Abstract
Reports of mecC methicillin-resistant Staphylococcus aureus (MRSA) strains have been published from several European countries. We describe the first six mecC MRSA isolates of human origin from Austria and report the application of a rapid PCR test. Candidate isolates (n = 295) received between 2009 and 2013 were investigated phenotypically by cefoxitin screening and streaking on ChromID MRSA plates. The presence of mecC was confirmed in six isolates from blood cultures, wound swabs and screening samples of four female and two male patients (age range 7–89 years) by an in-house PCR method and the new Genspeed MRSA test (Greiner Bio-One, Kremsmünster, Austria). The mecC MRSA were further characterized by whole genome sequencing, multilocus sequence and spa typing. Antimicrobial susceptibility testing was performed by Eucast disk-diffusion method and Vitek 2. The six mecC MRSA isolates were from two clonal lineages (CC130, including a new single-locus variant, and CC599) and four different spa types (t843, t1535, t3256, t5930). Analysis for virulence factor genes yielded lukED, eta, etd2 and edin-B (CC130 isolates) and tst, lukED, eta and sel (ST599 isolates). The Genspeed MRSA test identified mecC in all isolates whereas Vitek 2 failed to detect methicillin resistance in one isolate. The strains were susceptible to a wide range of non-β-lactam antibiotics. All patients were successfully treated or decolonized. mecC MRSA are present in Austria as colonizers but may also cause infections. Thus, laboratories must choose appropriate test methods such as cefoxitin screening and confirmation using molecular assays specifically targeting mecC.
Keywords: Austria, cefoxitin, Genspeed, mecC, MLST, MRSA, PCR, Staphylococcus aureus, whole genome sequencing
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) isolates carrying the mecA homologue mecC have been reported from all over Europe [1–8]. They may be detected phenotypically by routine cefoxitin screening and by PCR using specific primers; however, standard molecular diagnostic systems based on amplification of mecA fail to recognize these strains due to nucleic acid divergences between mecA and mecC. Published clinical data concerning mecC MRSA in humans include reports about colonization as well as skin and soft tissue infections [4], but also include fatal bacteremia [7] and osteomyelitis [9]. Thus, reliable detection of these strains in diagnostic microbiology routine is important [10].
The National Reference Centre for Antimicrobial Resistance and Nosocomial Infections at the Elisabethinen Hospital Linz receives bacterial isolates of human origin for identification, confirmation and typing from Austrian laboratories. Its strain collection contains over 5000 isolates of Staphylococcus spp. many of which have been extensively studied and typed using molecular methods [11–14]. We searched this strain collection for S. aureus carrying mecC using the conventional phenotypic approach followed by molecular confirmation with an in-house PCR method as well as one of the first commercially available systems also able to detect mecC, the Genspeed MRSA test (Greiner Bio-One, Kremsmünster, Austria). In addition, clinical and molecular typing data on four mecC-positive isolates detected as part of routine screening are presented, describing for the first time the presence of mecC MRSA in human samples from Austria.
Materials and Methods
Bacterial isolates, phenotypic and molecular antibiotic susceptibility testing and typing
Candidate S. aureus isolates (n = 295) that had tested negative for mecA and positive for femA using previously published primer sets between the years 2003 and 2012 were chosen from the strain collection [15,16]. Additionally, four strains received from Austrian laboratories in 2012–2013 for further testing regarding mecC were included in this study. Strains were subcultured overnight on trypticase soy agar containing 5% sheep’s blood (Oxoid, Wesel, Germany) at 36 ± 1°C in an aerobic atmosphere. Species identification of all isolates was done by matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry using the IVD MALDI Biotyper (Bruker Daltonik, Bremen, Germany) and the Vitek 2 system (bioMérieux, Marcy l’Etoile, France).
Susceptibility testing was performed according to the Eucast disk-diffusion method. All strains were screened phenotypically for methicillin susceptibility using cefoxitin 30 μg disks in quintuplicate. They were also inoculated onto ChromID MRSA agar plates (bioMérieux) that were read after 24 hours of incubation at 37°C. The broader antimicrobial susceptibility of mecC MRSA for a panel of substances (Table 1) was assessed by disk diffusion testing, and for selected substances, minimum inhibitory concentrations (MICs) were determined by gradient diffusion testing (Etest; bioMérieux) (Table 1). The susceptibility profiles of the mecC MRSA were also assessed using Vitek 2 Gram-positive antimicrobial susceptibility testing cards (bioMérieux).
TABLE 1.
Phenotypic and molecular typing data of six mecC methicillin-resistant Staphylococcus aureus isolates
| Isolate 4402/2009 |
Isolate 5127/2010 |
Isolate 5590/2012 |
Isolate 5625/2012 |
Isolate 5676/2012 |
Isolate 5752/2013 |
|
|---|---|---|---|---|---|---|
| Disk diffusion test | Diameter (mm) (category) | Diameter (mm) (category) | Diameter (mm) (category) | Diameter (mm) (category) | Diameter (mm) (category) | Diameter (mm) (category) |
| Cefoxitin | 18 (mean, R) | 16 (mean, R) | 21 (mean, R) | 18 (mean, R) | 17 (mean, R) | 20 (mean, R) |
| Gentamicin | 20 (S) | 22 (S) | 20 (S) | 22 (S) | 24 (S) | 22 (S) |
| Erythromycin | 26 (S) | 26 (S) | 24 (S) | 28 (S) | 30 (S) | 30 (S) |
| Clindamycin | 25 (S) | 26 (S) | 22 (S) | 26 (S) | 30 (S) | 30 (S) |
| Tetracycline | 25 (S) | 26 (S) | 22 (S) | 27 (S) | 25 (S) | 30 (S) |
| Fusidic acid | 30 (S) | 31 (S) | 26 (S) | 30 (S) | 30 (S) | 30 (S) |
| Trimethoprim/sulfa | 32 (S) | 31 (S) | 26 (S) | 34 (S) | 30 (S) | 30 (S) |
| Rifampicin | 31 (S) | 30 (S) | 27 (S) | 32 (S) | 30 (S) | 30 (S) |
| Gradient MIC test | MIC (mg/L) | MIC (mg/L) | MIC (mg/L) | MIC (mg/L) | MIC (mg/L) | MIC (mg/L) |
| Ceftaroline | 1 (S) | 1 (S) | 1 (S) | 1 (S) | 0.5 (S) | 0.5 (S) |
| Vancomycin | 1 (S) | 1 (S) | 2 (S) | 1 (S) | 1 (S) | 2 (S) |
| Teicoplanin | 1 (S) | 1 (S) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) |
| Tigecycline | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.125 (S) | 0.25 (S) |
| Linezolid | 0.5 (S) | 0.5 (S) | 1 (S) | 0.5 (S) | 0.5 (S) | 2 (S) |
| Daptomycin | 0.25 (S) | 0.125 (S) | 0.125 (S) | 0.125 (S) | 0.25 (S) | 0.25 (S) |
| Fosfomycin | 1 (S) | 1 (S) | 0.5 (S) | 1 (S) | 0.5 (S) | 2 (S) |
| Oxacillin | 4 | 4 | 2 | 8 | 4 | 2 |
| Cefoxitin | 16 | 32 | 16 | 16 | 32 | 16 |
| Typing | ||||||
| Multilocus sequence | 599 | 130 | SLV of 130 | 130 | 599 | 130 |
| Type | ||||||
| Clonal complex | 599 | 130 | 130 | 130 | 599 | 130 |
| spa Type | t5930 | t3256 | t1535 | t1535 | t5930 | t843 |
| Virulence factor gene | ||||||
| tst | + | − | − | − | + | − |
| lukED | + | + | + | + | + | + |
| eta | + | + | + | + | + | + |
| etd2 | − | + | + | + | − | + |
| edin-B | − | + | + | + | − | + |
| sel | + | − | − | − | + | − |
SLV, single locus variant; MIC, minimum inhibitory concentration.
All isolates showing a mean cefoxitin zone diameter <22 mm and/or growth on selective media underwent confirmatory PCR testing using a protocol published by Stegger et al. [17] to detect mecA, mecC and lukF-PV after extraction of bacterial DNA with InstaGene Matrix (BioRad, Hercules, CA, USA), with modified primer concentrations. Additionally, a commercially available rapid PCR system, the Genspeed MRSA (Greiner Bio-One, Kremsmünster, Austria) was evaluated. This system uses rapid bacterial lysis followed by conventional PCR and detection of mecA and mecC, including specific probes for S. aureus, via hybridization on a chip.
Isolates carrying mecC were further analyzed by pulsed-field gel electrophoresis of SmaI restriction fragments. They were also typed by amplification of the polymorphic X region of the protein A gene (spa) followed by sequencing and assignation of the spa type [18]. Whole genome sequencing of mecC-positive isolates was performed as previously described [19] to confirm their mecC gene status, to determine their multilocus sequence type (ST) and to identify virulence factor and antibiotic resistance genes. Nucleotide sequences have been deposited in the European short-read archive as ERR387183, ERR387184, ERR387185, ERR387090, ERR 490434 and ERR490433 for isolates 4402, 5127, 5590, 5625, 5675 and 5752, respectively.
Clinical information
Clinical information on patients with mecC MRSA was obtained from medical records retrospectively. Consent of the local ethics committee was provided (C-60-13).
Results
All strains were confirmed as Staphylococcus aureus with MALDI-TOF scores of >2.0. Phenotypic screening of the 295 isolates from the strain collection revealed two cefoxitin screening–positive isolates (0.7%) with mean diameters of 14 mm (range 13–14 mm) and 15 mm (all 15 mm), respectively. Fourteen isolates (4.7%), including the two that were cefoxitin screening positive, showed growth on ChromID MRSA agar. Cefoxitin diameters of the other 12 isolates were in the susceptible category (range 27–32 mm); thus, sensitivity of the ChromID MRSA agar for phenotypic methicillin resistance detection was 100% and specificity was 95.9%.
Further analysis by molecular testing was performed on these fourteen isolates. The two cefoxitin-resistant isolates were positive for mecC and negative for mecA by conventional PCR as well as with the new Genspeed MRSA test. In accordance with their susceptibility to cefoxitin, the other 12 isolates carried neither mecA nor mecC. Additionally, the four strains that were received in our capacity as reference laboratory were analyzed. All four isolates had a positive cefoxitin screening test with diameters of 17, 18, 20 and 21 mm, respectively. Presence of mecC and absence of mecA was confirmed with both PCR assays, and lukF-PV was not detected in any of the strains.
Analysis of spa sequencing data showed four different spa types among the six isolates: t5930 (n = 2), t1535 (n = 2), t843 (n = 1) and t3256 (n = 1). Whole genome sequencing of all isolates confirmed that each isolate encoded mecC within an SCCmec type XI. Sequence types derived from the genome sequences included three ST130, a single-locus variant of ST130 and two ST599. All six isolates were positive for the genes encoding leukocidin ED (lukED) (GenBank accession no. Y13225) and exfoliative toxin A (eta) (GenBank accession no. CAQ49592). The four isolates belonging to CC130 additionally carried the epidermal cell differentiation inhibitor B gene (edin-B) (GenBank accession no. AHC54577) and the exfoliative toxin D gene (etd2) (GenBank accession no. HF563069). The two ST599 isolates were also positive for the toxic shock syndrome toxin-1 (tst) (GenBank accession no. Q9F0L4) and the enterotoxin-L (sel) gene (GenBank accession no. CAI80052) (Table 1). Other than mecC and blaZ, no other acquired resistance genes were identified.
Analysis of SmaI pulsed-field gel electrophoresis patterns showed the strains to be unrelated, with the exception of isolates 5590/2012 and 5625/2012, which were isolated in the same area of Austria within 8 weeks of each other. The Dice coefficient for this pair was 85.9%, indicating possible relatedness.
Subsequently, the six mecC MRSA were subjected to antimicrobial susceptibility testing and yielded susceptible results for all non-β-lactam antibiotics (Table 1). Two isolates (5127/2010 and 5752/2013) had oxacillin MICs below the breakpoint usually used to infer methicillin resistance. Vitek 2 identified all six isolates correctly as S. aureus and gave a positive result for the cefoxitin screen in five isolates, whereas all isolates were given a susceptible result for oxacillin. One isolate (5752/2013) was identified as methicillin-susceptible Staphylococcus aureus (MSSA) with a negative cefoxitin screen also upon repetition of the test. The remaining susceptibility data of the AST GP card matched the results of manual testing.
Clinical data are summarized in Table 2, and the geographic location of the laboratories that provided the mecC MRSA strains as well as the year of detection are indicated in Fig. 1.
TABLE 2.
Clinical information of six mecC methicillin-resistant Staphylococcus aureus isolates
| Characteristic | Isolate 4402/2009 | Isolate 5127/2010 | Isolate 5590/2012 | Isolate 5625/2012 | Isolate 5676/2012 | Isolate 5752/2013 |
|---|---|---|---|---|---|---|
| Patient age/sex | 89/female | 54/female | 7/female | 72/female | 83/male | 70/male |
| Site of isolation | Blood culture | Multisite screen | Wound swab (outer ear) | Wound swab (leg ulcer) | Nose screen | Blood culture |
| Underlying disease | Myelodysplastic syndrome, sepsis | Diabetes, Eczema | Otitis externa | Ulcus cruris | Stroke | Peripheral arterial occlusion disease, infected leg ulcer |
| Therapy | Unknown | Octenidine/mupirocin | Topical ofloxacin | Topical silver-sulfa-diazine/povidone-iodine | Mupirocin | Clindamycin iv |
| Outcome | Recovered | Eradication | Recovered | Improvement | Eradication | Recovered |
| Risk factors | Unknown | Unknown | Pet rabbit (not screened) | Unknown | Unknown | Unknown |
| Geographic location | Upper Austria | Vienna | Vorarlberg | Vorarlberg | Salzburg | Upper Austria |
No contact could be established between patients.
FIG. 1.
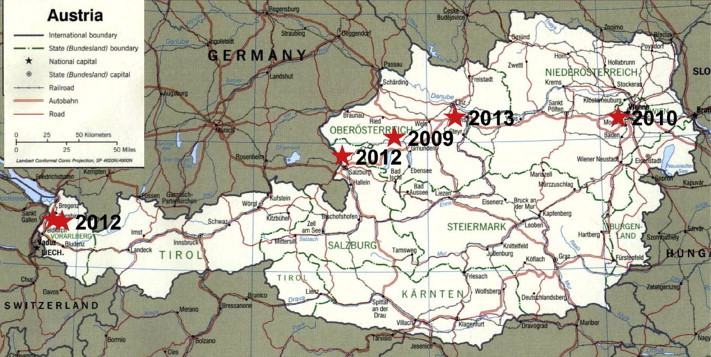
Location and year of isolation of six mecC methicillin-resistant Staphylococcus aureus isolates. Most patients lived in rural areas of Austria. (Map is a public domain file downloaded from http://www.mygeo.info/karten/austria_pol99.jpg.)
Discussion
To our knowledge, this is the first report concerning the detection of mecC MRSA in humans in Austria. We can confirm the presence of such strains back to the year 2009 resulting from the investigation of our strain collection. The only other data on mecC MRSA in Austria covers livestock- and wildlife-origin strains [20,21].
All mecC-positive isolates in this study were phenotypically recognizable as MRSA using cefoxitin in the Eucast disk-diffusion test, which has been shown to reliably detect these strains [22]. However, the built-in cefoxitin screen of the Vitek 2 GP AST card detected only five of the six isolates, which is in contrast to a recent study using the same system successfully on 62 mecC MRSA isolates [23]. However, the atypical Vitek 2 oxacillin-susceptible/cefoxitin-resistant profile of mecC MRSA as described by Cartwright et al. [23] was observed in the remaining five isolates. Isolate 5752/2013, which was misidentified as MSSA by Vitek 2, had the largest cefoxitin diameter (21 mm) of all isolates and thus was also challenging for disk-diffusion test reading. All isolates were universally susceptible to non-β-lactam antibiotics, which compares to data on mecC MRSA from other European countries [6,7,24].
Previous reports have shown reduced efficiency of growth on commercially available selective chromogenic media, also including ChromID agar [6], for ST130 mecC MRSA. In contrast, in our small sample also containing ST130 strains, all isolates grew well on ChromID agar. However, the cefoxitin MICs of our isolates were considerably higher than those reported by Cuny et al. [6] (all ≥16 mg/L). The published cefoxitin content of this medium is 4 mg/L [25,26], so efficient growth was not unexpected. Also, the observed false-positive rate of 4.1% is in line with published data [27].
Up to now, PCR methods for MRSA detection have successfully covered all known SCCmec types as a result of the high homology of the mecA gene. However, the newly discovered mecC element shares only 69% identity of its DNA sequence with mecA, leading to negative results using mecA-based PCR protocols and a need for additional primers to detect mecC. At the moment, not all commercially available assays cover this requirement. The Genspeed MRSA test that we used in this study can be completed in 75 minutes from culture; it can also be performed directly from swabs. It may be used as screening tool for colonization with S. aureus (MSSA) as well as mecA or mecC MRSA.
Four of the six mecC MRSA in our study belonged to CC130, which agrees with previously published data from other countries that also show CC130 to be predominant [1,2,7,17,24]. Analysis of whole genome sequences of six strains revealed that the CC130 isolates lacked pyrogenic toxin superantigen (PTSAg) encoding genes, such as the toxic shock toxin gene tst and staphylococcal enterotoxin genes. However they were all positive for exfoliative toxin genes (eta and edinB). The occurrence of mecC in MRSA isolates belonging to ST599 is relatively uncommon, but the two isolates in our study were positive for the PTSAg genes tst and sel, which matches the toxin gene profile of previously published ST599 mecC MRSA isolates [5,28]. Four different spa types were recovered in this study, all of which have previously been reported among mecC MRSA. t843 was the dominant spa type of mecC MRSA in a range of reports [4,7,17] and spa types t1535 and t3256 have also been described [7,24], whereas t5930 has so far only been mentioned in a single report from France [5].
The capability of mecC MRSA to cause clinical disease in a wide range of patients has been established by several reports. In a Spanish case series, most patients were elderly men who were colonized, but a patient with lethal sepsis was also described [7]. Petersen et al. [4] reported on 112 mecC MRSA isolates from Denmark, with equal sex distribution and a mean patient age of 51 years. Interestingly, the majority of the isolates were from infections, mostly skin and soft tissue infection, and not from colonization screening. In a recent report on the prevalence of human mecC MRSA in England, more than half of the isolates were identified from screening samples [24], whereas in a case series from Germany, most strains were isolated from wounds but also included was an isolate from nosocomial pneumonia [6]. A French case report described a patient with mediastinitis and sternal osteitis due to mecC MRSA [9]. In two of our six clinically diverse study patients, mecC MRSA were identified via routine screening, but in the remaining four patients, it was clearly associated with their clinical presentation.
Overall, mecC MRSA do not seem to be highly prevalent in Austria at the moment, which reflects the situation in most other European countries [10]. On the other hand, the frequency of mecC MRSA has increased significantly to a proportion of 2% of the total annual human MRSA cases in Denmark [4]. As mecC is carried on a mobile genetic element (SCCmec), it has the potential to spread into different lineages [29–31]. Thus, reliable detection and monitoring of mecC MRSA with appropriate phenotypic and molecular screening and confirmation methods is needed.
Conflict of interest
None declared.
Acknowledgements
The authors thank Lucia Berning, Magdalena Nimmervoll and Sarah Widhalm for excellent technical work and Harald Dirschmid (Institut für Pathologie, Landeskrankenhaus Feldkirch), Barbara Öhlinger (Institut für klinische Pathologie, Landeskrankenhaus Vöcklabruck), Jan Marco Kern (Division Medizinische Mikrobiologie, Universitätsinstitut für Medizinisch-Chemische Labordiagnostik, Paracelsus Medizinische Privatuniversität, Salzburger Landeskliniken) and Eva-Maria Zeitlberger (SMZ Ost Donauspital, Vienna) for their help in obtaining clinical data. This study was conducted as part of our routine work. The Genspeed MRSA kits used in this study were generously donated by Greiner Bio-One, Kremsmünster, Austria.
References
- 1.Garcia-Alvarez L., Holden M.T., Lindsay H., Webb C.R., Brown D.F., Curran M.D. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laurent F., Chardon H., Haenni M., Bes M., Reverdy M.E., Madec J.Y. MRSA harboring mecA variant gene mecC, France. Emerg Infect Dis. 2012;18:1465–1467. doi: 10.3201/eid1809.111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basset P., Prod’hom G., Senn L., Greub G., Blanc D.S. Very low prevalence of meticillin-resistant Staphylococcus aureus carrying the mecC gene in western Switzerland. J Hosp Infect. 2013;83:257–259. doi: 10.1016/j.jhin.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Petersen A., Stegger M., Heltberg O., Christensen J., Zeuthen A., Knudsen L.K. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19:E16–E22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- 5.Sabat A.J., Koksal M., Akkerboom V., Monecke S., Kriegeskorte A., Hendrix R. Detection of new methicillin-resistant Staphylococcus aureus strains that carry a novel genetic homologue and important virulence determinants. J Clin Microbiol. 2012;50:3374–3377. doi: 10.1128/JCM.01121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuny C., Layer F., Strommenger B., Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One. 2011;6:e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Garrote F., Cercenado E., Marin M., Bal M., Trincado P., Corredoira J. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraemia. J Antimicrob Chemother. 2013;69:45–50. doi: 10.1093/jac/dkt327. [DOI] [PubMed] [Google Scholar]
- 8.Schaumburg F., Kock R., Mellmann A., Richter L., Hasenberg F., Kriegeskorte A. Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J Clin Microbiol. 2012;50:3186–3192. doi: 10.1128/JCM.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barraud O., Laurent F., Francois B., Bes M., Vignon P., Ploy M.C. Severe human bone infection due to methicillin-resistant Staphylococcus aureus carrying the novel mecC variant. J Antimicrob Chemother. 2013;68:2949–2950. doi: 10.1093/jac/dkt274. [DOI] [PubMed] [Google Scholar]
- 10.Paterson G.K., Harrison E.M., Holmes M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krziwanek K., Luger C., Sammer B., Stumvoll S., Stammler M., Metz-Gercek S. PVL-positive MRSA in Austria. Eur J Clin Microbiol Infect Dis. 2007;26:931–935. doi: 10.1007/s10096-007-0391-4. [DOI] [PubMed] [Google Scholar]
- 12.Krziwanek K., Luger C., Sammer B., Stumvoll S., Stammler M., Sagel U. MRSA in Austria—an overview. Clin Microbiol Infect. 2008;14:250–259. doi: 10.1111/j.1469-0691.2007.01896.x. [DOI] [PubMed] [Google Scholar]
- 13.Krziwanek K., Metz-Gercek S., Mittermayer H. Methicillin-resistant Staphylococcus aureus ST398 from human patients, Upper Austria. Emerg Infect Dis. 2009;15:766–769. doi: 10.3201/eid1505.080326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krziwanek K., Metz-Gercek S., Mittermayer H. Trends in the occurrence of MRSA strains in Upper Austria from 2006 to 2009. Clin Microbiol Infect. 2011;17:920–923. doi: 10.1111/j.1469-0691.2010.03376.x. [DOI] [PubMed] [Google Scholar]
- 15.Ungeheuer J., Wilms S., Sietzen W. mecA-Gen bei Staphylokokken—vereinfachter Nachweis mit der PCR. Chemother J. 1994;1:31–35. [Google Scholar]
- 16.Kizaki M., Kobayashi Y., Ikeda Y. Rapid and sensitive detection of the femA gene in staphylococci by enzymatic detection of polymerase chain reaction (ED-PCR): comparison with standard PCR analysis. J Hosp Infect. 1994;28:287–295. doi: 10.1016/0195-6701(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 17.Stegger M., Andersen P.S., Kearns A., Pichon B., Holmes M.A., Edwards G. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251) Clin Microbiol Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y.W., Waddington M.G., Smith D.H., Manahan J.M., Kohner P.C., Highsmith L.M. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2000;38:1347–13451. doi: 10.1128/jcm.38.4.1347-1351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison E.M., Paterson G.K., Holden M.T., Morgan F.J., Larsen A.R., Petersen A. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. 2013;5:509–515. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loncaric I., Kubber-Heiss A., Posautz A., Stalder G.L., Hoffmann D., Rosengarten R. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J Antimicrob Chemother. 2013;68:2222–2225. doi: 10.1093/jac/dkt186. [DOI] [PubMed] [Google Scholar]
- 21.Loncaric I., Kubber-Heiss A., Posautz A., Stalder G.L., Hoffmann D., Rosengarten R. mecC-and mecA-positive meticillin-resistant Staphylococcus aureus (MRSA) isolated from livestock sharing habitat with wildlife previously tested positive for mecC-positive MRSA. Vet Dermatol. 2014;25:147–148. doi: 10.1111/vde.12116. [DOI] [PubMed] [Google Scholar]
- 22.Skov R., Larsen A.R., Kearns A., Holmes M., Teale C., Edwards G. Phenotypic detection of mecC-MRSA: cefoxitin is more reliable than oxacillin. J Antimicrob Chemother. 2014;69:133–135. doi: 10.1093/jac/dkt341. [DOI] [PubMed] [Google Scholar]
- 23.Cartwright E.J., Paterson G.K., Raven K.E., Harrison E.M., Gouliouris T., Kearns A. Use of VITEK 2 antimicrobial susceptibility profile to identify mecC in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2013;51:2732–2734. doi: 10.1128/JCM.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterson G.K., Morgan F.J., Harrison E.M., Cartwright E.J., Torok M.E., Zadoks R.N. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother. 2013;69:907–910. doi: 10.1093/jac/dkt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry J.D., Davies A., Butterworth L.A., Hopley A.L., Nicholson A., Gould F.K. Development and evaluation of a chromogenic agar medium for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2004;42:4519–4523. doi: 10.1128/JCM.42.10.4519-4523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra-Kumar S., Haccuria K., Michiels M., Ieven M., Poyart C., Hryniewicz W. Current trends in rapid diagnostics for methicillin-resistant Staphylococcus aureus and glycopeptide-resistant enterococcus species. J Clin Microbiol. 2008;46:1577–1587. doi: 10.1128/JCM.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malhotra-Kumar S., Abrahantes J.C., Sabiiti W., Lammens C., Vercauteren G., Ieven M. Evaluation of chromogenic media for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010;48:1040–1046. doi: 10.1128/JCM.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandendriessche S., Vanderhaeghen W., Soares F.V., Hallin M., Catry B., Hermans K. Prevalence, risk factors and genetic diversity of methicillin-resistant Staphylococcus aureus carried by humans and animals across livestock production sectors. J Antimicrob Chemother. 2013;68:1510–1516. doi: 10.1093/jac/dkt047. [DOI] [PubMed] [Google Scholar]
- 29.Spoor L.E., McAdam P.R., Weinert L.A., Rambaut A., Hasman H., Aarestrup F.M. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013;4(4) doi: 10.1128/mBio.00356-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nubel U., Dordel J., Kurt K., Strommenger B., Westh H., Shukla S.K. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 2010;6:e1000855. doi: 10.1371/journal.ppat.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monecke S., Coombs G., Shore A.C., Coleman D.C., Akpaka P., Borg M. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One. 2010;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]


