Abstract
We captured, ectoparasite-combed, and blood-sampled cave-roosting Madagascan fruit bats (Eidolon dupreanum) and tree-roosting Madagascan flying foxes (Pteropus rufus) in four single-species roosts within a sympatric geographic foraging range for these species in central Madagascar. We describe infection with novel Bartonella spp. in sampled Eidolon dupreanum and associated bat flies (Cyclopodia dubia), which nest close to or within major known Bartonella lineages; simultaneously, we report the absence of Bartonella spp. in Thaumapsylla sp. fleas collected from these same bats. This represents the first documented finding of Bartonella infection in these species of bat and bat fly, as well as a new geographic record for Thaumapsylla sp. We further relate the absence of both Bartonella spp. and ectoparasites in sympatrically sampled Pteropus rufus, thus suggestive of a potential role for bat flies in Bartonella spp. transmission. These findings shed light on transmission ecology of bat-borne Bartonella spp., recently demonstrated as a potentially zoonotic pathogen.
Author Summary
Bartonella spp. are bacteria that inhabit the red blood cells of both human and animal hosts. Among humans, Bartonella spp. are known to cause several febrile illnesses, including Carrion’s disease (Bartonella bacilliformis), trench fever (Bartonella quintana), and cat scratch fever (Bartonella henselae), all of which are transmitted via arthropod vectors—respectively sand flies, lice, and fleas. Bats are known to host multiple Bartonella spp., including some capable of infecting humans. Some bat species are also known to host obligate ectoparasites known as bat flies (Diptera: Hippoboscoidea), which also sometimes support Bartonella spp. infections. The role of bat flies and other bat ectoparasites as vectors for Bartonella spp. transmission has been suggested, but not fully explored. We demonstrate Bartonella spp. infection in one species of Madagascar fruit bat, which hosts bat flies, simultaneously with the absence of Bartonella in a fruit bat species of overlapping range that appears not to support these ectoparasites. In light of ongoing trends of zoonotic emergence of human diseases from bat reservoirs, further understanding of the transmission dynamics of bat-borne pathogens is paramount.
Introduction
The role of bats as reservoirs for viral pathogens—including several responsible for severe human disease—has received increasing attention in recent years [1]. The extent to which this pattern is mirrored by bats’ abilities to host and transmit other zoonotic agents, including bacteria, is less widely acknowledged. Bats have been confirmed as asymptomatic reservoirs for several species of gram-negative Bartonella bacteria in localities as wide-ranging as the United Kingdom [2], Kenya [3], Guatemala [4], Peru [5], Taiwan [6], Nigeria [7], and Puerto Rico [8]. Bartonella spp. infect erythrocytes and epithelial cells of predominantly mammalian hosts, and some are known to cause zoonotic disease (bartonellosis) in humans. Most recently, bats in the Northern Hemisphere have been implicated as hosts for the human pathogen, Bartonella mayotimonensis, although the mechanism of transmission between bats and humans remains unclear [9]. Bartonella spp. are frequently transmitted via arthropod vectors [10] and have been identified in several bat ectoparasites, including 19 bat fly species (Diptera: Hippoboscoidea: Nycteribiidae and Streblidae) [11,12]. However, the presence of Bartonella spp. within these arthropods may simply reflect their ingestion of host blood, and vector transmission of Bartonella between bats has yet to be confirmed via experimental trial or controlled field study. Nonetheless, phylogenetic analyses of global bat fly-Bartonella-bat associations demonstrate Bartonella spp. similarities across bat hosts and ectoparasites [12], suggesting that bat flies might play a vector role in transmission. To elucidate this relationship, we examined Bartonella prevalence in two sympatric Madagascar fruit bat species—one containing bat flies and fleas and one in which ectoparasites were conspicuously absent. Here we report the presence of closely related Bartonella genotypes in Madagascan fruit bats (Eidolon dupreanum) and their associated bat flies (Nycteribiidae) in Madagascar. We simultaneously report the absence of Bartonella spp. in bat fleas (Thaumapsylla sp.) of E. dupreanum, in addition to the concomitant absence of both ectoparasites and Bartonella in sympatric Madagascan flying foxes (Pteropus rufus).
Materials and Methods
Bat Capture and Sampling
In November 2013, 57 E. dupreanum and 32 P. rufus were mist-netted, sampled for pathogens, and live-released from four single-species roost sites in central Madagascar. E. dupreanum bats were captured from two cave roosts (Angavobe -18.918050S, 47.94360E; and Angavokely 18.932450 S, 47.7574170 E) and P. rufus bats netted from two tree roosts (Marovitsika -18.842180S, 48.033630E; and Ambakoana -18.511280S; 48.171120E) in the District of Moramanga. All four roost sites are within a 35km radius of one another and a 5km radius of neighboring human communities, distances well within the nightly foraging ranges of these flying foxes (Fig. 1) [13]. This highland region is dominated by savannah grassland interspersed with non-native plantation and mid-elevation (~1100m) humid forest. Both E. dupreanum and P. rufus feed on a range of fruits and nectars and are known to share feeding sites [13].
Fig 1. Sites of bat collection, showing numbers of bats collected.
Madagascar, 2013.
Upon capture, bats were thoroughly examined for ectoparasites, and all observed flies, fleas, and mites were removed and collected into vials of absolute ethanol with a comb (fleas) or tweezers (mites and bat flies). Blood (1.0ml) was collected from the brachial vein of adult bats (forearm >100mm) and robust juveniles (29 P. rufus, 47 E. dupreanum blood-sampled). Serum and blood cells were separated by centrifuging and stored in liquid nitrogen in the field, then transferred to -80°C freezers at the Institut Pasteur-Madagascar.
Ethics Statement
This study was carried out in strict accordance with guidelines posted by the American Veterinary Medical Association. All field protocols employed were pre-approved by the Princeton University Institutional Animal Care and Use Committee (IACUC Protocol # 1926), and every effort was made to minimize discomfort to animals.
Sample Processing and Molecular Analysis
Bat flies. Ectoparasite samples were processed at the University at Buffalo (Buffalo, NY, USA). Ectoparasite DNA was extracted from a subset of samples (19 bat flies and 6 fleas) using the Qiagen Animal Tissue kit (QIAGEN, Valencia, CA, USA). Ectoparasite voucher specimens were slide-mounted and identified using available taxonomic keys.
Blood pellets. Blood pellet samples were processed at the CDC’s Division of Vector-Borne Diseases (Fort Collins, CO, USA). DNA was extracted from blood samples using a Qiagen QIAamp tissue kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions.
Bartonella spp. assay. All DNA extractions (ectoparasites and blood) were examined for Bartonella spp. by conventional PCR targeting multiple genes employed in previous research: gltA, ftsZ, and nuoG genes for arthropod bartonellae, and gltA and ITS sequence for blood samples [3,11,12]. Only samples with sequences that unequivocally BLASTed to Bartonella spp. and nested within known Bartonella sequences by phylogenetic analysis were considered positive (RAxML 7.7) [14]. Samples positive by PCR with inconclusive sequence data were thus considered negative for Bartonella spp. in our analysis.
Statistical Analysis
We compared the frequency of bat fly (C. dubia) and bat flea (Thaumapsylla sp.) infections, as well as Bartonella spp. prevalence in both bat hosts and in ectoparasite arthropods. Differences were examined between species and across sampling sites using chi-squared and Fisher exact tests in the statistical program R [15]. We used a p-value threshold of 0.01 to assess whether observed ectoparasite burden and Bartonella spp. prevalence were independent of species and sampling site.
Results
Seven of 24 (29.2%) Eidolon dupreanum sampled from Angavobe cave and 20 of 23 (87%) E. dupreanum sampled from Angavokely cave were found to host Cyclopodia dubia (Nycteribiidae) bat flies. Ten of those 23 (43.5.1%) Angavokely E. dupreanum also hosted Thaumapsylla sp. fleas (Table 1). Both frequency of bat fly and flea hosting varied significantly by roosting site, via analysis by chi-squared tests of independence (bat fly: X2 = 13.768, df = 1, p = 0.0002; flea: X2 = 10.7863, df = 1, p = 0.001) and Fisher’s exact tests (bat fly: p = 8.828e-05; flea: p = 0.0002). Two of 2 (100%) bat flies processed from Angavobe and 15 of 17 (88.2%) bat flies processed from Angavokely were considered positive for Bartonella DNA by sequence, although all bat flies processed were Bartonella spp. positive by PCR alone. None of the six Thaumapsylla fleas processed were positive for any Bartonella target gene. The presence of Thaumapsylla sp. at the Angavokely site represents the first geographic record for Madagascar; this genus is known from Eidolon spp. elsewhere [16].
Table 1. Ectoparasites and Bartonella spp. in Madagascar fruit bats.
| Ectoparasite presence | Bartonella spp. prevalence | |||||
|---|---|---|---|---|---|---|
| Bat species | Locality | C. dubia | Thaumapsylla sp. | In bat host | In C. dubia | In Thaumapsylla sp. |
| E. dupreanum | Angavobe | 7/24 (29.2)* | 0/24 (0) † | 8/24 (33.3) | 2/2 (100) | — |
| Angavokely | 20/23 (87.0)* | 10/23 (43.5) † | 13/23 (56.5) | 15/17 (88.2) | 0/6 (0) | |
| P. rufus | Marovitsika | 0 (0) | 0 (0) | 0/12 (0) | — | — |
| Ambakoana | 0 (0) | 0 (0) | 0/17 (0) | — | — | |
*Between site differences statistically significant via chi-squared test for independence (X2 = 13.768, df = 1, p = 0.0002;) and Fisher’s exact test (p = 8.828e-05)
†Between site differences statistically significant via chi-squared test for independence (X2 = 10.7863, df = 1, p-value = 0.001) and Fisher’s exact test (p = 0.0002)
Table data indicate number positive/number sampled (%) for ectoparasite presence and Bartonella spp. prevalence (both in bat host and in hosted ectoparasites) for E. dupreanum and P. rufus.
Blood samples from eight of 24 (33.3%) Angavobe E. dupreanum and thirteen of 23 Angavokely (56.5%) E. dupreanum were positive for Bartonella DNA by PCR confirmed with sequence for one or more genes (Table 1). Bartonella spp. prevalence did not vary significantly between Angavobe and Angavokely roosting sites as indicated by a chi-squared test for independence (X2 = 1.7029, df = 1, p-value = 0.1919) and Fisher’s exact test (p-value = 0.1468). In Angavobe, bats demonstrated both singular infections with Bartonella spp. and with bat flies, as well as simultaneous co-infection with bat flies and Bartonella spp. (Fig. 2). In Angavokely, E. dupreanum individuals hosted every possible combination of bat fly/flea/Bartonella spp. infection and co-infection save for singular flea infestations in the absence of other pathogens (Fig. 2). It should be noted that, prior to processing, bats were housed together with others from the same sample site in wooden transport cages, and ectoparasite sharing among individuals was easily facilitated.
Fig 2. Venn-diagrams of infection/co-infection with bat flies, bat fleas, and Bartonella spp. across roosting sites for Eidolon dupreanum:
(A) Angavobe, N = 24; (B) Angavokely, N = 24. Both raw numbers of infected individuals and prevalence (%) are indicated. Note that all sampled P. rufus from both Marovitsika and Ambakoana were negative for all infections (i.e. bat flies, bat fleas, and Bartonella spp.).
No ectoparasites were recovered from either the 12 Pteropus rufus examined at the Marovitsika site or the 17 P. rufus sampled at the Ambakoana site (Table 1). As with ectoparasites, none of the 29 P. rufus samples (12 from Marovitisika, 17 from Ambakoana) were positive for Bartonella spp. by either molecular target (Table 1).
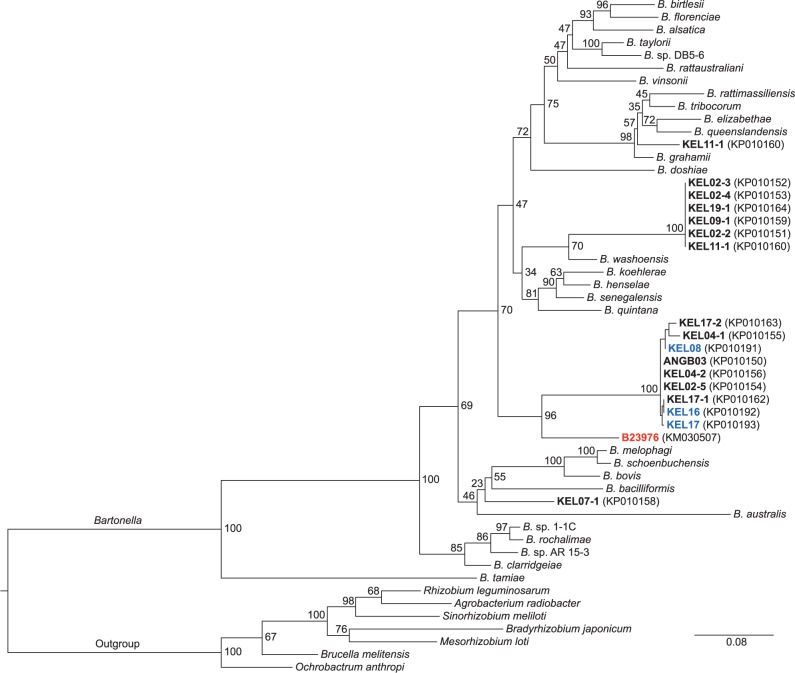
All Bartonella spp. sequences from E. dupreanum bats and associated C. dubia bat flies nested within or close to known major Bartonella lineages (Fig. 3) [17]. Although sequence data retrieved are insufficient to reach final Bartonella species identification, novel genotypes are present. Sequences (gltA) from sampled bats group with those retrieved from Cyclopodia bat flies.
Fig 3. Maximum likelihood phylogeny of representative gltA genes of Rhizobiales (ingroup: Bartonella spp.) (RAxML, GTR+G model, partitioned by codon position) (12).
KEL & ANGB—Madagascar samples. Blue: ex. Eidolon dupreanum (bat), Black: ex. Cyclopodia dubia (bat fly). RED comparative sequence ex. Cyclopodia dubia (bat fly) (9).
Discussion
The recent identification of bats as reservoirs for human pathogenic Bartonella mayotimonensis [9] validates further investigation of the zoonotic potential of Bartonella spp. in Chiropteran reservoirs. In Madagascar, insectivorous bats are known to roost in human residences, and both P. rufus and E. dupreanum are widely consumed as bushmeat, highlighting the extent of human-wildlife interface in the region [13].
In keeping with trends of persistent bacterial infection exhibited by bat-borne Bartonella elsewhere [3–5], we report high Bartonella spp. prevalence (57.4%) in a long-lived, cave-roosting E. dupreanum host (lifespan 10–20 years [13]). We correspondingly report no Bartonella infections in sympatric P. rufus, though our current sample size is too small to determine whether this absence is universal across the Madagascar population. Additionally, further study is needed to address whether these Bartonella spp. prevalence patterns are an artifact of phylogeny or ecology. The ability of Eidolon bats to serve as hosts for Bartonella spp. has now been documented in both tree-roosting [3] and cave-roosting environments—consistently in association with bat flies. P. rufus does not seem to host bat flies in Madagascar [18], although sampling has not been exhaustive enough to consider this absence a certainty. Tree-roosting Pteropus spp. are known to host bat flies throughout southeast Asia [19] and Australia [20], and investigation of Bartonella spp. infections in these populations will help address the relative influence of host genetic predisposition for Bartonella infection versus vector ecology.
In addition to pathogen prevalence in the host, we report Bartonella spp. infection in bat flies (Cyclopodia dubia) of E. dupreanum, simultaneous with Bartonella DNA absence in flea ectoparasites (Thaumapsylla sp.) of those same bats. Fleas are the confirmed vector for Bartonella henselae, the causative agent in cat scratch fever [21], and fleas of bats have been previously reported in association with Bartonella DNA [9,22]. In our study, both bat flies and fleas were host-specific and likely consumed host blood, although only flies tested positive for Bartonella spp., suggesting that the mechanisms by which arthropods host and transmit pathogens vary and impact their functionality as vectors. Sampling of flea ectoparasites was not extensive enough to assess the true extent of their ability, or lack of ability, to transmit Bartonella spp., and further experimental studies of the vector potential of both bat flies and fleas for Bartonella is warranted.
Finally, observed differences in the frequency of ectoparasite burden between sample sites for E. dupreanum indicated significantly higher rates of ectoparasite infection with both flies and fleas in Angavokely vs. Angavobe. These differences could result from ecological variation in both host density and/or climate between the two cave roosts. More extensive spatial sampling, in conjunction with climactic monitoring, in other E. dupreanum roost sites of varying size, temperature, and humidity across Madagascar will help elucidate habitat thresholds for ectoparasite invasion. In particular, sampling of E. dupreanum in reported tree roosts in central Madagascar will shed light on the extent to which roosting behavior limits bats’ abilities to support ectoparasites in this system [13]. If Bartonella spp. are, indeed, transmitted by bat fly vectors, such findings will have important implications for our understanding of the distribution, prevalence, and transmission dynamics of a potentially zoonotic pathogen.
Acknowledgments
We thank Jean-Michel Héraud and the Virology Unit of Institut-Pasteur Madagascar for laboratory space and logistical support in the field.
Data Availability
All relevant ecological data are within the paper and its Supporting Information files. All molecular sequences for Bartonella spp. files are available from the GenBank database (accession numbers: KP010160, KP010152, KP010153, KP010164, KP010159, KP010151, KP010163, KP010155, KP010191, KP010150, KP010156, KP010154, KP010162, KP010192, KP010193, KP010158, KM030507).
Funding Statement
Field work was supported via the following three small grants to CEB: [1] National Geographic Society Young Explorer's Grant to CEB (YEG #926-13, http://www.nationalgeographic.com/explorers/grants-programs/young-explorers/), [2] Princeton University Center for Health and Wellbeing Health Grand Challenge grant (http://www.princeton.edu/chw/education/grad-research-funding/graduate-research-funding-HGC/), [3] American Society of Mammalogists Grant-in-Aid of Research (http://www.mammalsociety.org/committees/grants-aid). Sample analysis was funded by a National Science Foundation Division of Environmental Biology grant (#:1050793) to KD. Publication fees were covered by a National Science Foundation grant (#: 23400 G0001 10004452) to APD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T (2006) Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19: 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ (2005) Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology 131: 489–496. [DOI] [PubMed] [Google Scholar]
- 3. Kosoy M, Ying Bai T, Lynch I V., Kuzmin M, Niezgoda R, et al. (2010) Bartonella spp. in bats, Kenya. Emerg Infect Dis 16: 1875–1881. 10.3201/eid1612.100601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, et al. (2011) Bartonella spp. in Bats, Guatemala. Emerg Infect Dis 17: 1269–1272. 10.3201/eid1707.101867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai Y, Recuenco S, Gilbert AT, Osikowicz LM, Gómez J, et al. (2012) Prevalence and diversity of Bartonella spp. in bats in Peru. Am J Trop Med Hyg 87: 518–523. 10.4269/ajtmh.2012.12-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin J-W, Hsu Y-M, Chomel BB, Lin L-K, Pei J-C, et al. (2012) Identification of novel Bartonella spp. in bats and evidence of Asian gray shrew as a new potential reservoir of Bartonella . Vet Microbiol 156: 119–126. 10.1016/j.vetmic.2011.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kamani J, Baneth G, Mitchell M, Mumcuoglu KY, Gutiérrez R, et al. (2014) Bartonella species in bats (Chiroptera) and bat flies (Nycteribiidae) from Nigeria, west Africa. Vector Borne Zoonotic Dis 14: 625–632. 10.1089/vbz.2013.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olival KJ, Dittmar K, Bai Y, Rostal MK, Lei BR, et al. (2014) Bartonella spp. in a Puerto Rican Bat Community. J Wildl Dis 51. [DOI] [PubMed] [Google Scholar]
- 9. Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis . Emerg Infect Dis 20 10.3201/eid2012.141026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB (2008) Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol 22: 1–15. 10.1111/j.1365-2915.2008.00713.x [DOI] [PubMed] [Google Scholar]
- 11. Billeter SA, Hayman DTS, Peel AJ, Baker K, Wood JLN, et al. (2012) Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology 139: 324–329. 10.1017/S0031182011002113 [DOI] [PubMed] [Google Scholar]
- 12. Morse SF, Olival KJ, Kosoy M, Billeter SA, Patterson BD, et al. (2012) Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect Genet Evol 12: 1717–1723. 10.1016/j.meegid.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 13. MacKinnon JL, Hawkins CE, Racey PA (2003) Pteropodidae, Fruit Bats, Fanihy, Angavo In: Goodman SM, Benstead JP, editors. The Natural History of Madagascar. The Universit of Chicago Press; pp. 1299–1302. [Google Scholar]
- 14. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 15.Team RC (2013) R: A language and environment for statistical computing. Available: http://www.r-project.org/.
- 16.Duchemin JB (2003) Biogéographie des puces de Madagascar. In French. Universite de Paris XII-Val de Marne.
- 17. Guy L, Nystedt B, Toft C, Zaremba-Niedzwiedzka K, Berglund EC, et al. (2013) A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella . PLoS Genet 9: e1003393 10.1371/journal.pgen.1003393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman SM (2013) Personal communication.
- 19. Olival KJ, Dick CW, Simmons NB, Morales JC, Melnick DJ, et al. (2013) Lack of population genetic structure and host specificity in the bat fly, Cyclopodia horsfieldi, across species of Pteropus bats in Southeast Asia. Parasites and Vectors 6: 231 10.1186/1756-3305-6-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Theodor O (1959) A revision of the genus Cyclopodia (Nycteribiidae, Diptera). Parasitology 49: 242–308. [DOI] [PubMed] [Google Scholar]
- 21. Chomel BB, Kasten RW, Chi B, Yamamoto K, Gurfield AN, et al. (1996) Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol 34: 1952–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reeves WK, Rogers TE, Durden LA, Dasch GA (2007) Association of Bartonella with the fleas (Siphonaptera) of rodents and bats using molecular techniques. J Vector Ecol 32: 118–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant ecological data are within the paper and its Supporting Information files. All molecular sequences for Bartonella spp. files are available from the GenBank database (accession numbers: KP010160, KP010152, KP010153, KP010164, KP010159, KP010151, KP010163, KP010155, KP010191, KP010150, KP010156, KP010154, KP010162, KP010192, KP010193, KP010158, KM030507).