Abstract
Prototheca zopfii associated with bovine mastitis and human protothecosis exists as two genotypes, of which genotype 1 is considered as non-infectious and genotype 2 as infectious. The mechanism of infection has not yet been described. The present study was aimed to identify genotype 2-specific immunodominant proteins. Prototheca proteins were separated using two-dimensional gel electrophoresis. Subsequent western blotting with rabbit hyperimmune serum revealed 28 protein spots. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis resulted in the identification of 15 proteins including malate dehydrogenase, elongation factor 1-alpha, heat shock protein 70, and 14-3-3 protein, which were previously described as immunogenic proteins of other eukaryotic pathogens.
Keywords: Bovine mastitis, immunodominant proteins, MALDI-TOF MS, proteomics, Prototheca, western blotting
Protothecosis refers to a rare infection caused by achlorophyllous unicellular algae of the genus Prototheca, which are reported to be saprophytic, ubiquitously distributed in the environment and closely related to Chlorella species [1,2]. Currently, among the six generally accepted species of Prototheca, Prototheca zopfii, Prototheca wickerhamii, Prototheca blaschkeae and Prototheca cutis have been associated with infections in cows, dogs, cats and, rarely, in humans. P. zopfii and P. blaschkeae are associated with antimicrobial-resistant bovine mastitis, which leads to heavy economic loss while P. zopfii appears to be frequently reported [3,4]. On the basis of biochemical, serological and phylogenetic analysis, P. zopfii has been divided into two genotypes, of which genotype 1 is considered to be non-pathogenic and genotype 2 is associated with bovine mastitis and human protothecosis [5–8]. Recently, differences at the proteomic level between these two genotypes of P. zopfii have been described [9–11]; however, the mechanism of infection has still not been described. Hence, the present study was carried out to identify the immunogenic proteins of P. zopfii genotype 2 using two-dimensional (2D) gel electrophoresis–western blotting with hyperimmune serum from experimentally immunized rabbits.
The type strain P. zopfii genotype 2 (SAG 2021), isolated from bovine mastitis [7], was utilized for extraction of protein [9]. In brief, cells cultured overnight were harvested by centrifugation at 1000 g for 5 minutes, reconstituted in lysis buffer (20 mM HEPES, pH 7.4, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 0.5% benzonase and 0.5% protease inhibitor cocktail tablet), sonicated and the supernatant was collected for further study. The specific rabbit primary polyclonal antibodies were raised in New Zealand White rabbits as described previously [2]. In brief, rabbits of body weight 2.5–3.5 kg were intradermally injected with 1.0 mL of emulsion containing equal volume of phosphate-buffered saline containing 107 cells/mL and Freund incomplete adjuvant (Sigma-Aldrich, Steinheim, Germany). Following 3 weeks of priming, the rabbits were boosted intravenously, biweekly three times with 107 viable cells of the homologous strain. Seven days after the last booster application and 10 weeks after priming, the rabbits were bled, serum was collected and stored at −80°C until further use. The immunization experiments with the rabbits were performed according to the German law on animal welfare and as approved by the German authorities (Regierungspräsidium Leipzig, Permission No. V 4/04).
Western blotting was carried out following the 2D separation of Prototheca proteins on 12% polyacrylamide gel after rehydration and isoelectrofocusing of 250 μg of whole cell lysate on an immobilized pH gradient strip of 7 cm and pI 3–10 (non-linear) (Immobilian drystrip; GE Healthcare, Munich, Germany). The separated proteins were then transferred to a nitrocellulose membrane (Trans Blot trans medium pure nitrocellulose membrane; Bio-Rad, Munich, Germany) using semi-dry transfer unit (80 mA per gel for 90 min) (GE Healthcare). Electrophoresis and blotting quality were checked by staining the membrane for 30 s in Ponceau S (1% Ponceau S, 0.1% acetic acid) followed by short destaining with Tris-buffered saline with Tween-20 (TBST). The membrane was incubated overnight in 1% skimmed milk powder (Carl Roth, Karlsruhe, Germany) in TBST, washed and incubated for 90 min with 1 : 100 diluted rabbit hyperimmune serum. The membrane was briefly rinsed with TBST and incubated for another 90 min with 1 : 2500 diluted goat anti-rabbit horseradish-peroxidase conjugated IgG-h-I (Biomol, Hamburg, Germany). Detection was carried out using 3,3′,5,5′-tetramethylbenzidine kit (Sigma Aldrich Chemie, Steinheim, Germany). The respective protein spots on a 2D gel performed in parallel and stained with Coomassie Brilliant Blue were identified after overlaying of the western blot image using Delta2D software version 4.0 (Decodon, Greifswald, Germany) [12].
Two-dimensional gel electrophoresis western blots from hyperimmune serum from two rabbits revealed 28 signals following analysis with Decodon software (Fig. 1). The corresponding protein spots were excised from the 2D gel and digested with trypsin. Protein identification was performed using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Ultraflex II TOF/TOF; Bruker Daltonics, Bremen, Germany), as described previously [9]. Protein identification was considered to be valid if more than two peptides matched and the MASCOT score was greater than or equal to the significance threshold (p <0.05). As a result, 15 proteins were successfully identified (Table 1). The identified proteins were found to be enzymes of energy metabolism and proteins involved in cellular transport and cell signalling including stress response.
Fig. 1.
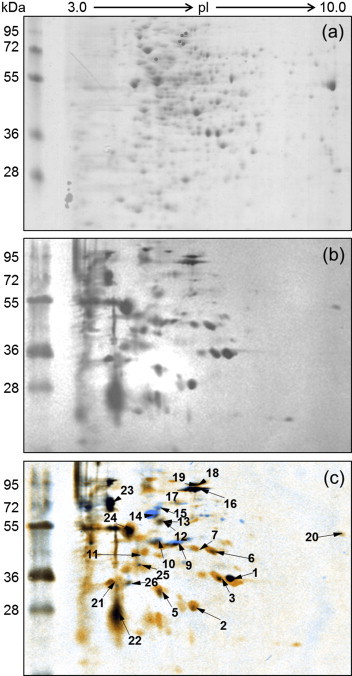
Two-dimensional gel electrophoresis (2DE) and western blot image of Prototheca cell lysate. (a) A representative 2DE (pH 3–10, non-linear, 7 cm) image showing the separation of Prototheca zopfii genotype 2 cell lysate. (b) A representative image of a western blot with serum from a rabbit intradermally challenged with live P. zopfii genotype 2 (SAG 2021). (c) Overlaid image of the two 2DE blots for identification of corresponding spots. The spots were numbered randomly.
Table 1.
List of identified proteins
| Spot ID | NCBI accession no. | Uni-Prot no. | Protein | Organism | MOWSE score | Molecular mass (Da) | pI | No. of peptides | Function |
|---|---|---|---|---|---|---|---|---|---|
| 1 | gi|317035255 | Malate dehydrogenase | Aspergillus niger | 105 | 32 405 | 7.71 | 5 | Energy metabolism | |
| 2 | gi|499658212 | Q3IXY7 | Serine recombinase | Rhodobacter sphaeroides | 87 | 57 111 | 7.12 | 14 | Site-specific recombinase |
| 4 | gi|294486349 | D6LCZ5 | Triosephosphate isomerase | Fusobacterium nucleatum | 110 | 27 607 | 5.51 | 7 | Energy metabolism (glycolysis) |
| 6 | gi|491508197 | Radical SAM protein | (Eubacterium) yurii | 105 | 36 228 | 6.36 | 13 | Enzyme superfamily, heterogeneous metabolic functions | |
| 7 | gi|307102699 | E1ZTB0 | Hypothetical protein CHLNCDRAFT_59826 | Chlorella variabilis | 92 | 38 692 | 6.28 | 3 | |
| 10 | gi|308475003 | E3MVY5 | Hypothetical protein | Caenorhabditis remanei | 94 | 62 721 | 9.21 | 17 | |
| 11 | gi|219117017 | B7FXA2 | Beta chain succinyl-coa synthetase synthetase | Phaeodactylum tricornutum | 102 | 48 047 | 5.06 | 7 | Energy metabolism (citric acid cycle) |
| 12 | gi|546319275 | R7QHD1 | S-adenosyl-l-homocysteine hydrolase, partial | Chondrus crispus | 113 | 54 526 | 5.31 | 7 | Enzyme of metabolism pathway |
| 15 | gi|145346182 | A4RWG3 | Heat shock protein 70 | Ostreococcus lucimarinus | 93 | 71 918 | 5.37 | 4 | Stress response, cell signalling |
| 18 | gi|547216904 | R5LKR2 | Sigma-70 region 2 | Eubacterium sp. | 86 | 18 444 | 5.97 | 10 | Ribosome biogenesis |
| 19 | gi|507081301 | Protein BolA | Citrobacter | 89 | 12 043 | 6.18 | 6 | Perhaps cell proliferation and cell cycle regulation (–> signalling) | |
| 20 | gi|224593225 | C0LL61 | Translation elongation factor-like protein (EF-1α) | Parachlorella kessleri | 159 | 34 932 | 7.88 | 10 | Protein biosynthesis |
| 22 | gi|74272601 | Q1WLZ5 | 14-3-3 protein | Chlamydomonas incerta | 112 | 29 683 | 4.9 | 4 | Diverse cell signalling |
| 23 | gi|116750623 | A0LN73 | Porphobilinogen deaminase | Syntrophobacter fumaroxidans MPOB | 92 | 33 376 | 7.04 | 8 | For synthase of porphyrin–> metabolism |
| 24 | gi|82793401 | ATP synthase subunit β | Plasmodium yoelii yoelii | 192 | 58 109 | 5.93 | 7 | Energy metabolism |
The MOWSE score (MOlecular Weight SEarch score) is calculated by –10 log (P), where P is the probability that the observed match is a random event. The identification is considered to be valid if the MOWSE score is greater than or equal to the significance threshold value (p <0.05) and more than two peptides match. The molecular mass was calculated from the identified protein sequence, and the pI (isoelectric point) was calculated from the identified protein sequence.
Among the identified proteins, malate dehydrogenase, elongation factor 1-alpha (EF-1α), heat shock protein 70 (Hsp70) and 14-3-3 protein have been described as immunoreactive proteins in serological studies with other eukaryotic pathogens [13–15]. The negative control serum appeared to possess antibodies against S-adenosyl-l-homocysteine hydrolase and EF-1α; however, the latter was earlier identified as an immunoreactive protein of Prototheca [16]. On the other hand, four proteins (malate dehydrogenase, S-adenosyl-l-homocysteine hydrolase, EF-1α, and Hsp70) were previously reported to be differentially expressed in P. zopfii genotype 2 [9]. Hence P. zopfii genotype 2 hyperimmune serum appeared to possess specific antibodies against two of the proteins, malate dehydrogenase and Hsp70, indicating a role of these two proteins in the immune reaction.
Hsp70 comprises a highly conserved protein family with housekeeping functions and stress-inducible chaperone functions when located intracellularly [17]. Extracellular Hsp70 in contrast, acts as a cytokine and activates immune cells. We can only speculate about the role of Hsp70 during infection. It should be noted that most of the identified proteins appear to be intracellular in nature, which supports the earlier speculations that the dead Prototheca cells induce a local immune response [18].
The absence of Prototheca-specific information in the public repositories resulted in the identification of proteins that are best described in other organisms [9]. De novo sequencing of the proteins might provide additional clues on the immunogenicity and virulence nature of Prototheca. The present study utilized serum from experimentally challenged rabbits; however, Prototheca infection in rabbit has not been reported yet. Hence, for a comprehensive description of genotype-specific antigens, which might serve as virulence factors, western blotting using serum of naturally infected animals should be carried out.
Transparency declaration
The authors state that they have no conflicts of interest.
Acknowledgements
We would like to acknowledge the assistance of the Bio-MS unit of the Core Facility BioSupraMol supported by the DFG. We would like to thank Michael Kühl for technical assistance.
References
- 1.Satoh K., Ooe K., Nagayama H., Makimura K. Prototheca cutis sp. nov., a newly discovered pathogen of protothecosis isolated from inflamed human skin. Int J Syst Evol Microbiol. 2010;60:1236–1240. doi: 10.1099/ijs.0.016402-0. [DOI] [PubMed] [Google Scholar]
- 2.Roesler U., Hensel A. Longitudinal analysis of Prototheca zopfii-specific immune responses: correlation with disease progression and carriage in dairy cows. J Clin Microbiol. 2003;41:1181–1186. doi: 10.1128/JCM.41.3.1181-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marques S., Silva E., Kraft C., Carvalheira J., Videira A., Huss V.A.R. Bovine mastitis associated with Prototheca blaschkeae. J Clin Microbiol. 2008;46:1941–1945. doi: 10.1128/JCM.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson G., Silva E., Marques S., Müller A., Carvalheira J. Algaemia in a dairy cow by Prototheca blaschkeae. Med Mycol. 2009;47:527–531. doi: 10.1080/13693780802566341. [DOI] [PubMed] [Google Scholar]
- 5.Jagielski T., Lagneau P. Protothecosis. A pseudofungal infection. J Mycol Med. 2007;17:261–270. [Google Scholar]
- 6.Möller A., Truyen U., Roesler U. Prototheca zopfii genotype 2—the causative agent of bovine protothecal mastitis? Vet Microbiol. 2007;120:370–374. doi: 10.1016/j.vetmic.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 7.Roesler U., Möller A., Hensel A., Baumann D., Truyen U. Diversity within the current algal species Prototheca zopfii: a proposal for two Prototheca zopfii genotypes and description of a novel species, Prototheca blaschkeae sp. nov. Int J Syst Evol Microbiol. 2006;56:1419–1425. doi: 10.1099/ijs.0.63892-0. [DOI] [PubMed] [Google Scholar]
- 8.Kishimoto Y., Kano R., Maruyama H., Onozaki M., Makimura K., Ito T. 26S rDNA-based phylogenetic investigation of Japanese cattle-associated Prototheca zopfii isolates. J Vet Med Sci. 2010;72:123–126. doi: 10.1292/jvms.09-0115. [DOI] [PubMed] [Google Scholar]
- 9.Murugaiyan J., Weise C., von Bergen M., Roesler U. Two-dimensional proteome reference map of Prototheca zopfii revealed reduced metabolism and enhanced signal transduction as adaptation to an infectious life style. Proteomics. 2013;13:2664–2669. doi: 10.1002/pmic.201300037. [DOI] [PubMed] [Google Scholar]
- 10.von Bergen M., Eidner A., Schmidt F., Murugaiyan J., Wirth H., Binder H. Identification of harmless and pathogenic algae of the genus Prototheca by MALDI-MS. Proteomics Clin Appl. 2009;3:774–784. doi: 10.1002/prca.200780138. [DOI] [PubMed] [Google Scholar]
- 11.Murugaiyan J., Ahrholdt J., Kowbel V., Roesler U. Establishment of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry database for rapid identification of infectious achlorophyllous green micro-algae of the genus Prototheca. Clin Microbiol Infect. 2012;18:461–467. doi: 10.1111/j.1469-0691.2011.03593.x. [DOI] [PubMed] [Google Scholar]
- 12.Berth M., Moser F.M., Kolbe M., Bernhardt J. The state of the art in the analysis of two-dimensional gel electrophoresis images. Appl Microbiol Biotechnol. 2007;76:1223–1243. doi: 10.1007/s00253-007-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludolf F., Patrocínio P.R., Corrêa-Oliveira R., Gazzinelli A., Falcone F.H., Teixeira-Ferreira A. Serological screening of the Schistosoma mansoni adult worm proteome. PLoS Negl Trop Dis. 2014;8:e2745. doi: 10.1371/journal.pntd.0002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaturvedi A.K., Weintraub S.T., Lopez-Ribot J.L., Wormley F.L. Identification and characterization of Cryptococcus neoformans protein fractions that induce protective immune responses. Proteomics. 2013;13:3429–3441. doi: 10.1002/pmic.201300213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L., Li F., Huang M., Lu J., Kong X., Wang S. Immunoproteomics based identification of thioredoxin reductase GliT and novel Aspergillus fumigatus antigens for serologic diagnosis of invasive aspergillosis. BMC Microbiol. 2012;12:11. doi: 10.1186/1471-2180-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques S. 2010. Protothecosis: agent characterization and pathogenesis. PhD Thesis. Porto. [Google Scholar]
- 17.Daugaard M., Rohde M., Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 18.Pérez J., Ginel P.J., Lucena R., Hervás J., Mozos E. Canine cutaneous protothecosis: an immunohistochemical analysis of the inflammatory cellular infiltrate. J Comp Pathol. 1997;117:83–89. doi: 10.1016/s0021-9975(97)80068-0. [DOI] [PubMed] [Google Scholar]


