger than those in the MATLOC cohort (61.5 vs 63.1 years, p = 0.0065). While cases were also younger in the DOCUMENT study (63.5 vs 64.7 years), this difference was not significant (p = 0.2501), indicating that the lower sensitivity was not particularly affected by age.
CONCLUSIONS
This multicenter study validates the previously determined 90% NPV in a population of 350 PSA screened men.16 In the current series an 88% NPV was achieved by correctly predicting the presence/absence of unsampled PCa after a histopathologically negative initial biopsy using an epigenetic assay. This epigenetic assay is a significant independent predictor that was the most valuable diagnostic aid of all evaluated risk factors in 2 independent trials. Negative findings of this assay could be used to decrease concern about unsampled cancer and effectively avoid unnecessary repeat biopsies.
Abbreviations and Acronyms
- DOCUMENT
Detection Of Cancer Using Methylated Events in Negative Tissue
- DRE
digital rectal examination
- GS
Gleason score
- HGPIN
high grade prostate intraepithelial neoplasia
- JHU
Johns Hopkins University
- LHMC
Lahey Hospital and Medical Center
- MATLOC
Methylation Analysis to Locate Occult Cancer
- NPV
negative predictive value
- PCa
prostate cancer
- PSA
prostate specific antigen
- UCLA
University of California-Los Angeles
- U.S
United States
References
- 1.Zlotta AR, Nam RK. To biopsy or not to biopsy—thou shall think twice. Eur Urol. 2012;61:1115. doi: 10.1016/j.eururo.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Pinsky PF, Crawford ED, Kramer BS, et al. Repeat prostate biopsy in the prostate, lung, colorectal and ovarian cancer screening trial. BJU Int. 2007;99:775. doi: 10.1111/j.1464-410X.2007.06708.x. [DOI] [PubMed] [Google Scholar]
- 4.Djavan B, Zlotta A, Remzi M, et al. Optimal predictors of prostate cancer on repeat prostate biopsy: a prospective study of 1,051 men. J Urol. 2000;163:1144. [PubMed] [Google Scholar]
- 5.Thompson IM, Tangen CM, Ankerst DP, et al. The performance of prostate specific antigen for predicting prostate cancer is maintained after a prior negative prostate biopsy. J Urol. 2008;180:544. doi: 10.1016/j.juro.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Troyer DA, Lucia MS, de Bruïne AP, et al. Prostate cancer detected by methylated gene markers in histopathologically cancer-negative tissues from men with subsequent positive biopsies. Cancer Epidemiol Biomarkers Prev. 2009;18:2717. doi: 10.1158/1055-9965.EPI-09-0068. [DOI] [PubMed] [Google Scholar]
- 7.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehrotra J, Varde S, Wang H, et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate. 2008;68:152. doi: 10.1002/pros.20675. [DOI] [PubMed] [Google Scholar]
- 9.Loeb S, Carter HB, Berndt SI, et al. Complications after prostate biopsy: data from SEER-Medicare. J Urol. 2011;186:1830. doi: 10.1016/j.juro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Neste L, Herman JG, Otto G, et al. The epigenetic promise for prostate cancer diagnosis. Prostate. 2012;72:1248. doi: 10.1002/pros.22459. [DOI] [PubMed] [Google Scholar]
- 12.Trock BJ, Brotzman MJ, Mangold LA, et al. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int. 2012;110:56. doi: 10.1111/j.1464-410X.2011.10718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa VL, Henrique R, Ribeiro FR, et al. Epigenetic regulation of Wnt signaling pathway in urological cancer. Epigenetics. 2010;5:343. doi: 10.4161/epi.5.4.11749. [DOI] [PubMed] [Google Scholar]
- 15.Dammann R, Schagdarsurengin U, Strunnikova M, et al. Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol Histopathol. 2003;18:665. doi: 10.14670/HH-18.665. [DOI] [PubMed] [Google Scholar]
- 16.Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol. 2013;189:1110. doi: 10.1016/j.juro.2012.08.219. [DOI] [PubMed] [Google Scholar]
- 17.Eichler K, Hempel S, Wilby J, et al. Diagnostic value of systematic biopsy methods in the investigation of prostate cancer: a systematic review. J Urol. 2006;175:1605. doi: 10.1016/S0022-5347(05)00957-2. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg DM, Fine J, Chappell R. Sample size for positive and negative predictive value in diagnostic research using case-control designs. Biostatistics. 2009;10:94. doi: 10.1093/biostatistics/kxn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancuso PA, Chabert C, Chin P, et al. Prostate cancer detection in men with an initial diagnosis of atypical small acinar proliferation. BJU Int. 2007;99:49. doi: 10.1111/j.1464-410X.2007.06544.x. [DOI] [PubMed] [Google Scholar]
- 20.Amin MM, Jeyaganth S, Fahmy N, et al. Subsequent prostate cancer detection in patients with prostatic intraepithelial neoplasia or atypical small acinar proliferation. Can Urol Assoc J. 2007;1:245. doi: 10.5489/cuaj.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Neste L, Bigley J, Toll A, et al. A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. BMC Urol. 2012;12:16. doi: 10.1186/1471-2490-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong M, Yang B, Livermore A, et al. Using the epigenetic field defect to detect prostate cancer in biopsy negative patients. J Urol. 2013;189:2335. doi: 10.1016/j.juro.2012.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 24.United States Preventive Services Task Force. . Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2008;149:185. doi: 10.7326/0003-4819-149-3-200808050-00008. [DOI] [PubMed] [Google Scholar]
- 25.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damaschke NA, Yang B, Bhusari S, et al. Epigenetic susceptibility factors for prostate cancer with aging. Prostate. 2013;73:1721. doi: 10.1002/pros.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]