Abstract
We have isolated cDNAs encoding PDE4A8 (phosphodiesterase 4 isoform A8), a new human cAMP-specific PDE4 isoform encoded by the PDE4A gene. PDE4A8 has a novel N-terminal region of 85 amino acids that differs from those of the related ‘long’ PDE4A4, PDE4A10 and PDE4A11 isoforms. The human PDE4A8 N-terminal region has diverged substantially from the corresponding isoforms in the rat and other mammals, consistent with rapid evolutionary change in this region of the protein. When expressed in COS-7 cells, PDE4A8 localized predominantly in the cytosol, but approx. 20% of the enzyme was associated with membrane fractions. Cytosolic PDE4A8 was exquisitely sensitive to inhibition by the prototypical PDE4 inhibitor rolipram (IC50 of 11 ± 1 nM compared with 1600 nM for PDE4A4), but was less sensitive to inhibition by cilomilast (IC50 of 101 ± 7 nM compared with 61 nM for PDE4A4). PDE4A8 mRNA was found to be expressed predominantly in skeletal muscle and brain, a pattern that differs from the tissue expression of other human PDE4 isoforms and also from that of rat PDE4A8. Immunohistochemical analysis showed that PDE4A8 could be detected in discrete regions of human brain, including the cerebellum, spinal cord and cerebral cortex. The unique tissue distribution of PDE4A8, combined with the evolutionary divergence of its N-terminus, suggest that this isoform may have a specific function in regulating cAMP levels in human skeletal muscle and brain.
Keywords: alternative mRNA splicing, cAMP, cilomilast, phosphodiesterase 4 isoform A (PDE4A), phosphoric diester hydrolyase, rolipram
INTRODUCTION
cAMP is the prototypical second messenger in mammalian cells and regulates numerous physiological processes, including glycogen storage in liver and muscle, smooth muscle tone, and many processes in the central nervous system, including learning, memory, mood and affect [1]. The cAMP-specific PDE4 (phosphodiesterase 4) enzymes, which are part of a larger family of cyclic nucleotide PDEs, hydrolyse cAMP and thus can regulate cAMP signalling pathways in cells [2–4]. Rolipram and other prototypical PDE4 inhibitors have cognitive-enhancing, anti-depressant, anti-inflammatory and smooth muscle relaxant properties in humans and other animals [2,3,5–9]. PDE4s can be distinguished from other PDE families by sequence identity in the catalytic region of the proteins [2,3,5,10] and by the presence of ‘signature’ regions of sequence called UCRs (upstream conserved regions) (UCR1 and UCR2), located in the N-terminal one-third of the proteins ([10,11], and reviewed in [5]).
Four different genes (PDE4A, PDE4B, PDE4C and PDE4D) encode the mammalian PDE4 isoforms, and each gene encodes multiple isoforms through the use of promoters unique to each isoform and alternative mRNA splicing (for reviews, see [2,3,5]). The various isoforms encoded by a single gene are divided into three groups: ‘long’ isoforms that contain both UCR1 and UCR2, ‘short’ isoforms that lack UCR1, but include UCR2, and ‘super-short’ isoforms that lack UCR1 and contain a truncated UCR2. The rat PDE4A5 (PDE4 isoform A5), PDE4A8, PDE4A10 and PDE4A11 isoforms are all long isoforms that vary from each other by containing unique blocks of amino acids at the N-terminal ends of their respective isoforms (Figure 1A) [12–15]. The rat PDE4A8 isoform is expressed almost exclusively in testis, in contrast with the rat PDE4A5, PDE4A10 and PDE4A11 isoforms, which are each expressed in a variety of tissues, including the brain and lung; however, each isoform has a distinct pattern of expression in these tissues [12–16]. The human forms of rat PDE4A5 (often called PDE4A4 in humans), PDE4A10 and PDE4A11 have been isolated and are strongly conserved between the two species, with the human and rat proteins being of near-identical length and having over 90% amino acid sequence identity [12,14,15]. The unique N-termini of these three human isoforms are strongly conserved when compared with their respective rat counterparts (PDE4A4 and PDE4A5 have 94 of 107 residues identical [12]; PDE4A10 has 39 of 46 residues identical [14], and PDE4A11 has 41 of 81 residues identical [15]). In addition, the super-short human [17] and rat [12,18] PDE4A1 isoforms are identical in the unique N-terminal regions, which are essential for their association with membranes [19–21].
Figure 1. Structure of PDE4A mRNAs and proteins.
(A) Schematic diagram of human PDE4A mRNA transcripts and their encoded proteins. The five dark stippled boxes indicate the unique N-terminal regions of the proteins encoded by the following cDNAs: PDE4A1 (GenBank® accession number U97584 [17]); PDE4A4 (GenBank® accession number L20965 [10]); PDE4A8 (GenBank® accession number AY593872; the present study); PDE4A10 (GenBank® accession number AF178570 [14]); PDE4A11 (GenBank® accession number AY618547 [15]). Also shown are regions of sequences that are highly conserved in other PDE4 isoforms, including the common long region, UCR1, UCR2 and the catalytic region. UCR1, UCR2 and the catalytic region are in turn separated by less conserved sequence regions, called LR (linker region) 1 and LR2. The C-terminal region is unique to PDE4A isoforms. The PKA phosphorylation site common to all long PDE4 isoforms is also indicated. (B) Alignment of the amino acid sequences of the N-terminal regions of the five human PDE4A proteins, as deduced from their cDNAs. Underlined regions indicate UCR1 and UCR2. Only the first 300 amino acids of the alignment are shown, as the remaining sequences of the four isoforms are all identical, as we and others have described previously [5]. (C) Alignment of the unique 5′-region of the human PDE4A8 cDNA with its corresponding genomic sequence (Genom; part of GenBank® accession number AC011548.4). Capital letters indicate regions of sequence identity. Dots indicate the 5′- and 3′-ends of exons. The physiological start codon is indicated in bold type. (D) Alignment of the nucleotide sequences of the unique 5′-regions of the human and rat PDE4A8 cDNAs. Vertical lines indicate sequence identities. Dots indicate gaps inserted by the program to improve the alignments. The physiological start codons and upstream in-frame stop codons are indicated in bold type. Also shown is the rat PDE4A8 amino acid sequence (GenBank® accession number L36467 [13]).
Given the generally strong sequence conservation of the mammalian PDE4 isoforms, we were intrigued when we isolated a cDNA clone for a novel human PDE4A isoform that has an N-terminal region with no amino acid similarity to its counterpart in other mammals. We now describe the cloning, sequence, expression and functional properties of this isoform, which we have called PDE4A8. Human PDE4A8 is a long PDE4A isoform with an N-terminal region (85 amino acids in length) that is distinct from the N-termini of human PDE4A5, PDE4A10 and PDE4A11. Although the nucleotide sequence of PDE4A8 has clear homology with that of rat PDE4A8, the amino acid sequences of the N-terminal regions of the isoforms from the two species are completely unrelated. We also report that human PDE4A8 is expressed at significant levels in several different regions of the brain, as well as in other tissues. We propose that the rapid evolutionary change in the PDE4A8 isoform represents its adaptation to the evolution of the function of the human brain.
EXPERIMENTAL
Isolation and analysis of PDE4A8 cDNA clones
Procedures were performed as described by Sambrook and Russell [22] unless specified otherwise. A human fetal brain cDNA library (Stratagene), cloned into the EcoRI site of the Lambda ZAP vector (Stratagene), was screened by hybridization using a probe corresponding to a portion of the human PDE4C catalytic region and 3′ sequences (the 1155 nt full-length insert of pPDE21 [10]; GenBank® accession number L20968). Hybridization was performed with a final wash of 0.3 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.3% SDS at 62°C. Sequencing was performed on both strands using an ABI 3700 sequencer (PerkinElmer). Alignments were generated with the Gap and Lineup programs of the GCG Wisconsin Package UNIX sequence software programs (Accelrys) using the standard defaults.
5′-RACE (rapid amplification of cDNA ends) and PCR cloning of the full ORF (open reading frame) of the human PDE4A8 cDNA
5′-RACE [22] was performed with human hippocampal Marathon-Ready cDNA (Clontech) and the Touch-Down PCR protocol was used (Clontech), with the following primers: RACE primer, 5′-CCATTCTCTGCCTCGAAGCTGTCGTTCTCGG-3′ (binds at nucleotide 326; all sequence co-ordinates refer to GenBank® accession number AY593872) and the nested PCR primer, 5′-CGAAGCTGTCGTTCTCGGCCCTGGTGATG-3′ (binds at nucleotide 314). The resulting PCR products were cloned into pcDNA3.1/V5-His-TOPO (Invitrogen) and sequenced. The sequence of the clone that extended the mRNA closest towards its 5′-end was used to design PCR primers for full-length cloning of the isoform.
To clone a cDNA that encoded the full-length PDE4A8 ORF, the following primers were used to amplify a human hippocampal cDNA library (Stratagene) by PCR: 5′-CGTCACGCCCCAGGAGAGGCAATAGGAGG-3′ (forward primer, binds at the first nucleotide) and 5′-GAGGGGAACAGGGACAGAGGTCTGGGG-3′ (reverse primer, binds at the 3′-untranslated region common to both PDE4A4 and PDE4A8). PCR was performed using Platinum High-Fidelity Taq polymerase (Invitrogen) in the presence of PCRx solution (0.5 × final concentration; Invitrogen). The resulting PCR product, ~3.0 kb in length, was cloned into pcDNA3.1/V5-His-TOPO and sequenced.
Semi-quantitative PCR analysis of PDE4A8 expression in tissues
The following primers were used to perform PCR on normalized human multiple tissue cDNA panels (Clontech): PDE4A8-specific forward primer (5′-CGTCACGCCCCAGGAGAGGCAATAGGAGG-3′, primes at the first nucleotide) and PDE4A-common reverse primer (5′-GAGGGTCTTGGTCGCGGCGCTTGCTG-3′, primes at nucleotide 949). PCR was performed with Platinum High-Fidelity Taq polymerase in the presence of PCRx solution (1 × final concentration) for 40 cycles. The amplification of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was performed as recommended by Clontech and allowed to proceed for 22 cycles. The resulting PCR products were analysed on a 1% (w/v) agarose gel and stained using ethidium bromide.
COS-7 cell culture
COS-7 cells [23] (A.T.C.C.) were cultured in 150 cm2 culture flasks (Corning) in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% (v/v) fetal bovine serum, 584 mg/l l-glutamine and 100 IU/ml penicillin/streptomycin at 37°C in 5% CO2/95% air. Cells were passaged at 70% confluence by rinsing with PBS, followed by incubation with trypsin/EDTA at 37°C for 3 min. Harvested cells were then resuspended in growth medium, centrifuged at 2000 g for 3 min, resuspended and seeded in fresh culture flasks.
Transient expression of PDE4A4 and PDE4A8 in COS-7 cells
Expression plasmids encoding human PDE4A4B and PDE4A8 were created by cloning the full ORF of PDE4A4B (GenBank® accession number L20965 [10,24]) or PDE4A8 into the pcDNA3 vector (Invitrogen), placing it under the control of the strong constitutive CMV (cytomegalovirus) intermediate-early gene promoter. The PDE4A8 protein was expressed with the VSV (vesicular-stomatitis virus) epitope at its C-terminus [14,15,25]. The plasmids were purified from Escherichia coli using the Wizard® Plus Maxiprep system (Promega). For transient transfections, COS-7 cells were seeded at a 1:3 ratio into culture flasks 24 h before transfection so that cells were ~60% confluent by the time of transfection. A transfection mixture containing 10 µg of plasmid DNA, 50 µl of PolyFect® transfection reagent (Qiagen) and 540 µl of serum-free DMEM was incubated at room temperature (25°C) for 10 min to allow complex formation. The growth medium was then refreshed, the DNA–PolyFect® reaction complex was added to the culture flasks, and the flasks were incubated for 24 h before use.
Generation of COS-7 cell lysates
Transfected cells (~90–100% confluent) were washed with PBS and harvested by using a cell scraper in lysis buffer [25 mM Hepes (pH 7.5), 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 30 mM sodium pyrophosphate and 10% (v/v) glycerol] containing Complete® protease inhibitor (Roche). Samples were then frozen on solid CO2, thawed and then manually disrupted with a Dounce homogenizer, followed by passage through a 26-gauge needle several times to ensure complete cell lysis. Cells were centrifuged at 15000 g for 5 min to remove any unbroken cells, and the resulting supernatant was frozen in solid CO2 and stored at −80°C until required. For experimentation, the protein concentration of whole-cell lysate from transfected and mock-transfected (vector only) cells was equalized (typically to 1 µg/µl).
Subcellular fractionation of COS-7 cells
Transfected cells (~90–100% confluent) were washed with PBS and harvested by using a cell scraper in KHEM buffer (50 mM KCl, 50 mM Hepes, pH 7.4, 10 mM EGTA and 1.92 mM MgCl2) containing 1 mM DTT (dithiothreitol) and protease inhibitors as described above. Samples were then manually disrupted with a Dounce homogenizer and syringe as described above. Fractionation of cells into their cytosolic and particulate fractions was achieved as described previously [26]. A low-speed centrifugation at 1000 g for 10 min yielded the P1 (particulate 1) fraction, which typically contained unbroken cells, nuclei and some fragments of the plasma membrane. The supernatant was then centrifuged further at 100000 g for 30 min at 4°C, from which the supernatant represented the high-speed S2 (cytosolic 2) fraction. The resulting pellet was resuspended in 500 µl of KHEM buffer and centrifuged again at 100000 g for 30 min at 4°C. The supernatant was discarded and the remaining pellet was retained as the P2 fraction, which typically contained mitochondria and fragments of endoplasmic reticulum, endosomes, lysosomes, Golgi apparatus and the plasma membrane. The particulate fractions were equated to the volume of the corresponding cytosolic fraction by resuspending in KHEM buffer. The fractions were frozen in solid CO2 and stored at −80°C until required for immunoblotting.
COS-7 cell treatment with IBMX (isobutylmethylxanthine) and forskolin
COS-7 cells were grown in six-well plates and transiently transfected at ~60% confluence as described above. The following day, the transfected cells (~90–100% confluent) were pre-treated with the non-selective PDE inhibitor IBMX (100 µM) (Sigma) for 2 min, followed by the addition of the adenylate cyclase activator forskolin (100 µM) (Sigma). Cells were then incubated at 37°C for 2.5, 5, 10, 15 or 20 min. COS-7 whole-cell lysates were then generated as described above. Untreated transfected COS-7 cells were used as controls. Where indicated, the same procedure was also performed with an additional 20 min pre-incubation step with the PKA (cAMP-dependent protein kinase) inhibitor H-89 (10 µM) (Calbiochem).
Antibodies
To develop antibodies specific to the unique N-terminus of PDE4A8, synthetic peptides (CGDERSRETPESDRAN and CRRLSSGPGLGWAQPE respectively) were conjugated to KLH (keyhole-limpet haemocyanin) and then injected into rabbits. Initial injections were performed with complete Freund’s adjuvant, and ‘booster’ injections were performed with incomplete Freund’s adjuvant, according to institutional animal care guidelines. Pre-immune and immune sera were both used at a dilution of 1:5000 for immunoblots and 1:1600 for immunohistochemistry respectively.
A ‘pan-4A’ human PDE4A-specific polyclonal antiserum, raised against the extreme C-terminal region that is unique to the PDE4A gene family and therefore capable of detecting all human PDE4A isoforms, was used as described previously [26]. The polyclonal antiserum, PS54-UCR1-A1, able to detect the conserved PKA consensus phosphorylation site in UCR1, of sequence RRESF, was also used as described previously [27]. The antibody against the VSV epitope [28] was used as described previously [14,15,25].
Immunohistochemistry
Human brain tissue was obtained at autopsy, according to a protocol approved by the IRB (institutional review board)/IEC (institutional ethics committee) of the University of Alabama at Birmingham. Sections of paraffin-embedded tissue (5 µm) were mounted on Bond-Rite slides (Richard-Allan Scientific) and heated at 60°C for 2 h. Paraffin was removed from the sections by three changes of xylenes and the sections were rehydrated through graded alcohols from absolute to 70% for 5 min each. The slides were rinsed with deionized water and high-temperature antigen retrieval was performed in 1 mM EDTA (pH 9) in a pressure cooker for 5 min. The slides were then allowed to cool for 20 min and transferred to 0.05 M Tris/HCl (pH 7.6), 0.15 M NaCl and 0.1% (v/v) Triton X-100. Endogenous peroxidases were quenched with an aqueous solution of H2O2 for 5 min. Biotin was blocked by treating the tissues with 10 µg/ml streptavidin (Jackson ImmunoResearch Laboratories) in PBS for 15 min, rinsing with Tris buffer, and applying 200 µg/ml biotin (Sigma–Aldrich) in PBE buffer (PBS with l% BSA, 1 mM EDTA and 0.15 mM sodium azide). Non-specific binding was blocked with 3% goat serum in PBE buffer on the sections for 20 min. Anti-PDE4A8 antibody, or pre-immune serum, was diluted in PBE buffer and applied to the sections for 1 h at room temperature. Slides were rinsed with Tris buffer after each step.
Immunodetection was performed with a biotinylated anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories) diluted 1:1000 in PBE buffer and avidin–HRP (horseradish peroxidase) (Signet Pathology Systems) for 20 min each. The chromagen used was 3,3′-diaminobenzidine (BioGenex). After 7 min, the slides were rinsed with water and lightly counterstained with Mayer’s haematoxylin. The sections were dehydrated through graded alcohols from 70% to absolute and three xylene baths for 5 min each. The coverslips were mounted with Permount.
Protein analysis
Protein concentration was determined using the Bradford method [29], with BSA as standard.
SDS/PAGE and Western blotting
Protein samples were loaded on 4–12% acrylamide gels (Invitrogen) and run at 180 V/gel for 1 h with cooling (1 × Mops running buffer) (Invitrogen). Whole-cell lysate samples were run at 5, 10 and 20 µl/well, protein amounts thus being 5, 10 and 20 µg/well. Fractionated samples were run at 20 µl/well, with protein amounts ranging from 1 to 50 µg/well. APDE4A standard was also used where indicated. Samples were resuspended in SDS sample buffer and boiled for 5 min before loading. The electrophoresed samples were transferred on to nitrocellulose membranes (Schleicher & Schuell) at 25 V for 2 h, or 10 V overnight (1 × transfer buffer with 20% methanol) (Invitrogen). Membranes were blocked with 5% non-fat dried skimmed milk powder (Marvel) in TBST [Tris-buffered saline containing 1% (v/v) Tween 20] at room temperature for 1 h before being immunoblotted with the appropriate antibody at room temperature for 1 h, or 4°C overnight. After washing membranes in TBST at room temperature for 1 h, labelled bands were identified using HRP-conjugated anti-rabbit IgG at room temperature for 1 h. All incubations were performed with gentle agitation. The Pierce enhanced chemiluminescence Western blotting kit was used for visualization. Band densities were calculated using Quantity One software (Bio-Rad).
PDE assays
PDE activity was determined using a two-step radioassay procedure as described previously [30].
Data analysis
Rolipram/cilomilast dose–response data were analysed using KaleidaGraph software (Abelbeck software), which also provided IC50 values. To define Km and Vmax values, data from PDE assays performed over a range of cAMP substrate concentrations were analysed by computer fitting to the hyperbolic form of the Michaelis–Menten equation using an iterative least-squares procedure (Ultra-Fit software, Biosoft).
RESULTS
Isolation of human PDE4A8 cDNAs
As part of an extensive search for novel PDE4 isoforms, we screened a human fetal brain library with a cDNA probe corresponding to a portion of the catalytic region of PDE4C [10]. This screen did not yield any PDE4C cDNAs, but did yield clones for PDE4A4 and several other PDE4 isoforms. Also obtained in the screen were multiple isolates of a novel clone, pPDE103. The sequence of pPDE103 was identical throughout its length with PDE4A4, PDE4A10, and PDE4A11, with the exception of a novel region of sequence at one end, clearly different from these previously described isoforms in this region (Figure 1A). To obtain an additional sequence corresponding to the unique region of pPDE103, we performed 5′-RACE on human hippocampal cDNA with primers based near the 5′-end of pPDE103. The 5′-RACE products extended the unique 5′-region of pPDE103 to include a start codon, which in turn was preceded by an in-frame stop codon, indicating the start of the ORF. The protein encoded by this ORF was a clearly a novel PDE4 isoform (Figure 1B).
Analysis of human genomic DNA sequences corresponding to PDE4A8
In silico analysis of human DNA sequences did not detect any cDNAs or ESTs (expressed sequence tags) corresponding to the unique 5′-end of our novel cDNA. However, BLAST searches of publicly available human genomic sequences demonstrated the presence of three contiguous blocks of sequence within the PDE4A gene on chromosome 19 (e.g. on the cosmid clone LLNLR-272E5; GenBank® accession number AC011548) that corresponded to the entire unique region of our new PDE4A cDNA (Figure 1C). Therefore we concluded that they were indeed exons specific to our new isoform. The sequence of these exons confirmed the start codon of the PDE4A8 cDNA and confirmed that the ORF upstream of the start codon was indeed blocked by a stop codon (Figure 1C).
The unique 5′-ends of the human and rat PDE4A8 cDNAs are conserved at the nucleotide level, but encode different proteins
Comparison of the novel sequence at the 5′-end of our new PDE4A cDNA with those of other mammals demonstrated that it had significant homology at the nucleotide level with that of rat PDE4A8 [13]; (Figure 1D). For this reason, we call our new human isoform PDE4A8, following the conventional nomenclature [31]. However, a number of insertions and other changes in the 5′-end completely alter the protein encoded by this region, compared with that of rat PDE4A8 (Figure 1D). The unique N-terminal region of rat PDE4A8 is 21 amino acids in length, out of a protein of 763 amino acids [13]. In contrast, the unique N-terminal region of human PDE4A8 is 85 amino acids in length, out of a protein of 864 amino acids (Figure 1B). These data demonstrate that the PDE4A8 N-terminus has undergone rapid change, which occurred after the divergence of humans and rodents. We were unable to find cDNAs encoding proteins homologous with the N-terminus of human PDE4A8 in the publicly available databases, including those derived from other primates. In contrast, searches of GenBank® revealed that the genomes of numerous mammals, including the mouse [32], bat, pig and dog, encode amino acid sequences strongly similar to that of the N-terminal region of rat PDE4A8 (results not shown).
Expression of human PDE4A8 mRNAs in tissues
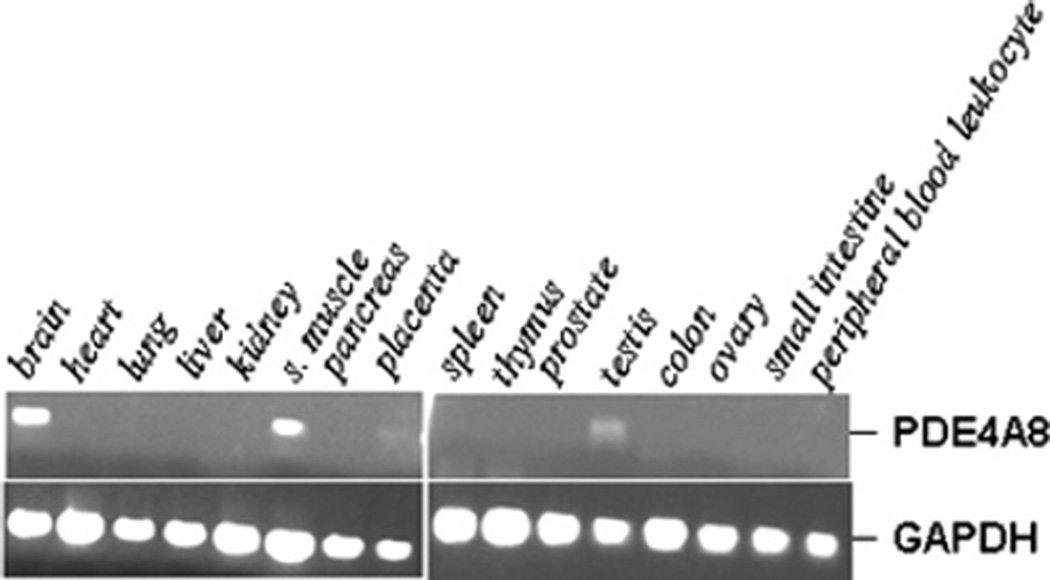
We have shown previously that rat PDE4A8 is present exclusively, or almost exclusively, in testis [13], whereas PDE4A4/PDE4A5, PDE4A10 and PDE4A11 are expressed in a variety of tissues, including the brain [12,14–16]. To determine the tissue distribution of human PDE4A8, semi-quantitative PCR was performed with a gene-specific primer set and a human multiple tissue cDNA panel. The highest levels of PDE4A8 mRNA were found in brain and skeletal muscle, although some expression in testis was also noted (Figure 2).
Figure 2. Tissue distribution of PDE4A8 mRNA.
The expression pattern of PDE4A8 was assessed by semi-quantitative PCR on human cDNA panels (Clontech). Each cDNA source was amplified for both PDE4A8 and GAPDH, as described in the Experimental section. The PCR products were resolved on 1% agarose gels and stained with ethidium bromide. s. muscle, skeletal muscle.
Expression of PDE4A8 protein in human tissues
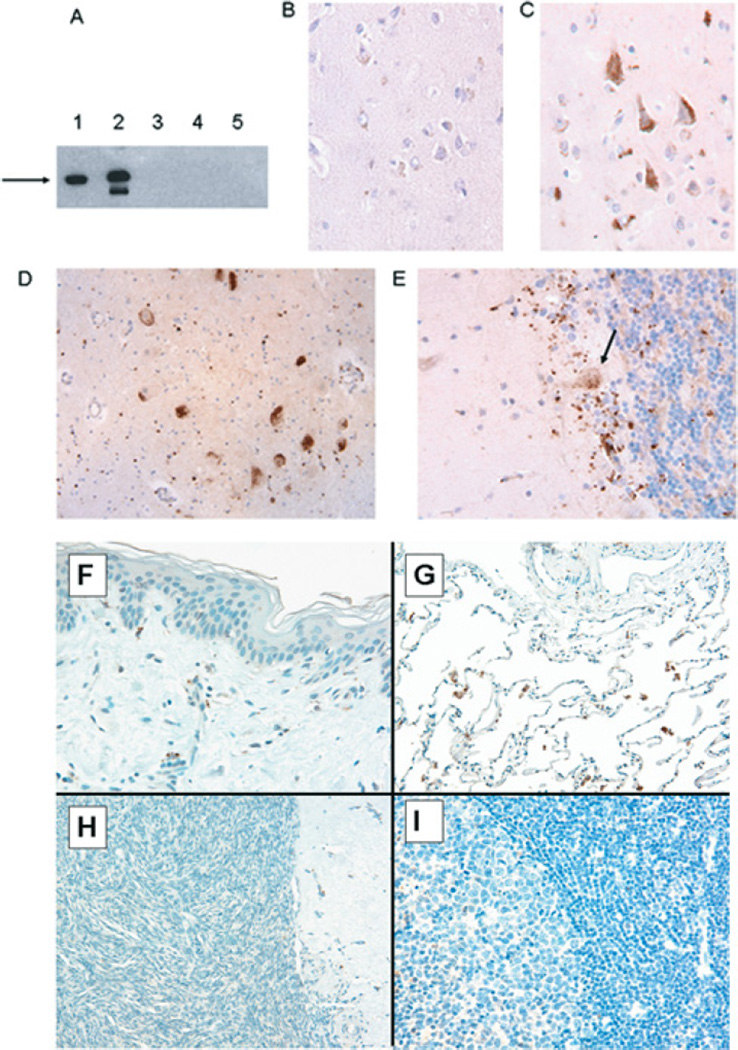
As our mRNA studies have demonstrated that PDE4A8 was expressed in the brain, we wished to determine the regional expression of PDE4A8 in human brain. For this purpose, rabbit polyclonal antibodies were generated against two different peptides, corresponding to discrete regions of the unique PDE4A8 N-terminus (see the Experimental section). These antibodies were shown to recognize recombinant PDE4A8 protein, as expressed in COS-7 cells (Figure 3A). They were then used in immunohistochemistry to detect PDE4A8 protein in tissues from selected regions of the human brain, obtained from autopsy specimens (Figures 3B–3E). These studies demonstrated the presence of PDE4A8 protein in pyramidal cells in cerebral cortex (Figure 3C) and also in a discrete population of large neurons, presumably sensory neurons, in the anterior horn of the spinal cord (Figure 3D). PDE4A8 was also detected in cells located at the base of the granular layer of the cerebellum, including Purkinje cells. The Purkinje cell staining tended to localize to the upper region of the main cell body (Figure 3E). These staining patterns were seen in tissues obtained from at least three of the four patients studied. Because banked autopsy specimens were used, not all regions of the brain were represented in the material available for study. No staining was seen in human lung, skin, ovary or lymph nodes (Figures 3F–3I) or uterine smooth muscle, which was consistent with the findings of the RT (reverse transcription)–PCR (see the previous section). These studies clearly demonstrated the presence of PDE4A8 in human brain, including regions involved in sensation, co-ordination and higher cognitive functions.
Figure 3. Tissue distribution of PDE4A8 protein.
(A) Immunoblotting of lysates from COS-7 cells transfected with a plasmid expressing PDE4A8. The recombinant protein contained a VSV epitope at its C-terminus. Lanes were immunoblotted as follows: 1, anti-VSV antibody; 2, anti-PDE4A8 antibody; 3, pre-immune serum; 4, anti-PDE4A8 antibody, cells transfected with vector only; and 5, anti-PDE4A8 antibody plus peptide used as immunogen (62.5 µg/ml). The recombinant protein migrated at 125 kDa (arrow). The less-intense band seen in lane 2 was thought to be a proteolytic fragment; we have observed such fragments before when PDE4 proteins are expressed at high levels in COS-7 cells [25]. (B) Immunohistochemistry of human cerebral cortex, using pre-immune serum. (C)–(I) show immunohistochemistry of human tissues, using the anti-PDE4A8 antibody. The tissues are as follows. (C) Cerebral cortex, showing several pyramidal neurons that stained strongly. (D) Spinal cord, showing a cluster of anterior horn neurons that stained strongly. (E) Cerebellar cortex; staining was seen in a diffuse band at the base of the Purkinje cell layer, including Purkinje cells (arrow). Staining of Purkinje cell bodies was most pronounced in a perinuclear pattern, extending into dendrites that project into the deeper portions of the molecular layer. (F) Skin, showing no staining. (G) Lung, showing staining of some rare scattered pulmonary macrophages, but no staining of alveolar or interstitial cells. (H) Ovarian parenchyma, showing no staining. (I) Germinal centre of a benign lymph node, showing no staining. All histological sections were at an original magnification of 400×.
Enzymatic properties of PDE4A8 expressed in COS-7 cells
We have used a COS-7 expression system to examine and compare the enzymatic properties of mammalian PDE4 isoforms, including their kinetic properties, phosphorylation by kinases and their susceptibility to various inhibitors [13–15,19,26]. Therefore human PDE4A8 was expressed in COS-7 cells under the control of the strong constitutive CMV intermediate-early gene promoter. Immunoblotting of COS-7 lysates with the pan-4A antibody, as described in the Experimental section, identified a single immunoreactive band of 125 ± 5 kDa in lysates prepared from PDE4A8-transfected COS-7 cells; lysates from vector-only transfectants were negative (Figure 3A). Immunoblotting PDE4A4 transfectants with the same antibody also gave a band of 125 ± 3 kDa, as we described previously [24,33].
PDE assays, performed as described in the Experimental section, with 1 µM cAMP as substrate, showed that cytosolic PDE4A8 exhibited a cAMP PDE activity of 265–273 pmol/min per mg of protein (n = 3), compared with 10–14 pmol/min per mg of protein (n = 3) in vector-only-transfected cells, suggesting that ~96% of total cytosolic PDE activity was due to recombinant PDE4A8. PDE4A8 activity was ~88 ± 8% (mean ± S.D.; n = 3) of that of PDE4A4 activity, when equal amounts of PDE4 protein were determined by immunoblotting.
Subcellular localization and activity of recombinant PDE4A8 in COS-7 cells
We have shown previously that specific PDE4 isoforms partition differently among various subcellular fractions when expressed in COS-7 cells [13–15,19,26]. To determine the subcellular fractionation of COS-7-expressed PDE4A8 and PDE4A4, transfected COS-7 cells were lysed and subcellular fractions were prepared, as described in the Experimental section. The fractions were then immunoblotted with the pan-4A antibody, to quantify the relative amounts of PDE4A8 protein in each fraction. PDE enzymatic activity was also assayed in each cellular fraction. The high-speed supernatant (S2) contained the bulk (81 ± 3%) of PDE4A8 immunoreactivity, with smaller fractions in the high-speed pellet (P2, 9 ± 2%) and low-speed pellet (P1, 10 ± 4%). Essentially identical proportions were obtained when PDE4A8 in the fractions was quantified using the PDE enzymatic assay (S2, 82 ± 3%; P2, 7 ± 2% and P1, 11 ± 2% respectively; all values are means ± S.D.; n = 3).
Inhibition of PDE4A8 by rolipram and cilomilast
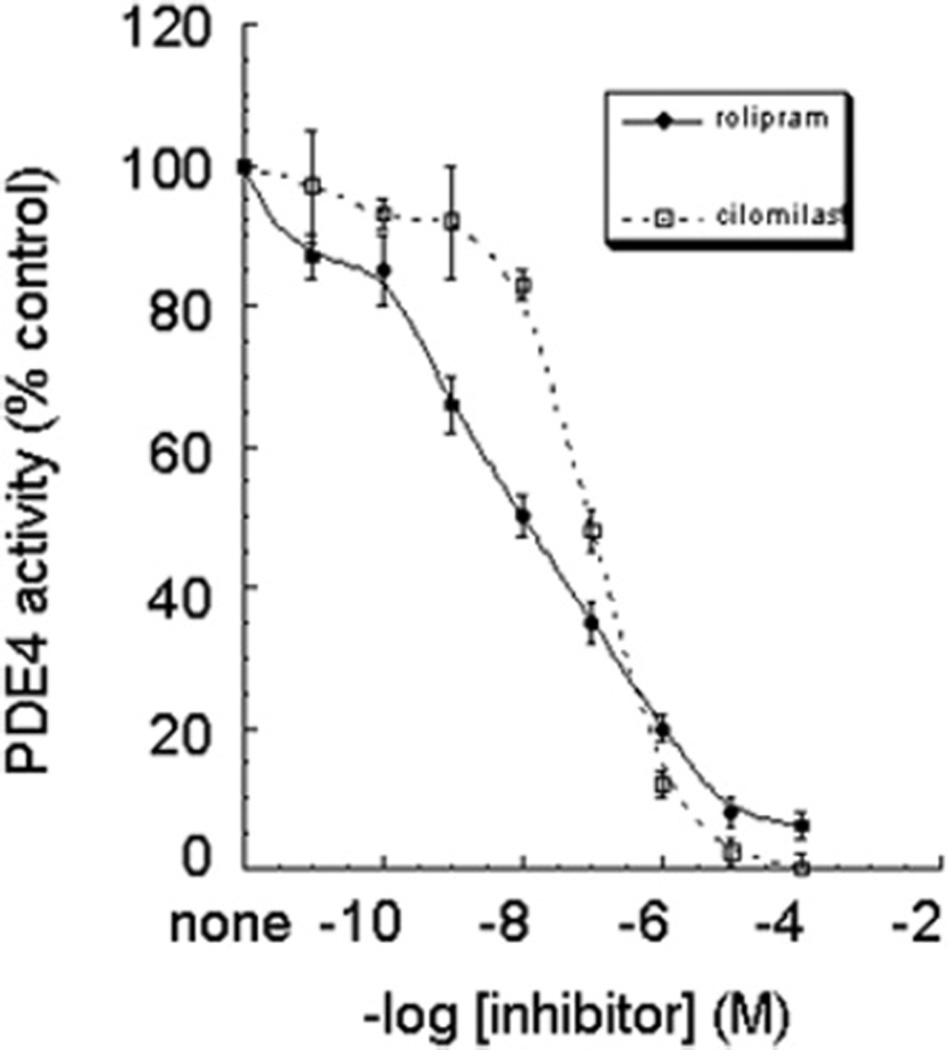
We next wished to determine the sensitivity of PDE4A8 to the prototypical selective PDE4 inhibitor rolipram, and also to cilomilast (Ariflo®, GlaxoSmithKline), another PDE4-selective inhibitor that has been tested in humans [34–36]. PDE assays, as described in the Experimental section, were performed on the S2 fraction of COS-7-expressed PDE4A8 with 1 µM cAMP as substrate and over a range of inhibitor concentrations (Figure 4). PDE4A8 was considerably more sensitive to rolipram than to cilomilast (IC50 for rolipram, 11 ± 1 nM, compared with 101 ± 7 nM for cilomilast; means ± S.D.; n = 3). Note that PDE4A8 is much more sensitive to inhibition by rolipram than is PDE4A4, which has an IC50 for rolipram of ~1.6 µM [24]. In contrast, PDE4A8 is less sensitive to cilomilast than is PDE4A4, with an IC50 of ~61 nM [15]. Thus the unique N-terminal regions of PDE4A isoforms can affect the conformation of the catalytic unit, leading to altered sensitivity to inhibition by various selective inhibitors.
Figure 4. Dose–response curves for the inhibition of PDE4A8 by rolipram or by cilomilast.
Lysates were prepared from COS-7 cells transfected to express PDE4A8. PDE activity was measured over a range of concentrations of inhibitor, as described in the text. Rolipram/cilomilast dose–response data were analysed using KaleidaGraph software to generate IC50 values (IC50 for rolipram, 11 ± 1 nM, and for cilomilast, 101 ± 7 nM; means ± S.D.; n = 3).
We noted significant differences between the slopes of the inhibition curves of rolipram and cilomilast. These differences are reminiscent of those seen for other PDE4 isoforms. It is understood that PDE4 isoforms can exist in different conformational states, in an equilibrium that can be affected by protein–protein interactions, including dimerization, as well as phosphorylation. It has been established that these conformational states can exhibit different sensitivity to certain, but not all, PDE4-selective inhibitors. Sensitivity to rolipram has been shown to be affected by PDE4 conformational status and in turn has been used to identify the presence of so-called high- and low-affinity PDE4 conformers. Thus, invariably, rolipram has a shallow slope of inhibition, because of the presence of different conformers with different affinities for this compound. In contrast, cilomilast does not discriminate between various conformers and therefore does not show the shallow slope typical of rolipram [2,5,15,33,37–39].
Phosphorylation of PDE4A8 by PKA
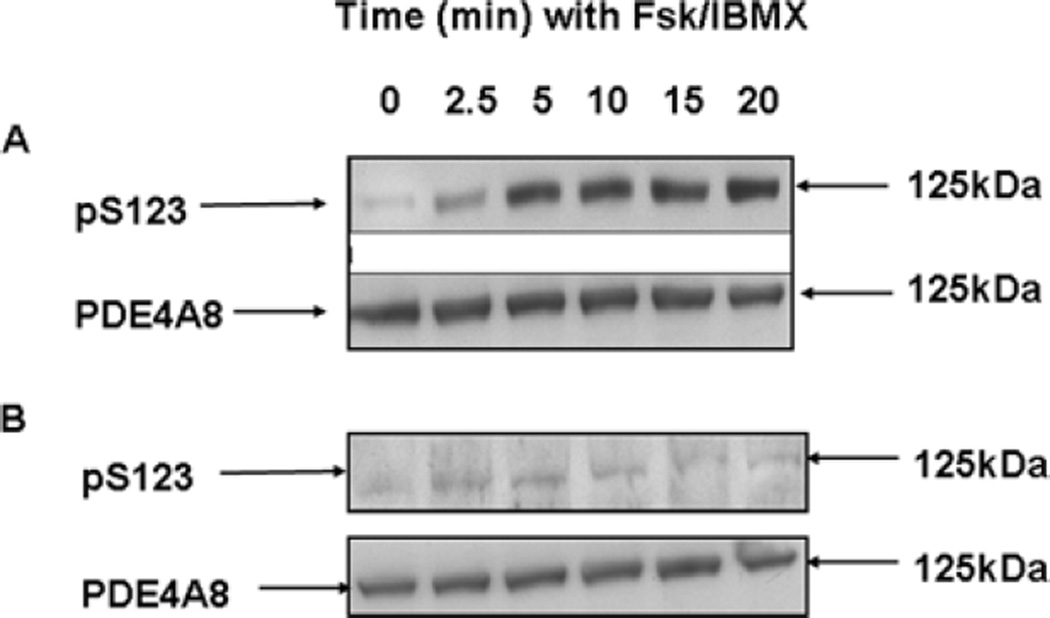
All long PDE4 isoforms can be activated by phosphorylation by PKA. PKA phosphorylation occurs at a serine residue located within a conserved PKA consensus phosphorylation site, of sequence RRESF, located at the very beginning of UCR1 (in PDE4A8, this site is Ser123; Figures 1A and 1B). We therefore studied PKA phosphorylation of PDE4A8, using methods that we have described for other PDE4 isoforms ([14,15,27], and see also the Experimental section). COS-7 cells transiently expressing PDE4A8 were treated with forskolin, a specific activator of adenylate cyclase, and the non-selective PDE inhibitor IBMX. Together, these agents raise intracellular cAMP levels sufficiently high to activate PKA in these cells [27]. Whole-cell lysates were prepared and immunoblotted with a phosphospecific antibody designed to recognize a UCR1-derived sequence that includes the PKA-phosphorylated serine residue, as described previously [27]. The data show that treatment of the cells with IBMX and forskolin produces a time-dependent phosphorylation of Ser123, as demonstrated by the appearance of a 125 kDa band in the immunoblots (Figure 5A).
Figure 5. Phosphorylation of PDE4A8 by PKA.
(A) COS-7 cells transfected to express PDE4A8 were treated with IBMX and forskolin (Fsk) to increase intracellular cAMP levels and activate PKA. At the indicated times after treatment, the cells were lysed and the lysates subjected to SDS/PAGE and immunoblotted. In the upper panel, they were immunoblotted with an antibody specific for the serine residue phosphorylated by PKA [i.e. Ser123 (pS123) in PDE4A8; Figures 1A and 1B]. In the lower panel, they were immunoblotted with the pan-4A antibody, to ensure that equal amounts of PDE4A8 protein were loaded on to each lane. (B) As in (A), but also with treatment with the PKA inhibitor H89. PDE4A8 migrated as a 125 kDa band. These results were typical of those obtained at least three times.
In order to confirm that PKA was indeed responsible for the phosphorylation that we observed, this experiment was repeated with the addition of the PKA inhibitor H89 (10 µM) before the administration of forskolin and IBMX. This treatment blocked the phosphorylation of Ser123, as manifested by the lack of appearance of the 125 kDa band upon immunoblotting with the phosphospecific antibody (Figure 5b).
DISCUSSION
One of the most remarkable aspects of PDE4 biology is the extreme diversity of PDE4 isoforms. Each of the four different PDE4 genes encodes multiple isoforms, each of which may differ in its enzymatic properties, tissue expression, protein partners or subcellular distribution [5,40]. The physiological and pharmacological consequences of this diversity is currently being investigated by numerous groups, but key to such efforts is to determine the full range of PDE4 isoforms that are present in mammals. In the present paper, we have described PDE4A8, a new isoform encoded by the human PDE4A gene that has a novel N-terminal region of 85 amino acids that is not present in any other PDE4 isoform. PDE4A8, like the other long PDE4A isoforms PDE4A4, PDE4A10 and PDE4A11 [10,14,15], contains both the UCR1 and UCR2 regulatory regions, which are located between the unique N-terminal region and the catalytic region (Figure 1A). The PDE4A4, PDE4A8, PDE4A10 and PDE4A11 mRNAs all have a common point of divergence (Figure 1A), which also corresponds to the major point of alternative mRNA splicing in transcripts from the human PDE4B and PDE4D genes [25,41], the mouse pde4a [32] gene, and the homologous dunce gene of Drosophila melanogaster ([10,25,42], and reviewed in [5]). These structural differences suggested that each of the PDE4A isoforms is generated by alternative mRNA splicing [10]. Since there is no region of sequence common to the 5′-ends of these isoforms, it is also likely that each is generated from a different transcriptional start site. Consistent with this interpretation, sequencing of the human PDE4A gene, located on chromosome 19, shows that the unique 5′-ends of each of these mRNAs is encoded by a different widely separated exon(s) [14,15,17].
Although the nucleotide sequences encoding the unique N-terminal regions of human and rat PDE4A8 are both unique and homologous, they encode completely different amino acid sequences. This result was unexpected, as almost all the PDE4 isoforms that we have identified to date have unique N-terminal amino acid regions that are conserved between these two species. To date, the one exception to this generalization is the PDE4B4 isoform, which is present in rodents, but not in humans [43].
Human PDE4A8 differs from other PDE4A isoforms in that it is expressed at high relative levels in skeletal muscle. In contrast, rat PDE4A8 is expressed almost exclusively in testis [13,44,45]. PDE4A8 is unlikely to be the only cAMP-hydrolysing PDE in skeletal muscle, as we and others have detected expression of PDE4 isoforms, including PDE4A10 [16], in skeletal muscle. In addition, PDE7A, which also has a high affinity for cAMP, is expressed at high relative levels in muscle [46]. It is likely that PDE4A8 plays an important role in the regulation of cAMP levels in skeletal muscle, although its exact physiological function(s) require further study.
We have also demonstrated in the present study that human PDE4A8 is expressed in brain. We and other groups have shown that PDE4A1, PDE4A4/PDE4A5 and PDE4A10 are also expressed in brain [12–16,20,47], but almost all of these data were obtained in rat or mouse. Human PDE4A8 is distinctive in that it is expressed in brain, whereas its rodent counterpart is not [13,45,47]. For this reason, we used immunohistochemistry to search for areas of the human brain that are enriched in PDE4A8 protein. We have shown that PDE4A8 is expressed in specific neuronal subpopulations in cortex, spinal cord and cerebellum. The physiological significance of these observations is as yet unclear, but they are certainly consistent with a physiological role in the functions performed by these regions of the brain.
The differences in PDE4A8 tissue expression between humans and rats is likely to be explained, at least in part, by differences in their promoters. Analysis of genomic DNA has shown that the promoter for each PDE4 isoform is located just upstream of the exon(s) encoding their unique N-termini [14,15,17,48,49]. For PDE4A8, these two regions diverge substantially between humans and rats (Figure 1D), presumably reflecting the evolutionary changes that also affect their coding regions.
PDE4A8, as expressed in COS-7 cells, is detected as a ~125 kDa band on immunoblots (Figures 3 and 5). As with all PDE4 isoforms analysed to date [5,14,26], this apparent size is larger than that predicted simply from the primary sequence alone, namely 96 kDa. Indeed, its apparent size is remarkably similar to that observed for PDE4A4, whose apparent and predicted sizes are 125 and 99.2 kDa respectively [24,26]. The inconsistency between the apparent and predicted size may be due to protein folding or to stretches of acidic amino acids reducing the amount of SDS bound to the protein [24]. Consistent with this observation, deletion studies have highlighted a region, rich in acidic amino acids, within the conserved PDE4A catalytic unit as the likely region causing this aberrant migration on SDS/PAGE [50].
The pattern of tissue expression and unique N-terminal region of PDE4A8 strongly suggest that it has specific properties related to cAMP signalling in brain and muscle. cAMP levels in muscle cells are regulated, at least in part, by β-adrenergic receptors linked to adenylate cyclase by G-proteins. Among the physiological targets of PKA in muscle cells are glycogen synthase and other enzymes that regulate energy metabolism. The transcription factor CREB (cAMP-response-element-binding protein) is also a physiological substrate of PKA in numerous cells, including brain and muscle, and mediates important functions, such as learning and memory, in the brain. These pathways are attractive candidates for the action of PDE4A8 in humans.
Acknowledgments
This work was supported by NIH (National Institutes of Health) grants R01-GM58553 (to G.B.B), 5U24CA086359-08 (to W. E.G.) and P30CA13148-36 (for core facilities), by MRC (Medical Research Council) grant G9504010 (to M.D.H.), the Leducq Foundation (to M.D.H.) and by European Commission grant 037189 (to M.D.H.). We thank Mary Barnette (GlaxoSmithKline) for Ariflo®.
Abbreviations used
- CMV
cytomegalovirus
- DMEM
Dulbecco’s modified Eagle’s medium
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HRP
horseradish peroxidase
- IBMX
isobutylmethylxanthine
- ORF
open reading frame
- P1 (etc.)
particulate 1 (etc.)
- PDE
phosphodiesterase
- PKA
cAMP-dependent protein kinase
- RACE
rapid amplification of cDNA ends
- S2
cytosolic 2
- TBST
Tris-buffered saline containing 1% (v/v) Tween 20
- UCR
upstream conserved region
- VSV
vesicular-stomatitis virus
Footnotes
The nucleotide sequence data reported for human PDE4A8 will appear in the DDBJ, EMBL, GenBank® and GSDB Nucleotide Sequence Databases under the accession number AY593872.
REFERENCES
- 1.Beavo JA, Brunton LL. Cyclic nucleotide research: still expanding after half a century. Nat. Rev. Mol. Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 2.Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 2003;278:5493–5496. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 3.Houslay MD, Adams DR. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolger GB, Conti M, Houslay MD. In: Cyclic Nucleotide Phosphodiesterases in Health and Disease. Beavo JA, Francis SH, Houslay MD, editors. Boca Raton: Taylor and Francis; 2007. pp. 99–129. [Google Scholar]
- 5.Houslay MD, Sullivan M, Bolger GB. The multienzyme PDE4 cyclic adenosine monophosphate-specific phosphodiesterase family: intracellular targeting, regulation, and selective inhibition by compounds exerting anti-inflammatory and antidepressant actions. Adv. Pharmacol. 1998;44:225–342. doi: 10.1016/s1054-3589(08)60128-3. [DOI] [PubMed] [Google Scholar]
- 6.Torphy TJ, Page C. Phosphodiesterases: the journey towards therapeutics. Trends Pharmacol. Sci. 2000;21:157–159. doi: 10.1016/s0165-6147(00)01478-4. [DOI] [PubMed] [Google Scholar]
- 7.Bach ME, Barad M, Son H, Zhuo M, Lu YF, Shih R, Mansuy I, Hawkins RD, Kandel ER. Age-related defects in spatial memory are correlated with defects in the late phase of hippocampal long-term potentiation in vitro and are attenuated by drugs that enhance the cAMP signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5280–5285. doi: 10.1073/pnas.96.9.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barad M, Bourtchouladze R, Winder DG, Golan H, Kandel E. Rolipram, a type IV-specific phosphodiesterase inhibitor, facilitates the establishment of long-lasting long-term potentiation and improves memory. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15020–15025. doi: 10.1073/pnas.95.25.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HT, Huang Y, Jin SL, Frith SA, Suvarna N, Conti M, O’Donnell JM. Antidepressant-like profile and reduced sensitivity to rolipram in mice deficient in the PDE4D phosphodiesterase enzyme. Neuropsychopharmacology. 2002;27:587–595. doi: 10.1016/S0893-133X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 10.Bolger G, Michaeli T, Martins T, St. John T, Steiner B, Rodgers L, Riggs M, Wigler M, Ferguson K. A family of human phosphodiesterases homologous to the dunce learning and memory gene product of Drosophila melanogaster are potential targets for antidepressant drugs. Mol. Cell. Biol. 1993;13:6558–6571. doi: 10.1128/mcb.13.10.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beard MB, Olsen AE, Jones RE, Erdogan S, Houslay MD, Bolger GB. UCR1 and UCR2 domains unique to the cAMP-specific phosphodiesterase family form a discrete module via electrostatic interactions. J. Biol. Chem. 2000;275:10349–10358. doi: 10.1074/jbc.275.14.10349. [DOI] [PubMed] [Google Scholar]
- 12.Bolger GB, Rodgers L, Riggs M. Differential CNS expression of alternative mRNA isoforms of the mammalian genes encoding cAMP-specific phosphodiesterases. Gene. 1994;149:237–244. doi: 10.1016/0378-1119(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 13.Bolger GB, McPhee I, Houslay MD. Alternative splicing of cAMP-specific phosphodiesterase mRNA transcripts: characterization of a novel tissue-specific isoform, RNPDE4A8. J. Biol. Chem. 1996;271:1065–1071. doi: 10.1074/jbc.271.2.1065. [DOI] [PubMed] [Google Scholar]
- 14.Rena G, Begg F, Ross A, MacKenzie C, McPhee I, Campbell L, Huston E, Sullivan M, Houslay MD. Molecular cloning, genomic positioning, promoter identification, and characterization of the novel cyclic AMP-specific phosphodiesterase PDE4A10. Mol. Pharmacol. 2001;59:996–1011. doi: 10.1124/mol.59.5.996. [DOI] [PubMed] [Google Scholar]
- 15.Wallace DA, Johnston LA, Huston E, MacMaster D, Houslay TM, Cheung YF, Campbell L, Millen JE, Smith RA, Gall I, et al. Identification and characterization of PDE4A11, a novel, widely expressed long isoform encoded by the human PDE4A cAMP phosphodiesterase gene. Mol. Pharmacol. 2005;67:1920–1934. doi: 10.1124/mol.104.009423. [DOI] [PubMed] [Google Scholar]
- 16.McPhee I, Cochran S, Houslay MD. The novel long PDE4A10 cyclic AMP phosphodiesterase shows a pattern of expression within brain that is distinct from the long PDE4A5 and short PDE4A1 isoforms. Cell. Signalling. 2001;13:911–918. doi: 10.1016/s0898-6568(01)00217-0. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan M, Rena G, Begg F, Gordon L, Olsen AS, Houslay MD. Identification and characterization of the human homologue of the short PDE4A cAMP-specific phosphodiesterase RD1 (PDE4A1) by analysis of the human HSPDE4A gene locus located at chromosome 19p13.2. Biochem. J. 1998;333:693–703. doi: 10.1042/bj3330693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RL, Takayasu H, Eberwine M, Myres J. Cloning and characterization of mammalian homologs of the Drosophila dunce+ gene. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3604–3608. doi: 10.1073/pnas.86.10.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakur Y, Pryde JG, Houslay MD. Engineered deletion of the unique N-terminal domain of the cyclic AMP-specific phosphodiesterase RD1 prevents plasma membrane association and the attainment of enhanced thermostability without altering its sensitivity to inhibition by rolipram. Biochem. J. 1993;292:677–686. doi: 10.1042/bj2920677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shakur Y, Wilson M, Pooley L, Lobban M, Griffiths SL, Campbell AM, Beattie J, Daly C, Houslay MD. Identification and characterization of the type-IVA cyclic AMP-specific phosphodiesterase RD1 as a membrane-bound protein expressed in cerebellum. Biochem. J. 1995;306:801–809. doi: 10.1042/bj3060801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huston E, Gall I, Houslay TM, Houslay MD. Helix-1 of the cAMP-specific phosphodiesterase PDE4A1 regulates its phospholipase-D-dependent redistribution in response to release of Ca2+ J. Cell Sci. 2006;119:3799–3810. doi: 10.1242/jcs.03106. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 24.Huston E, Pooley L, Julien P, Scotland G, McPhee I, Sullivan M, Bolger G, Houslay M. The human cyclic AMP-specific phosphodiesterase PDE-46 (HSPDE4A4B) expressed in transfected COS7 cells occurs as both particulate and cytosolic species which exhibit distinct kinetics of inhibition by the anti-depressant rolipram. J. Biol. Chem. 1996;271:31334–31344. doi: 10.1074/jbc.271.49.31334. [DOI] [PubMed] [Google Scholar]
- 25.Bolger GB, Erdogan S, Jones RE, Loughney K, Scotland G, Hoffmann R, Wilkinson I, Farrell C, Houslay MD. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem. J. 1997;328:539–548. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPhee I, Pooley L, Lobban M, Bolger G, Houslay MD. Identification, characterization and regional distribution in brain of RPDE-6 (RNPDE4A5), a novel splice variant of the PDE4A cyclic AMP phosphodiesterase family. Biochem. J. 1995;310:965–974. doi: 10.1042/bj3100965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKenzie SJ, Baillie GS, McPhee I, MacKenzie C, Seamons R, McSorley T, Millen J, Beard MB, Van Heeke G, Houslay MD. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in upstream conserved region 1 (UCR1) Br. J. Pharmacol. 2002;136:421–433. doi: 10.1038/sj.bjp.0704743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Marchmont RJ, Houslay MD. A peripheral and an intrinsic enzyme constitute the cyclic AMP phosphodiesterase activity of rat liver plasma membranes. Biochem. J. 1980;187:381–392. doi: 10.1042/bj1870381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beavo JA. Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physiol. Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 32.Olsen AE, Bolger GB. Physical mapping and promoter structure of the murine cAMP-specific phosphodiesterase pde4a gene. Mamm. Genome. 2000;11:41–45. doi: 10.1007/s003350010008. [DOI] [PubMed] [Google Scholar]
- 33.McPhee I, Yarwood SJ, Scotland G, Huston E, Beard MB, Ross AH, Houslay ES, Houslay MD. Association with the SRC family tyrosyl kinase LYN triggers a conformational change in the catalytic region of human cAMP-specific phosphodiesterase HSPDE4A4B: consequences for rolipram inhibition. J. Biol. Chem. 1999;274:11796–11810. doi: 10.1074/jbc.274.17.11796. [DOI] [PubMed] [Google Scholar]
- 34.Barnette MS, Christensen SB, Essayan DM, Grous M, Prabhakar U, Rush JA, Kagey SA, Torphy TJ. SB 207499 (Ariflo), a potent and selective second-generation phosphodiesterase 4 inhibitor: in vitro anti-inflammatory actions. J. Pharmacol. Exp. Ther. 1998;284:420–426. [PubMed] [Google Scholar]
- 35.Torphy TJ, Barnette MS, Underwood DC, Griswold DE, Christensen SB, Murdoch RD, Nieman RB, Compton CH. Ariflo™ (SB 207499), a second generation phosphodiesterase 4 inhibitor for the treatment of asthma and COPD: from concept to clinic. Pulm. Pharmacol. Ther. 1999;12:131–135. doi: 10.1006/pupt.1999.0181. [DOI] [PubMed] [Google Scholar]
- 36.Giembycz MA. Cilomilast: a second generation phosphodiesterase 4 inhibitor for asthma and chronic obstructive pulmonary disease. Expert Opin. Invest. Drugs. 2001;10:1361–1379. doi: 10.1517/13543784.10.7.1361. [DOI] [PubMed] [Google Scholar]
- 37.Houslay MD. PDE4 cAMP-specific phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:249–315. doi: 10.1016/s0079-6603(01)69049-4. [DOI] [PubMed] [Google Scholar]
- 38.Richter W, Conti M. The oligomerization state determines regulatory properties and inhibitor sensitivity of type 4 cAMP-specific phosphodiesterases. J. Biol. Chem. 2004;279:30338–30348. doi: 10.1074/jbc.M312687200. [DOI] [PubMed] [Google Scholar]
- 39.Rocque WJ, Tian G, Wiseman JS, Holmes WD, Zajac TI, Willard DH, Patel IR, Wisely GB, Clay WC, Kadwell SH, et al. Human recombinant phosphodiesterase 4B2B binds (R)-rolipram at a single site with two affinities. Biochemistry. 1997;36:14250–14261. doi: 10.1021/bi971112e. [DOI] [PubMed] [Google Scholar]
- 40.Bolger GB. Phosphodiesterase isoforms: an annotated list. In: Beavo JA, Francis SH, Houslay MD, editors. Cyclic Nucleotide Phosphodiesterases in Health and Disease. Boca Raton: CRC Press; 2007. pp. 19–31. [Google Scholar]
- 41.Cheung YF, Kan Z, Garrett-Engele P, Gall I, Murdoch H, Baillie GS, Camargo LM, Johnson JM, Houslay MD, Castle JC. PDE4B5, a novel, super-short, brain-specific cAMP phosphodiesterase-4 variant whose isoform-specifying N-terminal region is identical to that of cAMP phosphodiesterase-4D6 (PDE4D6) J. Pharmacol. Exp. Ther. 2007;322:600–609. doi: 10.1124/jpet.107.122218. [DOI] [PubMed] [Google Scholar]
- 42.Qiu YH, Chen CN, Malone T, Richter L, Beckendorf SK, Davis RL. Characterization of the memory gene dunce of Drosophila melanogaster. J. Mol. Biol. 1991;222:553–565. doi: 10.1016/0022-2836(91)90496-s. [DOI] [PubMed] [Google Scholar]
- 43.Shepherd M, McSorley T, Olsen AE, Johnston LA, Thomson NC, Baillie GS, Houslay MD, Bolger GB. Molecular cloning and subcellular distribution of the novel PDE4B4 cAMP-specific phosphodiesterase isoform. Biochem. J. 2003;370:429–438. doi: 10.1042/BJ20021082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farooqui SM, Al Bagdadi F, Houslay MD, Bolger GB, Stout R, Specian RD, Cherry JA, Conti M, O’Donnell JM. Surgically induced cryptorchidism-related degenerative changes in spermatogonia are associated with loss of cyclic adenosine monophosphate-dependent phosphodiesterases type 4 in abdominal testes of rats. Biol. Reprod. 2001;64:1583–1589. doi: 10.1095/biolreprod64.6.1583. [DOI] [PubMed] [Google Scholar]
- 45.Cherry JA, Davis RL. Cyclic AMP phosphodiesterases are localized in regions of the mouse brain associated with reinforcement, movement, and affect. J. Comp. Neurol. 1999;407:287–301. [PubMed] [Google Scholar]
- 46.Michaeli T, Bloom TJ, Martins T, Loughney K, Ferguson K, Riggs M, Rodgers L, Beavo JA, Wigler M. Isolation and characterization of a previously undetected human cAMP phosphodiesterase by complementation of cAMP phosphodiesterase-deficient Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:12925–12932. [PubMed] [Google Scholar]
- 47.D’Sa C, Eisch AJ, Bolger GB, Duman RS. Differential expression and regulation of the cAMP-selective phosphodiesterase type 4A splice variants in rat brain by chronic antidepressant administration. Eur. J. Neurosci. 2005;22:1463–1475. doi: 10.1111/j.1460-9568.2005.04321.x. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan M, Olsen AS, Houslay MD. Genomic organisation of the human cyclic AMP-specific phosphodiesterase PDE4C gene and its chromosomal localisation to 19p13.1, between RAB3A and JUND. Cell. Signalling. 1999;11:735–742. doi: 10.1016/s0898-6568(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 49.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 50.Johnston LA, Erdogan S, Cheung YF, Sullivan M, Barber R, Lynch MJ, Baillie GS, Van Heeke G, Adams DR, Huston E, Houslay MD. Expression, intracellular distribution and basis for lack of catalytic activity of the PDE4A7 isoform encoded by the human PDE4A cAMP-specific phosphodiesterase gene. Biochem. J. 2004;380:371–384. doi: 10.1042/BJ20031662. [DOI] [PMC free article] [PubMed] [Google Scholar]