Abstract
Background
Multidrug-resistant strains of Plasmodium vivax are emerging in Southeast Asia.
Methods
In vitro drug susceptibility and pvmdr1 genotype were determined in P. vivax field isolates from Indonesia and Thailand.
Results
Increased pvmdr1 copy number was present in 21% of isolates from Thailand (15/71) and none from Indonesia (0/114; P < .001). Compared with Indonesian isolates, the median IC50 of Thai isolates was lower for chloroquine (36 vs. 114 nmol/L; P < .001) but higher for amodiaquine (34 vs. 13.7 nmol/L; P = .032), artesunate (8.33 vs. 1.58 nmol/L; P < .001), and mefloquine (111 vs. 9.87 nmol/L; P < .001). In 11 cryopreserved Thai isolates, those with increased pvmdr1 copy number had a higher IC50 for mefloquine (78.6 vs. 38 nmol/L for single-copy isolates; P = .006). Compared with isolates with the wild-type allele, the Y976F mutation of pvmdr1 was associated with reduced susceptibility to chloroquine (154 nmol/L [range, 4.6–3505] vs. 34 nmol/L [range, 6.7–149]; P < .001) but greater susceptibility to artesunate (1.8 vs. 9.5 nmol/L; P = .009) and mefloquine (14 vs. 121 nmol/L; P < .001).
Conclusions
Amplification of pvmdr1 and single-nucleotide polymorphisms are correlated with susceptibility of P. vivax to multiple antimalarial drugs. Chloroquine and mefloquine appear to exert competitive evolutionary pressure on pvmdr1, similar to that observed with pfmdr1 in Plasmodium falciparum.
Malaria continues to exert a huge global burden both in public health and economic terms. Although Plasmodium falciparum infections have been the focus of research and control efforts, there is a growing awareness that the burden of, and morbidity and mortality associated with, Plasmodium vivax infections are also substantial [1-4]. Control strategies for both species have been confounded by the emergence of antimalarial drug resistance to multiple drugs. Although chloroquine resistant P. falciparum was first documented >50 years ago in Cambodia, the first cases of chloroquine-resistant P. vivax infection were not reported until 1989 from Papua New Guinea [5] and northern Papua, Indonesia [6]. In the last decade, chloroquine resistance has continued to emerge, with failure rates after monotherapy exceeding 70% in Papua [7-9]. Other reports suggest declining chloroquine efficacy across much of Asia and South America [1, 10].
Few studies have specifically addressed the clinical efficacy of other antimalarial regimens against P. vivax, although the available data suggest that in western Indonesia mefloquine retains clinical efficacy [11], whereas amodiaquine efficacy is waning, particularly in the eastern province of Papua [12] and Papua New Guinea [13]. The dearth of clinical studies of P. vivax reflects both the inherent difficulties associated with the in vivo test for relapsing malaria [14] and the difficulties in differentiating between reinfection, recrudescence, and relapse after treatment failure [15, 16].
To better define the drug-susceptibility profile of P. vivax, we recently developed a standardized in vitro assay [17] and validated it using isolates of P. vivax from Indonesia, where P. vivax is known to be highly resistant to chloroquine, and from Thailand, where strains remain susceptible to chloroquine [18]. The aim of the present study was to define the in vitro susceptibility profile of P. vivax for a wider range of antimalarial drugs and correlate this with pvmdr1 copy number and a polymorphism known to be prevalent in both Thailand and Indonesia [18].
METHODS
Field location and sample collection
Clinical isolates were collected between 2003 and 2007 from 2 sites, the first in Timika, southern Papua, Indonesia, and the other in Mae Sod on the western border of Thailand. In southern Papua, P. vivax demonstrates high-grade clinical resistance to chloroquine, with failure rates >65% on day 28 after chloroquine monotherapy [9]. In contrast,P. vivax in Mae Sod was clinically susceptible to chloroquine when last tested in 1999 [19]. The clinical efficacy of other monotherapies against P. vivax in either location is unknown.
Patients with symptomatic infections of pure P. vivax presenting to an outpatient facility were recruited into the study, and 5-mL blood samples were collected by venipuncture. After removal of host white blood cells using a CF11 column, 2 mL of packed infected red blood cells was divided as follows: 1 mL was cryopreserved in glycerolyte, 200 μL was spotted onto a filter paper, and 800 μL was used for the in vitro drug-susceptibility assay. Patients were treated with dihydroartemisinin-piperaquine (Indonesia) [20] or chloroquine (Thailand) [19] according to local guidelines but were not followed up routinely thereafter. The in vitro susceptibility to chloroquine for 124 isolates included in this study has been presented elsewhere [18].
In vitro drug-susceptibility assay
The antimalarial susceptibility of fresh field P. vivax isolates was measured using a modified World Health Organization microtest protocol, as described elsewhere [17]. Briefly, 200 μL of a 2% hematocrit blood-medium mixture made from McCoy’s 5A medium and 20% AB Rh+ human serum was added to each well of predosed drug plates. In Indonesia, each drug plate contained 11 serial concentrations of the antimalarials chloroquine (maximum concentration, 5910 nmol/L), amodiaquine (557 nmol/L), artesunate (93 nmol/L), mefloquine (338 nmol/L), and piperaquine (769 nmol/L). In Thailand only, chloroquine was tested from 2003 until 2006, after which all 5 drugs were assayed. A candle jar was used to mature the parasites at 37.5°C (22–42 h). Incubation was stopped when >40% of ring-stage parasites had matured to schizonts in the drug-free control well.
Thick blood films made from each well were stained with 5% Giemsa stain for 30 min and examined microscopically. Differential counts of 200 asexual parasites in the preincubation and test slides were classified into ring-stage parasites (ring-shaped trophozoites without pigment), mature trophozoites (single or double chromatin dot and hemazoin pigment visible), and schizonts. The number of schizonts (≥5 chromatin dots visible) per 200 asexual-stage parasites was determined for each drug concentration and normalized to the control well. The dose-response data were analyzed using nonlinear regression analysis (WinNonLin software; version 4.1; Pharsight), and the IC50 value was derived using a inhibitory sigmoid Emax model. In vitro data were only used from predicted curves for which the maximum (Emax) and minimum (E0) effect were within 15% of 100 or 0, respectively. Previous analysis has highlighted a significant stage specificity of drug activity, with chloroquine showing almost no activity against trophozoites stages in P. vivax infection [17]. For this reason, even though isolates were processed irrespective of the parasite staging in the initial culture, the analysis of geographical and molecular correlates of in vitro drug susceptibility was restricted to those isolates with predominantly ring-stage parasites initially (ring to trophozoite [RT] ratio >1).
In vitro drug-susceptibility assay in cryopreserved isolates
Because the number of Thai isolates with susceptibility data for drugs other than chloroquine was limited, a separate experiment was conducted using 11 cryopreserved isolates collected in Mae Sod in 2003 [21]. These comprised all 6 available isolates with pvmdr1 amplification and a random selection of 5 isolates with a single copy of pvmdr1. Cryopreserved isolates were thawed and set up in short-term culture, as described elsewhere [21].
Determination of pvmdr1 polymorphisms
Genomic DNA from blood spots and cryopreserved samples was extracted using a QIAamp DNA Mini Kit (Qiagen). Pvmdr1 single-nucleotide polymorphisms (SNPs) at 976 were detected using a DNA template mismatch primer method [18]. Polymerase chain reaction (PCR) was done in a total volume of 50 μL, containing 5 μL of 10× PCR buffer, 2.5 mmol/L MgCl2, 0.2 mmol/L each dNTP, 2 μmol/L Pvmdr976 forward primer, 1.5 μmol/LP vmdr 976 reverse primer, 1 μmol/L Pvmdr976 internal primer, 1.25 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), and 1 μL of genomic DNA. PCR was performed under the following conditions: 95°C for 10 min followed by 40 cycles of 94°C for 40 s, 55°C for 1 min, and 72°C for 2 min. The product was separated on 2% agarose gel.
The pvmdr1 gene copy number was estimated by a quantitative real-time SYBR Green PCR assay, as described elsewhere [18]. In brief, a single-copy gene coding for P. vivax aldolase (GenBank accession number AF247063) was used as a reference (normalizer) gene for estimating the pvmdr1 copy number. PCRs were performed in triplicate or quadruplet and contained 1× ABgene ABsolute QPCR SYBR Green Mix (catalog number AB-1166/a), 100 nmol/L ROX dye (passive reference dye), 1 μL of DNA template, and 75 nmol/L concentrations of each primer in a final volume of 25 μL. Cycling conditions were 95°C for 15 min followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s. Plasmids containing the aldolase gene fragment and 1 or 2 copies of the pvmdr1 gene fragments were used as controls. Fluorescence data were collected 3 times and averaged at the end of the annealing and extension steps. After the amplification cycles, a melting curve analysis was performed to confirm that the correct products were synthesized. The text report containing the threshold cycle values for every well was exported into Excel (Microsoft) and analyzed. Assessment of copy number was repeated at least twice for all isolates, and the repeatability coefficient determined was as 0.30 (viz., 95% of repeated estimates of pvmdr1 copy number were within 0.15 of the first).
Data and sequence analysis
Analysis was performed using SPSS for Windows (version 14; SPSS). The Mann-Whitney U test and Wilcoxon signed-rank test were used for nonparametric comparisons.
Ethics
Ethical approval for this study was obtained from the ethics committees of the National Institute of Health Research and Development, Ministry of Health, Indonesia; the Menzies School of Health Research, Darwin, Australia; and Mahidol University, Bangkok, Thailand.
RESULTS
In vitro susceptibility profile of Indonesian and Thai Isolates
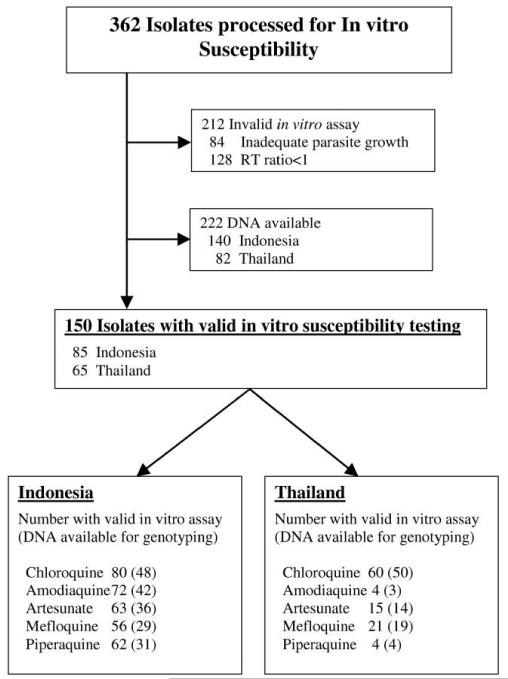
Between April 2003 and October 2007, 362 isolates were assayed for in vitro susceptibility (237 from Indonesia and 125 from Thailand). A total of 212 isolates were rejected owing to inadequate parasite growth (n = 84) or because the initial RT ratio was <1 (n = 128) (figure 1). Among the 150 isolates with acceptable data, the median percentage of ring-stage parasites in the initial culture was 84% (range, 50%–100%), with no significant difference in this proportion between Thai and Indonesian isolates.
Figure 1.
Selection of samples analyzed. RT ratio, ring to trophozoite ratio.
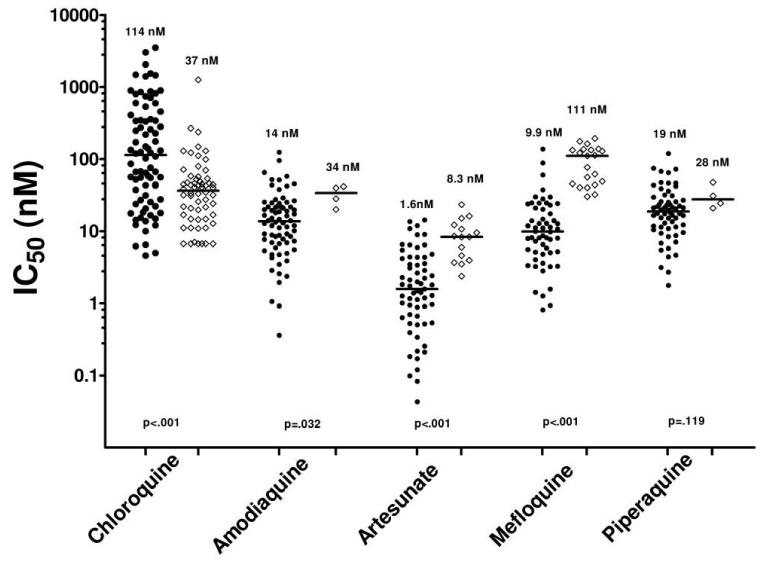
The median IC50 of P. vivax among Thai isolates was significantly lower than those among Indonesian isolates for chloroquine (36.7 vs. 114 nmol/L) but was higher for mefloquine (111 vs. 9.87 nmol/L), artesunate (8.33 vs. 1.58 nmol/L), and amodiaquine (34 vs. 13.7 nmol/L) (table 1 and figure 2).
Table 1. In vitro susceptibility of isolates from Thailand and Indonesia.
| Indonesian isolates | Thai isolates | ||||
|---|---|---|---|---|---|
| Drug | No. | IC50, nmol/L | No. | IC50, nmol/L | P |
| Chloroquine | 80 | 114(4.6–3506) | 60 | 37 (6.7–1264) | <.001 |
| Amodiaquine | 72 | 14(0.4–125) | 4 | 34 (20–42) | .032 |
| Artesunate | 63 | 1.6(0.04–14.3) | 15 | 8.3 (2.4–23) | <.001 |
| Mefloquine | 56 | 9.9(0.8–138) | 21 | 111 (30–194) | <.001 |
| Piperaquine | 62 | 19(1.8–120) | 4 | 28 (21–48) | .119 |
NOTE. Data for IC50 are median (range) values.
Figure 2.
In vitro drug-susceptibility profiles for Indonesian (circles) and Thai (diamonds) isolates. The nos. above each group and the horizontal bars indicate median values. P values are for the comparison between Indonesian and Thai isolates.
Prevalence of pvmdr1 amplification and Y976F point mutation
DNA was available for 222 (61%) of all isolates collected. The pvmdr1 copy number could be successfully quantified in 185 (83%) of these isolates and the 976 polymorphism identified in 211 (95%). In total, 15 (21%) of 71 isolates from Thailand had increased pvmdr1 copy number (13 with 2 copies, 2 with 3 copies), compared with 0 of 114 from Indonesia (P < .001). Conversely, the pvmdr1 976 mutant (Y976F) was present in 133 (96.4%) of 138 of Indonesian isolates, compared with 19 (26%) of 73 Thai isolates. In Thailand, all 15 of the isolates with increased copy number had the wild-type allele at codon 976, compared with 36 (67%) of 54 with a single copy of pvmdr1 (P = .007).
Correlation of pvmdr1 amplification and Y976F point mutation with in vitro susceptibilities in fresh field isolates
Compared with isolates with the wild-type allele, those with the pvmdr1 Y976F allele had reduced susceptibility to chloroquine (154 vs. 34.0 nmol/L; P < .001) but increased susceptibility to mefloquine (14.0 vs. 121 nmol/L; P < .001) and artesunate (1.8 vs. 9.52 nmol/L; P < .001) (table 2). The association between pvmdr1 mutation and IC50 remained after controlling for pvmdr1 amplification.
Table 2. Relationship of the pvmdr1 976 mutation and in vitro susceptibility of isolates from Thailand and Indonesia.
| Mutant | Wild type | ||||
|---|---|---|---|---|---|
| Drug, isolates | No. | IC50, nmol/L | No. | IC50, nmol/L | P |
| Chloroquine | |||||
| All | 58 | 154 (4.6–3506) | 40 | 34 (6.7–149) | <.001 |
| Indonesian | 47 | 274 (4.6–3506) | 1 | 103 | … |
| Thailand | 11 | 42.8 (6.7–239) | 39 | 33 (6.7–149) | .527 |
| Amodiaquine | |||||
| All | 43 | 14.0 (0.4–125) | 2 | 35 (28–42) | … |
| Indonesian | 42 | 13.7 (0.4–125) | 0 | … | … |
| Thailand | 1 | 20.1 | 2 | 35 (28–42) | … |
| Artesunate | |||||
| All | 41 | 1.80 (0.04–13.6) | 9 | 9.5 (3.7–16) | <.001 |
| Indonesian | 36 | 1.53 (0.04–13.6) | 0 | … | … |
| Thailand | 5 | 3.96 (2.4–5.97) | 9 | 9.5 (3.7–16) | .009 |
| Mefloquine | |||||
| All | 34 | 14.0 (0.9–138) | 14 | 121 (32–177) | <.001 |
| Indonesian | 29 | 12.8 (0.9–138) | 0 | … | … |
| Thailand | 5 | 49.2 (30–122) | 14 | 121 (32–177) | .064 |
| Piperaquine | |||||
| All | 31 | 20.9(1.8–120) | 4 | 28 (15–48) | .195 |
| Indonesian | 30 | 19.6(1.8–120) | 1 | 15.2 | … |
| Thailand | 1 | 21 | 3 | 31 (25–48) | … |
NOTE. Data for IC50 are median (range) values.
Because pvmdr1 amplification was absent in Indonesian isolates, the correlation with the in vitro response was restricted to Thai isolates. There was no significant difference between the chloroquine IC50 for 35 isolates with a single copy of pvmdr1 (median, 33.3 nmol/L [range, 6.7–239]) and that for the 12 isolates with pvmdr1 amplification (40.5 nmol/L [range, 11.1–99.4]; P = 772). The median mefloquine IC50 among single-copy Thai isolates was 77.0 nmol/L (range, 30.1-163 nmol/L), compared with 177 nmol/L and 137 nmol/L for the 2 Thai isolates with pvmdr1 amplification. Statistical comparison could not be made for the other drugs, because only 1 isolate with pvmdr1 amplification had a valid assay for artesunate (9.5 nmol/L), and none with pvmdr1 amplification had in vitro data on amodiaquine or piperaquine available. The median IC50 for isolates with a single copy of pvmdr1 was 8.3 nmol/L (range, 2.38–16.2 nmol/L) for artesunate, 28.2 nmol/L (range, 20.1–41.6 nmol/L) for amodiaquine, and 27.7 nmol/L (range, 20.9–47.6 nmol/L) for piperaquine.
Correlation between pvmdr1 amplification and in vitro susceptibility among thawed isolates
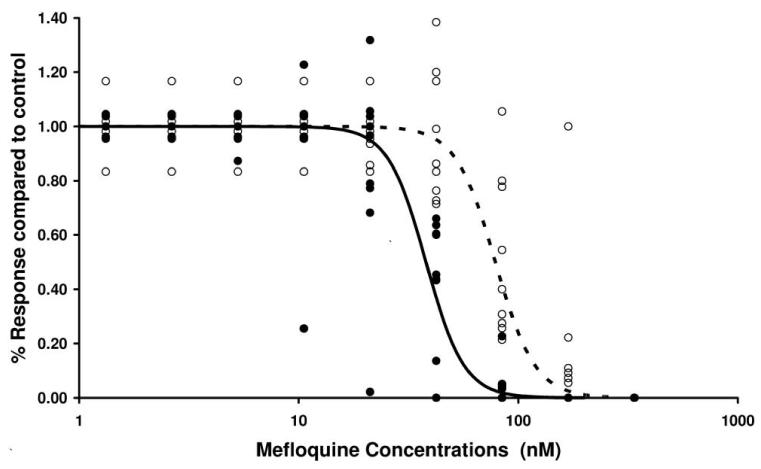
To investigate the role played by pvmdr1 amplification in drug susceptibility, 11 cryopreserved Thai isolates were thawed and assayed for in vitro susceptibility. Mefloquine was assayed for all isolates and artesunate for the 6 isolates for which there was sufficient sample to test a second drug. All assays undertaken were successful in generating IC50 data. The median IC50 for mefloquine among the 6 isolates with pvmdr1 amplification was 78.6 nmol/L (range, 56.5–188 nmol/L), compared with 38.0 nmol/L (range, 8.23–44.5 nmol/L) among the 5 isolates with a single copy of pvmdr1 (P = .006) (figure 3). The corresponding figures for artesunate were 12.01 nmol/L (range, 6.69–17.3 nmol/L) for the 2 isolates with increased pvmdr1 copy number and 2.57 nmol/L (range, 1.58–7.42 nmol/L) for the 4 isolates with a single copy of pvmdr1 (P = .165).
Figure 3.
In vitro susceptibility to mefloquine in thawed Thai isolates according to pvmdr1 genotype: single copy of pvmdr1 (black circles, solid line) vs. amplified pvmdr1 (white circles, dashed line). Data points represent assay wells for 11 isolates with 8 dilutions of mefloquine. The predicted population curves were calculated using the formula , where Y is the response compared with the control well, X is the mefloquine concentration, and IC50 and γ are the median values for the IC50 and slope, respectively, derived for each isolate by regression analysis.
DISCUSSION
Antimalarial resistance to most classes of antimalarial drugs has evolved, with the underlying resistance mechanisms derived mainly from mutations in the genes encoding target enzymes or transporters [22, 23]. In P. falciparum, the pfmdr1 gene on chromosome 5 encodes a P-glycoprotein pump that affects the intraparasitic concentrations of several important antimalarial drugs [24]. Point mutations in pfmdr1 are associated with decreased susceptibility to chloroquine and increased susceptibility to mefloquine, artesunate, and lumefantrine [25, 26]. Conversely, amplification of wild-type pfmdr1 is associated with reduced in vitro and in vivo susceptibility to structurally unrelated antimalarial drugs, such as mefloquine, artesunate, lumefantrine, and quinine [25, 27-29].
The molecular basis of antimalarial resistance in P. vivax has been less intensely studied. Although the pfcrt gene is the main determinant of chloroquine resistance in P. falciparum, polymorphisms of pvcrt, the orthologue in P. vivax, do not appear to affect susceptibility to chloroquine [18, 30]. However, our recent study suggested a role for the pvmdr1 gene, the orthologue of pfmdr1, with a tyrosine for phenylalanine substitution at position 976 (Y976F) associated with a 2-fold increase in chloroquine IC50 [18]. This article also noted the presence of gene amplification of pvmdr1 in Thai isolates. More recently, we have reported that the initial stage of the parasites before culture is a major confounding factor for in vitro parasite susceptibility [17] and that this varies considerably in isolates collected at different locations. Therefore, in the present study, we investigated the prevalence of pvmdr1 amplification and polymorphisms and their associated correlation with in vitro susceptibility in 2 geographically distinct sites, applying a more stringent assay restriction to isolates with a majority of ring-stage parasites before culture.
In Indonesia, high-grade resistance to chloroquine has emerged in P. vivax, and resistance in P. falciparum is mainly limited to chloroquine and sulfadoxine-pyrimethamine [9]. Before 2006, the mainstay of treatment for uncomplicated malaria in this region was chloroquine plus sulfadoxine-pyrimethamine; mefloquine was not available, and the use of the artemisinin derivatives was limited to clinical trials. On the western border of Thailand, P. vivax remained generally susceptible to chloroquine when last tested in 1999 [19], although in P. falciparum significant resistance has long been present to a wide range of drugs, including mefloquine, chloroquine, sulfadoxine-pyrimethamine, halofantrine, and lumefantrine [31]. In western Thailand, a combination of mefloquine and artesunate is used to treat P. falciparum, and chloroquine monotherapy is still recommended for P. vivax.
There were significant differences in the prevalence of pvmdr1 amplification and a SNP. In Papua, all isolates had single copies of the gene, with the Y976F allele present in 96.4% of isolates. In Thailand, on the other hand, the Y976F allele was present in only 26% of isolates, with pvmdr1 amplification present in 21%. The difference in allele frequencies was mirrored in the in vitro drug susceptibility profiles. In Indonesia, isolates were highly resistant to chloroquine but had low IC50 values for mefloquine, amodiaquine, and artesunate. Conversely, in Thailand, isolates demonstrated greater susceptibility to chloroquine but had reduced susceptibility to mefloquine, amodiaquine, and artesunate. These drug-susceptibility profiles, presented in table 1, concur with findings in other studies of Thai isolates [32].
When the in vitro response was correlated directly with molecular polymorphisms, the Y976F pvmdr1 mutant was associated with a 4-fold higher chloroquine IC50 (similar to our findings reported elsewhere [18]) but a 5-8-fold lower IC50 for artesunate and mefloquine (table 2). However, when only Thai isolates were selected, the correlation between chloroquine IC50 and the Y976F mutation was no longer significant, although the trend remained similar (table 2). It is possible that the current sample size no longer had the necessary power to determine a significant association, and a study with larger sample size may clarify the relationship between this mutation and drug susceptibility. Alternatively, the difference we reported in another study could have been attributable in part to the differences in the stage of the isolates between locations. In either case, although the 976 mutant may prove to be a useful population marker of emerging chloroquine-resistant P. vivax, alternative molecular mechanisms need to be evoked to account for the significant variation in IC50 values in wild-type pvmdr1 alleles.
The presence of pvmdr1 amplification in Thai but not Papuan isolates is intriguing, particularly because mefloquine is not a standard treatment for P. vivax in either area. Studies from Thailand recently showed that almost 70% of P. falciparum isolates from infections recurring after treatment with mefloquine had amplified pfmdr1 [33]. In practice, P. vivax and P. falciparum are often not discriminated between at diagnosis, and thus P. vivax infections are likely to be frequently treated with a mefloquine regimen. If the selective pressure on mdr1 amplification that is apparent in P. falciparum occurs in P. vivax, this would account for the high prevalence of pvmdr1 amplification present in Thailand even after inadvertent drug exposure. In Papua, where mefloquine has not been deployed, none of the isolates tested had increased pvmdr1 copy number. This hypothesis is supported by recent data showing a greater prevalence of pvmdr1 amplification among P. vivax isolates from Thailand than among isolates from other regions of Southeast Asia with less mefloquine exposure [34].
Unfortunately, apart from chloroquine, in vitro drug-susceptibility assays were available only for 2 field isolates with an increased pvmdr1 copy number. Interestingly, these isolates had a high IC50 for both mefloquine (177 and 137 nmol/L) and artesunate (9.5 nmol/L). To assess this further, 11 cryopreserved isolates were thawed, and the synchronous culture was tested against mefloquine and, when there was sufficient sample volume, artesunate. Despite the small numbers of isolates tested, pvmdr1 amplification was associated with a 2-fold increase in the median mefloquine IC50 (38–79 nmol/L; P = .006) and a 4.6-fold increase in the median artesunate IC50 (2.6–12.0 nmol/L; P = 17).
The association of a higher IC50 for both mefloquine and artesunate with increased pvmdr1 copy number suggests important similarities between the species with respect to the mechanism of multidrug resistance. Our results with P. vivax also suggest that chloroquine and mefloquine exert competitive evolutionary pressure on pvmdr1, as has been found elsewhere with P. falciparum. In Papua, chloroquine rather than mefloquine provides the main force for drug selection; hence, the observed high prevalence of single-copy pvmdr1 Y976F allele. Conversely, in Thailand, mefloquine has been used for many years for the treatment of falciparum malaria, and its widespread use has selected for amplification of pvmdr1, which occurs only in parasites with the wild-type allele at 976. Therefore, amplification of pvmdr1 may provide a counterbalance to chloroquine pressure, providing a plausible explanation for the low prevalence of Y976F and the retention of chloroquine-susceptible P. vivax in Thailand.
In conclusion, both a SNP and amplification of pvmdr1 are associated with variation in the in vitro susceptibility of P. vivax, similar to that associated with pfmdr1 in P. falciparum. Further studies are needed to confirm the correlation between such pvmdr1 polymorphisms and clinical response.
Acknowledgments
We are grateful to Lembaga Pengembangan Masyarakat Amungme Kamoro, the staff of the Rumah Sakit Mitra Masyarakat Hospital, and Dr. Paulus Sugiarto for their support in conducting this study and to Prayoga, Rosmini, and Yoshi Elvi for technical assistance. We also thank the Australian Red Cross Blood Transfusion Service for supplying serum samples for in vitro cultures.
Financial support: Wellcome Trust-National Health and Medical Research Council (NHMRC) International Collaborative Research Grants (Wellcome Trust GR071614MA; NHMRC ID 283321); Wellcome Trust-Mahidol University Oxford Tropical Medicine Research Programme; NHMRC (practitioner fellowship to N.A.); Wellcome Trust (career development award 074637 to R.P.).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- 2.Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007;23:533–9. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 4.Tjitra E, Anstey NM, Sugiarto P, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5:e136. doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet. 1989;2:1183–4. doi: 10.1016/s0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- 6.Baird JK, Basri H, Purnomo, et al. Resistance to chloroquine by Plasmodium vivax in Irian Jaya, Indonesia. Am J Trop Med Hyg. 1991;44:547–52. doi: 10.4269/ajtmh.1991.44.547. [DOI] [PubMed] [Google Scholar]
- 7.Tjitra E, Baker J, Suprianto S, Cheng Q, Anstey NM. Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob Agents Chemother. 2002;46:3947–53. doi: 10.1128/AAC.46.12.3947-3953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumawinata IW, Bernadeta, Leksana B, et al. Very high risk of therapeutic failure with chloroquine for uncomplicated Plasmodium falciparum and P. vivax malaria in Indonesian Papua. Am J Trop Med Hyg. 2003;68:416–20. [PubMed] [Google Scholar]
- 9.Ratcliff A, Siswantoro H, Kenangalem E, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and P. vivax to chloroquine and sulfadoxine-pyrimethamine in southern Papua, Indonesia. Trans R Soc Trop Med Hyg. 2007;101:351–9. doi: 10.1016/j.trstmh.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–83. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maguire JD, Krisin, Marwoto H, Richie TL, Fryauff DJ, Baird JK. Mefloquine is highly efficacious against chloroquine-resistant Plasmodium vivax malaria and Plasmodium falciparum malaria in Papua, Indonesia. Clin Infect Dis. 2006;42:1067–72. doi: 10.1086/501357. [DOI] [PubMed] [Google Scholar]
- 12.Hasugian AR, Purba HL, Kenangalem E, et al. Dihydroartemisinin-piperaquine versus artesunate-amodiaquine: superior efficacy and post-treatment prophylaxis against multidrug-resistant Plasmodium falciparum and Plasmodium vivax malaria. Clin Infect Dis. 2007;44:1067–74. doi: 10.1086/512677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marfurt J, Mueller I, Sie A, et al. Low efficacy of amodiaquine or chloroquine plus sulfadoxine-pyrimethamine against Plasmodium falciparum and P. vivax malaria in Papua New Guinea. Am J Trop Med Hyg. 2007;77:947–54. [PubMed] [Google Scholar]
- 14.Baird JK, Leksana B, Masbar S, et al. Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am J Trop Med Hyg. 1997;56:621–6. doi: 10.4269/ajtmh.1997.56.621. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Auliff A, Rieckmann K, Gatton M, Cheng Q. Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis. 2007;195:934–41. doi: 10.1086/512242. [DOI] [PubMed] [Google Scholar]
- 16.Imwong M, Snounou G, Pukrittayakamee S, et al. Relapses of Plasmodium vivax infection usually result from activation of heterologous hypnozoites. J Infect Dis. 2007;195:927–33. doi: 10.1086/512241. [DOI] [PubMed] [Google Scholar]
- 17.Russell B, Chalfein F, Prasetyorini B, et al. Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob Agents Chemother. 2008;52:1040–5. doi: 10.1128/AAC.01334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suwanarusk R, Russell B, Chavchich M, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS ONE. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luxemburger C, van Vugt M, Jonathan S, et al. Treatment of vivax malaria on the western border of Thailand. Trans R Soc Trop Med Hyg. 1999;93:433–8. doi: 10.1016/s0035-9203(99)90149-9. [DOI] [PubMed] [Google Scholar]
- 20.Ratcliff A, Siswantoro H, Kenangalem E, et al. Two fixed-dose artemisinin combinations for drug-resistant falciparum and vivax malaria in Papua, Indonesia: an open-label randomised comparison. Lancet. 2007;369:757–65. doi: 10.1016/S0140-6736(07)60160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosaisavee V, Suwanarusk R, Nosten F, et al. Plasmodium vivax: isotopic, PicoGreen, and microscopic assays for measuring chloroquine sensitivity in fresh and cryopreserved isolates. Exp Parasitol. 2006;114:34–9. doi: 10.1016/j.exppara.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plowe CV, Cortese JF, Djimde A, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–6. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 24.Wilson CM, Volkman SK, Thaithong S, et al. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–60. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- 25.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–9. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 26.Price RN, Cassar C, Brockman A, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price RN, Uhlemann AC, Vugt M, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–7. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–23. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura T, Carlton JM, Baird JK, et al. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J Infect Dis. 2001;183:1653–61. doi: 10.1086/320707. [DOI] [PubMed] [Google Scholar]
- 31.Nosten F, van Vugt M, Price R, et al. Effects of artesunate-mefloquine combination on incidence of Plasmodium falciparum malaria and mefloquine resistance in western Thailand: a prospective study. Lancet. 2000;356:297–302. doi: 10.1016/s0140-6736(00)02505-8. [DOI] [PubMed] [Google Scholar]
- 32.Chotivanich K, Udomsangpetch R, Chierakul W, et al. In vitro efficacy of antimalarial drugs against Plasmodium vivax on the western border of Thailand. Am J Trop Med Hyg. 2004;70:395–7. [PubMed] [Google Scholar]
- 33.Uhlemann AC, McGready R, Ashley EA, et al. Intrahost selection of Plasmodium falciparum pfmdr1 alleles after antimalarial treatment on the northwestern border of Thailand. J Infect Dis. 2007;195:134–41. doi: 10.1086/509809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imwong M, Pukrittayakamee S, Pongtavornpinyo W, et al. Gene amplification of Plasmodium vivax multidrug resistance 1 gene in Thailand, Laos, and Myanmar. Antimicrob Agents Chemother. 2008;52:2657–9. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]