Abstract
In France, approximately 1200 cases of Legionnaires disease (LD) are reported annually, and isolates are available for approximately 20% of cases identified since 2000. All Legionella pneumophila serogroup 1 (sg1) isolates are characterized by sequence-based typing at the National Reference Centre. LD cases caused by L. pneumophila sg1 reported from 2008 through 2012 were considered for the study. Our study objective was to describe cases according to their sequence type (ST). We also constructed multivariable modified Poisson regression models to estimate the incidence rate ratio (IRR) and to identify characteristics potentially associated with ST23 clones compared to ST1 and ST47 clones. We studied 1192 patients infected by ST1 (n = 109), ST23 (n = 236), ST47 (n = 123) or other STs (n = 724). The geographic distribution of the ST23 cases across the country was significantly different compared to other ST groups. This genotype was significantly associated with the absence of corticosteroid therapy compared to ST1 (IRR = 0.56; p 0.016). Concerning exposure, the ST23 genotype was significantly less associated with hospital-acquired infections compared to ST1 (IRR = 0.32; p 0.001), but it was more associated with infections acquired in hospitals and elderly settings compared with ST47. Finally, the ST23 genotype was less frequently associated with travel than other STs. Despite the large number of cases of ST23 infection, we did not identify any characteristics specific to this ST. However, we identified independent associations between ST1 and nosocomial transmission and steroid therapy. These findings should encourage further exploration, especially in terms of environmental diffusion, strain virulence and host factors.
Keywords: Legionella pneumophila, Legionnaires disease, molecular epidemiology, molecular typing, risk factors
Introduction
Legionnaires disease (LD) is usually acquired through inhalation of aerosolized water contaminated with Legionella spp., which is found ubiquitously in natural and man-made aqueous environments such as cooling towers, hot water systems or whirlpools [1]. Legionella pneumophila, especially serogroup 1 (sg1), is the major cause of LD, which remains a serious disease. Several host-related risk factors have been clearly identified [2–4], including older age, male sex, smoking, diabetes, chronic heart or lung disease, cancer or hematologic malignancy and steroid use or any immunosuppressive therapy.
The characterization of clinical isolates by molecular typing methods is essential for epidemiologic investigations, particularly for clusters and outbreaks. Thus, L. pneumophila sg1 isolates can be characterized by sequence-based typing (SBT) using isolates or by nested-PCR-based SBT using clinical samples [5,6]. The typing method for L. pneumophila sg1 was developed by the European Working Group for Legionella Infections (EWGLI was renamed the European Study Group for Legionella Infections, ESGLI, in September 2012). Both local and worldwide sequence types (STs), such as ST1, ST23, ST37, ST47 and ST62, have been identified [7–12]. In France, ST1- and ST47-associated LD represented 9% and 11% of culture-proven cases in 2008, respectively [7]. The epidemiologic characteristics of these two STs have previously been published: ST1 strains with the Paris pulsotype were more frequently identified among elderly patients, immunocompromised patients or women and had a high lethality rate (38%) compared to sporadic strains. In contrast, ST47 strains were significantly more frequently associated with smoking, but the case fatality rate was lower than with sporadic strains [13].
The present French epidemiologic report shows that the ST most frequently associated with LD is ST23. It accounted for approximately 20% of the strains isolated in France in 2008, but no specific host factors associated with this genotype have been reported thus far [7]. We thus characterized the epidemiologic and clinical features of 236 patients infected with ST23 and compared them to those of patients infected by two other predominant STs (ST1, ST47) and other nonpredominant STs.
Materials and Methods
Source of information
In France, all cases of LD are reported to the national health authority (Institut de Veille Sanitaire), which collects epidemiologic information through anonymous mandatory notification, including various risk factors and exposure routes for LD. The notification form allows the collection of information on current illness, the patient’s personal characteristics (sex, age and postcode of residence), disease outcome and factors known to be associated with LD. Moreover, exposure to any of the following settings during the 10-day period preceding symptom onset (i.e. the incubation period) is also recorded: hospital, elderly setting, thermal centre, whirlpool spa or travel site (campsite, hotel) [7].
Additionally, all corresponding Legionella isolates are sent to the NRCL (national reference centre for Legionella), where they are characterized by SBT [6].
Included cases
In our study, cases presenting clinical and radiologic signs of pneumonia combined with L. pneumophila sg1 isolation from a culture of sputum or bronchopulmonary secretions (bronchoalveolar lavage fluid or bronchial aspirate) were included. Cases of LD due to L. pneumophila sg1 reported from 2008 through 2012 were considered for the study. All patients lived in metropolitan France. The geographic distribution of cases (based on the postcode of residence) was used as a proxy for the geographic distribution of strains, and the patients were then classified into five regions according to their telephone area code (Fig. 1). Finally, on the basis of genotype distribution, for the purposes of the study, the population was divided into four groups: ST1, ST23, ST47 and other ST culture-confirmed cases.
FIG. 1.
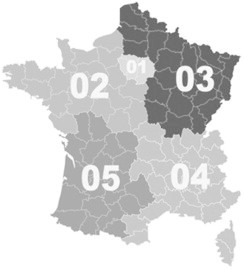
Division of metropolitan France into five regions to determine geographic distribution of cases.
Statistical analysis
We constructed multivariable modified Poisson regression models to estimate the incidence rate ratio (IRR) and to identify characteristics potentially associated with ST23 compared to the three other groups, considering a p value of ≤0.05 to be significant [14]. Analyses were performed by Stata 11.2 (StataCorp, College Station, TX, USA) software.
Results
From 2008 to 2012, a total of 6458 cases of LD were reported, and Legionella isolates were recovered in 1192 cases (18.5%), which were included in the study. ST23 accounted for the largest number of isolates (n = 236), representing 19.8% of all clinical strains. This rate did not change significantly from year to year over the study period. ST1 and ST47 L. pneumophila represented 9.1% and 10.3% of all isolates, respectively. ST23 isolates were found ubiquitously across France, but mainly in regions 3 and 4 (76.4%); this strain was also found in LD cases from other ST groups with only minor differences. On the other hand, ST1 cases occurred mostly in regions 1 and 4, and ST47 cases were mainly found in region 3 (Fig. 1; Table 1).
TABLE 1.
Epidemiologic characteristics of ST23 cases compared to ST1, ST47 other STs and all cases of Legionnaires disease due to Legionella pneumophila serogroup 1 in France, 2008–2012
| Characteristic | ST23 | ST1 | ST47 | Other ST | All |
|---|---|---|---|---|---|
| No. of subjects | 236 | 109 | 123 | 724 | 1192 |
| Male | 190 (80.5%) | 80 (73.4%) | 101 (82.1%) | 576 (79.6%) | 947 (79.5%) |
| Age (years) (mean/variable) | 59.1 | 63.3 | 60.6 | 58.7 | 59.4 |
| Death | 35 (16.1%) | 30 (30.9%) | 15 (13.2%) | 124 (18.6%) | 204 (18.6%) |
| Risk factor | |||||
| At least one | 172 (72.9%) | 82 (75.2%) | 85 (69.1%) | 540 (79.6%) | 879 (73.7%) |
| Cancer | 19 (8.1%) | 26 (23.9%) | 13 (10.6%) | 80 (11.1%) | 138 (11.6%) |
| Steroid therapy | 10 (4.2%) | 24 (22.0%) | 12 (9.8%) | 68 (9.4%) | 114 (9.6%) |
| Diabetes | 42 (17.8%) | 13 (11.9%) | 24 (19.5%) | 107 (14.8%) | 186 (15.6%) |
| Tobacco | 124 (52.5%) | 31 (28.4%) | 55 (44.7%) | 370 (51.1%) | 580 (48.7%) |
| Exposure | |||||
| Hospital-acquired infection | 7 (3.0%) | 34 (31.2%) | 3 (2.4%) | 45 (6.2%) | 89 (7.5%) |
| Travel | 29 (12.3%) | 12 (11.0%) | 21 (17.1%) | 142 (19.6%) | 204 (17.1%) |
| Elderly setting | 9 (3.8%) | 6 (5.5%) | 0 | 14 (1.9%) | 29 (2.4%) |
| Region | |||||
| 1 | 27 (11.4%) | 44 (40.4%) | 16 (13.0%) | 94 (13.0%) | 181 (15.2%) |
| 2 | 13 (5.5%) | 13 (11.9%) | 17 (13.8%) | 114 (15.8%) | 157 (13.2%) |
| 3 | 88 (37.3%) | 14 (12.8%) | 73 (59.4%) | 229 (31.6%) | 404 (33.9%) |
| 4 | 93 (39.4%) | 37 (33.9%) | 13 (10.6%) | 219 (30.3%) | 362 (30.4%) |
| 5 | 15 (6.4%) | 1 (0.9%) | 4 (3.3%) | 68 (9.4%) | 88 (7.4%) |
ST, sequence type.
Patients infected with L. pneumophila sg1 ST23 had a mean age of 59.1 years and were mostly men (Table 1). Of these patients, 72.9% had at least one of the risk factors associated with LD in 72.9%. This percentage was not significantly different from the corresponding percentages among patients infected by ST1, ST47 or other STs of L. pneumophila sg1. The rarest risk factors associated with ST23 LD were steroid therapy (4.2%) or cancer (8.1%). Conversely, smoking was the most frequently reported risk factor (n = 124; 52.5%). The percentage of cases with this risk factor was highest in the ST23 group compared to the other three groups. Moreover, ST23 cases were rarely hospital acquired (3.0%) or acquired in elderly settings (3.8%). Travel-associated cases represented 12.3% of the ST23 cases.
Multivariate analysis comparing the characteristics of ST 23 cases with those of other groups revealed five significant differences (Table 2). First, the geographic distribution of ST23 cases was significantly different than the geographic distribution of other STs (Table 2). Second, this genotype was significantly less associated with steroid therapy compared to ST1 (IRR = 0.56; p 0.016). This difference was not observed when comparing ST23 to ST47 or ST23 to other STs. Concerning exposure route, the ST23 genotype was significantly less associated with hospital-acquired infections compared to ST1 (IRR = 0.32; p 0.001), but it was more associated with hospital-acquired infections as well as with infections acquired in elderly settings compared to ST47. Finally, the ST23 genotype was less frequently associated with travel cases than other STs.
TABLE 2.
Multivariate analysis of epidemiologic characteristics of ST23 cases compared to ST1, ST47 and other STs
| Characteristic | ST23 vs. ST1 (n = 345) |
p | ST23 vs. ST47 (n = 359) |
p | ST23 vs. other STs (n = 960) |
p |
|---|---|---|---|---|---|---|
| IRR (95% CI) | IRR (95% CI) | IRR (95% CI) | ||||
| Male | 1.08 (0.92–1.26) | 0.35 | 1.02 (0.86–1.20) | 0.81 | 1.06 (0.80–1.40) | 0.67 |
| Age (years) (mean/variable) | 1.00 (0.99–1.0) | 0.81 | 1.00 (0.99–1.00) | 0.46 | 1.00 (0.99–1.00) | 0.85 |
| Risk factor | ||||||
| Cancer | 0.75 (0.55–1.03) | 0.07 | 0.93 (0.70–1.24) | 0.62 | 0.81 (0.53–1.24) | 0.33 |
| Steroid therapy | 0.56 (0.35–0.90) | 0.016* | 0.68 (0.43–1.09) | 0.11 | 0.56 (0.31–1.02) | 0.06 |
| Diabetes | 1.05 (0.91–1.21) | 0.55 | 0.99 (0.82–1.18) | 0.87 | 1.09 (0.82–1.45) | 0.58 |
| Tobacco | 1.04 (0.92–1.17) | 0.58 | 1.11 (0.95–1.31) | 0.19 | 0.99 (0.77–1.26) | 0.91 |
| Exposure | ||||||
| Hospital-acquired infection | 0.32 (0.16–0.63) | 0.001* | 1.49 (1.03–2.14) | 0.034* | 0.69 (0.34–1.41) | 0.31 |
| Travel | 0.97 (0.79–1.17) | 0.73 | 0.89 (0.70–1.13) | 0.34 | 0.66 (0.47–0.95) | 0.023* |
| Elderly setting | 0.95 (0.64–1.40) | 0.78 | 1.69 (1.31–2.20) | <10−3 | 1.54 (0.88–2.70) | 0.13 |
| Region | <10−3a | <10−3a,* | 0.003a | |||
| 1 | Ref. | Ref. | Ref. | |||
| 2 | 1.14 (0.71–1.81) | 0.60 | 0.68 (0.43–1.08) | 0.10 | 0.44 (0.24–0.80) | 0.007* |
| 3 | 1.85 (1.38–2.47) | <10−3* | 0.88 (0.67–1.14) | 0.33 | 1.12 (0.77–1.64) | 0.56 |
| 4 | 1.61 (1.20–2.16) | 0.001* | 1.43 (1.12–1.83) | 0.004* | 1.17 (0.80–1.70) | 0.42 |
| 5 | 1.95 (1.47–2.60) | <10−3 | 1.30 (0.94–1.76) | 0.11 | 0.73 (0.41–1.28) | 0.27 |
ST, sequence type; IRR, incidence rate ratio; CI, confidence interval; Ref., reference.
*Statistically significant.
Global test.
Discussion
In this study, we found a high rate of ST23 detection in LD; this ST represents 19.8% of cases with positive cultures in France between 2008 and 2012. This proportion is higher than those found in other countries, e.g. 6% in Belgium (2000–2010) [8], 4.7% in Japan (1980–2008) [10], 4.5% in Portugal (1987–2008) [15] and 0 observed in England and Wales (2000–2008) [11]. A compilation of European data (3200 isolates from 36 countries) showed that ST23 accounted for only 10.7% of clinical isolates, including French strains [16]. The results and statistical comparisons in the present work pertain to patients with culture-proven LD. The inclusion of only these patients could be a critical source of bias. Throughout the study period, considering all cases of non-culture-proven LD with complete allelic profiles determined by nested-PCR-based SBT (n = 70), the ST23 genotype was detected in 28.6% of patients (data not shown). This proportion is consistent with the number of ST23 culture-proven LD cases of our study.
It also appeared that 72.9% of the ST23-infected patients had at least one of the risk factors usually associated with LD acquisition. This overall percentage did not differ from those observed for cases of LD due to other ST; however, a more detailed analysis of the different risk factors showed that patients infected by ST23 strains were less likely to have received corticosteroid therapy than patients with ST1 (IRR = 0.56; p 0.016). Corticosteroids or other immunosuppressive therapies (e.g. anti-TNF-α) are increasingly used to treat systemic diseases such as rheumatoid arthritis, Crohn disease or cutaneous psoriasis. The use of these types of medications was shown to be associated with an increased relative risk of LD, leading to a 1.6- to 8-fold increase in the risk of LD compared to the general population [17]. The overall standardized incidence ratio was 13.1 for patients receiving TNF-α antagonists in the French population [18]. The frequency with which ST23-infected patients are receiving corticoids suggests that they are less likely to have chronic diseases requiring hospital admission. This finding is consistent with our observations, which revealed that ST23 infections were more often community acquired compared to ST1 infections. Indeed, the observed IRR was low (IRR = 0.32; p 0.001).
Comparing ST23 vs. ST1 cases regarding hospital exposure and corticosteroid therapy confirmed specific features related to ST1 strains that have already been reported [13]. In this study, 22% of ST1 patients received corticosteroids, and a high proportion of these patients were hospitalized (31.2%). Of course, we verified that there was no regional effect associated with these nosocomial infections (data not shown). The combination of environmental factors, strain virulence and host susceptibility may explain the observed differences among STs. Thus, concerning ST1 strains, we could hypothesize that they are either very well adapted to hospital water networks or are widely present in the environment. European data showed that ST1 accounted for 21.4% of environmental isolates, which is a relatively high percentage compared to other STs [16]. Other studies have also shown that ST1 was the most dominant ST in analysed strains from the environment in China (81/164, 49.4%) and in the United States (73/149, 49%) [19,20]. Moreover, in the Chinese study, ST1 accounted for 92.3% and 53.1% of the environmental sg1 isolates from potable water systems and cooling towers, respectively. This finding emphasises that ST1 strains are widely present in artificial water sources, but to our knowledge, these strains were never reported specifically in hospital water networks. Therefore, designing a ST1-specific PCR for rapid detection and identification in hospital environmental samples would be helpful. We could then hypothesize that this clone would be more ubiquitous and have lower virulence, infecting mainly immunocompromised patients. Finally, another approach would be for L. pneumophila ST1 to contain specific virulence determinants adapted for immunocompromised patients, who may also present specific host characteristics. The epidemiologic characteristics of ST1 cases could explain the higher case fatality rate observed with this clone (Table 1).
By contrast, both the ST47 and ST23 strains were rarely recovered from environmental samples [11,21]. European data indicated that ST47 and ST23 accounted for 0.4% and 1.5% of environmental isolates, respectively [16]. These rates suggest that these strains could have a peculiar virulence pattern because they are less disseminated in the environment and are less associated with patients receiving corticosteroids. Nevertheless, the ST23 genotype was more frequently associated with hospital-acquired infections and with infections acquired in elderly settings compared to ST47. We might therefore hypothesize that the source of infection could be different for these two clones.
Finally, although ST23 isolates could have developed a peculiar virulence pattern adapted to a larger population, we should have observed at least as many travel-associated cases in the ST23 group as we did in the other ST groups. In our study, this genotype was less frequently associated with travel cases than other STs. We unfortunately have no explanation for this difference. All of these observations suggest that ST23 could be a dominant infective clone in the global population of Legionella strains, as could ST1 and ST47. Nevertheless, there are still several unresolved issues, and further analysis will be necessary to determine the key host characteristics and virulence factors associated with susceptibility to these clones. Finally, this study reminds us that it is important to maintain clinician awareness of the need to collect sputum samples in order to document LD cases and to better understand the association between host characteristics and Legionella strains.
Conflict of Interest
None declared.
References
- 1.McDade J.E., Shepard C.C., Fraser D.W., Tsai T.R., Redus M.A., Dowdle W.R. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 2.Marston B.J., Lipman H.B., Breiman R.F. Surveillance for Legionnaires’ disease. Risk factors for morbidity and mortality. Arch Intern Med. 1994;154:2417–2422. [PubMed] [Google Scholar]
- 3.Chidiac C., Che D., Pires-Cronenberger S., Jarraud S., Campèse C., Bissery A. Factors associated with hospital mortality in community-acquired legionellosis in France. Eur Respir J. 2012;39:963–970. doi: 10.1183/09031936.00076911. [DOI] [PubMed] [Google Scholar]
- 4.Che D., Campese C., Santa-Olalla P., Jacquier G., Bitar D., Bernillon P. Sporadic community-acquired Legionnaires’ disease in France: a 2-year national matched case-control study. Epidemiol Infect. 2008;136:1684–1690. doi: 10.1017/S0950268807000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lück C., Fry N.K., Helbig J.H., Jarraud S., Harrison T.G. Typing methods for Legionella. Methods Mol Biol. 2013;954:119–148. doi: 10.1007/978-1-62703-161-5_6. [DOI] [PubMed] [Google Scholar]
- 6.Ginevra C., Lopez M., Forey F., Reyrolle M., Meugnier H., Vandenesch F. Evaluation of a nested-PCR-derived sequence-based typing method applied directly to respiratory samples from patients with Legionnaires’ disease. J Clin Microbiol. 2009;47:981–987. doi: 10.1128/JCM.02071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campese C., Bitar D., Jarraud S., Maine C., Forey F., Etienne J. Progress in the surveillance and control of Legionella infection in France, 1998–2008. Int J Infect Dis. 2011;15:30–37. doi: 10.1016/j.ijid.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Vekens E., Soetens O., De Mendonça R., Echahidi F., Roisin S., Deplano A. Sequence-based typing of Legionella pneumophila serogroup 1 clinical isolates from Belgium between 2000 and 2010. Euro Surveill. 2012;17:2030–2032. [PubMed] [Google Scholar]
- 9.Tijet N., Tang P., Romilowych M., Duncan C., Ng V., Fisman D.N. New endemic Legionella pneumophila serogroup I clones, Ontario, Canada. Emerg Infect Dis. 2010;16:447–454. doi: 10.3201/eid1603.081689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amemura-Maekawa J., Kura F., Helbig J.H., Chang B., Kaneko A., Watanabe Y. Characterization of Legionella pneumophila isolates from patients in Japan according to serogroups, monoclonal antibody subgroups and sequence types. J Med Microbiol. 2010;59(pt 6):653–659. doi: 10.1099/jmm.0.017509-0. [DOI] [PubMed] [Google Scholar]
- 11.Harrison T.G., Afshar B., Doshi N., Fry N.K., Lee J.V. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000–2008) Eur J Clin Microbiol Infect Dis. 2009;28:781–791. doi: 10.1007/s10096-009-0705-9. [DOI] [PubMed] [Google Scholar]
- 12.Lück C. Legionella pneumophila: genetic diversity of patients and environmental isolates. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2011;54:693–698. doi: 10.1007/s00103-011-1282-5. [DOI] [PubMed] [Google Scholar]
- 13.Ginevra C., Duclos A., Vanhems P., Campèse C., Forey F., Lina G. Host-related risk factors and clinical features of community-acquired legionnaires disease due to the Paris and Lorraine endemic strains, 1998–2007, France. Clin Infect Dis. 2009;49:184–191. doi: 10.1086/599825. [DOI] [PubMed] [Google Scholar]
- 14.Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 15.Chasqueira M.J., Rodrigues L., Nascimento M., Marques T. Sequence-based and monoclonal antibody typing of Legionella pneumophila isolated from patients in Portugal during 1987–2008. Euro Surveill. 2009;14(28) doi: 10.2807/ese.14.28.19271-en. [DOI] [PubMed] [Google Scholar]
- 16.Hilbi H., Jarraud S., Hartland E., Buchrieser C. Update on Legionnaires’ disease: pathogenesis, epidemiology, detection and control. Mol Microbiol. 2010;76:1–11. doi: 10.1111/j.1365-2958.2010.07086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tubach F., Ravaud P., Salmon-Céron D., Petitpain N., Brocq O., Grados F. Emergence of Legionella pneumophila pneumonia in patients receiving tumor necrosis factor-alpha antagonists. Clin Infect Dis. 2006;43:95–100. doi: 10.1086/508538. [DOI] [PubMed] [Google Scholar]
- 18.Lanternier F., Tubach F., Ravaud P., Salmon D., Dellamonica P., Bretagne S. Incidence and risk factors of Legionella pneumophila pneumonia during anti-tumor necrosis factor therapy: a prospective French study. Chest. 2013;144:990–998. doi: 10.1378/chest.12-2820. [DOI] [PubMed] [Google Scholar]
- 19.Qin T., Zhou H., Ren H., Guan H., Li M., Zhu B. Distribution of sequence-based types of Legionella pneumophila serogroup 1 strains isolated from cooling towers, hot springs, and potable water systems in China. Appl Environ Microbiol. 2014;80:2150–2157. doi: 10.1128/AEM.03844-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winchell J.M. Prevalence of sequence types among clinical and environmental isolates of Legionella pneumophila serogroup 1 in the United States from 1982 to 2012. J Clin Microbiol. 2014;52:201–211. doi: 10.1128/JCM.01973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginevra C., Forey F., Campèse C., Reyrolle M., Che D., Etienne J. Lorraine strain of Legionella pneumophila serogroup 1, France. Emerg Infect Dis. 2008;14:673–675. doi: 10.3201/eid1404.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]


