Abstract
Multi-Band Echo Planar Imaging (MB-EPI), a new approach to increase data acquisition efficiency and/or temporal resolution, has the potential to overcome critical limitations of standard acquisition strategies for obtaining high-resolution whole brain perfusion imaging using arterial spin labeling (ASL). However, the use of MB also introduces confounding effects, such as spatially varying amplified thermal noise and leakage contamination, which have not been evaluated to date as to their effect on cerebral blood flow (CBF) estimation. In this study, both the potential benefits and confounding effects of MB-EPI were systematically evaluated through both simulation and experimentally using a pseudo-continuous arterial spin labeling (pCASL) strategy. These studies revealed that the amplified noise, given by the geometry factor (g-factor), and the leakage contamination, assessed by the total leakage factor (TLF), have minimal impact on CBF estimation. Furthermore, it is demonstrated that MB-EPI greatly benefits high-resolution whole brain pCASL studies in terms of improved spatial and temporal signal-to-noise ratio efficiency, and increases compliance with the assumptions of the commonly used single blood compartment model, resulting in improved CBF estimates.
Keywords: arterial spin labeling (ASL), cerebral blood flow (CBF), leakage evaluation via acquired dummy slices (LEADS), multi-band echo planar imaging (MB-EPI), pseudo-continuous arterial spin labeling (pCASL), total leakage factor (TLF)
1. Introduction
Arterial spin labeling (ASL) perfusion imaging makes use of arterial blood water as an endogenous tracer to estimate tissue perfusion and evaluate tissue viability (Detre et al., 1992). The non-invasive and non-contrast enhanced characteristics of ASL imaging make it an attractive approach for both neuroscience research and clinical applications (Detre et al., 1998, Detre and Wang, 2002). High-resolution studies are desired in order to reduce partial volume effects on cerebral blood flow (CBF) quantification, increase the ability to identify small focal lesions (Bokkers et al., 2012) and improve perfusion quantification in small sub-cortical structures such as the hippocampus (Li et al., 2011, Li et al., 2013). However, obtaining high-resolution ASL based perfusion measurements is challenging due to the intrinsically low signal-to-noise ratio (SNR) and increased acquisition times when using standard acquisition strategies. Decreased perfusion SNR for high-resolution imaging is a consequence of multiple factors including; 1) increased in-plane resolution, 2) increased through-plane resolution and 3) the need for more slices to cover the same volume resulting in prolonged delay times between labeling and signal acquisition during which labeled spins experience longitudinal relaxation. The necessity of increasing the number of label/control image pairs for sufficient perfusion SNR greatly increases the total imaging acquisition time, thus limiting, if not prohibiting, the practice of acquiring high-resolution whole brain ASL perfusion data.
To overcome such challenges, different strategies have been previously proposed to increase perfusion SNR of ASL methods. Continuous arterial spin labeling (CASL) (Alsop and Detre, 1998), one of two major types of ASL methods, can provide better perfusion SNR than pulsed arterial spin labeling (PASL) (Kim and Tsekos, 1997, Wong, 2005), but requires a dedicated transmit/receive coil with the capability of continuous transmission of radiofrequency (RF) energy. More recently, an intermediate approach, pulsed- or pseudo-continuous arterial spin labeling (pCASL), has been shown to provide higher labeling efficiency than CASL without the need for a dedicated transmit/receive coil (Wu et al., 2007, Dai et al., 2008). The use of a body coil for RF transmission and a separate phased-array coil for signal reception makes pCASL amenable for applications on current clinical scanners. Therefore, pCASL has become the preferred ASL approach for whole brain ASL perfusion imaging (Alsop et al., 2014).
To increase the SNR of whole brain ASL imaging studies, 3D imaging methods have also been explored (Gunther et al., 2005, Dai et al., 2013). In this approach, a 3D slab, rather than a single slice, is excited by the RF excitation pulse. The excited spins are continuously refocused by using spin echo refocusing RF pulses between phase encoded slices and spatially encoded in plane with rapid k-space sampling strategies such as echo-planar imaging (EPI) (Gunther et al., 2005) or spiral (Dai et al., 2013) trajectories. Recently, to overcome the adverse T2 blurring effects of 3D acquisition on perfusion imaging, alternative imaging acquisition strategies have been proposed and demonstrated (Gai et al., 2011, Tan et al., 2011, Nielsen and Hernandez-Garcia, 2013).
In contrast to 3D imaging methods, 2D EPI does not suffer T2 blurring, but requires a longer total imaging acquisition time to cover the same volume. The imaging slices, typically acquired in an ascending slice order, experience widely varying post-labeling delay times, during which longitudinal relaxation of the labeled spins occurs. The total acquisition time is lengthened with increasing encoding matrices and slices as required in high-resolution whole brain studies. In such studies, the most inferior slices and superior slices are acquired at extremely short and long post-labeling delays, respectively. Short delay times have the potential problem of not allowing sufficient time for labeled spins to perfuse the target tissue and long delay times suffer from poor SNR. Particularly, the validity of the standard single-blood compartment model for CBF quantification can be compromised as the range of post-labeling delay times expands, which arises from using the T1 of the blood for the longitudinal relaxation decay correction of the perfusion signal (Wang et al., 2003).
Multi-band imaging, or simultaneous multi-slice imaging, provides an attractive and alternative solution to reduce the total acquisition time of high-resolution whole brain imaging with 2D EPI, especially when increased spatial or temporal resolution is desired (Moeller et al., 2010, Setsompop et al., 2012b). Multi-band EPI imaging (MB-EPI) uses multi-banded radiofrequency (RF) pulses to simultaneously excite multiple spatially distributed slices, where the superimposed signals acquired from the multiple slices are unwrapped via anti-aliasing reconstruction. The simultaneous acquisition of multiple slices can greatly reduce total imaging acquisition time for whole brain applications with EPI, and particularly has the potential to improve whole brain ASL perfusion studies where high in-plane and through-plane resolution is desired, necessitating the use of a large number of thin imaging slices to achieve the desired coverage. MB-EPI has been successfully demonstrated in functional magnetic resonance imaging (fMRI) (Moeller et al., 2010) and resting state fMRI (Feinberg et al., 2010, Koopmans et al., 2012, Smith et al., 2012), showing improved detection of resting sate networks, as well as diffusion-weighted imaging (DWI) (Feinberg et al., 2010, Setsompop et al., 2012a) for dramatic reductions in imaging time. As such, MB-EPI has become an essential data acquisition strategy for both fMRI and DWI in the Human Connectome Project (Van Essen et al., 2012).
MB-EPI has been recently demonstrated (Kim et al., 2013) and later compared to single-shot 3D GRASE (Feinberg et al., 2013) for PASL perfusion imaging in the brain using flow-sensitive alternating inversion recovery (FAIR) (Kim and Tsekos, 1997). By using a standard low imaging resolution, these studies found that perfusion signal differences between single-band (SB) and MB-EPI are minimal. While an important initial finding, these studies did not explore the potential of MB-EPI on ASL imaging in high-resolution whole brain applications where the slice dependent effects become pronounced. On the other hand, MB-EPI introduces confounding factors that may reduce the reliability of CBF estimation, including spatially varying noise amplification as characterized by geometry factor (g-factor) due to slice-GRAPPA (Generalized Autocalibrating Partially Parallel Acquisition) reconstruction (Robson et al., 2008) and leakage contamination resulting from imperfect anti-aliasing of simultaneously acquired multiple slices (Cauley et al., 2013, Xu et al., 2013). These potential confounding effects and their slice dependence were not directly and systematically investigated in the previous studies.
In this study, the potential benefits and confounding factors of MB-EPI for high-resolution whole brain ASL perfusion imaging are explored through both systematic experiments and theoretical simulations.
2. Materials and methods
2.1 MR imaging
Imaging studies were performed on a 3T Siemens Trio whole-body scanner (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany). The body coil was used for RF transmission, and the Siemens 32-channel phased array head coil for signal reception. High-resolution 3D anatomic images were acquired by using an MPRAGE (magnetization-prepared rapid acquisition with gradient echo) sequence with 1.0 mm isotropic resolution after a multi-plane scout localizer. These high-resolution images were used as the reference to prescribe imaging slices for the following ASL studies.
2.2 MB-EPI pCASL sequence
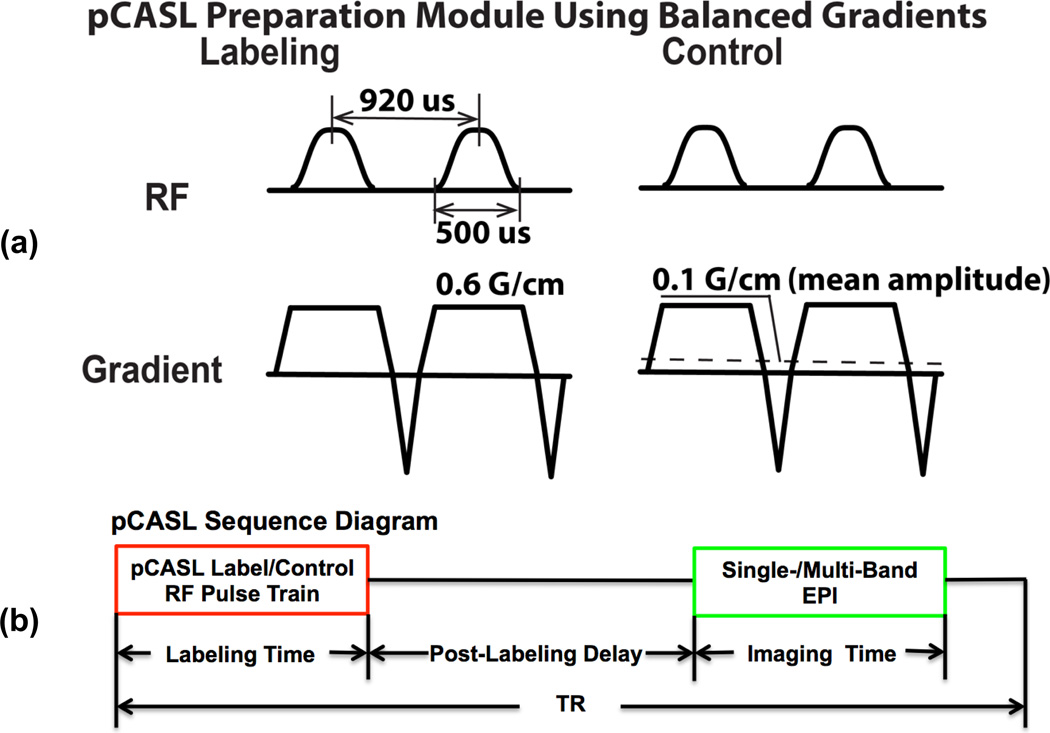
The pCASL preparation modules used the balanced gradient approach (Wu et al., 2007, Dai et al., 2008) (Figure 1a). The implemented pCASL EPI imaging sequence consisted of labeling/control RF pulse trains, post-labeling delay (PLD) and SB- or MB-EPI readout (Figure 1b). MB-EPI image slices are divided into a number of groups or bands equivalent to the MB factor (Figure 1c). Slices within each band can be collected in an ascending, descending or interleaved fashion, and slices across bands are acquired simultaneously in a spatially interleaved fashion. The applied MB-EPI method employed a blipped-CAIPI approach (Setsompop et al., 2012b) and aliasing constrained slice-GRAPPA algorithm to reduce leakage contamination (Cauley et al., 2013). In addition, a phase-scrambling strategy was applied for the MB RF pulses to reduce RF pulse peak power and/or duration (Goelman, 1997, Wong, 2012). The MB-EPI acquisition was preceded by a calibration scan for training the anti-aliasing reconstruction kernel without pCASL preparation. Additional EPI images could be optionally collected after the pCASL series with all RF pulses turned off for estimating thermal noise and calculating g-factor maps. Blipped CAIPI MB-EPI used a field of view (FOV) shift factor of 1/3, and MB-EPI reconstruction used a GRAPPA kernel size 5 for all studies.
Figure 1.
Implemented pCASL labeling/control preparation modules (a), single-/multi-band EPI pCASL sequence diagram (b), and illustration of imaging slice groups, shimming volume and labeling plane positions (c). Slice groups of MB-EPI with an acceleration factor 6 (MB6) are overlaid with the slice group matched by SB-EPI (in blue) in the comparison study 1. The time interval for Hanning-windowed hard pulses and RF pulse duration are 920 and 500 us, respectively. The maximal magnitude of slice-selective gradient is 0.6 G/cm, and the mean gradient amplitude for control preparation is 0.1 G/cm.
To minimize B0 off-resonance effects on pCASL labeling efficiency, EPI ghost artifacts and leakage contamination resulting from the EPI ghosts, Siemens’ advanced B0 shimming was applied in a targeted region covering both the imaging volume and the pCASL labeling plane (Figure 1c), in which B0 shimming was performed twice, one after the other, by using sequentially acquired B0 maps.
2.3 MB RF pulse manipulations and LEADS
To understand the impact of MB leakage contamination on CBF quantification, various studies were performed which involved manipulating components of the complex MB excitation RF pulse. Specifically, to estimate leakage signal contamination in a targeted slice, the MB RF pulse component exciting this specific slice was turned off (given zero amplitude) during imaging acquisition while all other simultaneously acquired slices were still excited, denoted as MB-RFout. With such an RF pulse manipulation, MB reconstruction of the aliased slices would reveal the total signal contamination from all other simultaneously excited slices within the unexcited specific slice (dummy slice). This measured total leakage contamination was used to estimate the total leakage factor (TLF) described below. In a similar fashion, experimental measurement of the traditional leakage factor (LF) (Xu et al., 2013) can be also obtained by only exciting a single slice in the MB acquisition and measuring the resulting contamination within the other (MB-factor)-1 slices which are not excited (i.e. dummy slices). Such an RF manipulation for MB imaging is denoted as MB-RFin. In contrast to both MB-RFout and MB-RFin, a full MB acquisition with all MB RF pulse components enabled will be referred to as MB-RFall. In general, the method of acquiring dummy slice(s) in an MB acquisition to experimentally determine leakage contamination will be referred to as LEADS, denoting Leakage Evaluation via Acquired Dummy Slice(s).
2.4 Total Leakage Factor (TLF)
In MB imaging, the resulting signals for a specific MB imaging slice mainly originate from the slice itself and superimposed amplified thermal nosie, but also contamination signals from signal leakage due to imperfect anti-aliasing of simultaneously acquired slices (Xu et al., 2013). To characterize the relative impact of this contamination on the signal of interest, the concept of the total leakage factor (TLF) is introduced,
| [1] |
where (STLC) is the total leakage contamination signal from all other simultaneously acquired slices within the targeted dummy slice from an MB-RFout acquisition, SNC is the signal of the targeted slice in the absence of contamination (NC, no contamination) and r represents spatial location. SNC can be acquired by using either an MB-RFin acquisition or a SB acquisition with MRI parameters matching those of the MB acquisition.
2.5 Subjects
Ten healthy volunteers were recruited for two comparison studies (please refer to sections 2.6 and 2.7, respectively): three males with ages 44 ± 21 years and two females with ages 25 ± 4 years for comparison study 1, and three males with ages 50 ± 19 years and two females with ages 26 ± 6 years for comparison study 2, expressed as mean ± S.D. (standard deviation). There were no significant differences between these two groups in terms of subject gender and age. Subjects were screened to assure their safety in the magnet and provided written informed consent prior to being studied according to a protocol approved by the local Institutional Review Board. All subjects were required to keep their eyes closed and remain awake during imaging sessions.
2.6 Comparison study 1
In this comparison study, low-resolution pCASL imaging was performed using the same MRI and pCASL parameters for both SB and MB, importantly including matching PLDs. To match PLDs between SB and MB imaging, the SB acquisition includes only slices matching the middle or middle superior slice group of whole brain MB acquisition. Perfusion signals between these two acquisitions were compared and the impact of the MB acceleration was assessed. MB pCASL imaging slice groups, as well as the slice group matched by those of SB are illustrated for MB imaging with MB factor 6 in Figure 1c.
The MB studies were performed with four different MB acceleration factors: 3, 4, 5, and 6. The previous described RF manipulations were performed using the middle group of imaging slices for MB 3 and 5 and the middle superior imaging slice groups for MB 4 and 6 as the dummy slices with MB-RFout and the excited slices with MF-RFin. For MB 3, 4 and 6, a total of 24 imaging slices were acquired, and for MB 5, a total of 25 imaging slices were acquired.
For this comparison study, the same EPI, RF pulse and pCASL parameters were used, including: TR/TE=3600/12 ms; phase encoding direction = anterior to posterior; FOV = 224 × 224 mm2; matrix size = 64 × 64; in-plane resolution = 3.5 × 3.5 mm2; slice thickness/gap= 5/1 mm; excitation RF pulse flip angle (FA)/duration/time bandwidth product (R)= 90 degree/4 ms/5.2; partial Fourier = 6/8; slice acquisition order = ascending; total number of perfusion imaging measurements = 60; total number of noise images = 200; labeling duration/PLD = 1.5 s/1.6 s.
A total of five data sets were acquired from each volunteer for each MB factor including SB pCASL, three MB pCASL imaging scans with the three MB RF pulse configurations (MB-RFin, MB-RFout, MB-RFall) and a second SB pCASL acquisition to evaluate perfusion signal variability across time due to either instrumental instability or physiological variations. For all but the second SB pCASL acquisition, 200 noise images were acquired following pCASL measurements by turning off all RF pulses within the sequence while keeping all other sequence parameters the same.
2.7 Comparison study 2
The second comparison study investigated the potential benefits of MB-EPI on high-resolution whole brain perfusion measurements in terms of CBF quantification, spatial and temporal perfusion SNRs and SNR efficiencies. A fair comparison was achieved for this study by utilizing MRI and pCASL parameters optimized individually for SB- and MB-EPI acquisitions. For this comparison, an MB-factor of 6 was applied for MB imaging.
MRI parameters common for both SB and MB imaging methods were: phase encoding direction = anterior to posterior; FA = 90 degree; FOV = 220 × 220 mm2; matrix size = 88 × 88; slice thickness/gap = 3.0/0.6 mm; partial Fourier = 6/8; the number of imaging slice = 36; slice acquisition order = ascending; total number of perfusion imaging measurements = 200; the number of noise images = 200; RF pulse time bandwidth product (R)= 5.2; and labeling duration = 1.5 s.
MRI parameters specific to SB-EPI pCASL imaging were: TR/TE = 4200/16 ms; RF pulse duration = 2.6 ms; PLD = 1.1 s; and imaging slice readout time = 44.5 ms. Specific MRI parameters for were: TR/TE = 3.4 s/18 ms; RF pulse duration = 4.0 ms; PLD = 1.6 s; and imaging slice readout time = 46.5 ms.
In this study, MB-RFin and MB-RFout scans were performed separately for each of the six bands in the MB 6 acquisition, resulting in six pairs of imaging series. To reduce the total study time, the acquisition time for each imaging series was about one minute with a total of 8 pairs of acquired label and control images.
2.8 Theoretical simulations
To theoretically evaluate the potential benefits of MB for whole brain pCASL imaging, numerical simulations were performed by using a single-blood compartment model (Wang et al., 2003) with T2* effect, using 90 and 35 ms for T2* for high and low imaging resolutions, respectively, estimated slice-wise g-factors, MRI parameters optimal for SB and MB, and previously reported longitudinal relaxation times of arterial blood: 1.66 s for T1 (Lu et al., 2004). T2* at 3T was estimated by using reported T2* values at 1.5T (Yang et al., 1999) and accounting for the inverse relationship between T2* and field strength and voxel size. Simulations were first performed with an assumed uniform GM perfusion across the brain, and later performed with slice-wise GM CBF as measured by high-resolution whole brain imaging acquisitions.
For low resolution, whole brain pCASL imaging (3.5 × 3.5 × 5 mm3), the same MRI parameters as those used in comparison study 1 were applied for the simulation except for the following: SB/MB TE = 12/13 ms; SB/MB RF duration = 2.6/4.0 ms; and SB/MB PLD = 1.36 s/1.6 s. The achievable minimal TRs were about 3.6 s and 3.2 s, and the slice acquisition times were about 29 ms and 30 ms for SB and MB, respectively. The use of 1.36 s for SB-EPI pCASL imaging is necessary to avoid intravascular artifacts (Dai et al., 2013), a finding also confirmed in our preliminary studies (data not shown).
For high-resolution whole brain perfusion imaging (2.5 × 2.5 × 3.0 mm3), MRI parameters used for the simulation were the same as those applied for comparison study 2.
2.9 TLF Evaluation
The imperfection of MB slice-GRAPPA anti-aliasing reconstruction can lead to local high TLFs across space (Cauley et al., 2013, Xu et al., 2013) while subject motion can cause the fluctuation of leakage contamination in imaging slices across time, both of which can affect perfusion temporal SNR. To accommodate the potential effects of subject bulk motion across time, the temporal averages of control images, and from MB-RFin and MB-RFout acquisitions, were used for TLF estimation. The voxel-wise TLF maps were calculated as the following and expressed in percentage:
| [2] |
and used to estiamte the mean TLF within grey matter. In equation [2], r and l represent the spatial location and the temporal index of the control image, respectively.
2.10 Perfusion image analysis and CBF quantification
Label and control images were first pair-wise subtracted and then averaged to obtain the mean perfusion-weighted image (PWI) signal. Subsequently, the grey matter (GM) CBF was estimated by using a single-blood compartment model (Wang et al., 2003):
| [3] |
where λ represents blood-tissue partition coeficient (assumed to be 0.9) (Roberts et al., 1996), ΔM mean perfusion-weighted imaging signal, α labeling efficiency (assumed to be 0.85) (Wu et al., 2007), M0 estimated proton density signal using the mean control image corrected for TR, T1b longitudinal relaxation time for arterial blood water, τ labeling time, and ω effecive slice-wise post-labeling delay time. The post-labeling delay time, ω, is further defined as:
| [4] |
where PLD represents norminal PLD defined in the pCASL EPI sequence, indexslice the chronological slice acquisition order for either SB or MB, and Tslice imaging slice acquisition time.
CBF differences between the first SB acquisiton (CBFSB1) and other acquisitions (CBFother) in comparison study 1 were evaluated as the following in percentage:
| [5] |
where r represents spatial location. The means of estiamted CBF differences within grey matter were obtained for each subjects.
2.11 Perfusion SNR and MB g-factor
Thermal noise was estimated from images of noise acquired immediately after the perfusion image acquistion within the same scan. Noise maps were generated by calculating standard deviations (stddev) across the 200 noise images on a voxel-wise basis;
| [6] |
where Snoise represents the signal within the noise image, σ thermal noise, n the index of noise image, N total number of noise images (i.e. 200) and r the spatial location.
For SB, the spatial mean of the noise map within grey matter was used as the estimate of thermal noise since the distribution across space is uniform in the absence of parallel imaging.
Perfusion imaging spatial SNR maps for SB and MB were then estiamted as follows:
| [7] |
| [8] |
where S(r)SB and S(r)MB represent SB and MB perfusion signals within grey matter, σSB and σ(r)MB estimated thermal noise levels for SB and MB, and Navg the number of temporal perfusion signal averages. To compare spatial perfusion SNRs between the first SB acquisition and other MB acquisitions in comparison study 1, SNR ratios (SNRMB/SNRSB1) were evalautred.
Temporal SNR (tSNR) of perfusion-weighted images was calculated as the ratio of the mean to the standard deviation of the perfusion signal S(r, n) across the ASL time series:
| [9] |
where n is the index of perfusion-weighted image, and N the total number of perfusion-weighted images.
The mean of perfusion imaging SNR within grey matter was used to estimate SNR effieciency as the following:
| [10] |
where SNReff is the perfusion SNR efficiency, and TRmin represents the achievable shortest repetition time. The g-factor of the MB acquisition was estimated by using the ratio of the MB noise map to the estimate of SB thermal noise:
| [11] |
and the mean g-factor within grey matter was then obtained.
2.12 Data processing and analysis
Post-processing operations, such as motion correction, segmentation and co-registration, were performed within SPM (Functional Imaging Laboratory, University College London). All image analysis, such as perfsuion SNR, g-factor estimation and theoretical simulation were all performed using scripts implemented in Matlab 7.1 (The MathWorks, Inc., Natick, Massachusetts).
All analysis and comparisons were performed only for GM. The GM masks were generated from the segmented images of MPRAGE acquisitions by using a probability threshold of 0.75 within SPM. White matter was not investigated because of the considerably longer PLDs required as a result of longer arterial transit times and lower perfusion SNR due to intrinsically lower perfusion levels. Group means and standard deviations of the different measures (e.g. SNR, CBF, TLF) were obtained for each comparison study. For TLFs and g-factors, the 90th percentiles were also calculated.
Paired two-tailed t-tests were performed to evaluate the existance of significant differences between the SB and MB pCASL imaging methods. Significance was achieved with p values less than or equal to 0.05.
3. Results
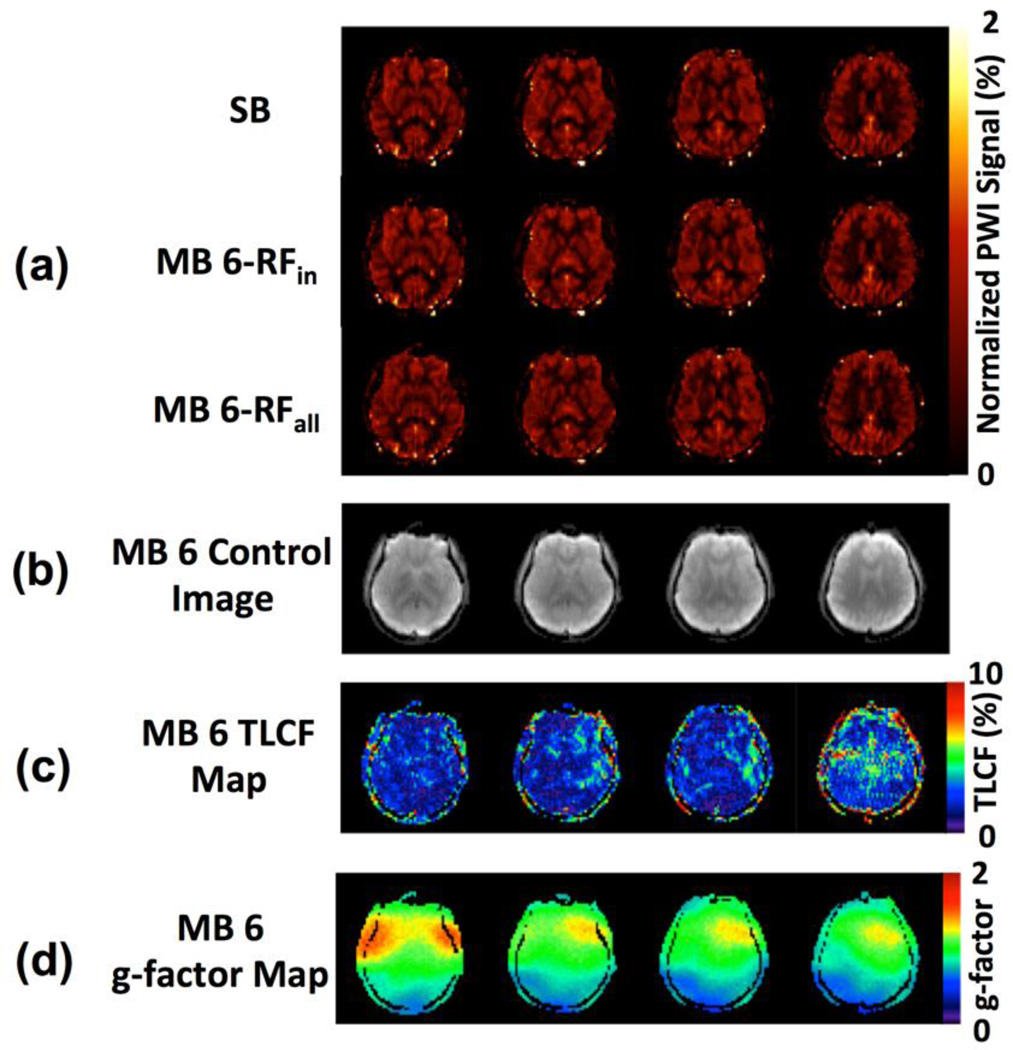
Results from comparison study 1 are summarized in Tables 1 and 2. Table 1 shows the CBF estimations and CBF differences relative to the first SB measurement (SB1). Table 2 shows the perfusion SNRs and the estimated g-factors and TLFs. One subject’s images from a pCASL study using SB (SB1, the first SB-EPI pCASL imaging measurement) and MB with MB factor 6 are shown in Figure 2. These data suggest that the measured perfusion signals are very similar and thus independent of the acquisition strategy (i.e. SB or MB with different reduction factors) when matched PLD and other imaging parameters are used (Figure 2a). With respect to the signal contamination resulting from the anti-aliasing of slices in MB, the spatial distribution of TLF varies both between and within slices as observed in Figure 2c. Figure 2d indicates that the estimated g-factors vary spatially. TLF and g-factor maps for other MB factors from the same subject are provided in the inline supplementary material, Figure A.1.
Table 1.
CBF analysis results for pCASL imaging using SB- and MB-EPI within matched imaging slices in the middle or middle superior brain regions. Results are presented as mean ± S.D. from five healthy voluteers.*
| MB Factor |
Mean GM CBF (mL/100 g/min) | ΔCBF (%) Relative to CBF by SB1 | |||||
|---|---|---|---|---|---|---|---|
| MB-RFall | MB-RFin | SB1 | SB2 | MB-RFall | MB-RFin | SB2 | |
| 3 | 67.9 ± 7.2 | 68.1 ± 6.1 | 67.7 ± 6.3 | 66.8 ± 4.6 | 0.3 ± 1.4 | 0.6 ± 0.8 | 1.2 ± 2.3 |
| 4 | 72.0 ± 6.8 | 71.8 ± 5.7 | 71.6 ± 6.8 | 71.6 ± 6.4 | 0.6 ± 0.9 | 0.4 ± 1.4 | −0.1 ± 2.6 |
| 5 | 65.2 ± 3.8 | 65.6 ± 2.5 | 65.0 ± 3.5 | 64.6 ± 4.4 | 0.4 ± 2.3 | 0.9 ± 1.8 | 0.7 ± 1.7 |
| 6 | 66.5 ± 5.7 | 65.5 ± 5.4 | 66.0 ± 5.3 | 65.4 ± 4.9 | 0.7 ± 2.2 | −0.7 ± 1.1 | 0.9 ± 1.8 |
CBF represents cerebral blood flow, GM grey matter, ΔCBF CBF difference, SB single-band, MB multi-band, S.D. standard deviation, MB-RFall indicates imaging results of MB-EPI with all MB RF pulse components on, MB-RFin with only turning on the RF pulse component exciting slices spatially matching those of SB-EPI in the middle or middle superior brain regions, SB1 and SB2 the first and second SB-EPI imaging scans.
Table 2.
Measured perfusion signal-to-noise ratio (SNR), g-factor and total leakage factor (TLF) for pCASL imaging using SB- and MB-EPI within matched imaging slices in the middle or middle superior brain regions. Results are presented as mean ± S.D. from five healthy voluteers.*
| MB Factor |
SNRMB/SNRSB1 | g-factor | TLF (%) | |||
|---|---|---|---|---|---|---|
| MB-RFall | MB-RFin | Mean | 90 percentile | Mean | 90 Percentile | |
| 3 | 0.95 ± 0.15 | 0.92 ± 0.13 | 1.12 ± 0.05 | 1.34 ± 0.06 | 1.3 ± 0.1 | 2.2 ± 0.2 |
| 4 | 0.89 ± 0.16 | 0.85 ± 0.10 | 1.15 ± 0.08 | 1.37 ± 0.10 | 1.6 ± 0.4 | 2.8 ± 0.4 |
| 5 | 0.86 ± 0.14 | 0.83 ± 0.15 | 1.18 ± 0.10 | 1.41 ± 0.07 | 2.8 ± 0.5 | 4.7 ± 0.6 |
| 6 | 0.82 ± 0.12 | 0.80 ± 0.14 | 1.22 ± 0.09 | 1.55 ± 0.06 | 3.6 ± 0.2 | 5.8 ± 0.2 |
SB represents single-band, MB multi-band, S.D. standard deviation, MB-RFall indicates imaging results of MB-EPI with all MB RF pulse components on, MB-RFin with only turning on the RF pulse component exciting slices spatially matching those of SB-EPI in the middle or middle superior brain region, and SB1 the first SB-EPI imaging scan.
Figure 2.
Images from a representative subject in comparison study 1 using SB- and MB-EPI with an acceleration factor 6: (a) normalized perfusion-weighed images (PWI) in the middle superior brain region by SB-EPI (top), MB-RFin (middle) and MB-RFall (bottom); (b) control images acquired in the MB-RFall acquisition; (c) total leakage factor (TLF) map, and (d) g-factor map. MB-RFin refers to acquisition when only turning on the RF pulse component exciting slices which match the SB-EPI acquisition, and MB-RFall refers to standard without turning off any MB-EPI excitation RF pulse components. The presented PWIs were normalized PWIs by using mean control images as the reference.
For all applied MB factors, paired two-tailed t-tests indicated no significant differences between MB (MB-RFall or MB-RFin) and SB (SB1 or SB2) with respect to CBF estimates. The differences between MB and SB with respect to CBF were also comparable (Table 1). Perfusion SNRs for the MB methods were consistently and significantly lower than those for SB, and TLFs increase consistently and significantly with the increase of MB factor (Table 2). The highest group mean and 90th percentile of TLFs for MB factor 6 were: 3.6 ± 0.2% and 5.8 ± 0.2%, respectively.
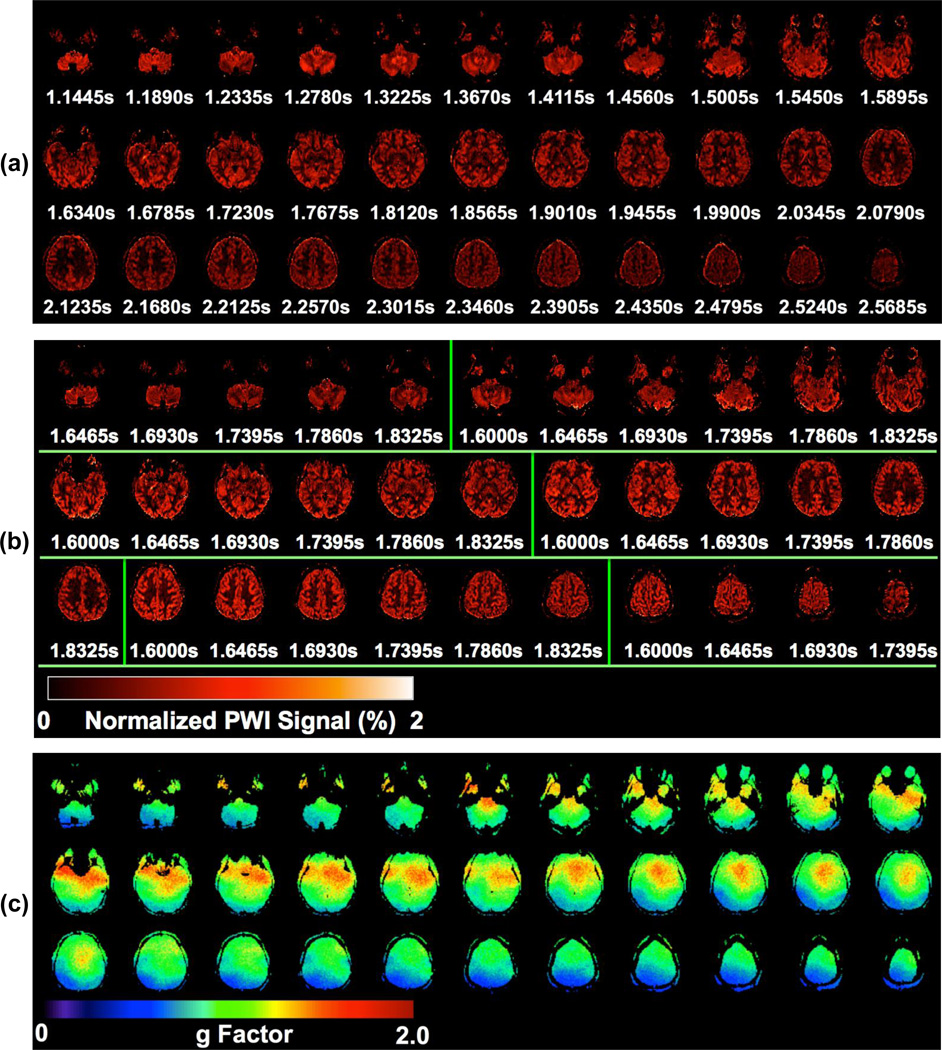
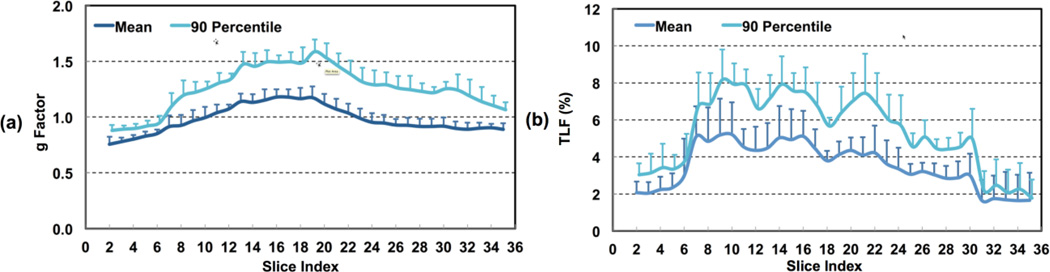
Figures 3 and 4 show a typical subject’s data from comparison study 2, including PWIs, g-factor maps, mean control images from MB-RFin and MB-RFout acqusitions and TLF maps. CBF maps of SB and MB6 from the same subject are presented in the online supplementary material, Figure A.2. G-factor slice-wise means and 90th percentiles along with TLFs averaged across all subjects are provided in Figure 5. The group mean g-factors varied across slices with values less than 1.0 in the inferior and superior brain regions and larger than 1.0 in the middle part of the brain.
Figure 3.
Images of a representative subject from comparison study 2 where SB- and MB-EPI with an acceleration factor 6 were evaluated for high-resolution whole brain pCASL imaging: normalized perfusion-weighted images (PWIs) for (a) SB and (b) MB, and (c) g-factor maps for MB6. Results are presented for 33 out of total 36 imaging slices. The 33 PWIs for MB6 shown in (b) are divided into 6 slice groups representing the MB slice bands by using overlaid vertical green lines. The presented PWIs were normalized PWIs by using mean control images as the reference. Under each PWI in (a) and (b), the effective PLD (in second) is displayed.
Figure 4.
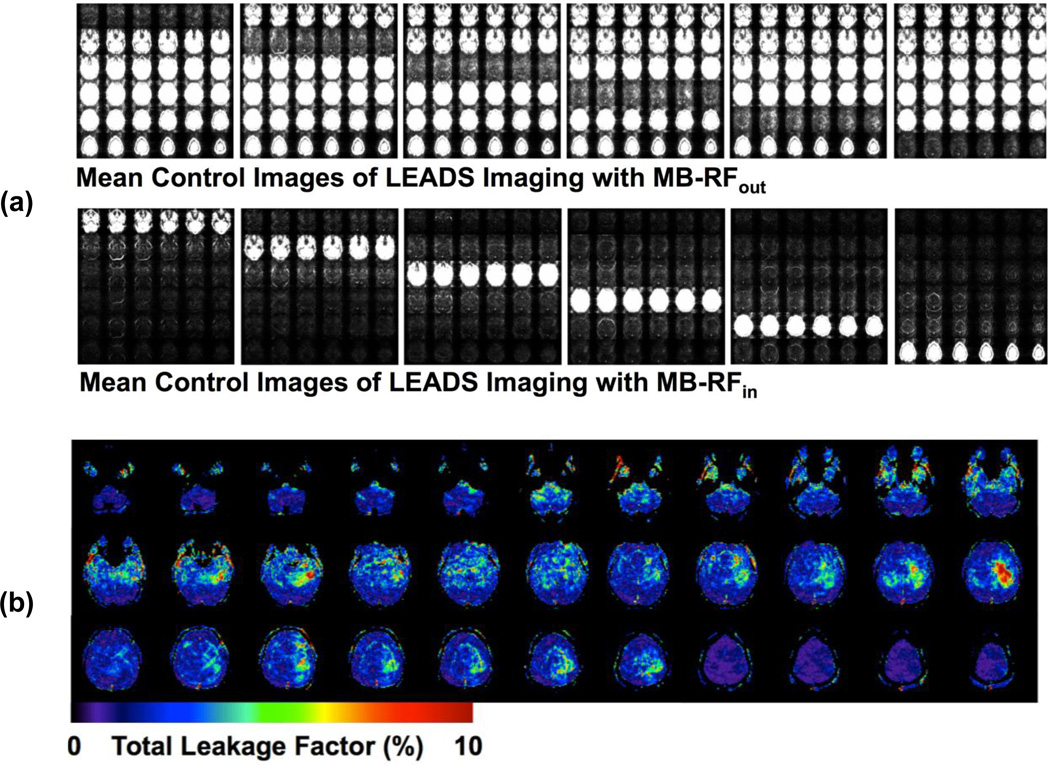
One subject’s imaging results of leakage evaluation studies using LEADS method from comparison study 2: (a) mean control images of the pCASL acquisition and (b) total leakage factor (TLF) maps for MB6.
Figure 5.
Slice-wise analysis results of g-factor (a) and total leakage factor (TLF) for high-resolution whole brain pCASL imaging using MB-EPI with MB acceleration factor 6 from five healthy volunteers. Error bars represent standard errors.
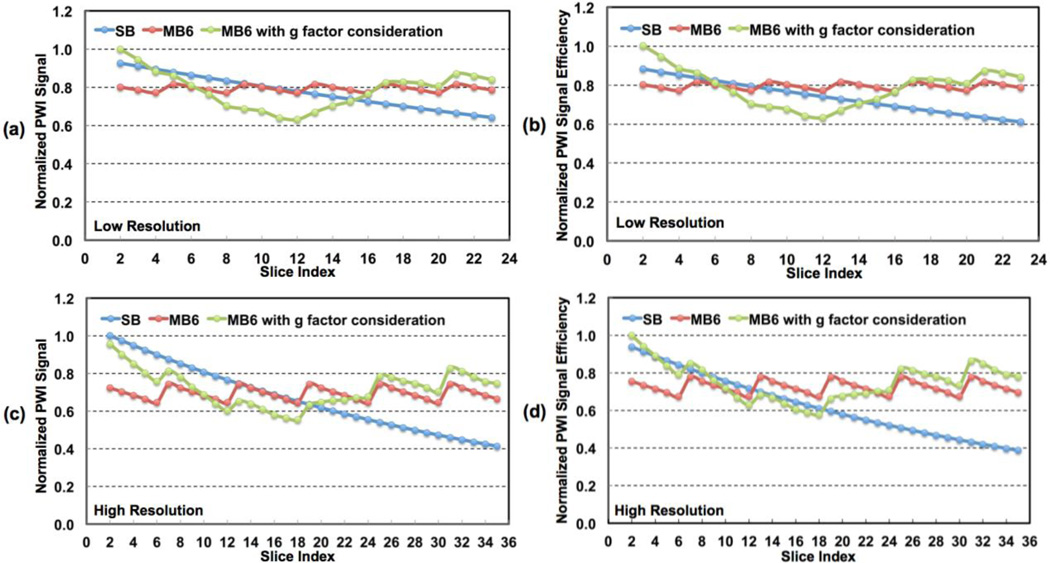
The group means of estimated 90th percentile g-factors have similar across-slice patterns but higher estimated values, and can be as high as about 1.6 in the middle part of the brain. Similar to the observations in comparison study 1, TLF varied across space with higher levels in the inferior middle part of the brain and lower levels in the inferior and superior brain regions. The group mean 90th percentile TLF was as high as 8%. Simulation results of normalized PWI signals and PWI signal efficiencies for both low- and high-resolution studies, assuming a uniform CBF across all slices, are presented in Figure 6. Slice dependent values are shown for SB and MB6 with and without the inclusion of experimentally obtained slice dependent g-factors. The SB acquisition shows monotonically decreasing PWI signals and signal efficiencies with increasing slice number as expected due to longitudinal relaxation of the labeled blood. The simulations for MB pCASL imaging shows a saw-tooth patterns oscillating about a constant mean PWI value due to the fact that labeled blood decays only over a period 1/MB-factor times of the SB acquisition and across the slices of a single band not the slices across the entire head. The 9th and 13th slices of SB and MB6 possess the same effective PLDs for low- and high-resolution imaging studies, respectively, and therefore the same perfusion
Figure 6.
Theoretical simulation results for perfusion-weighted image (PWI) signals and signal efficiencies for (a, b) low- and (c, d) high-resolution whole brain perfusion imaging using pCASL. Simulations were performed with assumed uniform CBF across all slices. SB and MB6 represent SB and MB with MB factor 6, respectively. The simulation results of MB6 including g-factor penalty reflect perfusion SNRs and SNR efficiencies by MB relative to those of SB.
SNRs with the chosen MRI parameters: nominal 1.36 s and 1.6 s PLDs with 29 ms and 30 ms slice acquisition times for SB and MB in low-resolution imaging, and nominal 1.1 s and 1.6 s PLDs with 44.5 ms and 46.5 ms slice acquisition times for SB and MB in high-resolution imaging, respectively. Simulation results using the smoothed experimentally determined slice dependent CBF values (Figure 7a) are presented in Figure 7c. When using the experimentally determined slice-dependent CBF values, the results for both the SB and MB simulations (Figure 7c) are similar to those achieved experimentally (Figure 7d).
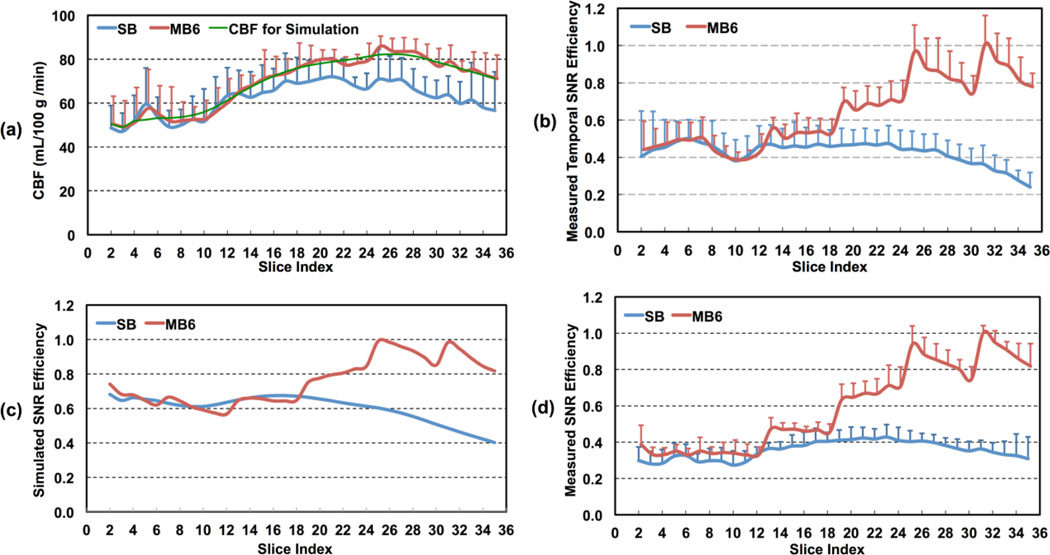
Figure 7.
Slice-wise analysis results for high-resolution whole brain pCASL imaging using SB- and MB-EPI with MB acceleration factor 6 from five healthy subjects: a) CBF measurements using SB- and MB-EPI, and smoothed CBF values used for theoretical simulation; b) estimated temporal perfusion SNR efficiencies; c) spatial perfusion SNR efficiencies from theoretical simulation; and d) measured spatial perfusion SNR efficiencies. Error bars represent standard errors.
Slice-wise CBF estimations from SB and MB, smoothed CBF values used for theoretical simulation, and measured SB and MB spatial and temporal SNR efficiencies from comparison study 2 are presented in Figure 7. Spatial and temporal SNR efficiencies were normalized to the highest achieved in the MB acquisition. The perfusion SNR and temporal SNR efficiencies in Figures 7b and 7d are shown to greatly increase in the superior slices of the brain for MB.
4. Discussion
Although MB-EPI has been recently demonstrated (Kim et al., 2013) and compared to single-shot 3D GRASE for brain perfusion imaging using FAIR (Feinberg et al., 2013), we explored the potential benefit of MB-EPI in high-resolution whole brain pCASL imaging for the first time. We have demonstrated the impact of MB on ASL imaging both theoretically and experimentally, taking into account both the beneficial characteristics (improved imaging efficiency) and confounding effects (amplified thermal noise and leakage contamination).
4.1 TLF and LEADS
To characterize the signal leakage contamination resulting from imperfect slice un-aliasing during MB reconstruction, the leakage factor (LF), was originally proposed (Xu et al., 2013). The LF provides an estimate of the residual aliasing from one slice into other simultaneously acquired slices. To determine LF, different methods have been suggested and demonstrated based on a single, representative SB acquisition followed by simulation either using a Frequency Modulation/Monte Carlo (FMMC) (Moeller et al., 2012) method or a Linear System Leakage Approach (LSLA) (Cauley et al., 2013). However, the LF does not directly reflect the impact of the total leakage contamination from all other simultaneously acquired imaging slices on a single slice of interest, and therefore, TLF, as a more meaningful metric, was proposed to better faciliate the understanding of the impact of the signal contamination versus the desired signal. The TLF can be estimated using previous methods (Moeller et al., 2012, Cauley et al., 2013). However, calculating TLF from a single non-MB acquisition by summing together LF estimates is complicated by the complex nature of the signals as well as the absence of MB RF pulses. MB RF pulses introduce additional effects (e.g. MB RF side-lobe effects) that can be only realized through LEADS. In addition, TLF can be simply quantified through LEADS using the existing online reconstruction system.
In the evaluation of TLF, the reference for deriving the fractional contamination described in Equation 1 should ideally be SB images acquired with the same acquisition parameters as the MB study since SB images do not contain spatially-varying amplified thermal noise. However, our study indicates that the difference between TLFs calculated with MB-RFin and SB as the reference was quite small (much less than 1%). Therefore, images from MB-RFin acquisition were used as the reference for TLF evaluation. In addition, to avoid potential influence of temporal perfusion variability on TLF evaluation, the control images, instead of the label images, from the pCASL imaging series were used. Furthermore, to accommodate the potential effects of subject bulk motion across time, the temporal mean control images were used in TLF evaluation (Equation 2).
4.2 Confounding effects of MB-EPI
As demonstrated in the current study, particularly in comparison study 1 (Table 2), MB introduces confounding effects, which can have a negative impact on perfusion imaging quality. First, MB suffers from amplified thermal noise due to coil-sensitivity-based slice-GRAPPA reconstruction, as reflected by the spatially varying g-factor maps. This effect was found to increase with MB factor, and could result in lower perfusion SNR. Second, MB-EPI introduces leakage contamination resulting from the imperfect anti-aliasing reconstruction, as evaluated by TLF, which also increases with MB factor.
The spatially varying g-factors shown in Figure 3c highlight the fact that the amplified thermal noise is also spatially varying throughout the volume. At first, even within a single imaging slice, there exist areas with g-factors less than 1.0, indicating increased perfusion SNR, and areas with g-factors larger than 1.0, suggesting decreased perfusion SNR. Across imaging slices from the inferior to superior brain regions, the mean g-factors were non-uniform as well, giving g-factors less than 1.0 in the inferior and superior imaging slices. The observed g-factors less than 1.0 in the inferior and superior brain regions is mainly due to the calculation of the weighting factors for fitting missing lines in k-space on a least-squares basis in MB reconstruction (Robson et al., 2008). Therefore, with SB as the reference, MB can provide either an increase or decrease in perfusion SNR. Importantly, as observed in comparison study 1 (Table 1), the comparable CBF measurements between MB-RFin and SB suggest that the MB reconstruction itself has little effect on CBF estimations.
Similar to g-factor, the estimated TLFs are also spatially varying (Figures 2c and 4b). Particularly, as indicated by the right TLF image on the second row in Figure 4b, due to the imperfection of MB anti-aliasing reconstruction, high total leakage contamination can occur in local brain regions. Excessive subject motion can induce the fluctuation of leakage signal within these areas, reducing perfusion temporal SNR. To avoid such an adverse effect, the control of subject motion will become more important for perfusion imaging using MB-EPI. However, such effects were found statistically insignificant with respect to CBF estimates. Although MB-RFall CBF measurements were consistently higher than SB across all MB factors, no significant differences were found between SB and MB, and CBF differences between MB-RFall and SB scan 1 were all less than 1% (Table 1). Furthermore, CBF differences between MB-RFall and SB scan 1 were very comparable to those between SB scan 1 and SB scan 2 (Table 1). Therefore, results indicate that with similar pCASL acquisition parameters (i.e. matched PLDs in comparison study 1), there is no statistical difference between MB and SB perfusion measurements.
One result of comparison study 1 worth noting is that CBF differences between MB and SB were much smaller than estimated TLFs (Tables 1 and 2); CBF differences between MB-RFall and SB scan 1 were all less than 1%, while TLF were above 1%. This may be because the leakage contamination contains a significant amount of signals from the skull and subcutaneous fat as a result of EPI ghosting and the hyperintense cerebrospinal fluid (CSF). The paired subtraction nature of ASL calculation tends to remove these contaminating non-perfusion signals. The analysis of mean perfusion-weighted images acquired using MB-RFin with 30 averages further confirmed that the leakage contamination in the difference images was negligible (data not shown). Using imaging acquisition methods better immune to the ghost artifacts (Wang et al., 2013) for acquiring the MB calibration scans may improve MB reconstruction by reducing leakage contamination resulting from EPI ghosts. Applying background suppression techniques (Ye et al., 2000, Garcia et al., 2005) will also help to reduce leakage contamination originating from hyperintense CSF signals. On the other hand, these results suggest that even assuming the artificial perfusion signal originating from leakage contamination has the same level as measured TLFs (the worst case scenario), the contributions of leakage contamination will be less than 6% of total measured perfusion signals.
4.3 PLD for MB-EPI
In contrast to the ascending slice acquisition order used for SB, the spatially interleaved slice acquisition characteristics of MB requires a proper selection of PLD to avoid potential intravascular artifacts in brain regions where long arterial transit times exist. Recently, whole brain ASL imaging studies with normal healthy volunteers have demonstrated that arterial transit times do not monotonically increase from the inferior to superior brain regions and that the visual cortex has the longest arterial transit time (Gunther et al., 2005, Dai et al., 2013). Such results provided a basis for proper selection of PLD for the whole brain ASL studies with either 3D methods or MB-EPI. In the current studies, conservatively long PLDs equal to 1.1 s and 1.6 s were used for the high-resolution whole brain SB and MB pCASL acquisitions, respectively. These PLDs were chosen based on the consideration of slice acquisition times and orders, and successfully avoided intravascular artifacts as confirmed by study results (Figures 2a and 3a). For SB, 1.1 s was used for PLD to avoid intravascular artifacts observed in the inferior middle brain regions (data not shown). It is worth noting that these selected PLDs can only be appropriate for pCASL imaging with healthy volunteers, and properly selected longer PLDs should be used for the applications with patient populations (Alsop et al., 2014).
Due to the interleaved slice acquisition order of MB, there exist obvious perfusion signal oscillations across the slice groups (Figure 3b). Such perfusion signal oscillations are also reflected in slice-wise perfusion SNR efficiencies from both theoretical simulation and experimental results (Figures 6 and 7b–7d), but not observed in measured slice-wise CBF values (Figure 7a). The resolved oscillations of the perfusion signal in the CBF estimates indicate that the longitudinal decay of labeled blood can be properly corrected by the single-blood compartment model for the range of delay times experienced within each band (i.e. 1.6–1.9 s) and thus do not affect CBF quantification.
4.4 CBF quantification model and MB-EPI
By using the single-blood compartment model for CBF quantification, the estimated CBF values by MB were comparable to those by SB in the inferior brain region, but higher than those by SB in the middle and superior brain regions. This observed discrepancy in CBF quantification most likely is a result of the imperfect single-blood compartment model and not a result of leakage contamination or intravascular artifacts.
The single-blood compartment model assumes that the perfusion signals decay at the longitudinal relaxation rate of arterial blood, and that most of the labeled blood water spins remain in the vascular space before perfusion signal acquisition. However, due to the greatly prolonged imaging acquisition times required for SB in the high-resolution studies, labeled blood spins have more chance to exchange extravascularly into the brain parenchyma, thus their signals will decay at the shorter longitudinal relaxation rate of brain tissue. With increasing signal decay occurring in the brain tissue, poorer perfusion SNR results while the correction assuming a single longitudinal relaxation time becomes less reliable. The end result is increasingly underestimated brain perfusion in late acquired imaging slices, which are those located most superiorly. On the contrary, compared to SB, although longer initial PLD of 1.6 s was used, the effective PLDs for the superior imaging slices are greatly reduced due to dramatically shortened total imaging acquisition time for MB (about 280 ms for MB versus about 1.6 s for SB). For MB, fewer spins exit the vasculature during the effective PLDs, resulting in a decrease in perfusion signal decay and an increase in the resulting CBF estimation. Therefore, it is reasonable to postulate that MB can better satisfy the assumption of the single-blood compartment model, and should provide more reliable CBF quantification in high-resolution whole brain imaging. While further validation may be required, support for this statement can be found in the results presented in this study
As indicated by the results of comparison study 1, SB and MB provided very comparable CBF estimations within matched imaging slices when using the same MRI acquisition parameters (Table 1). In additional studies, these results were confirmed to hold for the high-resolution acquisitions (data not shown). The higher CBF estimations for superior imaging slices in MB is not a result of intravascular signals as a PLD of 1.6 s was used, which avoided such artifacts as demonstrated in the presented PWI maps (Figure 3a). It is also clear that the distribution of measured TLFs across slices (Figure 5b) is not consistent with the observed CBF differences between SB and MB (Figure 7a). Furthermore, the observed increases of perfusion SNR efficiency for MB-EPI pCASL imaging by both theoretical simulation and experimental results are not correlated with the measured TLFs across slices. These results further support the claim that leakage contamination is not the reason for the observed discrepancy between SB and MB CBF quantification.
4.5 Benefits of MB-EPI for pCASL imaging
The major obstacle to obtaining high-resolution whole brain ASL imaging is insufficient perfusion SNR. In addition, perfusion SNR efficiency has been presented as an ideal metric to evaluate different ASL imaging techniques and imaging readouts (Wong et al., 1998). Therefore, the benefit of MB-EPI for pCASL imaging was evaluated based on a comparison of perfusion SNR efficiency with SB. This comparison was ensured to be meaningful by using MRI parameters optimal for each imaging approach in comparison study 2.
To explore the potential benefits of MB for high-resolution whole brain ASL imaging, theoretical simulations were performed with an assumed uniform perfusion across the brain (Figure 6). Based on the argument that MB ASL may better support the single-blood compartment model (see the section CBF quantification model and MB-EPI), the theoretical simulations were also performed with smoothed, slice-wise, CBF estimations from the MB6 studies in comparison study 2 (Figure 7c).
Simulation results indicated that MB can greatly benefit high-resolution but not typically acquired low-resolution whole brain pCASL imaging in terms of perfusion SNR efficiency, especially in the middle part of the brain where thermal noise level was elevated (Figure 6). The reduction of total imaging time for low-resolution is only about 400 ms, resulting in a 5.6% efficiency gain for MB relative to SB, while for high-resolution imaging, total imaging time of MB is about 800 ms shorter than that of SB, giving an 11.1% efficiency gain. In addition, the prolonged effective PLD in SB due to increased slice acquisition times in high-resolution imaging causes larger perfusion signal decay than in low-resolution imaging. This loss in signal is most pronounced in the superior slices. The combination of these effects, increased efficiency in slice acquisition and more consistent signal decay due to narrower range of effective PLDs across the brain, enables MB to achieve perfusion SNR efficiency comparable to SB in the middle part of the brain for high-resolution studies. Due to the reduction of g-factors (less than 1.0), together with an increase in imaging efficiency, MB was able to provide similar SNR efficiency in the inferior brain regions. In the most superior regions of the brain, however, MB provides much higher perfusion SNR efficiency for both low-resolution and high-resolution imaging.
Such a benefit of MB for high-resolution whole brain perfusion imaging was corroborated by the results of the comparison study 2 (Figures 7b and 7d). The measured spatial perfusion SNR efficiency agrees with those from theoretical simulation using smoothed, slice-wise CBF estimations by MB (Figure 7c). Without considering time efficiency, measured perfusion SNRs of MB were lower than, comparable to and higher than those of SB in the inferior, middle and superior brain regions, respectively (data not shown), which were similar to those from theoretical simulation.
Although the distribution of g-factor within imaging slices is not uniform and there exist subject-dependent variation for estimated slice-wise g-factors, such a simulation can provide valuable insights into how MB can benefit high-resolution whole brain pCASL imaging in terms of perfusion SNR efficiency. For the resolution (2.5 × 2.5 × 3.0 mm3) and the number of imaging slices (36 slices) used for the current high-resolution whole brain study, the reduction in imaging acquisition time by applying higher MB factor (e.g. 7 or 8) will be quite small. For example, when using MB 7 for 35 slices or MB 8 for 40 slices (the number of imaging slices has to be integer times of MB factor for MB imaging), the imaging acquisition time will be only about 46.5 ms shorter than that of current study using MB 6 for 36 slices. This indicates that the increases of spatial and temporal SNR efficiencies due to reduced imaging acquisition time by applying higher MB factors for the current imaging resolution will be small even when the level of thermal noise is assumed the same as that of MB 6. The benefits of MB-EPI for whole brain perfusion imaging with higher in-plane resolution, thinner imaging slices and larger MB factors can be expected, but should be further validated by experiments since both thermal noise amplification and TLF depend on imaging resolution and MB factor.
Compared to SB, MB provided comparable temporal perfusion SNR efficiency in the inferior brain region and higher in the middle and superior parts of the brain for high-resolution whole brain pCASL imaging (Figure 7b). It is expected that temporal SNR efficiency can be further increased for MB when background suppression methods are applied (Ye et al., 2000, Garcia et al., 2005). It is worth noting that with the tremendous reduction in total imaging slice acquisition time, MB becomes much more amenable to background suppression techniques, and therefore is superior to SB-EPI in this respect. The benefits and impact of MB to the application of background suppression and increasing spatial resolutions in pCASL studies of the brain are under investigation.
5. Conclusions
Compared to SB-EPI, MB-EPI can improve high-resolution whole brain pCASL perfusion imaging by providing improved spatial and temporal perfusion SNR efficiencies for late acquired imaging slices, and provide data which is more amenable to the assumptions implicit in the single-blood compartment model for CBF quantification.
Supplementary Material
Highlights.
MB leakage contamination has minimal impact on CBF quantification in pCASL imaging
MB improves SNR efficiency for high-resolution pCASL imaging using 2D EPI
MB-EPI better supports the single-blood compartment model for CBF quantitation
Acknowledgments
This work was supported by grants P41 RR008079, P41 EB015894, P30 NS076408 and Human Connectome Project (1U54 MH091657) from the 16 Institutes and Centers of the National Institutes of Health that support the NIH Blueprint for Neuroscience Research. Research reported in this publication was also supported by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ASL
Arterial Spin Labeling
- CBF
Cerebral Blood Flow
- EPI
Echo Planar Imaging
- FA
Flip Angle
- FAIR
Flow-sensitive Alternating Inversion Recovery
- FOV
Field Of View
- GRAPPA
GeneRalized Autocalibrating Partially Parallel Acquisitions
- GRE
Gradient Recalled Echo
- LEADS
Lead Evaluation via Acquired Dummy Slices
- MB
Multi-Band
- MPRAGE
Magnetization Prepared Rapid Acquisition Gradient Echo
- MR
Magnetic Resonance
- MRI
Magnetic Resonance Imaging
- PASL
Pulsed Arterial Spin Labeling
- pCASL
Pseduo-Continuous Arterial Spin Labeling
- ROI
Region Of Interest
- SB
Single-Band
- S.D.
Standard Deviation
- TE
Echo Time
- TLF
Total Leakage Factor
- TR
Repetition Time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alsop DC, Detre JA. Multisection cerebral blood flow MR imaging with continuous arterial spin labeling. Radiology. 1998;208:410–416. doi: 10.1148/radiology.208.2.9680569. [DOI] [PubMed] [Google Scholar]
- Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, Macintosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC, Zaharchuk G. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2014 doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokkers RP, Hernandez DA, Merino JG, Mirasol RV, van Osch MJ, Hendrikse J, Warach S, Latour LL, National Institutes of Health Stroke Natural History I. Whole-brain arterial spin labeling perfusion MRI in patients with acute stroke. Stroke; a journal of cerebral circulation. 2012;43:1290–1294. doi: 10.1161/STROKEAHA.110.589234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauley SF, Polimeni JR, Bhat H, Wald LL, Setsompop K. Interslice leakage artifact reduction technique for simultaneous multislice acquisitions. Magn Reson Med. 2013 doi: 10.1002/mrm.24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med. 2008;60:1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Shankaranarayanan A, Alsop DC. Volumetric measurement of perfusion and arterial transit delay using hadamard encoded continuous arterial spin labeling. Magn Reson Med. 2013;69:1014–1022. doi: 10.1002/mrm.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detre JA, Alsop DC, Vives LR, Maccotta L, Teener JW, Raps EC. Noninvasive MRI evaluation of cerebral blood flow in cerebrovascular disease. Neurology. 1998;50:633–641. doi: 10.1212/wnl.50.3.633. [DOI] [PubMed] [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- Detre JA, Wang J. Technical aspects and utility of fMRI using BOLD and ASL. Clin Neurophysiol. 2002;113:621–634. doi: 10.1016/s1388-2457(02)00038-x. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Beckett A, Chen L. Arterial spin labeling with simultaneous multi-slice echo planar imaging. Magn Reson Med. 2013;70:1500–1506. doi: 10.1002/mrm.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PloS one. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai ND, Talagala SL, Butman JA. Whole-brain cerebral blood flow mapping using 3D echo planar imaging and pulsed arterial tagging. J Magn Reson Imaging. 2011;33:287–295. doi: 10.1002/jmri.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Duhamel G, Alsop DC. Efficiency of inversion pulses for background suppressed arterial spin labeling. Magn Reson Med. 2005;54:366–372. doi: 10.1002/mrm.20556. [DOI] [PubMed] [Google Scholar]
- Goelman G. Two methods for peak RF power minimization of multiple inversion-band pulses. Magn Reson Med. 1997;37:658–665. doi: 10.1002/mrm.1910370506. [DOI] [PubMed] [Google Scholar]
- Gunther M, Oshio K, Feinberg DA. Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med. 2005;54:491–498. doi: 10.1002/mrm.20580. [DOI] [PubMed] [Google Scholar]
- Kim SG, Tsekos NV. Perfusion imaging by a flow-sensitive alternating inversion recovery (FAIR) technique: application to functional brain imaging. Magn Reson Med. 1997;37:425–435. doi: 10.1002/mrm.1910370321. [DOI] [PubMed] [Google Scholar]
- Kim T, Shin W, Zhao T, Beall EB, Lowe MJ, Bae KT. Whole brain perfusion measurements using arterial spin labeling with multiband acquisition. Magn Reson Med. 2013;70:1653–1661. doi: 10.1002/mrm.24880. [DOI] [PubMed] [Google Scholar]
- Koopmans PJ, Boyacioglu R, Barth M, Norris DG. Whole brain, high resolution spin-echo resting state fMRI using PINS multiplexing at 7 T. NeuroImage. 2012;62:1939–1946. doi: 10.1016/j.neuroimage.2012.05.080. [DOI] [PubMed] [Google Scholar]
- Li X, Sarkar SN, Purdy DE, Spence JS, Haley RW, Briggs RW. Anteroposterior perfusion heterogeneity in human hippocampus measured by arterial spin labeling MRI. NMR in biomedicine. 2013;26:613–621. doi: 10.1002/nbm.2898. [DOI] [PubMed] [Google Scholar]
- Li X, Spence JS, Buhner DM, Hart J, Jr, Cullum CM, Biggs MM, Hester AL, Odegard TN, Carmack PS, Briggs RW, Haley RW. Hippocampal dysfunction in Gulf War veterans: investigation with ASL perfusion MR imaging and physostigmine challenge. Radiology. 2011;261:218–225. doi: 10.1148/radiol.11101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Clingman C, Golay X, van Zijl PC. Determining the longitudinal relaxation time (T1) of blood at 3.0 Tesla. Magn Reson Med. 2004;52:679–682. doi: 10.1002/mrm.20178. [DOI] [PubMed] [Google Scholar]
- Moeller S, Xu J, Auerbach EJ, Yacoub E, Ugurbil K. Signal Leakage(L-Factor) as a Measure for Parallel Imaging Performance Among Simultaneously Multi-Slice (SMS) Excited and Acquired Signals. Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2012. 2012:Abstract 0519. [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JF, Hernandez-Garcia L. Functional perfusion imaging using pseudocontinuous arterial spin labeling with low-flip-angle segmented 3D spiral readouts. Magn Reson Med. 2013;69:382–390. doi: 10.1002/mrm.24261. [DOI] [PubMed] [Google Scholar]
- Roberts DA, Rizi R, Lenkinski RE, Leigh JS., Jr Magnetic resonance imaging of the brain: blood partition coefficient for water: application to spin-tagging measurement of perfusion. J Magn Reson Imaging. 1996;6:363–366. doi: 10.1002/jmri.1880060217. [DOI] [PubMed] [Google Scholar]
- Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med. 2008;60:895–907. doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Cohen-Adad J, Gagoski BA, Raij T, Yendiki A, Keil B, Wedeen VJ, Wald LL. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. NeuroImage. 2012a;63:569–580. doi: 10.1016/j.neuroimage.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med. 2012b;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Hoge WS, Hamilton CA, Gunther M, Kraft RA. 3D GRASE PROPELLER: improved image acquisition technique for arterial spin labeling perfusion imaging. Magn Reson Med. 2011;66:168–173. doi: 10.1002/mrm.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E Consortium WU-MH. The Human Connectome Project: a data acquisition perspective. NeuroImage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Vu J, Yacoub ES, Ugurbil K, Deshpande V. High Resolution T2-Weighted Imaging with Whole Brain Coverage at 7 Tesla Using Multiband Slice Accelerated Spin Echo. Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2013. 2012:Abstract 2677. [Google Scholar]
- Wang J, Alsop DC, Song HK, Maldjian JA, Tang K, Salvucci AE, Detre JA. Arterial transit time imaging with flow encoding arterial spin tagging (FEAST) Magn Reson Med. 2003;50:599–607. doi: 10.1002/mrm.10559. [DOI] [PubMed] [Google Scholar]
- Wong E. Optimized Phase Schedules for Minimizing Peak RF Power in Simultaneous Multi-Slice RF Excitation Pulses. Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2012. 2012:Abstract 2209. [Google Scholar]
- Wong EC. Quantifying CBF with pulsed ASL: technical and pulse sequence factors. J Magn Reson Imaging. 2005;22:727–731. doi: 10.1002/jmri.20459. [DOI] [PubMed] [Google Scholar]
- Wong EC, Buxton RB, Frank LR. A theoretical and experimental comparison of continuous and pulsed arterial spin labeling techniques for quantitative perfusion imaging. Magn Reson Med. 1998;40:348–355. doi: 10.1002/mrm.1910400303. [DOI] [PubMed] [Google Scholar]
- Wu WC, Fernandez-Seara M, Detre JA, Wehrli FW, Wang J. A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous arterial spin labeling. Magn Reson Med. 2007;58:1020–1027. doi: 10.1002/mrm.21403. [DOI] [PubMed] [Google Scholar]
- Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, Ugurbil K. Evaluation of slice accelerations using multiband echo planar imaging at 3 T. NeuroImage. 2013;83:991–1001. doi: 10.1016/j.neuroimage.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wen H, Mattay VS, Balaban RS, Frank JA, Duyn JH. Comparison of 3D BOLD functional MRI with spiral acquisition at 1.5 and 4.0 T. NeuroImage. 1999;9:446–451. doi: 10.1006/nimg.1998.0422. [DOI] [PubMed] [Google Scholar]
- Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magn Reson Med. 2000;44:92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.