SUMMARY
Glycogen is the major mammalian glucose storage cache and is critical for energy homeostasis. Glycogen synthesis in neurons must be tightly controlled, due to neuronal sensitivity to perturbations in glycogen metabolism. Lafora disease (LD) is a fatal, congenital, neurodegenerative epilepsy. Mutations in the gene encoding the glycogen phosphatase laforin result in hyperphosphorylated glycogen that forms water-insoluble inclusions called Lafora bodies (LBs). LBs induce neuronal apoptosis and are the causative agent of LD. The mechanism of glycogen dephosphorylation by laforin and dysfunction in LD is unknown. We report the crystal structure of laforin bound to phosphoglucan product, revealing its unique integrated tertiary and quaternary structure. Structure-guided mutagenesis combined with biophysical and biochemical analyses reveal the basis for normal function of laforin in glycogen metabolism. Analyses of LD patient mutations define the mechanism by which subsets of mutations disrupt laforin function. These data provide fundamental insights connecting glycogen metabolism to neurodegenerative disease.
INTRODUCTION
Glycogen, the major glucose storage molecule in animals, plays an essential role in energy metabolism throughout the body. The brain is the organ most susceptible to decreases in glucose availability (Dinuzzo et al., 2014; Fryer and Brown, 2014). During the past 20 years, the perceived role of brain glycogen has shifted from an emergency energy supply to a dynamic participant in brain metabolism (Dinuzzo et al., 2014; Fryer and Brown, 2014; Swanson, 1992). While neuronal glycogen was thought to be limited to embryonic neurons, adult neurons express both glycogen synthase and glycogen phosphorylase, and they produce low levels of glycogen (Duran et al., 2014; Lovatt et al., 2007; Pfeiffer-Guglielmi et al., 2003; Saez et al., 2014; Vilchez et al., 2007). However, glycogen synthesis in neurons must be tightly controlled, because both over-accumulation and aberrant accumulation induce neuronal apoptosis (DePaoli-Roach et al., 2010; Duran et al., 2014; Turnbull et al., 2011; Valles-Ortega et al., 2011; Vilchez et al., 2007).
Glycogen is a branched polymer of glucose units joined by α-1,4-glycosidic linkages, formed by glycogen synthase, and branches occurring every 12–14 units via α-1,6-glycosidic branches, created by branching enzyme (Roach et al., 2012). Branches within glycogen are evenly distributed, resulting in a spherical structure with exposed non-reducing chain ends. This unique organization allows cells to store up to 55,000 glucose units in a water-soluble form that can be rapidly released during bursts of metabolic energetics. Mutations in the Epilepsy, Progressive Myoclonus 2A (EPM2A) gene encoding the glycogen phosphatase laforin cause Lafora disease (LD) (Minassian et al., 1998; Serratosa et al., 1999). Laforin mutations result in hyperphosphorylated glycogen with decreased branching and increased glucose chain length that eventually forms an insoluble glucan inclusion called a Lafora body (LB) (Nitschke et al., 2013; Tagliabracci et al., 2011). Notably, it was recognized that LBs share a similar morphology with plant amylopectin, the major constituent of starch (Yokoi et al., 1968). Starch also contains branched polymers of glucose, but it is water-insoluble due to its longer glucose chains of 20–25 glucose units that are branched in clusters and form helices that exclude water (Blennow and Engelsen, 2010; Gallant et al., 1997). Recent studies demonstrated that LBs possess longer glucose chains than glycogen and they contain monoester-bound phosphate, both molecular features that resemble plant starch rather than human glycogen (Nitschke et al., 2013; Tagliabracci et al., 2011). Indeed, LD is one of a family of glycogen storage diseases and offers a unique window into glycogen metabolism and associated diseases.
LD is a fatal, autosomal recessive, neurodegenerative disorder and one of the five major progressive myoclonic epilepsies (Berkovic et al., 1991; Minassian et al., 2001). LD patients typically present with an epileptic event in the second decade of life that invariably progresses to intractable and fatal seizures within 5–10 years (Berkovic et al., 1991; Minassian et al., 2001). Disease symptoms result from neuronal apoptosis driven by the accumulation of the cytoplasmic, hyperphosphorylated, water-insoluble LB inclusions, which are normally prevented by the action of laforin (Duran et al., 2014; Turnbull et al., 2011; Valles-Ortega et al., 2011; Vilchez et al., 2007). These inclusions occur throughout the body, but neurons are acutely sensitive to polyglucosan-mediated toxicity (DePaoli-Roach et al., 2010; Duran et al., 2014; Valles-Ortega et al., 2011; Vilchez et al., 2007).
Laforin is the founding member of the glucan phosphatase family and dephosphorylates glycogen both in vitro and in vivo (Gentry et al., 2007; Tagliabracci et al., 2008; Tagliabracci et al., 2007; Worby et al., 2006). Laforin possesses a carbohydrate binding module (CBM) family 20 domain followed by a dual specificity phosphatase (DSP) domain (Minassian et al., 1998; Serratosa et al., 1999; Wang et al., 2002). Highlighting its basic biological importance, laforin orthologs are found in all vertebrates as well as several unicellular eukaryotes (Gentry et al., 2007; Gentry and Pace, 2009). While having a critical role in normal glycogen metabolism and aberrant LB formation, the mechanism of glycogen dephosphorylation by laforin is not known. Further, LD mutations are distributed throughout the primary sequence of laforin, leaving open the critical question of the mechanism(s) by which mutations in laforin lead to LB formation.
Plants utilize a cyclic process of reversible phosphorylation by glucan dikinases and glucan phosphatases for efficient starch degradation (Silver et al., 2014; Streb and Zeeman, 2012). Phosphorylation of starch outer glucans results in solubilization, thereby allowing degradation by starch hydrolyzing amylases, and subsequent dephosphorylation by the glucan phosphatases Starch EXcess 4 (SEX4) and Like Sex Four2 (LSF2). All known glucan phosphatases are members of the Protein Tyrosine Phosphatase (PTP) superfamily within the Dual-Specificity Phosphatases (DSPs) clade (Gentry et al., 2009; Gentry et al., 2007; Tonks, 2006). The DSP domain is an ~150 amino acid domain that is <10% identical among the 65 human DSPs. The heterogeneous DSPs all utilize a cysteine residue at the base of the active site within the conserved Cx5R catalytic motif to perform nucleophilic attack on the phosphorus atom of the substrate (Tonks, 2006). Differences within the DSP domain and active site allow for different members of the DSP family to specifically dephosphorylate proteinaceous substrates, glucans, lipids, or nucleic acids (Moorhead et al., 2009; Tonks, 2006).
Each glucan phosphatase possesses unique features that enable it to bind and dephosphorylate phosphorylated glucans. We recently demonstrated the molecular basis for plant glucan phosphatase function (Meekins et al., 2013; Meekins et al., 2014; Vander Kooi et al., 2010). SEX4 possesses a DSP and CBM domain followed by a carboxy-terminal (CT)-motif. In SEX4, the DSP and CBM share an extensive interdomain interface that forms a continuous binding pocket to engage a hexasaccharide. Conversely, LSF2 possesses only a DSP domain and CT-motif and utilizes two Secondary Binding Sites (SBSs) to engage glucan substrates. Laforin possesses CBM and DSP domains in the reverse orientation as SEX4, it lacks a CT motif, and it possesses a unique inter-domain linker suggesting a novel physical basis for the specific activity of laforin versus glycogen.
Despite glycogen metabolism being a mainstay in textbooks, work on Lafora disease has yielded a number of recent breakthroughs regarding glycogen biology including the discovery of both glycogen dephosphorylation as well as putative therapies for glycogen storage diseases. Laforin is critical for regulating the phosphorylation state of glycogen, which is necessary to maintain glycogen homeostasis. Understanding the mechanism(s) underlying human disease mutations in laforin is critical for understanding the molecular basis for LD and normal glycogen metabolism. Herein, we report the structure of laforin bound to phospho-glucan product. The structure reveals the unique tertiary and quaternary architecture of laforin, providing a platform for understanding its causative role in LD. We define the basis for subsets of mutations that selectively disrupt laforin substrate engagement and specific activity. These data provide a key connection between this critical fundamental biological process and human neurodegenerative disease.
RESULTS
Crystal Structure of Laforin Bound to Maltohexaose
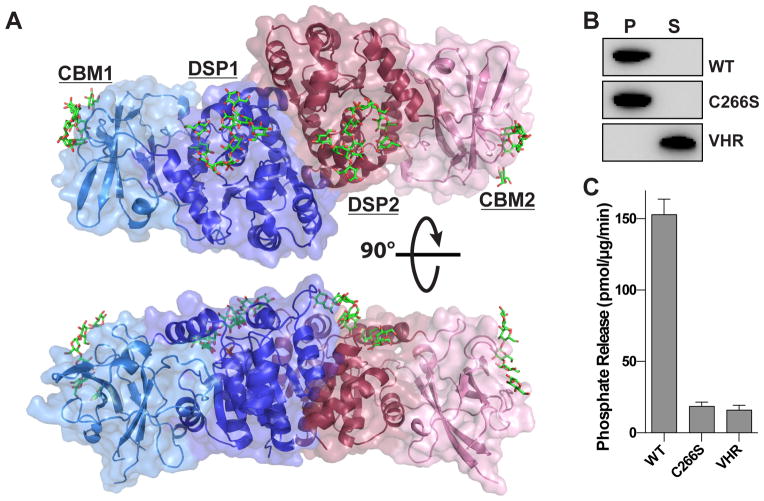
The structure of human laforin bound to maltohexaose and phosphate was determined using tungstate single-wavelength anomalous diffraction (SAD) phasing and refined to a resolution of 2.4 Å (Figure 1A and Table 1). There are two laforin molecules in the asymmetric unit, which are superimposable and each possesses a CBM and DSP domain. Laforin is the only known phosphatase in the human proteome that contains CBM and DSP domains, and the structure displays unique tertiary and quaternary structure. The N-terminal CBM domain is intimately associated with the DSP domain, forming an integrated tertiary structure. Furthermore, the two laforin molecules form an antiparallel dimer mediated entirely by the DSP domain (Figure 1A). Multiple maltohexaose molecules are bound to the laforin CBM and DSP domains, along with a single phosphate at the DSP active site. Maltohexaose is a soluble oligosaccharide composed of six glucose moieties with α-1,4-glycosidic linkages, and serves as a model for the linear portions of glycogen.
Figure 1. Crystal structure of human laforin bound to maltohexaose and phosphate.
(A) Structure of human laforin bound to maltohexaose (green) and phosphate (orange). The laforin structure is an anti-parallel dimer with one molecule in blue and the other in red. Each molecule contains a CBM and DSP domain yielding a tetramodular CBM1-DSP1-DSP2-CBM2 structure. Maltohexaose chains are bound at the CBM and DSP domains with a single phosphate molecule located at the base of the catalytic site.
(B) Glycogen co-sedimentation assay to measure glycogen binding. Recombinant laforin wild-type (WT), laforin C266S, and VHR were incubated with glycogen bound to Con A agarose beads, glycogen was pelleted by centrifugation, and proteins in the pellet (P) and supernatant (S) were separated by SDS-PAGE and visualized by Western analysis. Glycogen-bound proteins are in the pellet and unbound proteins are in the supernatant.
(C) Specific activity of laforin WT, inactive mutant laforin C266S, and human VHR against glycogen. 100 ng of protein was incubated with 45 μg glycogen for 20 minutes. Each bar is the mean ± SEM of six replicates, P < 0.05.
Table 1.
Data collection and refinement statistics
| Native | Tungstate | |
|---|---|---|
| Data Collection | ||
| Beamline | APS 22-ID | APS 22-ID |
| Wavelength | 1.0000 | 1.2148 |
| Space group | P43212 | P43212 |
| Unit cell parameters (a, b, c) | 78.71, 78.71, 321.12 | 78.68, 78.68, 321.10 |
| Unique reflections | 37,585 | 38,389 |
| Completeness (%) | 92.3 (92.0) | 99.8 (100.0) |
| Resolution (Å) | 20.0–2.4 (2.49–2.40) | 20.0–3.0 (3.11–3.00) |
| Rmerge (%) | 9.7 (51.6) | 10.5 (26.4) |
| Redundancy | 3.6 (2.8) | 3.8 (3.6) |
| I/σ(I) | 11.6 (2.5) | 12.3 (5.7) |
| Refinement | ||
| Resolution limits (Å) | 20.00–2.40 | |
| No. reflections/no. to compute Rfree | 33896/3582 | |
| R/Rfree | 16.6/21.1 | |
| No. protein residues | 624 | |
| No. glucose/phosphate | 30/2 | |
| No. of water | 143 | |
| RMSD Bond, Å | 0.008 | |
| RMSD Angle, ° | 1.54 | |
| Ramachandran favored/outlier (%) | 98.24/0.0 | |
| MolProbity score | 0.92 (100th percentile) | |
Laforin is the only known human phosphatase that can dephosphorylate glucan substrates (Gentry et al., 2007; Sherwood et al., 2013; Worby et al., 2006). To confirm the function of the crystallized laforin construct, we tested the ability of the catalytically active protein to both bind and dephosphorylate glycogen. Laforin readily bound to glycogen, whereas the prototypical human DSP protein phosphatase vaccinia H1-related (VHR/ DSP3) did not (Figure 1B). Additionally, laforin readily dephosphorylated glycogen (Figure 1C). This activity was specific, since mutation of the catalytic nucleophile (C266S) lacked specific activity. Additionally, VHR also lacked glycogen phosphatase activity. Thus, the crystallized laforin construct represents an active specific glycogen phosphatase.
This structure opens new avenues for understanding laforin function in glycogen dephosphorylation and the effect of LD disease mutations. A significant number of LD mutations are localized to regions of the protein involved in physical interactions, including the DSP active site, CBM binding site, CBM-DSP interface, buried regions within individual domains, and the DSP-DSP dimer interface. Thus, the laforin structure allows us to probe the mechanism(s) by which LD patient mutations affect laforin function.
Maltohexaose and Phosphate Bound at the Laforin Active Site
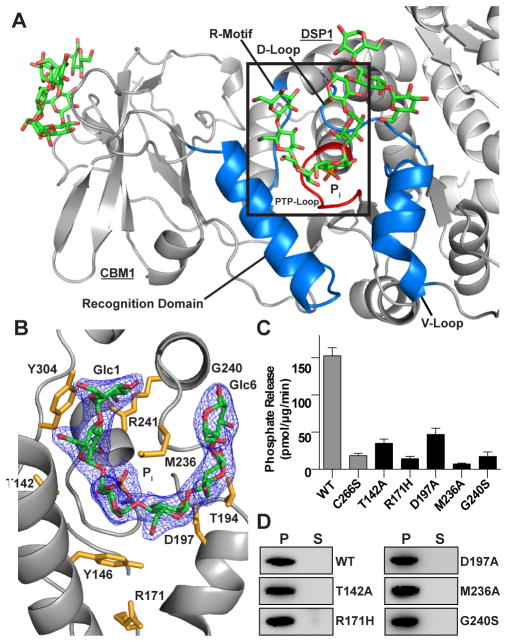
A bound maltohexaose chain and phosphate are tightly integrated into the laforin DSP domain active site, with a contact area of 655 Å2. The active site is broad, with a cross section ~17 Å wide and 10 Å deep, and is formed by residues from different portions of the DSP domain whose variability among DSP family members yields unique substrate specificities (Figure 2A). The laforin DSP domain is structurally most similar to the plant glucan phosphatases LSF2 and SEX4, with an R.M.S.D. of 2.0 and 2.1Å (Figure S1), respectively. The electron density of maltohexaose in the laforin active site allows unambiguous assignment of glucan chain orientation, labeled Glc1-Glc6 from the nonreducing to reducing end (Figure 2B). Seven conserved laforin residues make extensive interactions with the maltohexaose glucose moieties (Figure 2B). M236 fully engages the concave surface of the maltohexaose chain. T142, Y146, T194, D197, R241, N267, and Y304 interact with the convex surface, which involves many hydrogen bonds as well as van der Waals interactions. Each of these interacting residues is strictly conserved in laforin vertebrate orthologs (Figure S2A). At the base of the active site is the PTP-loop Cx5R motif (HCNAGVGR, residues 265–272). The cysteine within this motif sits at the base of the catalytic cleft and functions as a nucleophile during catalysis. The O3 and O2 of Glc3 are positioned 2.7 Å and 3.8 Å from the phosphate, respectively, with the O6 at a distance of 7.7 Å pointed towards solvent (Figure S3).
Figure 2. Maltohexaose bound at the laforin DSP domain active site.
(A) Ribbon diagram of one laforin molecule with maltohexaose (green), phosphate (orange), backbone (grey), DSP domain motifs (blue), active site loop (red), and the second molecule (light grey).
(B) Inset from A, showing a 2Fo-Fc electron density map (1.3 σ) with maltohexaose (green) and phosphate (orange). Glucose moieties (Glc) are numbered from non-reducing to reducing end. Side chains of interacting residues are colored yellow. See also Figure S3.
(C) Specific activity of laforin WT and C266S (from Figure 1) compared to DSP active site mutants. Each bar is the mean ± SEM of six replicates, P < 0.05.
(D) Glycogen co-sedimentation assay of WT and DSP active site mutants.
To investigate the role of these residues in laforin function, we generated alanine mutations of M236, T142, and D197. Additionally, we tested multiple LD mutations in the DSP domain that are localized to the DSP active site, including R171H and G240S (Figure 2B). We first tested the effect of these mutations on laforin stability and folding by measuring protein stability using differential scanning fluorimetry (DSF). None of the mutations showed a significant decrease in stability (Table S1). After establishing that the laforin point mutants did not destabilize the protein, we tested the mutants’ phosphatase activity against glycogen. In all cases, we found a significant decrease in specific activity, ranging from 69–95% decrease compared to wild-type (WT) (Figure 2C). M236A was the most deleterious to laforin activity, consistent with its observed extended interaction surface with the bound carbohydrate.
Since the laforin DSP independently engages a glucan chain, a loss of glucan phosphatase activity may indicate a decrease in glucan binding. To assess binding to glycogen, we incubated laforin with glycogen coupled to Concanavalin A (Con A) Agarose (GE Healthcare), washed the agarose beads, performed a low speed spin, and used Western analysis to determine if laforin was bound to glycogen in the pellet or remained in the supernatant. We found that the laforin DSP mutants pelleted with glycogen similarly to WT (Figure 2D). Thus, mutations in the DSP domain active site channel do not dramatically decrease glycogen binding, but specifically impact the ability of laforin to dephosphorylate glycogen.
Laforin CBM-Glucan Interactions
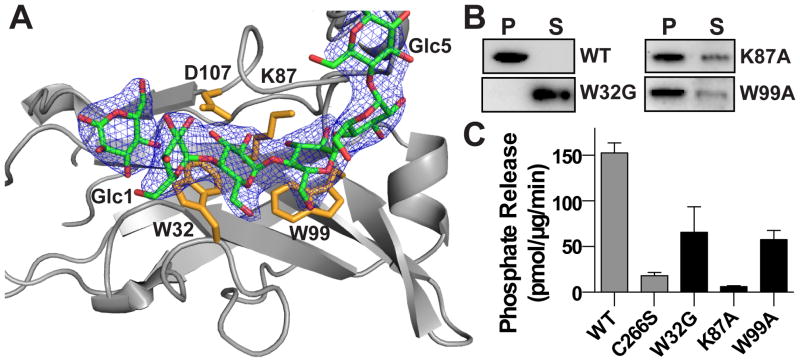
A separate maltohexaose chain is bound to the canonical carbohydrate binding site in the laforin CBM domain (Figure 3A). Aromatic and hydrophilic residues within the CBM engage the maltohexaose, with a contact area of 327 Å2, via a network of stacking interactions and hydrophilic residues including the sidechains of W32, K87, W99, and D107 and backbone of G103. The tryptophan residues interact with one face of the glucose units. W32, K87, and W99 are strictly conserved and D107 is highly conserved in all laforin orthologs (Figure S2B).
Figure 3. Maltohexaose bound at the laforin CBM domain.
(A) Close-up of the laforin CBM domain with bound maltohexaose (green) 2Fo-Fc electron density map (1.0 σ) with CBM residues (yellow). Glucose moieties (Glc) are numbered from nonreducing to reducing end.
(B) Glycogen co-sedimentation assay of WT and CBM domain mutants.
(C) Specific activity of laforin WT and C266S (from Figure 1) compared to CBM domain mutants. Each bar is the mean ± SEM of six replicates, P < 0.05.
To define the contribution of the CBM to laforin activity, we mutated K87 and W99 to alanine, as well as assessed the LD patient mutation W32G. W32G showed reduced stability with the Tm decreased by 5.1 °C, indicating that it contributes to protein stability (Table S1). Indeed, the structure identifies a number of key core hydrophobic residues including F5, Y86, and F88 that are mutated in LD and likely destabilize the protein. In contrast, we found the other mutant proteins had equivalent stability to WT protein. To determine how these mutations impact laforin binding to glycogen, we employed the Con A-glycogen experiment. We found that W32G was the most deleterious, being found entirely in the supernatant fraction. K87A and W99A also exhibited decreased glycogen binding by ~40% (Figure 3B). The CBM mutations resulted in a 57–96% decrease in glycogen phosphatase activity (Figure 3C). Thus, LD and CBM mutations that affect CBM domain stability and/or direct glucan engagement lead to loss of enzymatic activity. These data indicate the importance of glycogen engagement by the CBM domain for laforin activity.
Laforin Architecture and Dynamics
While each domain is clearly important for laforin function, the structure reveals a unique tertiary architecture with tightly integrated CBM and DSP domains, with a contact area of 1154 Å2. The relative orientation of the CBM and DSP domains is dramatically different than that observed in the plant glucan phosphatase SEX4 (Figure S1). Most strikingly, the two carbohydrate binding sites are not proximal, as was seen in SEX4 (Figure S1). Thus, we next probed the relationship and coupling between the protein domains.
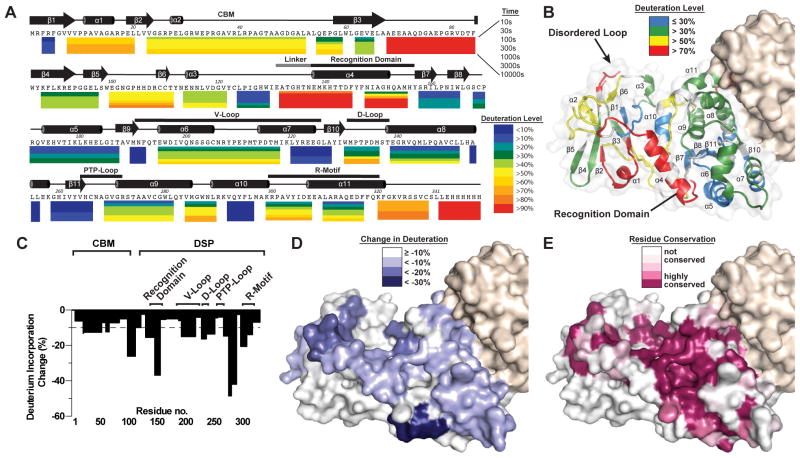
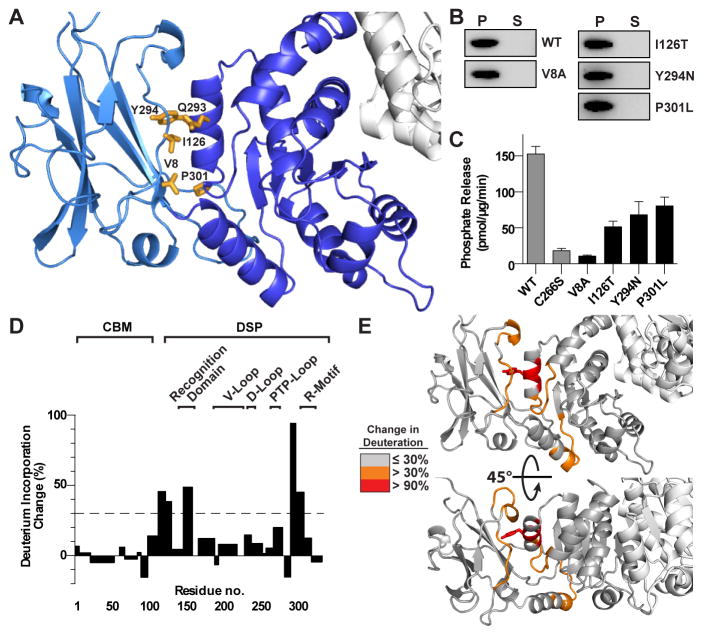
We employed deuterium exchange mass spectrometry (DXMS) to elucidate the conformational dynamics of laforin in solution, and the effect of binding to its physiological substrate glycogen in terms of changes in tertiary and quaternary structure. Optimization of the assay allowed 100% coverage of C-terminally His6-tagged laforin WT and allowed us to assess the change in peptide mass with increasing time (Figure S4A). Strong differences in protection were observed for different regions of the protein (Figure 4A and 4B). Among the most protected regions, defined as <20% deuteration from 10 s to 1,000 s, are residues 122–129 in the CBM domain and 290–296 in the DSP domain. These are residues in the complementary surfaces of the CBM-DSP domain interface, indicating that the observed tertiary structure is stable in solution and suggesting that there is minimal structural rearrangement of this region. Additionally, residues 218–227 and 252–256, both located at the dimer interface in the DSP domain, exhibit <20% deuteration throughout the experiment, consistent with stable dimer association in solution.
Figure 4. Deuterium exchange mass spectrometry (DXMS) analysis of laforin architectural dynamics.
(A) Laforin dynamics from DXMS expressed as percent deuteration of laforin peptides with the lowest deuteration in blue to highest in red, as indicated. Each colored bar below the primary sequence represents percent deuteration at one of seven timepoints. Secondary structure elements are depicted above the primary sequence with the CBM domain and DSP motifs labeled.
(B) Surface mapping of the percent deuterium exchange of each peptide at 300 s was mapped onto the laforin structure. Residues 69–79 were 100% deuterated throughout the DXMS experiment, and these residues were disordered and not modeled in the structure (labeled disordered loop).
(C) Maximal percent change in deuteration of laforin ± glycogen versus the laforin primary sequence. The dotted line designates 10% decrease. See also Figure S4B.
(D) Surface mapping depicting the regions that exhibit decreased deuteration in the presence of glycogen.
(E) Surface model of laforin illustrating amino acid conservation using the sequences from Figure S2A and S2B.
Regions that are highly accessible to solvent, defined as greater than or equal to 50% deuterated after 300 s in D2O, are primarily within surface exposed regions (Figure 4B). The region that is the most highly solvent accessible, exhibiting >90% deuteration at all time points, is residues 66–84 of the CBM domain that corresponds to the disordered loop not modeled in the crystal structure (Figure 4A). The CBM-DSP domain linker and DSP recognition motif also displayed >70% deuteration at 300 s (Figure 4A and 4B) yet other peptides did not display high levels of deuteration, suggestive of local dynamics but not global conformational changes.
Next, we utilized DXMS to evaluate structural changes in laforin in the presence of its endogenous substrate glycogen. In the presence of glycogen, we observed significant substrate-induced decreases in the exchange rate within both the CBM and DSP domain (Figures 4C and S4B). Three peptides within the CBM exhibited 13–27% decreases of deuteration in the presence of glycogen: residues 21–52, residues 60–65, and residues 98–111. The identified oligosaccharide-binding residues W32, W99, and D107 lie within these regions (Figure 4A). Likewise, regions within the DSP domain surrounding the active site also showed significant protection, decreases of 13–45%, including within the recognition domain (residues 130–145, 146–155), V-loop (residues 193–217), D-loop (residues 228–236), PTP loop (267–281), and R-motif (308–317). Changes were restricted to the surface of the laforin structure encompassing the active site cleft with little to no change on the opposite face (Figure 4D). Peptides involved in the CBM-DSP domain interface and the dimerization interface did not exhibit significant changes in the presence of glycogen. Indeed, there were no significant increases in deuteration in any region of laforin, suggesting that there is not an increase in protein dynamics associated with substrate binding. Intriguingly, the CBM and DSP-bound maltohexaose chains are positioned in a parallel orientation, i.e. the non-reducing and reducing ends are oriented in the same direction. When residue conservation is mapped onto the surface of the structure, very high levels of conservation are observed for the active site and intervening region to the CBM binding site (Figure 4E). These regions closely resemble the less deuterated regions in the presence of glycogen, suggestive of a physical path for cooperative binding of glycogen.
The CBM and DSP Domain Interaction is Critical for Function and Impacts Lafora Disease
Given the extensive interdomain interface, stable dynamics, and conservation we hypothesized that the observed CBM/DSP orientation exhibited in the laforin tertiary structure is critical for its function (Figure 5A). Indeed, many of the residues that participate in interdomain interactions are mutated in LD, including I126T, Q293L, Y294N, and P301L. We generated an alanine mutation of V8, as well as analyzed the LD mutants I126T, Y294N, and P301L, all of which are located at the interdomain interface. P301L displayed a decreased Tm, while all other mutant proteins were stable and well folded with no significant difference in Tm compared to WT protein (Table S1). We investigated the glycogen binding of these mutations using a Con A-glycogen assay and found that each mutant pellets with glycogen, indicating that glycogen binding is not dramatically affected (Figure 5B). Next, we tested the glycogen phosphatase activity of each mutant and found that V8A, I126T, Y294N, and P301L significantly decreased the glycogen phosphatase activity of laforin by 47–93% (Figure 5C). Thus, mutations within the CBM-DSP interface significantly decrease glycogen phosphatase activity without diminishing glycogen binding.
Figure 5. The laforin CBM-DSP interface is necessary for laforin function.
(A) Close up of the CBM (light blue) DSP (dark blue) domain interface. Residues that comprise the CBM-DSP interface are yellow.
(B) Glycogen co-sedimentation assay of WT and CBM-DSP interface mutants.
(C) Specific activity of laforin WT and C266S (from Figure 1) compared to CBM-DSP interface mutants. Each bar is the mean ± SEM of six replicates, P < 0.05.
(D) Structural dynamics of the laforin mutant Y294N with a bar graph depicts maximal percent change in deuteration between WT and Y294N, positive values depict increased deuteration in Y294N and negative values depict decreases. The dotted line designates 30% change in deuteration.
(E) Surface mapping of regions that exhibit changes deuteration between WT and Y294N.
To further explore the structural dynamics of the CBM-DSP interface, we performed DXMS analysis of the LD patient mutation laforin Y294N. Strikingly, the Y294N mutation significantly increased the solvent accessibility of the CBM/DSP interface (Figures 5D, 5E and S4C). Specifically, residues 290–296 and 297–307 were 95% and 45% more deuterated in laforin Y294N than in WT, respectively. These residues surround Y294 and include the DSP domain residues that comprise the CBM-DSP interface (Figure 5E). Further, CBM domain residues comprising the opposite side of the interface also exhibit increased deuteration compared to WT with residues 113–121 and 122–129 increasing 50% and 39%, respectively. While the CBM/DSP interface exhibited increased deuteration, there was no significant change in deuteration of peptides comprising the dimer interface. These results indicate that the unique tertiary structure with specific CBM/DSP orientation is required for laforin activity and provides an explanation for the role that mutations of these interfacial residues play in LD.
Laforin Dimerization
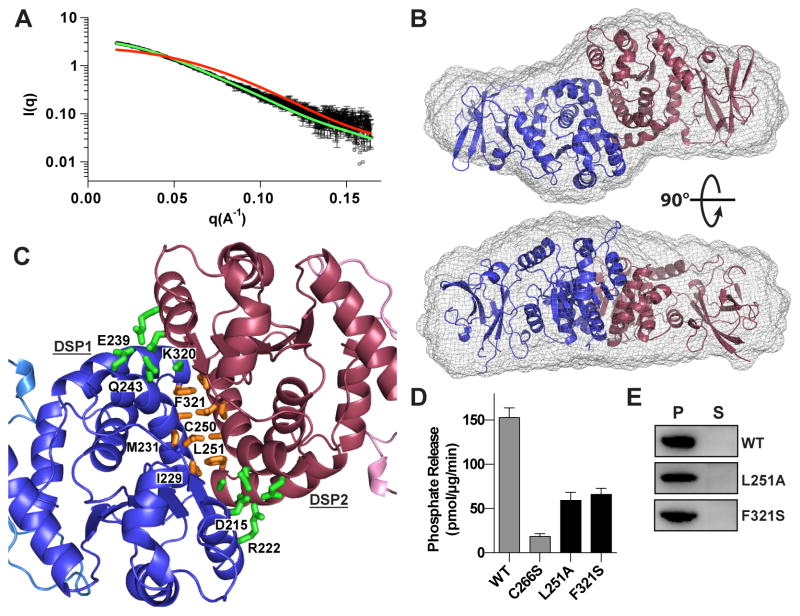
The physical and functional role of laforin oligomerization has been controversial (Dukhande et al., 2011; Koksal and Cingolani, 2011; Liu et al., 2006; Liu et al., 2009; Sanchez-Martin et al., 2013). In situ and in vivo experiments have demonstrated that laforin is able to both dimerize and multimerize, but different models have been proposed for the extent to which oligomers form and which domains contribute. The structure reveals that laforin is an anti-parallel dimer mediated exclusively by the DSP domains, with an extended 906 Å2 interface and high shape complementarity (Sc=0.81) (Figure 1A).
To examine the structure of laforin in solution, we employed inline size exclusion chromatography-small angle X-ray scattering (SEC-SAXS). Laforin eluted from SEC as a single species (Figure S5A) with molecular size and shape parameters Rg = 33.9 ± 0.2 Å and Dmax = 119 Å. Rg was invariant across a twenty five-fold concentration range as the protein eluted from the column (33.7 ± 0.7 Å for the entire range) indicating that the protein was not exchanging between oligomeric states. The experimental scattering curve demonstrated strong agreement with the theoretical profile calculated for the laforin dimer (predicted Rg = 31.2 Å and Dmax = 121 Å) observed in the crystal structure (χ2 = 1.1) (Figures 6A, S5B, and S5C). In contrast, the theoretical scattering profile for a single copy of laforin monomer (predicted Rg = 21.2 Å and Dmax = 78 Å) showed a marked deviation from the experimental data (χ2 = 27.0). An ab initio reconstruction of the average molecular envelope revealed an extended bilobal shape that closely matched the crystal structure of laforin (Figure 6B). The excluded volume indicated that the molecular weight of the protein was 72 kDa, matching the theoretical molecular weight of the laforin dimer (74 kDa) and further confirming that laforin forms a stable dimer in solution. Thus, the solution scattering data demonstrate that the tertiary and quaternary structure of laforin observed in the crystal structure represent the native conformation of the protein in solution.
Figure 6. Laforin quaternary structure is critical for its function.
(A) Scattering curve of laforin C266S (black) with curves computed from the crystallographic structure for monomer (red) and dimer (green).
(B) Ab initio model of laforin C266S with the computed surface (gray mesh) overlayed with the crystal structure.
(C) The laforin DSP domain dimer interface with interfacial hydrophilic (green) and hydrophobic interacting residues (gold). Residues from DSP1 are labeled.
(D) Specific activity of WT and C266S (from Figure 1) compared to dimer interface mutants. Each bar is the mean ± SEM of six replicates, P < 0.05.
(E) Glycogen co-sedimentation assay of WT and dimer interface mutants.
Closer examination of the dimerization interface demonstrates that it contains a core hydrophobic region comprised of I229, M231, L251, C250, and F321 and outer hydrophilic regions containing D215, R222, E239, Q243, and K320 (Figure 6C). In particular, F321 is buried at the interface and is mutated in the LD patient mutation F321S.
To determine the effect of mutations at the laforin dimeric interface on function we next tested the effect of F321S or L251A mutations. Both mutations significantly affected laforin stability, resulting in >5°C decreases in protein stability (Table S1). Mutation at the dimer interface also significantly decreased glycogen phosphatase activity, with observed activity decreases of 56–62%, but without global effects on glycogen binding (Figure 6D and 6E).
Laforin Substrate Binding Cooperativity
To examine the oligomeric state of the F321S LD mutation, we employed analytical ultracentrifugation (AU). Consistent with SEC and SAXS solution measurements, WT laforin exists as a stable dimer in solution with a single peak observed at 79 ± 6 kDa (Figure 7A and S6A). Strikingly, the F321S mutation disrupts dimerization, existing as a single species of 41 ± 6 kDa, consistent with the monomeric molecular weight of laforin (Figure 7B, S6B).
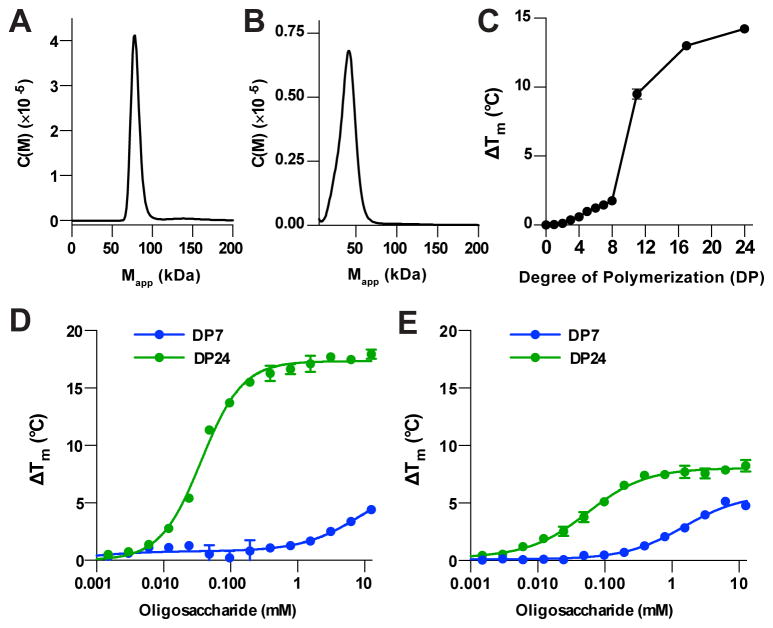
Figure 7. Cooperative binding to glucan substrates.
(A) Sedimentation velocity experiments reveal that laforin WT exists as a dimer in solution.
(B) Sedimentation velocity experiments reveal that laforin F321S exists primarily as a monomer in solution.
(C) Thermal shift assay with 1 mM oligosaccharides reveals that laforin C266S is more stabilized in the presence of increasing degree of polymerization (DP).
(D) Concentration dependent increase in laforin C266S melting temperature (ΔTm) with maltohexaose (DP6) maltodextrin (DP24).
(E) Concentration dependent increase in laforin F321S melting temperature (ΔTm) with maltohexaose (DP6) maltodextrin (DP24).
The dimeric form of laforin is structurally organized with CBM binding interfaces and DSP active sites aligned. We next tested the role of dimerization in substrate binding by measuring the binding of laforin to varying lengths of oligosaccharides using a thermal shift assay. We first tested the degree of stabilization in the presence of malto-oligosaccharides from maltose of degree of polymerization 2 (DP2) through DP24. We saw little effect with DP2-DP4 oligosaccharides and a minimal length-dependent increase in Tm for DP5-DP8 with each additional glucose moiety yielding an ~0.3 °C ΔTm (Figure 7C). Strikingly, there was a dramatic increase in stabilization when using longer oligosaccharides, with a 24-mer yielding a ΔTm increase of ~14 °C. Further, when the concentration dependence was measured, a shorter oligosaccharide showed a broad transition with two binding events, whereas the longer oligosaccharide showed a single high affinity binding with a Hill coefficient of 1.41, demonstrating striking cooperativity in binding (Figure 7D). We next assessed the effect of laforin mutant protein for cooperative binding. F321S exhibited a significantly decreased degree of stabilization by glucan substrate and notably loss of cooperativity, with a Hill coefficient of 0.95 (Figure 7E). Overall, these data provide a mechanism for laforin dimerization in its function, and provide a basis for understanding the effect of LD mutations that disrupt dimerization.
DISCUSSION
The role of glycogen dephosphorylation is an emerging area of research that directly impacts our understanding of normal glycogen metabolism as well as dysfunction in the fatal human epilepsy Lafora disease. The data presented here provide fundamental insights into how laforin functions in glycogen dephosphorylation and uncovers novel features of the laforin protein that provide a basis to understand disease mechanisms. The domain orientation in laforin is unique and distinct from the other known glucan phosphatases with the carbohydrate binding regions of the CBM and DSP domains distant from each other. In contrast, SEX4 employs a continuous binding pocket that spans both the CBM binding site and DSP active site allowing it to bind a single maltohexaose chain (Meekins et al., 2014). The laforin dimer produces a unique tetramodular architecture with CBM-DSP-DSP-CBM domains aligned and engaging multiple maltohexaose chains. Structure-guided and patient mutations are described that result in loss of functional interactions in the CBM binding site, DSP active site, CBM/DSP interface, and dimerization interface. Our data demonstrate that this unique tertiary and quaternary structure allows for engagement of glycogen by the CBM domain, cooperative substrate binding by the dimer, engagement of the phosphorylated glucan at the DSP active site, and finally catalysis.
This model has direct implications for our understanding of the source and position-specific function of glycogen phosphorylation on both glycogen superstructure and LB formation, which is currently being explored. Some propose that phosphate is introduced by a glycogen synthase side reaction, while others suggest specific incorporation by a yet unidentified human glycogen kinase (Nitschke et al., 2013; Tagliabracci et al., 2011). In plants, regulated phosphorylation of starch is better characterized. Glucan, water dikinase (GWD) specifically phosphorylates the C6 position that targets phosphoglucan, water dikinase (PWD) to phosphorylate C3 hydroxyls. These phosphorylation events induce steric strain and disrupt the helical structure of starch, and allow hydrolytic enzymes to processively degrade starch (Blennow and Engelsen, 2010; Silver et al., 2014; Streb and Zeeman, 2012). Complementary to the dikinases, the plant glucan phosphatases possess distinct preference for dephosphorylating glucose positions. SEX4 preferentially dephosphorylates the C6 position, while LSF2 exclusively dephosphorylates the C3 position (Kotting et al., 2009; Meekins et al., 2013; Meekins et al., 2014; Santelia et al., 2011). This position-specific activity was shown to be due to orientation-specific binding of the glucan substrates in the active site with C3-hydroxyls in the active site of LSF2 and C6-hydroxls for SEX4 (Meekins et al., 2013; Meekins et al., 2014). Glycogen can be phosphorylated at the C2, C3, and C6 position (Nitschke et al., 2013). The laforin structure shows preferential binding of the glucan with C3/C2 oriented towards the catalytic center, suggesting that it will preferentially dephosphorylate C3 or C2 positions (Figure S3). Indeed, the C6 hydroxyls are pointed away from the laforin active site into the solvent. As with SEX4, laforin likely will be able to act on all positions to some extent, but would require the opposite glucan orientation to accommodate dephosphorylation of the C6 position and thus is expected to have significantly reduced activity level for the C6 position.
Currently, the position of phosphorylation with respect to α-1,6 branchpoints is unresolved. It has been reported that C6 monophosphates may be selectively elevated in LBs and predominate near the center of glycogen molecules, thereby affecting branching (Nitschke et al., 2013). Intriguingly, an additional portion of an oligosaccharide chain is observed in the crystal structure of laforin near the DSP active site and a single ordered glucose moiety is observed near the CBM binding site. This is likely a consequence of the high concentration of carbohydrate used in crystallization, but it may suggest the possibility of preferential binding to specific glycogen regions, particularly branch points. The laforin structure demonstrates that a shallow and wide binding pocket (10 Å × 17 Å) is a conserved feature of glucan phosphatases from plants to animals and an architectural feature that would accommodate a branched glycogen chain. The ability of laforin to bind and localize to specific sub-regions within glycogen is an area of on-going research that will require temporal and quantitative comparisons of wild type and mutant proteins in the context of physiological substrates. These results will further define the role of phosphorylation/ dephosphorylation in LB formation and disease progression.
The physical and physiological relevance of laforin dimerization has been a point of debate in the field (Dukhande et al., 2011; Liu et al., 2006; Liu et al., 2009; Sanchez-Martin et al., 2013). The crystal structure, along with solution-based measurements using complementary techniques, establishes that laforin exists as an antiparallel dimer mediated by the DSP domain. Notably, laforin does not possess a CT-motif, which is required for stability and function in plant glucan phosphatases. Instead, the laforin dimer interface partially overlaps with the region where the CT-motif binds to the DSP domain (Figure S1). Furthermore, we demonstrate that the LD mutation F321S disrupts laforin dimerization and diminishes glycogen phosphatase activity and cooperative substrate binding. Dimerization is a common theme regulating the activity of transmembrane receptor-like PTPs (RPTPs). A helix-turn-helix wedge motif of one RPTP obstructs the active site of the dimer partner, possibly allowing a ligand-dependent manner to modulate phosphatase activity (Tonks, 2006, 2013). Conversely, dimerization of cytoplasmic PTPs is much less common and DSPs are typically regarded as monomeric enzymes. The human phosphoinositide lipid phosphatase myotubularin related protein 2 (MTMR2) and the Vaccinia virus gene H1 (VH1) are both DSPs that dimerize. The laforin DSP domain forms its dimer interface, while both MTMR2 and VH1 form dimers via ancillary domains. MTMR2 forms a dimer via its C-terminal coiled-coil domain, while the VH1 dimer interface is formed by a swapped N-terminal α-helix (Berger et al., 2003; Koksal and Cingolani, 2011; Phan et al., 2007).
LD offers a unique window into both normal cellular glycogen metabolism and disease implications when the process is perturbed. Recently, a study using a novel glycogen detection method demonstrated that neurons contain low levels of glycogen, and this glycogen protects neurons from hypoxic stress in vitro and in vivo (Saez et al., 2014). Yet, glycogen synthesis in neurons must be tightly controlled, because over-accumulation induces neuronal apoptosis (Duran et al., 2014; Valles-Ortega et al., 2011; Vilchez et al., 2007). Intriguingly, insoluble starch-like accumulations also accumulate in neurons during human aging. Corpora amylacea are insoluble, glucan-based aggregates that form in brain, eye, and peripheral nervous system neurons of individuals thirty years old and greater (Cavanagh, 1999; Sinadinos et al., 2014). The function of corpora amylacea as well as their impact on cellular homeostasis and involvement of laforin is unknown, yet recent evidence suggests that corpora amylacea may constitute an important factor in age-related neurological decline (Sinadinos et al., 2014).
In addition to impacting neuronal processes, perturbations in glycogen metabolism are a feature of glycogen storage diseases, diabetes, cardiac pre-excitation syndromes, and cancer (Light, 2006; Roach, 2002; Roach et al., 2012; Ros and Schulze, 2012). Given the central importance of glycogen to cellular homeostasis, glycogen (de)phosphorylation may impact other aspects of human disease that are yet unexplored. The role of regulated carbohydrate phosphorylation in diverse biological systems by glucan phosphatases, glucan dikinases, and yet to be discovered enzymes opens new avenues for understanding novel aspects of glycogen metabolism as well as human diseases.
EXPERIMENTAL PROCEDURES
Please refer to Supplemental Experimental Procedures for additional experimental details.
Cloning, Protein Expression, and Protein Purification
H. sapiens (Hs) laforin residues 1–328 was expressed from pET28b (Novagen) as an N-terminal His6 tag fusion. Wild-type and mutant proteins were expressed in BL21-CodonPlus E. coli cells and purified using IMAC and SEC.
Crystal Structure Determination
High quality single native and tungstate co-crystals were obtained and data collected on the 22-ID beamline of SER-CAT at the Advanced Photon Source, Argonne National Laboratory to 2.4 Å and 3.0 Å, respectively (Table 1).
Phosphatase Assays
We utilized a phosphatase assay that employs malachite green to quantify inorganic phosphate release from rabbit muscle glycogen, assays were performed six times or more (Sherwood et al., 2013; Tagliabracci et al., 2008; Tagliabracci et al., 2007).
Glycogen Binding Assays
Glycogen binding assays were performed using Concanavalin A (Con A) Agarose beads (Sigma) as described in Supplemental Experimental Procedures.
Hydrogen Deuterium Exchange Mass Spectrometry (DXMS)
DXMS experiments were performed as previously described (Hsu et al., 2009; Pantazatos et al., 2004). 134 high-quality peptides with 100% coverage of laforin were obtained. Exchange of laforin with and without glycogen was performed three independent times, with all samples prepared in triplicate.
Size-Exclusion Chromatography Small - Angle X-ray Scattering (SEC-SAXS)
SEC-SAXS experiments were performed at the 18-ID beamline of BioCAT, Advanced Photon Source, Argonne National Laboratory using an inline Superdex 75 10/300 column (GE). Guinier analysis demonstrated that the system is monodisperse with particles that have a radius of gyration (Rg) = 33.9 ± 0.2 Å (Figure S5B). Pair-distance distribution function (P(r)) demonstrated a well-defined Dmax = 119 Å and real space Rg (35.0 Å) consistent with Guinier analysis (Figure S5C).
Analytical Ultracentrifugation (AU)
Sedimentation velocity experiments were conducted using a Beckman XL-I analytical ultracentrifuge (Beckman-Coulter, Fullerton, CA). Protein was concentrated to 5 μM in size exclusion buffer.
Differential Scanning Fluorimetry (DSF)
Thermal stability experiments were carried out using a CFX96 Real-Time PCR system (BioRad). 2 μM of protein was combined with 5X SYPRO Orange Protein Gel Stain (Invitrogen) with or without maltoligosaccharides and stability assessed from 20 to 90 °C at 1 °C/50 s in triplicate.
Supplementary Material
Figure S1, related to Figure 2. Structure comparison of glucan phosphatases laforin, SEX4 and LSF2.
Ribbon diagrams of the glucan phosphatases laforin, Starch EXcess4 (SEX4), and Like Sex Four2 (LSF2) with a star indicating the DSP domain active site. The laforin dimer interface is spatially positioned similar to the SEX4 and LSF2 CT-motif.
Figure S2, related to Figure 2 and Figure 3. Alignments of laforin orthologs.
(A) Alignment of the DSP domain of laforin vertebrate orthologs. Secondary structure elements are depicted above the primary sequence and DSP motifs are labeled. The catalytic triad residues D235, C266, and R272 are highlighted in red and residues interacting with maltohexaose are in yellow.
(B) Alignment of the CBM domain of laforin vertebrate orthologs. Secondary structure elements are depicted above the primary sequence. Critical residues for glycogen binding are highlighted in blue.
Figure S3, related to Figure 2. The laforin catalytic triad.
Close-up of the laforin DSP domain active site, showing a ribbon diagram of maltohexaose (green) and phosphate (orange) with 2Fo-Fc electron density map (1.3 σ). Glucose moieties in the maltohexaose chain are numbered from non-reducing to reducing end. Side chains of the catalytic triad are displayed, D235 (yellow), S266 (C266 in WT laforin, teal), and R272 (yellow). The O2 and O3 hydroxyls of Gl3 are pointing towards the active site with the O6 hydroxyl in solvent.
Figure S4, related to Figure 4. Deuterium exchange mass spectrometry (DXMS) peptide maps and peptide graphs.
(A) Sequence coverage map of identified pepsin-digested laforin peptides. A total of 134 high-quality peptides were detected. Solid blue lines indicate the 27 peptides utilized in the analysis. Dashed lines indicate peptides that were not used but were examined. Laforin primary sequence is above the corresponding peptides.
(B) Line graphs depicting peptides that exhibited a > 10% change in deuterium incorporation in the presence versus absence of glycogen. Data shown as green represents percentages of deuterium incorporation in glycogen-free laforin and blue represents glycogen-bound laforin. Percent deuterium incorporation is shown in a time-dependent manner.
(C) Line graphs depicting peptides that exhibited a > 10% change in deuterium incorporation for laforin Y294N versus laforin WT. Data shown as red represents percentages of deuterium incorporation for Y294N and green represents WT. Percent deuterium incorporation is shown in a time-dependent manner.
Figure S5, related to Figure 6. Size exclusion chromatography-small angle x-ray scattering (SEC-SAXS) analysis of laforin.
(A) Size exclusion chromatography profile of laforin C266S loaded onto a Superdex 75 10/300 GL (GE Healthcare) reveals that laforin elutes as a single species.
(B) Guinier plot of the low q region of the x-ray scattering data allows measurement of Rg = 33.9 ± 0.2 Å for laforin.
(C) The distance distribution P(r) function of SAXS scattering data of laforin.
Figure S6, related to Figure 7. Analytical ultracentrifugation analysis of laforin.
(A) Laforin WT sedimentation coefficient distribution as determined by sedimentation velocity analytical ultracentrifugation.
(B) Laforin F321S sedimentation coefficient distribution as determined by sedimentation velocity analytical ultracentrifugation.
Table S1: Tm of laforin and laforin mutants
Acknowledgments
We thank Drs. Mike Begley, Carolyn Worby, Skip Waechter, Carlos Roma-Mateo, Pablo Sanchez-Martin, and Yvonne Fondufe-Mittendorf for assistance and advice as well as Drs. Carol Beach, Jing Chen, and Martin Chow of the UK COBRE Center for Molecular Medicine Proteomics and Protein Analytical Core. This study was supported by NIH Grants R01NS070899 (M.S.G.), P20GM103486 (M.S.G., C.W.V.K), R01AI081982 (S.L. and V.V.W.), R01GM020501 (S.L. and V.V.W.), and R01AI101436 (S.L. and V.V.W); KSEF Grants KSEF-2268-RDE-014 and KSEF-2971-RDE-017 (M.S.G.); Mizutani Foundation for Glycoscience Award (M.S.G.); NSF Grant IIA-1355438 (M.S.G.); University of Notre Dame Office of Research (L.M.H.); and SAF2011-27442 grant from the Spanish Ministry of Economy (P.S.).
Footnotes
ACCESSION NUMBERS
The atomic coordinates and structure factors are available in the Protein Data Bank deposited with the PDB code 4RKK.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online.
AUTHOR CONTRIBUTIONS
M.S.G. and C.W.V.K. conceived the experiments, analyzed data, and wrote the paper. C.W.V.K. collected X-ray diffraction data and determined the crystal structure. M.R. conceived experiments, purified protein, generated and optimized crystals, generated mutants, performed DSF and phosphatase assays, analyzed data, and generated figures. M.K.B. generated mutants, performed assays, analyzed DXMS data, and generated figures. B.K.W. and H.S. performed DXMS experiments under the supervision of S.L. and V.V.W. A.R.S. generated constructs, purified proteins, performed assays, analyzed DXMS data, and generated figures. M.W.P., S.C., and C.W.V.K. performed SAXS experiments and analysis. L.M.H. performed and analyzed AUC data. S.H. generated mutants and performed binding experiments. P.S. provided constructs and analyzed data. T.M.B., B.C.P., K.D.A., D.A.M., B.D.T., V.V.D., and A.O.T. generated constructs, purified proteins, performed crystallization screens, and performed assays.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berger P, Schaffitzel C, Berger I, Ban N, Suter U. Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc Natl Acad Sci U S A. 2003;100:12177–12182. doi: 10.1073/pnas.2132732100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovic SF, So NK, Andermann F. Progressive myoclonus epilepsies: clinical and neurophysiological diagnosis. J Clin Neurophysiol. 1991;8:261–274. [PubMed] [Google Scholar]
- Blennow A, Engelsen SB. Helix-breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010;15:236–240. doi: 10.1016/j.tplants.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev. 1999;29:265–295. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J Biol Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinuzzo M, Mangia S, Maraviglia B, Giove F. Does abnormal glycogen structure contribute to increased susceptibility to seizures in epilepsy? Metabolic brain disease. 2014 doi: 10.1007/s11011-014-9524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukhande VV, Rogers DM, Roma-Mateo C, Donderis J, Marina A, Taylor AO, Sanz P, Gentry MS. Laforin, a dual specificity phosphatase involved in Lafora disease, is present mainly as monomeric form with full phosphatase activity. PLoS One. 2011;6:e24040. doi: 10.1371/journal.pone.0024040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J, Gruart A, Garcia-Rocha M, Delgado-Garcia JM, Guinovart JJ. Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum Mol Genet. 2014;23:3147–3156. doi: 10.1093/hmg/ddu024. [DOI] [PubMed] [Google Scholar]
- Fryer KL, Brown AM. Pluralistic roles for glycogen in the central and peripheral nervous systems. Metabolic brain disease. 2014 doi: 10.1007/s11011-014-9516-5. [DOI] [PubMed] [Google Scholar]
- Gallant DJ, Bouchet B, Baldwin PM. Microscopy of starch: evidence of a new level of granule organization. Carbohydrate Polymers. 1997;32:177–191. [Google Scholar]
- Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glycogen metabolism of the organism. BMC Evol Biol. 2009;9:138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S, Kim Y, Li S, Durrant ES, Pace RM, Woods VL, Jr, Gentry MS. Structural insights into glucan phosphatase dynamics using amide hydrogen-deuterium exchange mass spectrometry. Biochemistry. 2009;48:9891–9902. doi: 10.1021/bi9008853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksal AC, Cingolani G. Dimerization of Vaccinia virus VH1 is essential for dephosphorylation of STAT1 at tyrosine 701. J Biol Chem. 2011;286:14373–14382. doi: 10.1074/jbc.M111.226357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, et al. STARCH-EXCESS4 Is a Laforin-Like Phosphoglucan Phosphatase Required for Starch Degradation in Arabidopsis thaliana. Plant Cell. 2009;21:334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light PE. Familial Wolff-Parkinson-White Syndrome: a disease of glycogen storage or ion channel dysfunction? Journal of cardiovascular electrophysiology. 2006;17(Suppl 1):S158–S161. doi: 10.1111/j.1540-8167.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Wu C, Liu Y, Zheng P. Dimerization of Laforin is required for its optimal phosphatase activity, regulation of GSK3beta phosphorylation, and Wnt signaling. J Biol Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Wu C, Zheng P. Deletions and missense mutations of EPM2A exacerbate unfolded protein response and apoptosis of neuronal cells induced by endoplasm reticulum stress. Hum Mol Genet. 2009;18:2622–2631. doi: 10.1093/hmg/ddp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovatt D, Sonnewald U, Waagepetersen HS, Schousboe A, He W, Lin JH, Han X, Takano T, Wang S, Sim FJ, et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins DA, Guo HF, Husodo S, Paasch BC, Bridges TM, Santelia D, Kotting O, Vander Kooi CW, Gentry MS. Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell. 2013;25:2302–2314. doi: 10.1105/tpc.113.112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS. Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc Natl Acad Sci U S A. 2014;111:7272–7277. doi: 10.1073/pnas.1400757111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian BA, Andrade DM, Ianzano L, Young EJ, Chan E, Ackerley CA, Scherer SW. Laforin is a cell membrane and endoplasmic reticulum-associated protein tyrosine phosphatase. Ann Neurol. 2001;49:271–275. [PubMed] [Google Scholar]
- Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- Nitschke F, Wang P, Schmieder P, Girard JM, Awrey DE, Wang T, Israelian J, Zhao X, Turnbull J, Heydenreich M, et al. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 2013;17:756–767. doi: 10.1016/j.cmet.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL., Jr Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci U S A. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer-Guglielmi B, Fleckenstein B, Jung G, Hamprecht B. Immunocytochemical localization of glycogen phosphorylase isozymes in rat nervous tissues by using isozyme-specific antibodies. J Neurochem. 2003;85:73–81. doi: 10.1046/j.1471-4159.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- Phan J, Tropea JE, Waugh DS. Structure-assisted discovery of Variola major H1 phosphatase inhibitors. Acta Crystallogr D Biol Crystallogr. 2007;63:698–704. doi: 10.1107/S0907444907014904. [DOI] [PubMed] [Google Scholar]
- Roach PJ. Glycogen and its Metabolism. Current Molecular Medicine. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros S, Schulze A. Linking glycogen and senescence in cancer cells. Cell Metab. 2012;16:687–688. doi: 10.1016/j.cmet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Saez I, Duran J, Sinadinos C, Beltran A, Yanes O, Tevy MF, Martinez-Pons C, Milan M, Guinovart JJ. Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2014;34:945–955. doi: 10.1038/jcbfm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin P, Raththagala M, Bridges TM, Husodo S, Gentry MS, Sanz P, Roma-Mateo C. Dimerization of the glucan phosphatase laforin requires the participation of cysteine 329. PLoS One. 2013;8:e69523. doi: 10.1371/journal.pone.0069523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Kotting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, et al. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011;23:4096–4111. doi: 10.1105/tpc.111.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- Sherwood AR, Paasch BC, Worby CA, Gentry MS. A malachite green-based assay to assess glucan phosphatase activity. Anal Biochem. 2013;435:54–56. doi: 10.1016/j.ab.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DM, Kotting O, Moorhead GB. Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 2014;19:471–478. doi: 10.1016/j.tplants.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Sinadinos C, Valles-Ortega J, Boulan L, Solsona E, Tevy MF, Marquez M, Duran J, Lopez-Iglesias C, Calbo J, Blasco E, et al. Neuronal glycogen synthesis contributes to physiological aging. Aging cell. 2014;13:935–945. doi: 10.1111/acel.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Zeeman SC. Starch metabolism in Arabidopsis. The Arabidopsis book / American Society of Plant Biologists. 2012;10:e0160. doi: 10.1199/tab.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson RA. Physiologic coupling of glial glycogen metabolism to neuronal activity in brain. Can J Physiol Pharmacol. 1992;70(Suppl):S138–144. doi: 10.1139/y92-255. [DOI] [PubMed] [Google Scholar]
- Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for lafora disease. J Biol Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci U S A. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. Febs J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J, Depaoli-Roach AA, Zhao X, Cortez MA, Pencea N, Tiberia E, Piliguian M, Roach PJ, Wang P, Ackerley CA, et al. PTG Depletion Removes Lafora Bodies and Rescues the Fatal Epilepsy of Lafora Disease. PLoS Genet. 2011;7:e1002037. doi: 10.1371/journal.pgen.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, et al. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO molecular medicine. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. From the Cover: Structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci U S A. 2010;107:15379–15384. doi: 10.1073/pnas.1009386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J Biol Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, Dixon JE. Laforin: A dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Austin J, Witmer F, Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol. 1968;19:15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 2. Structure comparison of glucan phosphatases laforin, SEX4 and LSF2.
Ribbon diagrams of the glucan phosphatases laforin, Starch EXcess4 (SEX4), and Like Sex Four2 (LSF2) with a star indicating the DSP domain active site. The laforin dimer interface is spatially positioned similar to the SEX4 and LSF2 CT-motif.
Figure S2, related to Figure 2 and Figure 3. Alignments of laforin orthologs.
(A) Alignment of the DSP domain of laforin vertebrate orthologs. Secondary structure elements are depicted above the primary sequence and DSP motifs are labeled. The catalytic triad residues D235, C266, and R272 are highlighted in red and residues interacting with maltohexaose are in yellow.
(B) Alignment of the CBM domain of laforin vertebrate orthologs. Secondary structure elements are depicted above the primary sequence. Critical residues for glycogen binding are highlighted in blue.
Figure S3, related to Figure 2. The laforin catalytic triad.
Close-up of the laforin DSP domain active site, showing a ribbon diagram of maltohexaose (green) and phosphate (orange) with 2Fo-Fc electron density map (1.3 σ). Glucose moieties in the maltohexaose chain are numbered from non-reducing to reducing end. Side chains of the catalytic triad are displayed, D235 (yellow), S266 (C266 in WT laforin, teal), and R272 (yellow). The O2 and O3 hydroxyls of Gl3 are pointing towards the active site with the O6 hydroxyl in solvent.
Figure S4, related to Figure 4. Deuterium exchange mass spectrometry (DXMS) peptide maps and peptide graphs.
(A) Sequence coverage map of identified pepsin-digested laforin peptides. A total of 134 high-quality peptides were detected. Solid blue lines indicate the 27 peptides utilized in the analysis. Dashed lines indicate peptides that were not used but were examined. Laforin primary sequence is above the corresponding peptides.
(B) Line graphs depicting peptides that exhibited a > 10% change in deuterium incorporation in the presence versus absence of glycogen. Data shown as green represents percentages of deuterium incorporation in glycogen-free laforin and blue represents glycogen-bound laforin. Percent deuterium incorporation is shown in a time-dependent manner.
(C) Line graphs depicting peptides that exhibited a > 10% change in deuterium incorporation for laforin Y294N versus laforin WT. Data shown as red represents percentages of deuterium incorporation for Y294N and green represents WT. Percent deuterium incorporation is shown in a time-dependent manner.
Figure S5, related to Figure 6. Size exclusion chromatography-small angle x-ray scattering (SEC-SAXS) analysis of laforin.
(A) Size exclusion chromatography profile of laforin C266S loaded onto a Superdex 75 10/300 GL (GE Healthcare) reveals that laforin elutes as a single species.
(B) Guinier plot of the low q region of the x-ray scattering data allows measurement of Rg = 33.9 ± 0.2 Å for laforin.
(C) The distance distribution P(r) function of SAXS scattering data of laforin.
Figure S6, related to Figure 7. Analytical ultracentrifugation analysis of laforin.
(A) Laforin WT sedimentation coefficient distribution as determined by sedimentation velocity analytical ultracentrifugation.
(B) Laforin F321S sedimentation coefficient distribution as determined by sedimentation velocity analytical ultracentrifugation.
Table S1: Tm of laforin and laforin mutants