Abstract
Clostridium difficile causes antibiotic-associated diarrhoea and pseudomembranous colitis. The main virulence factors of C. difficile are the toxins A (TcdA) and B (TcdB). A third toxin, called binary toxin (CDT), can be detected in 17% to 23% of strains, but its role in human disease has not been clearly defined. We report six independent cases of patients with diarrhoea suspected of having C. difficile infection due to strains from toxinotype XI/PCR ribotype 033 or 033-like, an unusual toxinotype/PCR ribotype positive for CDT but negative for TcdA and TcdB. Four patients were considered truly infected by clinicians and were specifically treated with oral metronidazole. One of the cases was identified during a prevalence study of A−B−CDT+ strains. In this study, we screened a French collection of 220 nontoxigenic strains and found only one (0.5%) toxinotype XI/PCR ribotype 033 or 033-like strain. The description of such strains raises the question of the role of binary toxin as a virulence factor and could have implications for laboratory diagnostics that currently rarely include testing for binary toxin.
Keywords: A−B−CDT+ strains, binary toxin, diagnostic, PCR ribotype 033, toxinotype XI, Clostridium difficileI
Introduction
Clostridium difficile is responsible for 15% to 25% of antibiotic-associated diarrhoea and for more than 95% of pseudomembranous colitis. This Gram-positive spore-forming bacterium is the major cause of hospital-acquired diarrhoea [1,2]. Since 2003, many countries have reported outbreaks of severe C. difficile infections (CDI). This trend is assumed to be due in part to the emergence and rapid dissemination of an epidemic clone of C. difficile named NAP1/BI/027 [3]. Other changes have been observed in the epidemiology of CDI: (a) community cases of CDI have increased and have been described in people previously considered to be at low risk [4,5], (b) another clone, named 078, involved in severe CDI has been described [6], (c) C. difficile has been recognized as a pathogen or commensal in numerous animals [7,8] and (4) detection of C. difficile in food products has been reported [4,9].
The main virulence factors of C. difficile are two large clostridial toxins, toxins A (TcdA) and B (TcdB), encoded by the genes tcdA and tcdB, respectively. These two genes are located within a locus of pathogenicity (PaLoc) with three accessory genes: tcdC, tcdR and tcdE. Sequencing of the PaLoc has indicated a series of genetic polymorphisms. On the basis of PCR–restriction fragment length polymorphism of this locus, C. difficile strains are currently divided into 31 toxinotypes (or toxin variant strains) that are characterized by insertions, deletions and sequence mutations compared to the reference strain, VPI 10463 (toxinotype 0) [10,11]. Among the 31 toxinotypes, only the toxinotype XI strains do not produce TcdA and TcdB (A−B− strains) related to an absence of the tcdB gene and the presence of a large deletion in the 5′ region of the tcdA gene [10].
A third toxin, called binary toxin or CDT, was isolated for the first time by Popoff et al. [12] in a patient with severe pseudomembranous colitis. This toxin is detected in 17% to 23% of C. difficile strains in nonoutbreak situations [13–15]. This toxin is encoded by two genes, cdtA and cdtB, located on the CDT locus (CdtLoc), which is separated from the PaLoc on the C. difficile chromosome [12,16]. Strains carrying the CdtLoc belonged to specific toxinotypes, e.g. toxinotypes III, IV, V and XI, or, more rarely, to strains for which the PaLoc is absent [17]. The role of binary toxin in the pathophysiology of CDI remains unclear [18].
Five nonepidemiologically related C. difficile strains, characterized by the absence of tcdB gene but the presence of the binary toxin genes, and suspected to belong to the toxinotype XI (A−B−CDT+), were sent to the National Reference Center (NRC) for C. difficile in Paris, France, between December 2011 and February 2013. The objectives of this study were to confirm the toxinotype of those strains, to describe the clinical features of the five patients in whom these strains were isolated and to estimate the prevalence of such strains that produce binary toxin but not large clostridial toxins.
Materials and methods
Characterization of five strains with atypical toxin-encoding gene content received by the NRC and analysis of clinical data
Between December 2011 and February 2013, five strains with atypical toxin-encoding gene content were sent to the NRC for C. difficile in Paris. Three of five strains were identified as B−CDT+ using the molecular method Xpert® C. difficile (Cepheid, Sunnyvale, CA, USA) and were sent by the biologists to the NRC for confirmation of this particular result and to exclude a genetic drift in tcdB. The two other strains were characterized as A−B−CDT+ by the NRC performing routine PCR targeting tcdA, tcdB, cdtA and cdtB genes on DNA extracted from C. difficile cultures.
DNA extraction from colonies of C. difficile was performed with the InstaGene Matrix kit® (Bio-Rad Laboratories, Hercules, CA, USA). Amplification by PCR of tcdA, tcdB, tcdC, cdtA and cdtB genes coding for toxin A, toxin B, TcdC and the binary toxin, respectively, was performed using primers described elsewhere [13,19]. The toxinotypes were determined according to the method described by Rupnik et al. [19]. Briefly, amplification of A2 and A3 fragments of tcdA and B1, B2 and B3 fragments of tcdB was performed as described. Fragment A3 was digested using EcoRI to determine the toxinotype. PCR ribotyping was performed as described by Bidet et al. [20].
In vitro toxin production was tested by inoculating two to five colonies into brain–heart infusion broth (Oxoid, Hampshire, England, UK) that was incubated 5 days under an anaerobic atmosphere. The supernatant was filtered (0.22 μm pore size), and the filtrate was inoculated on MRC-5 cells. Toxin detection was also tested by C. diff Quik Chek complete® (Alere, Orlando, FL, USA) performed directly on colonies. Clinical data were reviewed and analyzed for the five patients harbouring these strains.
Screening of a A−B− strain collection
To estimate the frequency of A−B−CDT+ strains, 220 consecutive nontoxinogenic strains (A−B−) isolated in Paris (North of France) (n = 84) and Montpellier (South of France) (n = 136) between July 2011 and April 2013 were screened. All strains were isolated from patients with diarrhoea who were suspected of having CDI, who were admitted in Saint Antoine (Paris) or Arnaud de Villeneuve (Montpellier) hospitals and strains were characterized as nontoxinogenic strains by toxigenic culture (TC). The in vitro determination of C. difficile isolates ability to produce toxins (TC) was performed either by inoculating supernatant from a 5-day brain–heart infusion broth (Oxoid) on MRC-5 cells (Paris) or by using ImmunoCard® Toxins A&B (Meridian Bioscience, Cincinnati, OH, USA) directly on colonies as recommended by the manufacturer (Montpellier).
Absence of the PaLoc was confirmed as described elsewhere by obtaining an amplification of about 700 bp using primers lok1 and lok3 anchored outside the targeting PaLoc [21]. In case of a negative result (i.e., absence of the PaLoc was not confirmed), amplification by PCR of tcdA, tcdB, tcdC, cdtA and cdtB genes, toxinotyping and PCR ribotyping were performed as described above [13].
Results
Characteristics of five A−B−CDT+ strains and associated clinical data
Amplification of B1, B2 and B3 fragments of tcdB was confirmed as negative for the five strains (data not shown). Only the 3′ region (A2 and A3 fragments) of the tcdA gene was amplified for all the strains. Binary toxin genes and a 39 bp deletion in tcdC were identified in the five isolates. They were confirmed to be toxinotype XI strains after enzymatic restriction. Three isolates were defined as toxinotype XIa (EcoRI restriction pattern of A3 PCR fragment was of type 5d) and two isolates as XIb (EcoRI restriction pattern of A3 PCR fragment was of type 8) (Fig. 1). The five strains showed either a similar or slightly different PCR ribotyping banding pattern as reference strains (R11402, 542 and CD219), suggesting that the five strains belonged to the same lineage (Fig. 2). They were characterized as PCR ribotype 033 (or 033-like) isolates. The strains tested negative for their in vitro toxin production either by the cytotoxicity assay on MRC-5 cells or the C. diff Quik Chek complete® assay.
FIG. 1.
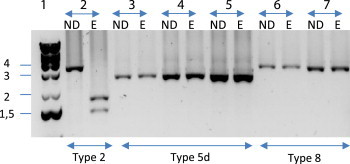
Restriction patterns obtained for A3 amplified fragment of tcdA gene for Clostridium difficile A−B−CDT+ strains isolated from patients compared with reference strains. ND, unrestricted; E., EcoRI digestion. Lanes: 1, DNA ladder (kb); 2, PCR ribotype 027; 3, 542 (reference strain for toxinotype XIa); 4, CD219 (reference strain for toxinotype XIa); 5, strain isolated from patient 1; 6, R11402 (reference strain for toxinotype XIb); 7, strain isolated from patient 2.
FIG. 2.
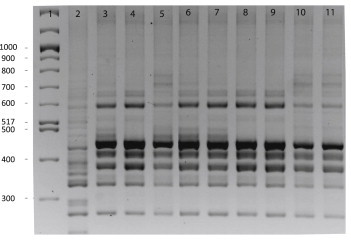
PCR ribotype of Clostridium difficile A−B−CDT+ strains isolated from patients compared with reference strains. Lanes: 1, DNA ladder (bp); 2, PCR ribotype 027; 3, R11402 (reference strain; toxinotype XIb); 4, patient 2; 5, patient 4; 6, strain isolated in prevalence study; 7, 542 (reference strain; toxinotype XIa); 8, patient 1; 9, CD219 (reference strain; PCR ribotype 033; toxinotype XIa); 10, patient 3; 11, patient 5.
Clinical and biological data of the five patients were analyzed (Table 1). The five patients were hospitalized in French hospitals in different areas, and their infections were considered to be epidemiologically unrelated. All five patients were symptomatic (diarrheic stools) and had risk factors for CDI (antibiotics n = 5, age >65 years n = 3). Four of five patients harbouring toxinotype XI strains were considered truly infected by their treating physician and were specifically treated with oral metronidazole. The clinical presentation of one patient was atypical because diarrhoea was probably the manifestation of ileal CDI, as the patient had previously undergone a total colectomy (Table 1, patient 1). One patient was not treated for CDI, but symptomatic treatment was started and diarrhoea resolved in the following days (patient 3).
TABLE 1.
Clinical and biological data of the six patients harbouring A−B−CDT+ strains
| Patient no. | Diagnostic | Toxinotype | Date of admission (dd/mm/yy) | Sampling date (dd/mm/yy) | Age (years) | Gender | Ward/hospital | Location (city) | Main reason for admission | White blood cell count | Origin of diarrhoea | Type of diarrhoea | Salmonella, Shigella, Campylobacter, Yersinia | Previous antibiotics | Specific treatment for CDI | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B−CDT+ (Xpert C. difficile Assay, Cepheid) | XIa | 02/12/2011 | 29/12/2011 | 70 | Male | Vascular surgery/Hôpital Henri Mondor | Créteil (North of France) | Surgical site infection after aortobifemoral prosthetic bypass | 7.8 × 109/L | HC-CDI | Watery | Absent | Imipenem, gentamicin | MTZ po, 500 mg 3 times a day | Diarrhoea resolved/no recurrence |
| 2 | B−CDT+(Xpert C. difficile Assay, Cepheid) | XIb | 05/10/2012 | 07/10/2012 | 81 | Female | Emergency and internal medicine, infectious disease/Hôpital André Mignot | Versailles (North of France) | Pneumonia | 2.9 × 109/L | HC-CDI | Watery and mucoid | Absent | Piperacillin and tazobactam | MTZ po 500 mg 3 times a day | Diarrhoea resolved/no recurrence |
| 3 | A−B−CDT+ (NRC, PCR on colonies) | XIa | 27/11/2012 | 03/12/2012 | 89 | Male | Long-term care/Hôpital de Bourg-en-Bresse | Bourg-en-Bresse (Centre of France) | Vomiting and repetitive falls, pneumonia | 6.4 × 109/L | HC-CDI | Bloody | Absent | Amoxicillin and clavulanic acid | No∗ (symptomatic treatment for diarrhoea) | Diarrhoea resolved |
| 4 | A−B−CDT+ (NRC, PCR on colonies) | XIb | 03/11/2012 | 04/12/2012 | 56 | Male | Nephrology/Hôpital Pellegrin | Bordeaux (South of France) | Acute renal failure and pneumonia | 13 × 109/L | HC-CDI | PMC | ND | Amoxicillin and clavulanic acid | MTZ po 250 mg 3 times a day | Diarrhoea resolved |
| 5 | B−CDT+ (Xpert C. difficile Assay, Cepheid) | XIa | 04/02/2013 | 05/02/2013 | 44 | Male | Hematology/Institut Paoli Calmette | Marseille (South of France) | Febrile diarrhoea | 25 × 109/L | CO-HC-CDI | Unknown | ND | Ticarcillin and clavulanic acid (+cancer chemotherapy) | MTZ po 500 mg 3 times a day | Death not related to CDI |
| 6 | A−B−CDT+ (prevalence study) | XIb | 10/09/2012 | 18/09/2012 | 73 | Male | Hepatologygastroenterology/Hôpital Saint Antoine | Paris (North of France) | Worsening of general conditions with hepatocellular carcinoma | 5.2 × 109/L | HC-CDI | Unknown | Absent | Unknown | No | Death not related to CDI |
ND, not done; HC, health care associated; CDI, C. difficile infection; CO, community onset; PMC, pseudomembranous colitis; MTZ, metronidazole; NRC, national reference centre.
Clostridium difficile has been isolated only by the NRC during EUropean, multicentre, prospective biannual point prevalence study of Clostridium difficile Infection in hospitalized patients with Diarrhoea Euclid study. C. difficile testing had not been requested by the physician nor done by the laboratory. Because this study was noninterventional, the result was not immediately transmitted to the physician.
Prevalence of A−B−CDT+ strains
Among the 220 studied strains, an amplification was obtained with primers lok1 and lok3, confirming the absence of the PaLoc, for 219 strains (99.5%). No amplification could be obtained for tcdA and tcdB. The binary toxin genes were also absent. These 219 strains were confirmed as nontoxigenic strains (A−B−CDT−). Absence of amplification with primers lok1 and lok3 was observed for one strain (0.5%). Further amplifications for tcdA and tcdB showed the presence of the 3′ region of tcdA only. This strain harboured a 39 bp deletion in the tcdC gene. Binary toxin genes were present, and the strain was characterized as toxinotype XIb. This strain belonged to the same PCR ribotype (033/033-like) as the five toxinotype XI strains previously identified in our laboratory (Fig. 2). Clinical data for the patient hospitalized in Hôpital Saint Antoine, Paris, and harbouring this strain were sparse; no specific treatment was initiated despite clinical symptoms, probably because the strain was reported as nontoxigenic by TC to the physician (Table 1, patient 6).
Discussion
We report here the six first strains of toxinotype XI isolated in patients suspected of having CDI in France. Toxinotype XI strains produce only binary toxin but not the common large clostridial toxins TcdA and TcdB. Toxinotype XI strains are atypical because they exhibit major rearrangement of the PaLoc. Only portion of the 3′ end region of PaLoc is present, bearing part of the sequence of tcdA and tcdC, and the 5′ region covering tcdB, tcdR and tcdE is deleted [10]. Rupnik et al. [22] described heterogeneity in the A3 region of tcdA that leads to distinguish two subtypes, XIa and XIb. So far, toxinotype XI is the only C. difficile toxinotype to be positive for binary toxin but negative for TcdA and TcdB production (A−B−CDT+) (http://www.mf.uni-mb.si/tox/images/Table1_Toxinotypes-characteristics.pdf).
Uncertainties still remain about the clinical significance of toxinotype XI in the pathogenesis of CDI because the role of binary toxin as a virulence factor is still controversial [18]. This toxin is produced by the highly virulent NAP1/BI/027 clone, which has caused severe outbreaks in North America and Europe, and by the emerging 078 clone, suggesting that it may serve as an additional virulence factor and may act in synergy with the large clostridial toxins [3]. A few clinical and epidemiological studies that compared data from infections with strains producing binary toxin in addition to TcdA and TcdB and infections with strains producing only TcdA and TcdB suggested that there could be a correlation between the production of binary toxin and the severity of CDI [23–25]. A case–control study conducted in 2005 compared the clinical presentation of 26 patients infected with strains producing binary toxin in addition to toxins A and B to 42 controls infected with strains producing only toxins A and B. Using univariate analysis, diarrhoea due to a binary toxin–positive strain was more often community acquired (p = 0.017) and associated with abdominal pain (p = 0.07) than diarrhoea due to binary toxin–negative strains. Diarrhoea was more often the cause of hospitalization in cases than in controls (p = 0.003) [23]. A more recent study reported that patients infected with C. difficile harbouring genes for toxins A and B and binary toxin had higher case-fatality rates than patients infected with C. difficile harbouring genes for toxin A and B without binary toxin [24]. Another study has shown that the presence of the binary toxin gene in C. difficile isolates was the only independent predictive factor for recurrent CDI [25]. The effects of binary toxin in animal models gave some conflicting results. In the rabbit ileal loop model, inoculation of supernatants from culture of binary toxin–positive strains that produced neither TcdA nor TcdB leads to enterotoxic response, suggesting that binary toxin may act as a virulence factor. Nevertheless, in the hamster model of ileocolitis, these strains produced no symptoms despite colonization [26]. Recent in vitro experiments have shown that binary toxin induces redistribution of microtubules and formation of long microtubule-based protrusions at the surface of intestinal epithelial cells. The CDT-induced microtubule protrusions form a dense mesh work at the cell surface, which wrap and embed bacterial cells, thereby largely increasing the adherence of C. difficile [27]. Finally, the importance of binary toxin was demonstrated using isogenic toxin mutants (A−B−CDT+) of NAP1/027/BI strains producing only binary toxin inoculated to the hamsters [28]. Three of nine hamsters inoculated with this A−B−CDT+ died.
PCR ribotype 033 strains have been reported in animals. In Australia, this PCR ribotype is the second most common strain isolated in cattle and calves [29]. However, very few cases of human CDI due to toxinotype XI have been described in the literature. Among the five cases reported by Geric et al. [17], three were from asymptomatic patients and one was from a symptomatic patient; no clinical information was available for the remaining case. In the present report, C. difficile strains were isolated from six symptomatic patients with several risk factors for CDI, including advanced age and previous broad-spectrum antibiotic treatment. Four of six patients were successfully treated with metronidazole. These data support the conclusion that diarrhoea in four patients was really due to C. difficile.
Only a few studies have estimated the prevalence of toxinotype XI strains. In this study, we found a prevalence of 0.5% (1/220). In a European study, 411 clinical C. difficile isolates from 38 hospitals in 14 European countries were characterized by toxinotyping. A total of 354 isolates (86.1%) were toxigenic. Among the toxigenic isolates, 268 (75.7%) were from toxinotype 0; 86 strains (24.3%) belonged to nine variant toxinotypes, but none was of toxinotype XI [15]. A large collection (5000 isolates) of C. difficile isolates from the United States and other sources over a 20-year period was examined for the presence of binary toxin genes among strains that do not produce TcdA and TcdB. Eight isolates have been reported as A−B− but CDT+ [17]. Among these eight isolates, five had a truncated PaLoc characteristic of toxinotype XI (including two toxinotype XIa and three toxinotype XIb isolates). However, most of the toxinotype XI strains isolated in that study were recovered from asymptomatic patients. All these data indicate that the prevalence of strains from toxinotype XI in humans seems very low. However, we hypothesize that the prevalence is likely underestimated. Indeed, the microbiological diagnosis of CDI in France is mostly based on the detection of TcdA or TcdB in stool samples, and clinical laboratories do not specifically look for binary toxin. As a consequence, strains from toxinotype XI would be likely reported as nontoxigenic isolates when tested in a clinical laboratory. Since 2009, nucleic acid amplifications assays have been increasingly used for the diagnosis of CDI. Most of the assays only target the tcdA or tcdB genes of C. difficile. Among these tests, the Xpert® C. difficile assay simultaneously detects three targets, including tcdB, binary toxin genes and the deletion in position 117 within the tcdC gene. This assay was initially developed in order to presumptively identify CDI due to the epidemic strain NAP1/BI/027 [3,30]. In recent years, this assay has been widely used in many laboratories in France for the routine diagnosis of CDI. Three cases of toxinotype XI that we report here were detected in clinical laboratories using the Xpert C. difficile assay and were suspected only because of the presence of binary toxin gene. It is likely that the increasing use of this test will enable a better recognition of toxinotype XI and therefore of its potential role in clinical disease.
In conclusion, the present study suggests that C. difficile strains producing only binary toxin seem to be pathogenic despite the lack of TcdA and TcdB. A result of C. difficile testing that would be only positive by PCR for binary toxin should be taken into account and may reveal a toxinotype XI strain. However, the prevalence of toxinotype XI seems low in France.
Conflict of interest
None declared.
References
- 1.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A. Multistate point-prevalence survey of health care–associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rupnik M., Wilcox M.H., Gerding D.N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 3.McDonald L.C., Killgore G.E., Thompson A., Owens R.C., Kazakova S.V., Sambol S.P. An epidemic, toxin gene–variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 4.Hensgens M.P.M., Keessen E.C., Squire M.M., Riley T.V., Koene M.G.J., de Boer E. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect. 2012;18:635–645. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox M.H., Mooney L., Bendall R., Settle C.D., Fawley W.N. A case–control study of community-associated Clostridium difficile infection. J Antimicrob Chemother. 2008;62:388–396. doi: 10.1093/jac/dkn163. [DOI] [PubMed] [Google Scholar]
- 6.Goorhuis A., Bakker D., Corver J., Debast S.B., Harmanus C., Notermans D.W. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- 7.Avbersek J., Janezic S., Pate M., Rupnik M., Zidaric V., Logar K. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe. 2009;15:252–255. doi: 10.1016/j.anaerobe.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Riley T.V., Adams J.E., O'Neill G.L., Bowman R.A. Gastrointestinal carriage of Clostridium difficile in cats and dogs attending veterinary clinics. Epidemiol Infect. 1991;107:659–665. doi: 10.1017/s0950268800049359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckert C., Burghoffer B., Barbut F. Contamination of ready-to-eat raw vegetables with Clostridium difficile in France. J Med Microbiol. 2013;62(pt 9):1435–1438. doi: 10.1099/jmm.0.056358-0. [DOI] [PubMed] [Google Scholar]
- 10.Geric Stare B., Rupnik M. Clostridium difficile toxinotype XI (A−B−) exhibits unique arrangement of PaLoc and its upstream region. Anaerobe. 2010;16:393–395. doi: 10.1016/j.anaerobe.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Rupnik M. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev. 2008;32:541–555. doi: 10.1111/j.1574-6976.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- 12.Popoff M.R., Rubin E.J., Gill D.M., Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert C., Coignard B., Hebert M., Tarnaud C., Tessier C., Lemire A. Clinical and microbiological features of Clostridium difficile infections in France: the ICD-RAISIN 2009 national survey. Med Mal Infect. 2013;43:67–74. doi: 10.1016/j.medmal.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Bauer M.P., Notermans D.W., van Benthem B.H.B., Brazier J.S., Wilcox M.H., Rupnik M. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377(9759):63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 15.Barbut F., Mastrantonio P., Delmee M., Brazier J., Kuijper E., Poxton I. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect. 2007;13:1048–1057. doi: 10.1111/j.1469-0691.2007.01824.x. [DOI] [PubMed] [Google Scholar]
- 16.Perelle S., Gibert M., Bourlioux P., Corthier G., Popoff M.R. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geric B., Johnson S., Gerding D.N., Grabnar M., Rupnik M. Frequency of binary toxin genes among Clostridium difficile strains that do not produce large clostridial toxins. J Clin Microbiol. 2003;41:5227–5232. doi: 10.1128/JCM.41.11.5227-5232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerding D.N., Johnson S., Rupnik M., Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2013;5(1) doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupnik M., Avesani V., Janc M., von Eichel-Streiber C., Delmee M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidet P., Barbut F., Lalande V., Burghoffer B., Petit J.C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 21.Braun V., Hundsberger T., Leukel P., Sauerborn M., von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996;181:29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 22.Rupnik M., Brazier J.S., Duerden B.I., Grabnar M., Stubbs S.L. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology. 2001;147(pt 2):439–447. doi: 10.1099/00221287-147-2-439. [DOI] [PubMed] [Google Scholar]
- 23.Barbut F., Decre D., Lalande V., Burghoffer B., Noussair L., Gigandon A. Clinical features of Clostridium difficile–associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J Med Microbiol. 2005;54(pt 2):181–185. doi: 10.1099/jmm.0.45804-0. [DOI] [PubMed] [Google Scholar]
- 24.Bacci S., Mølbak K., Kjeldsen M.K., Olsen K.E.P. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis. 2011;17:976–982. doi: 10.3201/eid1706.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart D.B., Berg A., Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg. 2013;17:118–124. doi: 10.1007/s11605-012-2056-6. [DOI] [PubMed] [Google Scholar]
- 26.Geric B., Carman R.J., Rupnik M., Genheimer C.W., Sambol S.P., Lyerly D.M. Binary toxin-producing, large clostridial toxin-negative Clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J Infect Dis. 2006;193:1143–1150. doi: 10.1086/501368. [DOI] [PubMed] [Google Scholar]
- 27.Schwan C., Stecher B., Tzivelekidis T., van Ham M., Rohde M., Hardt W.-D. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehne S.A., Collery M.M., Kelly M.L., Cartman S.T., Cockayne A., Minton N.P. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight D., Thean S., Putsathit P., Fenwick S., Riley T.V. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol. 2013;79:2630–2635. doi: 10.1128/AEM.03951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldenberg S.D., Dieringer T., French G.L. Detection of toxigenic Clostridium difficile in diarrheal stools by rapid real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2010;67:304–307. doi: 10.1016/j.diagmicrobio.2010.02.019. [DOI] [PubMed] [Google Scholar]


