Abstract
Prostate cancer is a heterogeneous disease and thus, it is important to understand whether among the heterogeneous collection of cell types, androgen-deprivation insensitive cells exist prior to hormonal manipulation. We established several LNCaP subclones with distinct insensitivities to androgen deprivation from a parental LNCaP cell line. In the resulting clones, the sensitivity to androgen-deprivation negatively correlated with their PSA expression levels. In two of these clones, an androgen insensitive clone, LNCaP-cl1, and an androgen sensitive clone, LNCaP-cl5, the DNA copy number differed significantly, indicating that these clones contain genetically distinct cells. LNCaP-cl1 had higher PSA expression but lower invasiveness and tumor growth potential than LNCaP-cl5. The expression levels of two genes that are known to be regulated by miR-21, an androgen-regulated microRNA, Sprouty1 (SPRY1) and Jagged1 (JAG1) were significantly lower in LNCaP-cl1 than in LNCaP-cl5. Knocking down SPRY1 in LNCaP cells enhanced PSA expression and cell proliferation. JAG1 administration in LNCaP cells enhanced cell invasion and JAG1 knockdown in PC3 cells suppressed cell invasion and tumor formation. These results indicated that the expression differences in SPRY1 and JAG1 may contribute to the phenotypic differences between the LNCaP-cl1 and LNCaP-cl5 clones. In tissue samples, SPRY1 expression levels were significantly lower in prostate cancer patients with PSA recurrence after surgical treatment (P = 0.0076) and JAG1 expression levels were significantly higher in Gleason sum (GS) 8–9 disease than in GS 5–6 (P = 0.0121). In summary a random population of LNCaP cells comprises a heterogeneous group of cells with different androgen-deprivation sensitivities and potential for invasiveness.
Keywords: PROSTATE CANCER, ANDROGEN SENSITIVE, MARKER
A central question in cancer biology is whether treatment-resistant cells exist prior to the initiation of a given treatment. Androgen deprivation therapy has been the mainstay in the treatment of advanced metastatic prostate cancer. However, most patients ultimately relapse after a period of initial response to this approach and progress to castration resistant prostate cancer (CRPC). Unfortunately, the mechanisms contributing to castration resistant progression are not fully elucidated. It has been proposed that prostate cancer contains a heterogeneous mixture of cells that vary in their dependence on androgens for growth and survival and that treatment with androgen ablation therapy provides selective pressure and alters the relative concentration of these cells, thereby leading to the outgrowth of CRPC [Craft et al., 1999]. The existence of androgen-deprivation insensitive cells prior to hormonal manipulation may be indicative of the potential response to this therapeutic approach and therefore could predict patient prognosis as well as provide a better understanding of treatment resistance.
The LNCaP cell line is a widely used model for in vitro prostate cancer research due to its sensitivity to androgen-deprivation. Although acute androgen ablation results in growth arrest without inducing apoptosis in this cell line, long-term androgen ablation transforms a subpopulation to androgen-deprivation insensitive clones [van Steenbrugge et al., 1991; Pflug et al., 1999; Inoue et al., 2006, 2008]. It has also been reported that the low-passage number LNCaP cells grow slowly in an androgen-deprivation sensitive manner, whereas high-passage cells can grow aggressively and also become androgen-deprivation insensitive [Lin et al., 1998; Denmeade et al., 2003]. These results led to the hypothesis that LNCaP cells contain subclones with different genetic compositions and that androgen-deprivation insensitive cells exist prior to them being challenged with androgen deprivation. To confirm this hypothesis, we established LNCaP subclones using limiting dilution without specific selection media. Upon analyzing the individual “clones,” it was evident that their PSA expression levels and in vitro androgen sensitivities were different at the same passage numbers. Surprisingly, the LNCaP clone with higher PSA expression and in vitro androgen-deprivation insensitivity had lower invasiveness. By comparing gene expression patterns between these clones, we could identify genes that could serve as biomarkers for detecting aggressive and potentially androgen-derivation insensitive prostate cancers. These results provide findings that: (1) an androgen-deprivation insensitive or invasive clone existed prior to any challenge; (2) this clone has a unique genetic and protein signature; and (3) the characterization of this signature may provide for a way to identify the existence of the capability of resistant to a therapy prior to its use.
MATERIALS AND METHODS
CELL LINES
LNCaP and PC3 cells were purchased from the American Type Culture Collection and cultured under typical conditions in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS). LNCaP cells at their 25th passage number were used for establishing LNCaP clones. In a 10-cm dish 1 × 104 cells were plated and grown for 40 days in normal medium. Utilizing limiting dilution in which single cells were placed into individual wells, fifty colonies were selected, of which 22 subclones continued to grow and were established. For androgen-depleted conditions, cells were cultured in phenol red-free RPMI 1640 (Invitrogen) supplemented with 10% charcoal-stripped fetal bovine serum (CSFBS) (Hyclone). R1881 (Perkin Elmer) was used as a synthetic androgen and Bicalutamide (Toronto Research Chemicals) was used as an anti-androgen. Cell numbers were counted and cell diameters were measured using Cellometer Auto T4 (Nexcelom Bioscience).
CELL PROLIFERATION ASSAY
In a 96-well plate, 3 × 103 cells were plated in 100 μl of medium, incubated for the indicated time period, after which 20 μl of CellTiter 96 Aqueous One Solution (Promega) was added. After an additional 2 h of incubation at 37°C, the absorbance at 490 nm of each well was measured. To compare the insensitivity to androgen deprivation of each LNCaP clone, five of these clones were grown in normal and androgen-depleted medium and their cell proliferation rates were examined. All experiments were performed three times in triplicate. Epidermal growth factor (EGF; Sigma–Aldrich), extracellular-signal-regulated kinase (ERK) inhibitor PD98059 (Enzo Lifescience) and JAG1 recombinant protein (1277-JG, R&D systems) were used in the concentrations as indicated in the Results section.
MATRIGEL INVASION ASSAY
In vitro tumor cell invasion was measured with BD Biocoat Matrigel Invasion Chambers (BD). The upper chambers were filled with 1 × 105 cells in serum-free medium containing 0.1% BSA and the lower chambers were filled with normal medium containing 10% FBS. The numbers of cells that invaded through the matrix were counted after 48 h of incubation for the PC3 cells and 72 h incubation for the LNCaP cells. All experiments were performed in triplicate.
XENOGRAFT TUMOR FORMATION ASSAY
Five million LNCaP or PC3 cells were suspended in 200 ml of media and subcutaneously injected into the right flank of 6-week-old SCID mice (LNCaP) or nude mice (PC3). The LNCaP cells were inoculated in combination with 100 μl of Matrigel (BD). Tumor volumes were measured with a caliper using the formula, a × b2 × 0.52 where a is the largest diameter and b is the largest diameter perpendicular to a. These experiments were approved by the Johns Hopkins Institutional Animal Care and Use Committee.
RNA ISOLATION AND REAL-TIME PCR
Total RNA was isolated using an RNeasy Mini Kit (QIAGEN). After quantification, 1 μg total RNA was transcribed into first strand cDNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc.). Quantitative real-time PCR was carried out with the CFX96 Real-Time PCR Detection System using iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.). PCR primers were 5’-cacagcctgtttcatcctga-3’ (forward) and 5’-aggtccatgaccttcacagc-3’ (reverse) for PSA, 50-cctgctaaaggatgcctgaa-3’ (forward) and 5’-gaggtacaacccacctccaa-3’ (reverse) for SPRY1, 5’-gaacccgatcaaggaaatca-3’ (forward) and 5’-gagctcagcaagggaacaag-3’ (reverse) for JAG1, and 5’-gaatataatcccaagcggtttg-3’ (forward) and 5’-acttcacatcacagctcccc-3’ (reverse) for TATA-binding protein (TBP). For the microRNA analysis, total RNA was isolated using a miRNeasy Mini Kit (QIAGEN). TaqMan MicroRNA assay systems (Applied Biosystems) were used for measuring miR-21 (000397) and RUN24 (001001) expression levels. Analysis and fold differences were determined using the comparative threshold method. All experiments were performed in triplicate.
DNA COPY NUMBER VARIATION AND DNA MICROARRAY ANALYSIS
For DNA copy number variation analysis, total DNA was isolated from LNCaP-cl1 and -cl5 using DNeasy Blood and Tissue Kit (QIAGEN). DNA copy number was compared using the Human SNP array 6.0 (Affymetrix). For DNA microarray analysis, total RNA was isolated from LNCaP-cl1 and -cl5 and gene expression differences were analyzed using the Human Genome U133 Plus2.0 (Affymetrix) array. These data were analyzed using Genotyping Console v3.0 and Affymetrix Expression Console v1.0, respectiely.
WESTERN BLOT ANALYSIS
Cells were lysed with RIPA buffer (Thermo Scientific) with 1 mM PMSF (Sigma), 1 mM DTT (Sigma), and Halt Protease Inhibitor Cocktail (Thermo Scientific). Protein (30 μg) was electrophoresed on Mini-PROTEAN TGX precast gels (Bio-Rad) and blotted with antibodies for AR (1:500, rabbit polyclonal, N-20, Santa Cruz), PSA (1:500, goat polyclonal, C-19, Santa Cruz), SPRY1 (1:500, mouse monoclonal, RR-15, Santa Cruz), JAG1 (1;500, rabbit polyclonal, H-144, Santa Cruz), N-cadherin (1:1,000, rabbit polyclonal, #4061, Cell Signaling), and β-actin (1:5,000, mouse monoclonal, clone AC15, Sigma).
RNA INTERFERENCE
Cells were plated at 2–3 × 105 per well in six well plates and transfected after incubation for 24 h. ON-TARGET plus SMART pool siRNA for SPRY1 (J27339) JAG1 (J011060) and negative control siRNA (D1810), miRIDIAN Mimic hsa-miR-21 (C300492) and miRIDIAN Mimic negative control (CN1000), lentiviral shRNA vector pGIPZ-shJAG1 (V2LHS-255871) were all purchased from Thermo Scientific. Cells were transiently transfected with each siRNA (50 nM) or miRNA (50 nM) using DharmaFECT (Thermo Scientific) and stably transfected with scrambled shRNA (pGIPZ-shCtr) and pGIPZ-shJAG1 using Trans-Lentiviral Packaging Kit (Thermo Scientific) according to the manufacturer's protocol. Stably transfected cells were grown in 1 μg/ml puromycin (Invitrogen).
DUAL-LUCIFERASE REPORTER ASSAYS
Cells were plated at 1.5 × 105 per well in 24-well plates and were transiently co-transfected with 250 ng of pGL3-PSAP [Hu et al., 2009] and 5 ng of pTK-RL using Lipofectamine LTX reagent (Invitrogen). After 48 h of incubation in media containing FBS, the luciferase activity of the cell lysate was measured using the Dual-Luciferase Reporter Assay System (Promega) with a microplate reader (BMG Labtech) in triplicate.
CLINICAL SAMPLES
Prostate tissue specimens from 70 clinically localized prostate cancers (27 without recurrence and 43 with recurrence) used in this study were collected and frozen at the time of radical prostatectomy at the Johns Hopkins Hospital from 1993 to 2007 [Shiraishi et al., 2011]. Samples with large volume tumors were selected for obtaining high quality and high quantity RNA. Tissue-Tek (Sakura Finetek) embedded frozen tissue blocks were manually trimmed to enrich the content of target tissue lesions prior to sectioning and RNA extraction. For tumor samples, tumor cells comprised more than 70% of the tissue content (calculated by averaging the % tumor content in the first and last sections) in all cases. PSA recurrence is defined as a PSA increase of greater than 0.2 ng/ml after radical prostatectomy. The use of de-identified surgical specimens for molecular analysis was approved by the Johns Hopkins Medicine Institutional Review Boards.
STATISTICAL ANALYSIS
Differences between each group are compared by paired t-test. Differences in xenograft tumor volumes were compared by ANOVA. Statistical tests were two-sided and P values <0.05 were considered to be statistically significant.
RESULTS
ANDROGEN-INSENSITIVE CLONES EXIST WITHIN A POPULATION OF LNCaP CELLS PRIOR TO SELECTION
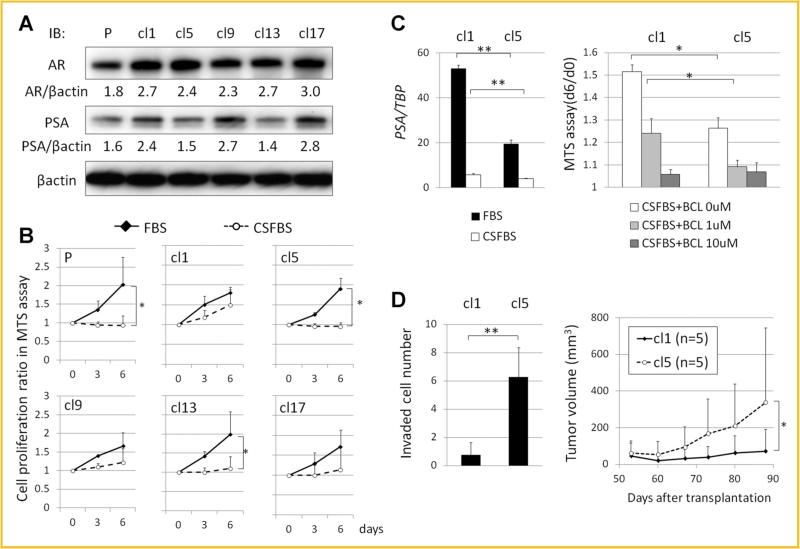
LNCaP clones were established by limiting dilution as described in the Materials and Methods section. The morphologies of these clones were not significantly different from each other. The expression levels of AR and PSA in the parental LNCaP (LNCaP-P) and randomly selected LNCaP clones (LNCaP-cl1, -cl5, -cl9, -cl13, and -cl17) grown in normal medium containing FBS were compared by Western blotting. AR/β-actin ratios were 1.8, 2.7, 2.4, 2.3, 2.7, 3.0, and PSA/β-actin ratios were 1.6, 2.4, 1.5, 2.7, 1.4, and 2.8 in LNCaPP, cl1, -cl5, -cl9, -cl13, and -cl17, respectively. Among the LNCaP clones, PSA expression levels were higher in LNCaP-cl1, -cl9, and -cl17, and lower in LNCaP-cl5 and -cl13 without significant differences in AR expression levels (Fig. 1A). The androgen sensitivities were compared between these clones by growing them in normal and androgen depleted medium. Cell proliferation rates in normal medium were not different among the clones. However, androgen deprivation significantly suppressed cell proliferation of LNCaP-P, -cl5, and -cl13 but not that of LNCaP-cl1, -cl9, and -cl17 (Fig. 1B). These results indicated that among the LNCaP clones established, the sensitivity to androgens were different and negatively correlated with PSA expression.
Fig. 1.
Establishment and characterization of LNCaP clones. A: AR and PSA expression levels in parental LNCaP (P) and LNCaP clones (cl1, cl5, cl9, cl13, and cl17) grown in normal medium containing FBS by immunoblotting. Expression ratios of AR/β-actin and PSA/β-actin calculated by ImageJ 1.47t software (http://imagej.nih.goc/ij) are shown. B: Cell proliferation ratios of parental LNCaP and LNCaP clones in normal (FBS) and androgen-depleted medium (CSFBS) on days 3 and 6 relative to day 0 by MTS assays. C: PSA mRNA expression levels relative to TBP of LNCaP-cl1 and -cl5 in FBS and CSFBS (left). Cell proliferation of LNCaP-cl1 and -cl5 in CSFBS under androgen receptor antagonism by 0, 5, and 10 μM bicalutamide (BCL) (right). D: Invaded cell numbers of LNCaP-cl1 and -cl5 in Matrigel invasion assays after 72 h incubation (left) and sequential changes in tumor volumes of LNCaP-cl1 and -cl5 in xenograft in vivo tumor formation assays at indicated days after transplantation (n = 5 each) (right). (*P < 0.05, **P < 0.005).
THE LNCaP CLONE WITH HIGHER AR ACTIVITY AND HIGHER ANDROGEN-INSENSITIVITY IS LESS INVASIVE AND HAS LOWER IN VIVO TUMOR GROWTH POTENTIAL
AR expression levels in both clones LNCaP-cl1 and -cl5 were similar while PSA expression levels were higher in LNCaP-cl1 than in LNCaP-cl5. As PSA is an androgen regulated gene, this suggested that the AR activity was different between these clones, causing the differences in their androgen insensitivity. To test this possibility, AR function was evaluated by dual-luciferase reporter assay. AR activity appeared to be higher in LNCaP-cl1 than in LNCaP-cl5, although the difference was not statistically significant (Supplement Fig. S1A). In contrast, PSA expression levels were significantly higher in LNCaP-cl1 than in LNCaP-cl5 even after androgen deprivation (Fig. 1C). Moreover, the cell proliferation of LNCaP-cl1 without androgen was partially and completely suppressed by the administration of 5 and 10 mM of bicalutamide, respectively (Fig. 1C). These results indicate that the androgen-insensitivity of LNCaP-cl1 is associated with its higher AR activity. Next, to compare the aggressiveness of LNCaP-cl1 and -cl5, Matrigel invasion assays and in vivo tumor formation assays were performed. Surprisingly, the numbers of cells that invaded as well as the in vivo tumor growth rate were significantly higher in LNCaP-cl5 than in LNCaP-cl1 (Fig. 1D). Taken together, LNCaP-cl1 had higher AR activity and androgen insensitivity but lower invasiveness and in vivo tumor growth potential.
ANDROGEN-SENSITIVE AND -INSENSITIVE CLONES EXHIBIT DNA COPY NUMBER VARIATIONS
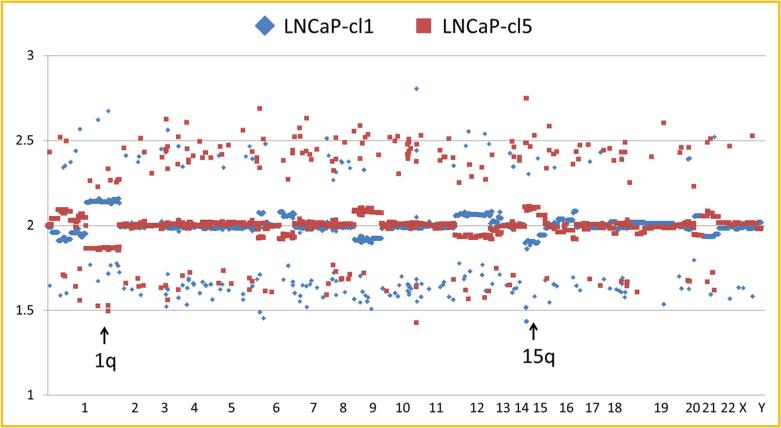
Since LNCaP clones with different androgen sensitivities and aggressiveness were identified from a population of LNCaP cells, it is possible that the observed differences were caused by genetic differences. To answer the question whether these clones are genetically distinct, DNA copy number variation was analyzed in the LNCaP-cl1 and -cl5. Indeed, several differences were observed that included gene copy numbers 2.15 ± 0.04 in LNCaP-cl1 and 1.87 ± 0.04 in LNCaP-cl5 at 1q region (P < 0.0001), 1.91 ± 0.09 in LNCaP-cl1 and 2.10 ± 0.12 in LNCaP-cl5 at 15q region (P < 0.0001), and also at 1p, 6q, 9p, and 12q (Fig. 2). To discern the effects of copy number variation on gene expression in these clones, DNA microarray analysis was performed. Some genes highly expressed in LNCaP-cl1 were located in the 1q region (RGS2, NBPF10, or FAM5C) and some genes that were highly expressed in LNCaP-cl5 were located in the 15q region (C15orf21, ADAMTSL3, CYP11A1, SH3GL3, or SH3GL3), indicating that gene expression differences at least, to some extent, were associated with gene copy number variants (Supplement Table S1). In this microarray data, the cl1/cl5 ratio of AR expression was 1.10 and PSA expression was 2.13 consistent with the observed AR and PSA protein expression differences.
Fig. 2.
DNA copy number variants between LNCaP-cl1 and -cl5. Blue (cl1) and red (cl5) squares show the copy number differences at each SNP marker on all the chromosomes in human SNP array 6.0. Arrows show 1p (cl1>cl5) and 15q (cl1<cl5) lesions.
CELL SIZE AND THE EXPRESSION LEVELS OF SPRY1 AND JAG1 WERE SIGNIFICANTLY DIFFERENT BETWEEN ANDROGEN-SENSITIVE AND -INSENSITIVE CLONES
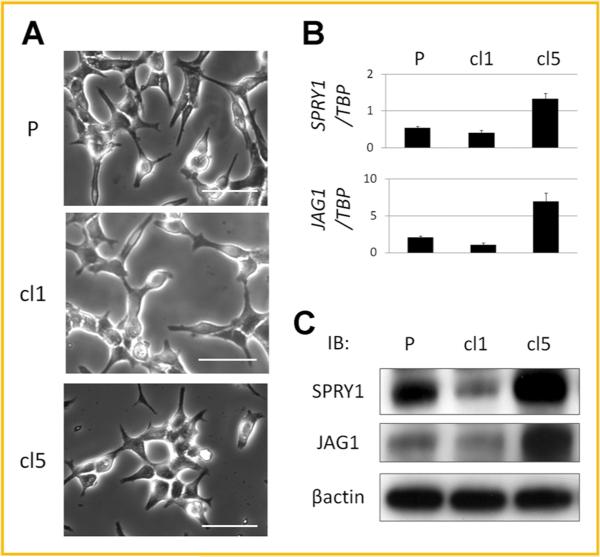
Among the LNCaP clones, cellular morphology was not apparently different when examined by light microscopy (Fig. 3A). However, after detaching the cells with trypsin, the cell diameters of LNCaP-P, -cl1, and -cl5 were 18.4 3.4, 21.4 5.0, and 18.0 ± 5.4 μm, respectively. Those of LNCaP-cl1 were significantly larger than those of LNCaP-cl5 (P < 0.0001). To gain additional insight, we examined the gene expression profiles using DNA microarrays and focused on two genes, SPRY1 (cl5/cl1 ratio = 5.67) and JAG1 (cl5/cl1 ratio = 5.27) that were significantly down-regulated in LNCaP-cl1 compared to LNCaP-cl5. SPRY1 and JAG1 mRNA and protein expression levels in parental LNCaP-P, -cl1, and -cl5 were validated by real-time PCR (Fig. 3B) and Western blotting (Fig. 3C), respectively. SPRY1 expression was moderate in LNCaP-P, low in LNCaP-cl1 and high in LNCaP-cl5.
Fig. 3.
Cell shapes and expression levels of SPRY1 and JAG1 in LNCaP-P, -cl1, and -cl5. A: The cell shapes of LNCaP-P, -cl1, and -cl5 (bars: 50 μm), B: SPRY1 and JAG1 mRNA expression levels relative to TBP by real-time PCR. C: SPRY1 and JAG1 protein expression levels by Western blotting.
miR-21, AN ANDROGEN-REGULATED microRNA, IS HIGHLY EXPRESSED IN THE ANDROGEN INSENSITIVE CLONE AND NEGATIVELY REGULATES SPRY1 AND JAG1 EXPRESSION
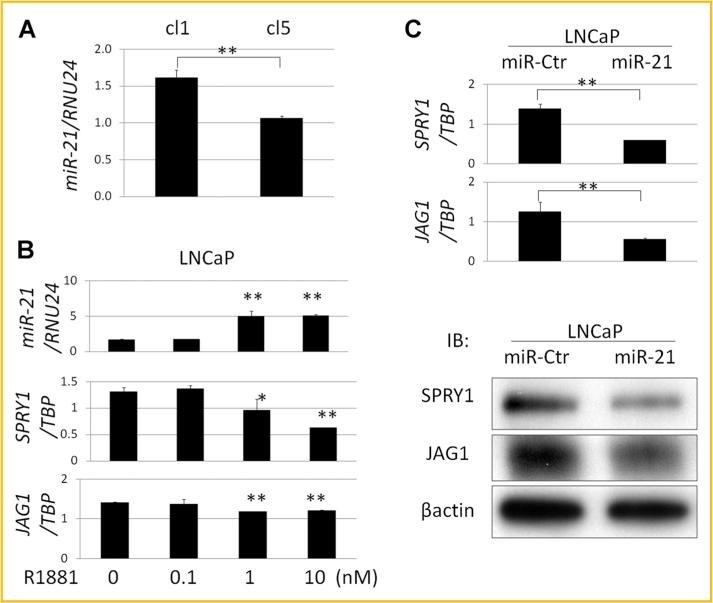
It has previously been reported that in prostate cancer, SPRY1 is regulated by miR-21 [Darimipourain et al., 2011] a microRNA that is regulated by androgens and associated with prostate cancer or disease progression [Ribas et al., 2009]. Using Target Scan (http://www.targetscan.org/), miR-21binding sites in the 3’UTR were found in SPRY1 and in JAG1 (Supplement Fig. S1B). The expression levels of miR-21 relative to the internal control microRNA RNU24 were significantly higher in LNCaP-cl1 than in LNCaP-cl5 (Fig. 4A). Furthermore, miR-21 expression levels were enhanced by treating the cells with 1–10 nM R1881 while the expression levels of SPRY1 and JAG1 were suppressed (Fig. 4B). Transfection of miR-21 into LNCaP cells significantly suppressed the expression levels of SPRY1 and JAG1 (Fig. 4C). Taken together, these results indicated that miR-21 negatively regulates SPRY1 and JAG1 which might explain the inverse correlation between AR activation and expression levels of SPRY1 and JAG1 in LNCaP-cl1 and -cl5.
Fig. 4.
Regulation of SPRY1 and JAG1 by miR-21. A: miR-21 expression levels relative to RNU24 in LNCaP-cl1 and -cl5 by real-time PCR. B: miR-21, SPRY1, and JAG1 mRNA expression changes under the stimulation with 0, 0.1, 1, and 10 nM of synthetic androgen (R1881). C: SPRY1 and JAG1 mRNA (upper) and SPRY1 and JAG1 protein (lower) expression levels of LNCaP cells transfected with a negative control microRNA (miR-Ctr) and hsa-miR-21 (miR-21). (*P < 0.05, **P < 0.005).
SPRY1 NEGATIVELY REGULATES AR ACTIVATION AND EGF-INDUCED ANDROGEN-INSENSITIVE CELL PROLIFERATION POTENTIATED BY THE ERK PATHWAY
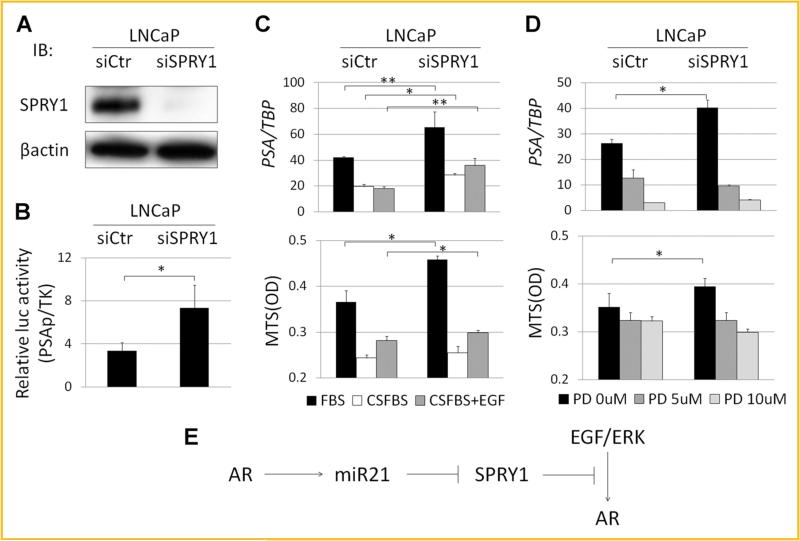
To examine the functions of SPRY1 in LNCaP cells, its expression was knocked down using an siRNA. SPRY1 protein expression levels were significantly reduced by knocking down SPRY1 (Fig. 5A). However, AR activity, evaluated by a luciferase reporter assay, was significantly up-regulated by knocking down SPRY1 (Fig. 5B). Moreover, SPRY1 silencing resulted in enhanced PSA expression and cell proliferation in normal medium (Fig. 5C). Although PSA expression in androgen-depleted medium was slightly enhanced by the knocking down of SPRY1, cell proliferation was not changed. SPRY1 is a known negative regulator of receptor tyrosine kinase (RTK)-induced ERK signal [Hanafusa et al., 2002]. The EGF receptor is an RTK and EGF expression levels are higher in LNCaP-cl1 than in LNCaP-cl5 (cl1/cl5 ratio = 2.82; Supplement Table S1). Therefore, we examined the changes in PSA expression and cell proliferation as a result of SPRY1 knockdown by stimulating cells with EGF in androgen-depleted medium. PSA expression was significantly enhanced by SPRY1 knockdown under the stimulation of EGF. Cell proliferation was also enhanced by EGF stimulation and more significantly by SPRY1 knockdown (Fig. 5C). To confirm the association between SPRY1 and the ERK pathway, we used an ERK inhibitor, PD98059. As expected, inhibiting ERK signaling significantly suppressed the up-regulation of PSA as well as the cell proliferation induced by SPRY1 knockdown in a dose dependent manner (Fig. 5D). ERK phosphorylation was higher in LNCaP-cl1 than in LNCaP-cl5, which was induced by the SPRY1 knockdown and EGF stimulation in LNCaP cells (Supplement Fig. S2A). Taken together, these data strongly indicated that SPRY1 negatively regulates AR activation and EGF-induced PSA expression and cell proliferation of LNCaP cells.
Fig. 5.
SPRY1 suppresses the EGF/ERK/AR pathway. A: SPRY1 protein expression levels by Western blotting in LNCaP cells transfected with a negative control (siCtr) and SPRY1 targeting (siSPRY1) siRNA. B: AR activation of LNCaP-siCtr and -siSPRY1 evaluated by dual luciferase reporter assay using pGL3-luc containing PSA promoter (PSAp) and pTK-RL (TK). C: PSA mRNA expression levels relative to TBP (upper) and cell proliferation in MTS assays (lower) of LNCaP-siCtr and -siSPRY1 cells in normal medium (FBS), androgen-depleted medium without EGF (CSFBS), and androgen-depleted medium with 2 ng/ml EGF (CSFBS + EGF). D: PSA mRNA expression levels relative to TBP (upper) and cell proliferation in MTS assays (lower) of LNCaP-siCtr and -siSPRY1 cells in normal medium with 0, 5, and 10 μM of ERK inhibitor PD98059 (PD). Cells were incubated for 72 h. (*P < 0.05, **P < 0.005). E: AR regulates miR-21. miR-21 negatively regulate SPRY1. SPRY1 negatively regulate the EGF/ERK/AR pathway.
JAG1 IS ASSOCIATED WITH PROSTATE CANCER CELL INVASION AND IN VIVO PROLIFERATION
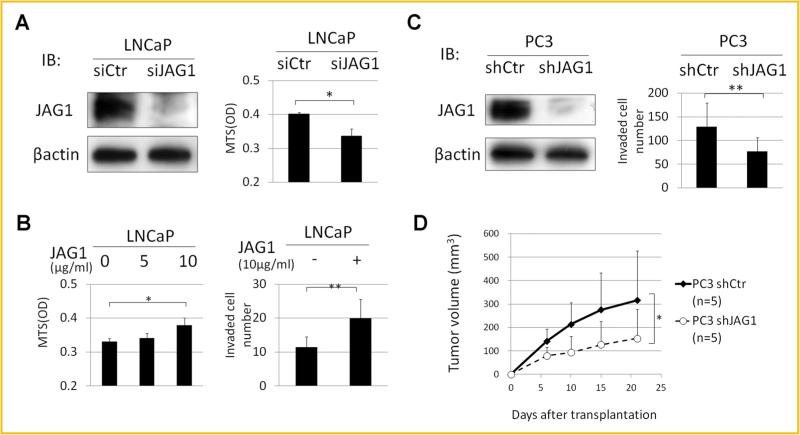
JAG1 is one of several Notch receptor ligands that appears to play an important role in cancer cell invasion [Ranganathan et al., 2011]. In LNCaP cells, knocking down JAG1 expression significantly suppressed cell proliferation (Fig. 6A), while administration of JAG1 recombinant protein significantly enhanced cell proliferation in a dose dependent manner (Fig. 6B). Moreover, in Matrigel invasion assays, JAG1 recombinant protein administration also enhanced cell invasion significantly (Fig. 6B). These results indicated that JAG1 is positively associated with proliferation and invasion of LNCaP cells. JAG1 is also highly expressed in the more invasive PC3 cells. Thus, to further examine the association of JAG1 with cell invasion and in vivo tumor growth potential, we performed stable knockdown of JAG1 using a JAG1-specific shRNA. Knocking down JAG1 in PC3 cells significantly suppressed cell invasion (Fig. 6D) without affecting cell proliferation (data not shown). Moreover, in vivo tumor growth of PC3-shJAG1 was significantly lower than that observed with PC3 treated with a scrambled shRNA (PC3-shCtr; Fig. 6D). N-cadherin is reported to be associated with prostate cancer invasion or metastasis [Amano et al., 2010]. The N-cadherin expression levels in LNCaP-cl5 were higher than in LNCaP-cl1, correlating with the JAG1 expression levels. When the JAG1 was knocked down in PC3 cells, the expression levels of N-cadherin were significantly down-regulated (Supplement Fig. S2B). These results indicated that JAG1 regulates N-cadherin and is positively associated with cell invasion and in vivo tumor growth of PC3 cells. Therefore, JAG1 might be associated with the higher invasive and in vivo tumor growth potential of LNCaP-cl5.
Fig. 6.
JAG1 enhances cell invasiveness. A: JAG1 protein expression levels of LNCaP cells transfected with negative control (siCtr) and JAG1 targeting (siJAG1) siRNAs by Western blotting (left). Their cell proliferation in MTS assays after 72 h incubation (right). B: Cell proliferation of LNCaP cells under the administration of 0, 5, and 10 μg/ml JAG1 recombinant protein (left). Numbers of invaded LNCaP cells without or with 10 μg/ml JAG1 recombinant protein in Matrigel invasion assays after 72 h incubation (right). C: JAG1 protein expression levels of PC3 cells transfected with pGIPZ Non-silencing control (shCtr) and pGIPZ-shJAG1 (shJAG1) by Western blotting (left). Numbers of invaded PC3-shCtr and -shJAG1 cells in Matrigel invasion assays after 48 h incubation (right). D: Sequential changes in tumor volumes of PC3-shCtr and -shJAG1 xenograft tumor at indicated days after transplantation (n = 5 each). (*P < 0.05, **P < 0.005).
SPRY1 AND JAG1 mRNA EXPRESSION IN HUMAN PROSTATE CANCERS
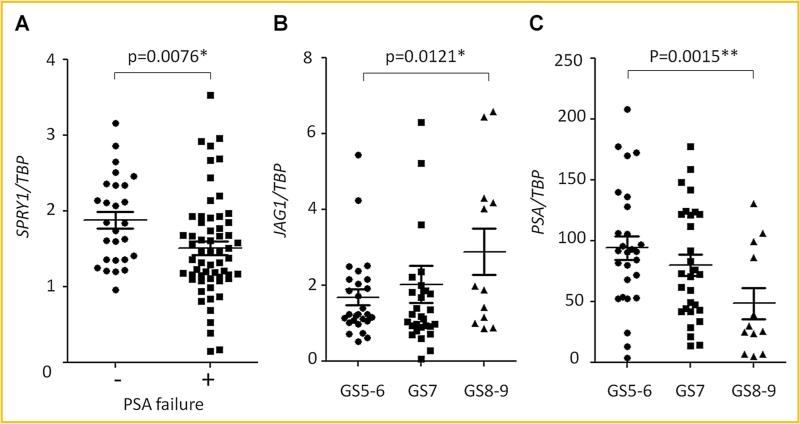
In order to establish the clinical relevance of the observed results, we next examined SPRY1 and JAG1 mRNA expression in clinical prostate cancer specimens. Of the 70 samples that were accumulated with follow-up information for up to 14 years, 43 patients had PSA recurrence and 27 patients did not. The median duration from surgery to PSA recurrence was 2 (1–8) years. The median follow-up duration in patients without PSA recurrence was 6 (1–14) years. The numbers in each Gleason sum (GS) group were 27, 31, and 12 in GS5–6, 7, and 8–9, respectively. In patients without and with PSA recurrence, the mean ages were 54.8 ± 6.9 and 59.4 ± 6.9 years (P = 0.004), the mean pre-operative serum PSA levels were 8.1 ± 7.1 and 12.8 ± 8.9 ng/ml (P = 0.011) and the mean GSs were 6.5 ± 0.9 and 7.1 ± 0.9 (P = 0.007), respectively. SPRY1 mRNA expression was significantly higher in the prostate cancer tissues of patients without a PSA recurrence than patients that had a PSA recurrence of their disease (P = 0.0076) (Fig. 7A) and the JAG1 mRNA expression was significantly higher in the GS8–9 tumors than in the GS5–6 tumors (P = 0.0121) (Fig. 7B). Interestingly, PSA mRNA levels were significantly lower in GS8–9 than in GS5–6 (P = 0.0015) (Fig. 7C), suggesting that SPRY1 and JAG1 mRNA expression levels in prostate cancer may be potential tissue biomarkers to differentiate aggressive prostate cancer.
Fig. 7.
SPRY1 and JAG1 mRNA expression levels in PC tissues. A: SPRY1 mRNA expression levels by real time PCR in prostate cancer tissues of patients without (–) and with (+) PSA recurrence. B:JAG1 mRNA expression levels by real time PCR in prostate cancer tissues with Gleason sum score (GS) 5–6, 7, and 8–9 diseases. C:JAG1 mRNA expression levels by real time PCR in PC tissues with Gleason sum score (GS) 5–6, 7, and 8–9 diseases. (*P < 0.05, **P < 0.005).
DISCUSSION
Most prostate cancer lesions are multifocal and cancer cells of high and low Gleason grade co-exist together within a prostate [Arora et al., 2004]. It has been reported that these multiple and independent histologic foci of cancer are genetically distinct [Macintosh et al., 1998; Mehra et al., 2007]. Although it is now widely accepted that CRPC is a phenotypically heterogeneous group of diseases [Shah et al., 2004], molecular analyses showed that multiple metastases in the same patient were clonally related, indicated that CRPC is genetically monoclonal [Mehra et al., 2008; Liu et al., 2009]. These findings suggest that CRPC arise from the selective advantage of individual clones during cancer progression. However, this process of clonal evolution may also represent the consequence of androgen deprivation, which may differentially target cells of varying levels of insensitivity to androgen deprivation or malignant potential [Shen and Abate-Shen, 2010].
LNCaP cells were originally established from a lymph node metastasis from a prostate cancer patient receiving androgen deprivation therapy. Although the tumors were derived from CRPC tissues, LNCaP cells harbor insensitivity to androgen deprivation which may reflected the well characterized androgen receptor mutation that exists within the line. These studies do suggest though that androgen sensitive and insensitive cells co-exist in LNCaP cell line [Horoszewicz et al., 1983]. Various kinds of androgen-insensitive LNCaP subclones have been established by long-term androgen deprivation [van Steenbrugge et al., 1991; Pflug et al., 1999; Inoue et al., 2006, 2008]. However, to the best of our knowledge, there are no reports showing that these androgen insensitive cells pre-existed before the hormone deprivation. In the present study, the LNCaP clones were established without androgen deprivation. These clones have different levels of AR and PSA, and differ in their insensitivity to androgen deprivation. Moreover, the DNA copy numbers in some regions were different between these LNCaP clones. These results would suggest that these LNCaP clones contained genetically different types of cells and the prevalence of these cells could account for the differences in their phenotypes and gene expression patterns. However, a majority of the genes differentially expressed between these clones are on the regions where no DNA copy number differences were identified. These genes are suggested to be regulated by other means such as translational, post-translational, or epigenetic changes. These studies were performed in media containing either intact and charcoal stripped FBS. While the purpose of the study was to utilize the charcoal stripping of the FBS as a means to remove androgens from the media, the process of charcoal stripping does remove other hormones and growth factors. The elimination of these other entities is a limitation to the studies outlined.
SPRY was originally identified in Dtosophila as a negative regulator of fibroblast growth factor (FGF) signaling during tracheal development [Hacohen et al., 1998]. Subsequent studies have shown SPRY to be a general inhibitor of growth factor-induced RTK signaling pathways involved in Dtosophila development and organogenesis [Casci et al., 1999; Reich et al., 1999]. While Dtosophila has only one SPRY gene, at least four SPRY homologues (SPRY1–4) are found in humans [de Maximy et al., 1999]. SPRY also inhibits growth factor-induced cell response by inhibiting the RTK-dependent Ras/mitogen-activated protein (MAP) kinase signaling pathway [Hanafusa et al., 2002]. In this study, we found that SPRY1 negatively regulates AR activation. To the best of our knowledge, this is the first report demonstrating an association between SPRY1 and AR activation. It was previously reported that activation of RTK and MAP kinase are associated with castration-resistant progression of prostate cancer via AR activation without androgens [Feldman and Feldman, 2001]. ERK phosphorylation was up-regulated by EGF stimulation and SPRY1 knockdown. ERK inhibition suppressed cell proliferation and PSA up-regulation induced by SPRY1 knockdown. These results indicated that negative regulation of EGF/ERK pathway was one of the mechanisms for SPRY1 to inactivate AR function and suppress cell proliferation. However, to elucidate the association between AR and ERK signaling needs further studies.
Notch signaling is involved in a variety of cellular processes, such as cell fate specification, differentiation, proliferation, and survival. It has been reported that the Notch signaling network is frequently de-regulated in various kinds of human malignancies [Ranganathan et al., 2011]. Four Notch receptors (NOTCH1–4) and five ligands (JAG-1, -2, DLL-1, -3, -4) have been described and binding of ligand induces cleavage of the receptor, and its intracellular domain translocates into the nucleus and mediates the transcription of target genes [Wang et al., 2008]. JAG1 has been reported to positively regulate prostate cancer cell growth, migration, and invasion [Zhang et al., 2006; Wang et al., 2010]. Recent studies have suggested that Notch can drive cancer progression through the repression of E-cadherin [Chen et al., 2010]. N-cadherin has been shown to be important for prostate cancer progression [Tanaka et al., 2010] and the E- to N-cadherin switch in primary tumors predictive of recurrence and prostate cancer-related death [Gravdal et al., 2007]. In our study, knocking down JAG1 in PC3 cells down-regulated N-cadherin, and suppressed cell invasion. These results indicated that N-cadherin up-regulation may be one of the mechanisms by which JAG1 enhances cell invasion. Further studies are needed for elucidating the association between JAG1 and N-cadherin.
The expression levels of two genes SPRY1 and JAG1 were higher in LNCaP-cl5 than in LNCaP-cl1 and miR-21 regulates both of these genes. mRNA and protein expression levels of these genes were down-regulated by the transfection of miR-21 (Fig. 4C). These results along with identification of candidate miR-21 binding sites in the 3’UTR of the SPRY1 and JAG1 mRNAs indicate that miR-21 regulation of SPRY1 and JAG1 may be mediated directly by miR-21 binding to sequences in their 3’UTRs. Androgen regulated miR-21 and both AR activity and miR-21 expression were higher in LNCaP-cl1 than in LNCaP-cl5. Thereafter, the expression levels of SPRY1 and JAG1 were lower in LNCaP-cl1 than in LNCaP-cl5 (Supplement Fig. S3). There were no significant differences between the proliferation rates of LNCaP-cl1 and -cl5 in normal medium. In LNCaP cells SPRY1 negatively and JAG1 positively regulated cell proliferation. Interestingly, ERK phosphorylation levels were regulated by SPRY1 and were higher in LNCaP-cl1 than in LNCaP-cl5. AR activity was negatively regulated by SPRY1 and higher in LNCaP-cl1 than in LNCaP-cl5 (Supplement Fig. S1A). N-cadherin expression levels were regulated by JAG1 and higher in LNCaP-cl5 than in LNCaP-cl1 (Supplement Fig. S2B). It is suggested that SPRY1 and JAG1 compensate each other and therefore do not impact cell proliferation. However, SPRY1 negatively regulates PSA expression and JAG1 positively regulates cell invasion. This may explain the differences in their insensitivity to androgen deprivation and invasiveness (Supplement Fig. S3).
The phenomenon of epithelial to mesenchymal transition (EMT) occurs when epithelial cells undergo several morphological changes and assume a mesenchymal phenotype that includes decreased adhesion, increased production of extracellular matrix components, increased migration, increased resistance to apoptosis and invasiveness. EMT is a prerequisite for the tumor cells to cross the basement membrane, enter into circulation and result in distant metastases [Thiery, 2003]. PSA is specifically expressed in prostate epithelium and would be typically down-regulated during an EMT transition [Pretlow et al., 1991; Magklara et al., 2000]. Notch signaling appears to play an important role in regulating EMT [Ranganathan et al., 2011]. In the genes differentially expressed between LNCaP-cl1 and -cl5, it was reported that UGT2B17 (cl1/cl5 ratio = 3.11) was associated with androgen synthesis [Nadeau et al., 2011] and TWIST1 (cl5/cl1 ratio = 2.59) was an important gene regulating EMT [Mani et al., 2008]. These genes might contribute to their phenotypic differences as well. Taken together, LNCaP-cl1 and -cl5 have “epithelial” and “mesenchymal” phenotypes, respectively. Our study using LNCaP clones indicate that the prevalence of cells harboring these different phenotypes might be associated with androgen-dependence or invasiveness of prostate cancer.
It was previously reported that SPRY1 is down-regulated in a subset of prostate cancer tissues when compared with normal prostate tissues [Kwabi-Addo et al., 2004]. In addition, down-regulation of SPRY1 was associated with biochemical recurrence following radical prostatectomy [Fritzsche et al., 2006]. JAG1 expression was reported to be higher in high grade and metastatic prostate cancer tissues [Santagata et al., 2004]. In our study, SPRY1 mRNA expression levels in prostate cancer tissues were negatively correlated with PSA recurrence. However, JAG1 mRNA expression levels did not correlate with PSA recurrence (data not shown). On the other hand, JAG1 mRNA expression levels were higher in high GS disease. Interestingly, PSA mRNA expression levels were lower in high GS disease, inversely correlating with the expression pattern of JAG1. In every patient, the expression levels of SPRY1, JAG1, and PSA mRNA did not correlate with one another by Pearson's correlation coefficient analysis (data not shown), suggesting that in addition to miR-21, other factors may regulate SPRY1 and JAG1 in the clinical setting.
There are two models suggested for the castration-resistant progression of prostate cancer. The “adaptation” model proposes that CRPC arises through genetic or epigenetic conversion of previously androgen-dependent cells during conditions of androgen deprivation, while the “clonal selection” model suggests that emergence of castration-resistance reflects the proliferation of a previously quiescent population ofrare CRPC cells within an otherwise androgen-dependent tumor [Isaacs and Coffey, 1981]. Studies of the limiting dilution and fluctuation analyses of an androgen-dependent xenograft [Craft et al., 1999] or the onset of castration-resistance in TRAMP mice [Gingrich et al., 1997] provide evidence to support the “clonal selection” model. In addition, analysis of localized prostate cancer tumors suggests that rare AR mutations can be detected prior to androgen deprivation therapy [Gaddipati et al., 1994; Tilley et al., 1996]. Furthermore, the finding CRPC cells such as CARNs (castration-resistant Nkx3.1-expressing cells) represent a cell of origin for prostate cancer in which the rare CRPC population might also correspond to putative cancer stem cells [Wang et al., 2009]. In these reports, increased AR activity prior to androgen deprivation may be selected during prostate cancer progression. Our study demonstrates that LNCaP cells are collections of cells with distinct AR activities which are correlated with their androgen insensitivity and consistent with this theory. Surprisingly however, LNCaP cells with low AR activity had a higher level of invasiveness. CRPC cells with high AR activity could be treated by new drugs to block androgen receptor activity (MDV3100) or steroidal synthesis (abiraterone, GTx-758, or TAK-700) [Attard et al., 2009; Lassi and Dawson, 2009]. It has been reported that androgen deprivation therapy enhanced expression levels of some EMT markers in androgen-dependent prostate cancer cells [Tanaka et al., 2010; Sun et al., 2011]. Our results indicate that androgen-sensitive but aggressive prostate cancer cells with higher EMT markers exist prior to hormonal manipulation. Taken together, these findings provided a rationale for the usefulness of EMT marker to predict the prognosis of prostate cancer as well as the potential of EMT blockade therapy for the treatment of CRPC.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donald Vindivich, David Yeater, and Dana Wodzenski in the Department of Urology, at the Johns Hopkins University School of Medicine for the technical assistance with this project. This work was supported by grants from the National Cancer Institute (U54CA143803 and R01CA143299).
Footnotes
Conflict of interest: None.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web-site.
REFERENCES
- Amano J, Masuyama N, Hirota Y, Tanaka Y, Igawa Y, Shiokawa R, Okutani T, Miyayama T, Nanami M, Ishigai M. Antigen-dependent internalization is related to rapid elimination from plasma of humanized anti-HM1. 24 monoclonal antibody. Drug Metab Dispos. 2010;38:2339–2346. doi: 10.1124/dmd.110.035709. [DOI] [PubMed] [Google Scholar]
- Arora R, Koch MO, Eble JN, Ulbright TM, Li L, Cheng L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–2366. doi: 10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- Attard G, Reid AH, Olmos D, de Bono JS. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–4940. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Chen J, Imanaka N, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102:351–360. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft N, Chhor C, Tran C, Belldegrun A, DeKernion J, Witte ON, Said J, Reiter RE, Sawyers CL. Evidence for clonal outgrowth of androgen-independent prostate cancer cells from androgen-dependent tumors through a two-step process. Cancer Res. 1999;59:5030–5036. [PubMed] [Google Scholar]
- Darimipourain M, Wang S, Ittmann M, Kwabi-Addo B. Transcriptional and post-transcriptional regulation of Sprouty1, a receptor tyrosine kinase inhibitor in prostate cancer. Prostate Cancer Prostatic Dis. 2011;14:279–285. doi: 10.1038/pcan.2011.33. [DOI] [PubMed] [Google Scholar]
- de Maximy AA, Nakatake Y, Moncada S, Itoh N, Thiery JP, Bellusci S. Cloning and expression pattern of a mouse homologue of drosophila sprouty in the mouse embryo. Mech Dev. 1999;81:213–216. doi: 10.1016/s0925-4773(98)00241-x. [DOI] [PubMed] [Google Scholar]
- Denmeade SR, Sokoll LJ, Dalrymple S, Rosen DM, Gady AM, Bruzek D, Ricklis RM, Isaacs JT. Dissociation between androgen responsiveness for malignant growth vs. expression of prostate specific differentiation markers PSA, hK2, and PSMA in human prostate cancer models. Prostate. 2003;54:249–257. doi: 10.1002/pros.10199. [DOI] [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- Fritzsche S, Kenzelmann M, Hoffmann MJ, Muller M, Engers R, Grone HJ, Schulz WA. Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr Relat Cancer. 2006;13:839–849. doi: 10.1677/erc.1.01190. [DOI] [PubMed] [Google Scholar]
- Gaddipati JP, McLeod DG, Heidenberg HB, Sesterhenn IA, Finger MJ, Moul JW, Srivastava S. Frequent detection of codon 877 mutation in the androgen receptor gene in advanced prostate cancers. Cancer Res. 1994;54:2861–2864. [PubMed] [Google Scholar]
- Gingrich JR, Barrios RJ, Kattan MW, Nahm HS, Finegold MJ, Greenberg NM. Androgen-independent prostate cancer progression in the TRAMP model. Cancer Res. 1997;57:4687–4691. [PubMed] [Google Scholar]
- Gravdal K, Halvorsen OJ, Haukaas SA, Akslen LA. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin Cancer Res. 2007;13:7003–7011. doi: 10.1158/1078-0432.CCR-07-1263. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, Bova GS, Luo J. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Yoshida T, Shimizu Y, Kobayashi T, Yamasaki T, Toda Y, Segawa T, Kamoto T, Nakamura E, Ogawa O. Requirement of androgen-dependent activation of protein kinase Czeta for androgen-dependent cell proliferation in LNCaP Cells and its roles in transition to androgen-independent cells. Mol Endocrinol. 2006;20:3053–3069. doi: 10.1210/me.2006-0033. [DOI] [PubMed] [Google Scholar]
- Inoue T, Leman ES, Yeater DB, Getzenberg RH. The potential role of purine-rich element binding protein (PUR) alpha as a novel treatment target for hormone-refractory prostate cancer. Prostate. 2008;68:1048–1056. doi: 10.1002/pros.20764. [DOI] [PubMed] [Google Scholar]
- Isaacs JT, Coffey DS. Adaptation versus selection as the mechanism responsible for the relapse of prostatic cancer to androgen ablation therapy as studied in the Dunning R-3327-H adenocarcinoma. Cancer Res. 1981;41:5070–5075. [PubMed] [Google Scholar]
- Kwabi-Addo B, Wang J, Erdem H, Vaid A, Castro P, Ayala G, Ittmann M. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004;64:4728–4735. doi: 10.1158/0008-5472.CAN-03-3759. [DOI] [PubMed] [Google Scholar]
- Lassi K, Dawson NA. Emerging therapies in castrate-resistant prostate cancer. Curr Opin Oncol. 2009;21:260–265. doi: 10.1097/CCO.0b013e32832a1868. [DOI] [PubMed] [Google Scholar]
- Lin MF, Meng TC, Rao PS, Chang C, Schonthal AH, Lin FF. Expression of human prostatic acid phosphatase correlates with androgen-stimulated cell proliferation in prostate cancer cell lines. J Biol Chem. 1998;273:5939–5947. doi: 10.1074/jbc.273.10.5939. [DOI] [PubMed] [Google Scholar]
- Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, Nelson WG, Yegnasubramanian S, Luo J, Wang Y, Xu J, Isaacs WB, Visakorpi T, Bova GS. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintosh CA, Stower M, Reid N, Maitland NJ. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 1998;58:23–28. [PubMed] [Google Scholar]
- Magklara A, Scorilas A, Stephan C, Kristiansen GO, Hauptmann S, Jung K, Diamandis EP. Decreased concentrations of prostate-specific antigen and human glandular kallikrein 2 in malignant versus nonmalignant prostatic tissue. Urology. 2000;56:527–532. doi: 10.1016/s0090-4295(00)00621-x. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- Mehra R, Tomlins SA, Yu J, Cao X, Wang L, Menon A, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau G, Bellemare J, Audet-Walsh E, Flageole C, Huang SP, Bao BY, Douville P, Caron P, Fradet Y, Lacombe L, Guillemette C, Levesque E. Deletions of the androgen-metabolizing UGT2B genes have an effect on circulating steroid levels and biochemical recurrence after radical prostatectomy in localized prostate cancer. J Clin Endocrinol Metab. 2011;96:E1550–E1557. doi: 10.1210/jc.2011-1049. [DOI] [PubMed] [Google Scholar]
- Pflug BR, Reiter RE, Nelson JB. Caveolin expression is decreased following androgen deprivation in human prostate cancer cell lines. Prostate. 1999;40:269–273. doi: 10.1002/(sici)1097-0045(19990901)40:4<269::aid-pros9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Pretlow TG, Pretlow TP, Yang B, Kaetzel CS, Delmoro CM, Kamis SM, Bodner DR, Kursh E, Resnick MI, Bradley EL., Jr. Tissue concentrations of prostate-specific antigen in prostatic carcinoma and benign prostatic hyperplasia. Int J Cancer. 1991;49:645–649. doi: 10.1002/ijc.2910490503. [DOI] [PubMed] [Google Scholar]
- Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: A little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- Reich A, Sapir A, Shilo B. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development. 1999;126:4139–4147. doi: 10.1242/dev.126.18.4139. [DOI] [PubMed] [Google Scholar]
- Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, Mendell JT, Lupold SE. MiR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, Rubin MA, Aster JC. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi T, Terada N, Zeng Y, Suyama T, Luo J, Trock B, Kulkarni P, Getzenberg RH. Cancer/testis antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, Chen D, Seo K, Modrusan Z, Gao WQ, Settleman J, Johnson L. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: Implications for androgen-deprivation therapy. Cancer Res. 2011;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kono E, Tran CP, Miyazaki H, Yamashiro J, Shimomura T, Fazli L, Wada R, Huang J, Vessella RL, An J, Horvath S, Gleave M, Rettig MB, Wainberg ZA, Reiter RE. Monoclonal antibody targeting of N-cadherin inhibits prostate cancer growth, metastasis and castration resistance. Nat Med. 2010;16:1414–1420. doi: 10.1038/nm.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Tilley WD, Buchanan G, Hickey TE, Bentel JM. Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res. 1996;2:277–285. [PubMed] [Google Scholar]
- van Steenbrugge GJ, van Uffelen CJ, Bolt J, Schroder FH. The human prostatic cancer cell line LNCaP and its derived sublines: An in vitro model for the study of androgen sensitivity. J Steroid Biochem Mol Biol. 1991;40:207–214. doi: 10.1016/0960-0760(91)90184-7. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Sarkar FH. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer Res. 2008;28:3621–3630. [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V, Hay N, Sarkar FH. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Ahmed F, Banerjee S, Li Y, Sarkar FH. Down-regulation of Jagged-1 induces cell growth inhibition and S phase arrest in prostate cancer cells. Int J Cancer. 2006;119:2071–2077. doi: 10.1002/ijc.22077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.