Abstract
Background
Increased pfmdr1 copy number is associated with reduced susceptibility to structurally unrelated antimalarial drugs. We assessed how administration of different antimalarial drugs altered pfmdr1 polymorphism in parasites from patients who experienced treatment failure.
Methods
In studies conducted on the northwestern border of Thailand, amplifications and single-nucleotide polymorphisms in pfmdr1 were compared before and after antimalarial drug treatment.
Results
Intrahost changes in pfmdr1 copy number were observed in 20% (26/132) of patients with recurrent infections. Among infections that recrudesced after mefloquine-containing regimens, increases in pfmdr1 copy number occurred in 68% (95% confidence interval [CI], 46%–85%), and decreases occurred in 2% (95% CI, 0.4%–11%) of isolates; corresponding proportions after artemether-lumefantrine were 25% (2/8) and 11% (2/19); after quinine, 50% (1/2) and 40% (4/10); and after artemisinins alone, 0% (0/10) and 19% (3/16) of isolates (overall P < .001).
Conclusions
Intrahost selection based on pfmdr1 copy number occurs frequently in parasite populations within individual patients. Amplification confers multidrug resistance but probably imposes a significant fitness cost to the parasites.
Antimalarial drug resistance poses a major threat to the control of malaria. In Plasmodium falciparum infection, resistance has evolved to all classes of antimalarial drugs with the exception of the artemisinin derivatives. Resistance mechanisms underlying this mainly involve mutations in the genes encoding target enzymes or transporters [1, 2]. Despite tight control of its use, significant resistance to mefloquine (MFQ) emerged within 6 years after it was first deployed in Thailand in 1984 [3]. MFQ resistance is largely mediated by a P-glycoprotein pump (Pgh), which is encoded by the pfmdr1 gene [4, 5] on chromosome 5. Pgh affects the intraparasitic concentrations of several important antimalarial drugs. Point mutations in pfmdr1 are associated with resistance to chloroquine and are negatively associated with resistance to MFQ [6, 7]. Amplification of wild-type pfmdr1 is associated with reduced susceptibility in vitro to structurally unrelated antimalarial drugs, such as MFQ, artesunate (ART), lumefantrine, and quinine (QU) [5, 8]. In vivo, increased pfmdr1 copy number accounts for more than half of the treatment failures after MFQ, MFQ plus artesunate (MAS), and artemether-lumefantrine (AL) [5, 8].
Over the past 20 years, prospective studies of antimalarial drug efficacy have been conducted on the northwestern border of Thailand. We have utilized samples taken during these studies from patients who experienced treatment failure to investigate how administration of different antimalarial drugs selects for point mutations and amplification in pfmdr1.
SUBJECTS, MATERIALS, AND METHODS
Study sites
From December 1990 to August 2002, patients with P. falciparum infection were recruited into a series of large chemotherapeutic studies conducted by the Shoklo Malaria Research Unit (SMRU) [9-17]. The majority of patients in these investigations were part of clinical trials of antimalarial drugs, whereas the remainder were treated in SMRU outpatient clinics and identified on presentation with clinical symptoms. Patients came from 3 sites in Tak Province on the northwestern border of Thailand: Maela, Shoklo (both camps for displaced Karen people; 60 and 120 km to the north of the Thai provincial town of Mae Sot, respectively), and Mawker Tai (an area hosting migrant workers from neighboring Burma; 60 km to the south of Mae Sot). The area is one of low and unstable transmission of P. falciparum and P. vivax malaria [18, 19]. All P. falciparum and most P. vivax infections are symptomatic. Patients were included in these trials on the condition that they or their accompanying relatives provided fully informed consent. The project was part of clinical drug studies that were individually approved by the ethics committees of the Faculty of Tropical Medicine (Mahidol University, Bangkok), the Karen Refugee Committee (Mae Sot), and the Oxford Tropical Research Ethics Committee (United Kingdom). Molecular studies were approved by the Wandsworth Local Research Ethics Committee (United Kingdom).
Drug treatments
Antimalarial treatment was classified into 5 groups: MFQ monotherapy (25 mg/kg) (the MFQ group), MFQ (25 mg/kg) plus 3 days of artesunate (12 mg/kg) (the MAS3 group), AL (4 or 6 doses) (the AL group), ART-based monotherapy with an ART derivative (artemether or artesunate 12 mg/kg over 7 days) (the ART group), and QU with or without doxycycline over 7 days (the QU group).
Collection of blood samples
Fingerprick blood samples were obtained from patients with acute, uncomplicated falciparum malaria. Cases of malaria were confirmed by microscopy, and individuals were treated with a variety of antimalarial regimens depending on age, pregnancy status, and random allocation in the various chemotherapy trials at the time of recruitment. A small quantity (30 μL) of whole blood from these patients was spotted onto 3MM filter paper (Whatman), airdried, and stored in plastic bags at room temperature until DNA extraction. Parasite counts were measured by microscopy on presentation with symptoms and then assessed daily until negative and weekly until day 42 or 63 in nonpregnant patients or for the entire length of gestation in pregnant women. A second blood spot was obtained from those patients who became slide positive again within the follow-up period or who presented at a later date to one of the SMRU clinics. Primary and recurrent infection sample pairs were selected randomly from the 5 treatment groups. For cases in which there were both recrudescences and new infections, equal numbers of both were chosen.
Genotyping of P. falciparum by polymerase chain reaction (PCR)
Genetic comparison of P. falciparum in acute and recurrent infections was performed at the SMRU laboratory based in Mae Sot. Parasite DNA was extracted from blood spots using PrepMan Ultra (Applied Biosystems) or the saponin lysis/chelex extraction method [5]. Three genetic loci, which exhibit repeat-number polymorphisms, were amplified by nested PCR from the DNA of each isolate. These were the genes encoding the merozoite surface proteins 1 and 2 and the gene encoding the glutamate rich protein, located on chromosomes 9, 2, and 10, respectively. Primers and amplification conditions for all 3 loci have been described elsewhere [20]. PCR products were detected by electrophoresis of 8 μL from each reaction on 1.7% agarose gels. The PCR products were visualized by ethidium-bromide staining and UV fluorescence and were sized against a 100-bp molecular-weight marker (Life Technologies). Clonality of infection was determined according to multiple banding at any 1 of the 3 loci assessed.
Quantitation of pfmdr1 copy number and detection of polymorphisms
pfmdr1 copy number was assessed by quantitative PCR (ABI Sequence Detector 7700; Applied Biosystems) using the methods described elsewhere [5]. All reactions were performed in triplicate and were rejected if they did not conform to exponential kinetics. Copy number is reported as a continuous variable. Previous studies defined the repeatability coefficient of the copy number assay as 0.64 (viz., 95% of repeated estimates of pfmdr1 copy number would be expected to be within ±0.32 of the first). Single-nucleotide polymorphisms (SNPs) in pfmdr1 and pfcrt were detected by nested PCR–restriction fragment–length polymorphism methods, also as described elsewhere [21, 22]. All PCRs and digests included, as positive and negative controls, DNA of laboratory strains 3D7, HB3, 7G8, and Dd2, respectively.
Statistical analyses
Data were analyzed using SPSS for Windows (version 14; SPSS). The Mann-Whitney U test or the Kruskal-Wallis method was used for nonparametric comparisons, and Student’s t test or 1-way analysis of variance was used for parametric comparisons. For categorical variables, percentages and corresponding 95% confidence intervals (CIs) were calculated using Wilson’s method. Proportions were examined using the χ2 test with Yates’s correction or Fisher’s exact test. Paired proportions were compared using McNemar’s test. The relationship between the copy numbers of the pre- and posttreatment isolates was assessed on ordinal variables by rounding copy numbers to the nearest integer. For polyclonal infections, the assessment of copy number generates a figure representing an average of the copy numbers of genes in individual parasite clones weighted by their respective proportions. Therefore, to determine the variation in copy number attributable to the selection of resistant parasites from polyclonal infections, we also assessed pre- and posttreatment isolates’ copy number as a continuous variable. Linear regression analysis was used to determine the relationship between the initial copy number and the change between the copy number of the pre- and posttreatment isolates.
RESULTS
Between December 1992 and June 2002, a total of 1289 paired isolates were identified from patients whose parasitemia reappeared within the follow-up period. In total, 230 pairs (18%) were subjected to molecular analysis of pfmdr1 and genotyping of merozoite antigens (table 1), of which 79 (34%) came from pregnant women (29 after MFQ, 33 after ART, and 17 after QU). pfmdr1 copy number was successfully determined in 199 pairs (87%), and SNPs in 169 (73%); these proportions did not differ significantly between treatment groups or between pregnant and nonpregnant patients. Polyclonal infections were detected for 73 (32%) pretreatment isolates and for 52 (23%) posttreatment isolates (P = .04).
Table 1. Classification according to 3-loci genotyping (merozoite surface proteins 1 and 2 and the gene encoding the glutamate rich protein) of the 230 parasite isolates subjected to pfmdrl genotyping in the 5 treatment groups.
| Total | Mefloquine | Mefloquine plus artesunate | Artemether-lumefantrine | Artemisinin monotherapya | Quinine | Total |
|---|---|---|---|---|---|---|
| Novel infections | 1 | 21 | 18 | 14 | 1 | 55 |
| Recrudescent | 33 | 40 | 32 | 29 | 17 | 151 |
| Recrudescent + novel infections | 5 | 6 | 0 | 4 | 0 | 15 |
| Indeterminate | 3 | 5 | 0 | 1 | 0 | 9 |
| Total | 42 | 72 | 50 | 48 | 18 | 230 |
Artesunate or artemether at 12 mg/kg over 7 days.
pfmdr1 copy number
After MAS3 treatment, increased copy number in the primary isolate was significantly associated with PCR-confirmed recrudescences; PCR-confirmed recrudescences occurred for 81% (26/32) of the primary isolates with an increased pfmdr1 copy number, compared with 44% (10/23) for those with single copies (relative risk, 1.9 [95% CI, 1.1–3.1]; P = .009).
PCR-confirmed recrudescences
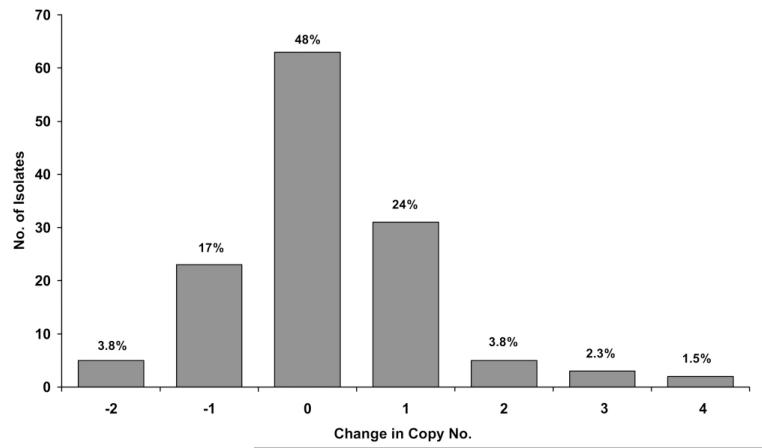
Among the 151 paired isolates from patients with PCR-confirmed recrudescences, copy number could be assessed in both the primary and the recurrent infection isolates in 132, of which 93 (70%) had a primary isolate with a single pfmdr1 copy. Detection of intrahost changes in copy number were frequent, occurring in 52% (69/132) of patients, of which 41 (59%) were increases in copy number, and 28 (41%) were decreases (figure 1). In total, amplification from a single to >1 copy number occurred in 12% (16/132) of patients, and deamplification from multiple to a single copy number occurred in 7.5% (10/132). The change in copy number from the pre- to posttreatment isolate differed significantly between treatment groups (table 2). Although 68% (13/19) of isolates from patients with recrudescent infections after MFQ or MAS3 had an increase in pfmdr1 copy number, this occurred in only 25% (2/8) of patients after AL (P = .09) and in 0% (0/10) of patients after ART (P < .001). These proportions were similar when selecting only monoclonal infections.
Figure 1.
Histogram of change in copy no. (rounded to nearest integer) between the posttreatment isolate and the pretreatment isolate (shown as the former minus the latter), in polymerase chain reaction–confirmed recrudescent infections (n = 132).
Table 2. Change in pfmdrl copy no. (CN) between pretreatment and posttreatment isolates after polymerase chain reaction–confirmed recrudescence.
| Pretreatment CN, posttreatment CN | Mefloquine | Mefloquine plus artesunate | Artemether-lumefantrine | Artemisinin monotherapya | Quinine |
|---|---|---|---|---|---|
| Single | |||||
| Single | 3 (33) | 3 (30) | 6 (75) | 10 (100) | 1 (50) |
| Increased | 6 (67) | 7 (70) | 2 (25) | 0 (0) | 1 (50) |
| Total | 9 | 10 | 8 | 10 | 2 |
|
| |||||
| Increased | |||||
| Increased | 22 (100) | 25 (96) | 17 (89) | 13 (81) | 6 (60) |
| Single | 0 (0) | 1 (3.8) | 2 (11) | 3 (19) | 4 (40) |
| Total | 22 | 26 | 19 | 16 | 10 |
NOTE. Data are no. (%) of isolates.
Artesunate or artemether at 12 mg/kg over 7 days.
There was also a significant difference between treatment groups in the rates at which recurrent isolates showed a decrease in pfmdr1 copy number, although by definition this analysis was confined to pairs in which the primary isolate had an increased pfmdr1 copy number. Only 2% (1/48) of isolates from patients experiencing treatment failure after MFQ or MAS3 deamplified from multiple to a single copy number, compared with 19% (3/16) after ART (P = .045), 40% (4/10) after QU (P = .002), and 11% (2/19) after AL (P = .19).
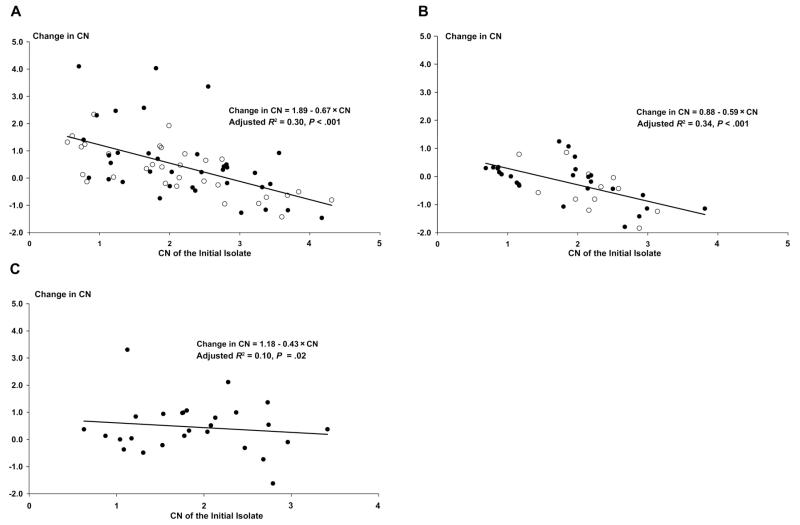
When copy number was quantified as a continuous variable, the mean change between the primary and recurrent isolate was +0.58 (95% CI, 0.1–1.04) after MAS3, +0.46 (95% CI, 0.09–0.83) after AL, +0.32 (95% CI, 0.01–0.66) after MFQ, −0.16 (95% CI, −0.45 to 0.13) after ART, and −0.46 (95% CI, −0.97 to 0.05) after QU (overall P = .029). The change in copy number was correlated with the copy number of the primary isolate (Rs = −0.48; P < .001), and this was apparent for each treatment group (figure 2). Hence, isolates with a low pretreatment copy number had the greatest increase after recrudescence, whereas those starting with a high copy number were more likely to decrease. In a linear regression model, there was no significant difference in the change in copy number between MFQ and MAS3 nor between ART and QU. After combining treatment groups with long terminal elimination half-life (MFQ + MAS3), there was a significant difference in the linear regression model with treatment regimens that were eliminated rapidly (ART + QU) (P < .001).
Figure 2.
Change in copy no. (CN; posttreatment isolate minus pretreatment isolate) in polymerase chain reaction–confirmed recrudescent infections. A, Mefloquine (white circles) and mefloquine plus 3 days of artesunate (black circles). B, Artemisinin monotherapy (white circles) and quinine (black circles). C, Artemether-lumefantrine.
In patients treated with MFQ or MAS3, the change in copy number between primary and recurrent isolates was correlated with the time to recrudescence (R = 0.3; P = .01); hence, infections with a low copy number in the primary isolate had the greatest increase after recrudescence, whereas those infections starting with a high copy number were more likely to decrease. However, patients with an initially higher copy number also tended to recrudesce earlier (R = −0.3; P = .01). After controlling for the copy number of the initial isolate, there was no effect of the change in copy number on the time to recrudescence, age of the patient, clonality of infection, or initial parasitemia.
New infections
Of the 55 paired isolates from reinfections, copy number could be assessed in both primary and recurrent isolates in 48 (87%) (table 3). Overall, 70% (19/27) of patients with isolates with a single pfmdr1 copy number were reinfected with isolates with an increased copy number, compared with 19% (4/21) of patients with isolates with increased copy number being reinfected with isolates with a single copy number (P = .003). However, after stratifying by treatment group, individual group numbers were small, and the difference did not reach significance.
Table 3. Population prevalence of copy no. in the pretreatment and reinfecting (posttreatment) isolates.
| Category | Mefloquine | Mefloquine plus artesunatea | Artemether-lumefantrineb | Artemisinin monotherapyc,d |
|---|---|---|---|---|
| Pretreatment | ||||
| Single | 1 (100) | 13 (68) | 7 (41) | 6 (55) |
| Increased | 0 (0) | 6 (32) | 10 (59) | 5 (45) |
|
| ||||
| New infections | ||||
| Single | 0 (0) | 7 (37) | 2 (12) | 3 (27) |
| Increased | 1 (0) | 12 (63) | 15 (88) | 8 (73) |
|
| ||||
| Total | 1 | 19 | 17 | 11 |
NOTE. Data are no. (%) of isolates. Paired proportions for each treatment group were compared using McNemar’s test to determine the rates of change in copy no. (single to amplified vs. amplified to single).
P = .15.
P = .06.
P = .38.
Artesunate or artemether at 12 mg/kg over 7 days.
SNPs
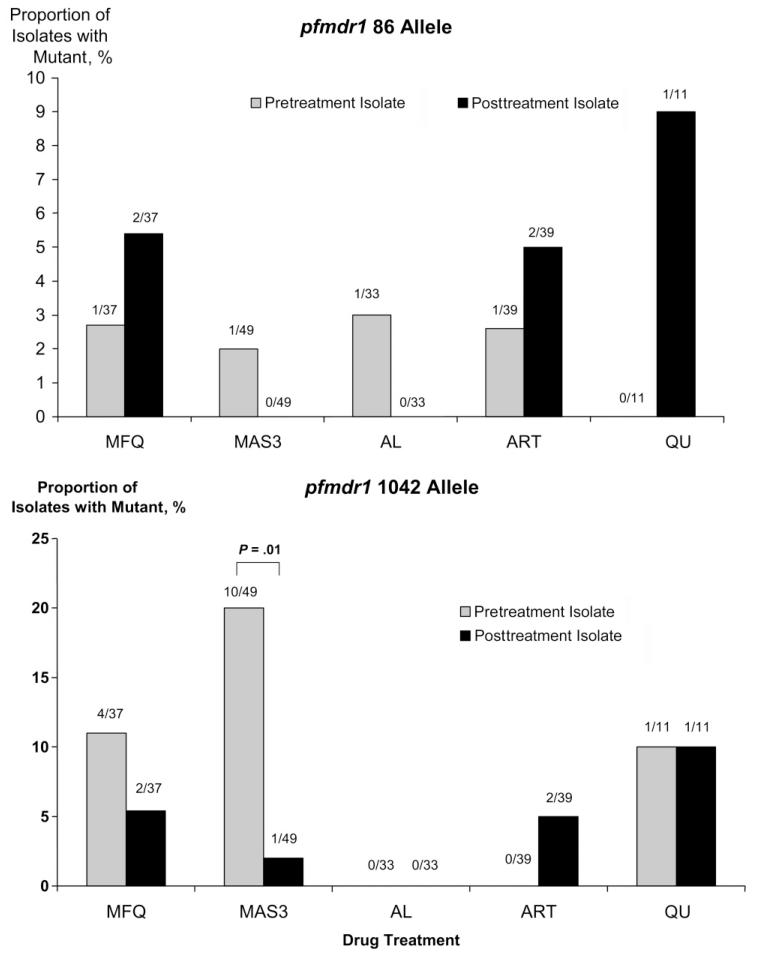
pfmdr1 SNPs were detected at codon 86 in 2.4% (4/169) of pretreatment isolates, at codon 184 in 32% (39/121), at codon 1034 in 0.6% (1/169), and at codon 1042 in 8.9% (15/169). After MAS3, all (10/10) of the 1042D alleles present in the pretreatment isolate had reverted to 1042N (wild type), whereas the reverse process (1042N to 1042D) occurred only in 2.6% (1/39) (P = .01) (figure 3). No significant difference was observed after any of the other treatments or for SNPs at codons 86, 184, or 1034.
Figure 3.
Proportion of isolates with mutant pfmdr1 alleles in pre- and posttreatment isolates after antimalarial treatment (including reinfections and recrudescences). AL, artemether-lumefantrine; ART, artemisinin monotherapy; MAS3, mefloquine plus 3 days of artesunate; MFQ, mefloquine monotherapy; QU, quinine.
DISCUSSION
Our results demonstrate that pfmdr1 copy number often changes during the course of a recurrent infection after antimalarial therapy. Overall, 54% (38/71) of primary isolates with a single pfmdr1 copy number had an increased copy number in the recurrent isolate, and this figure did not differ when selecting only single clone infections according to 3-loci genotyping. There are 3 possible explanations for these findings. First, there could be expansion of an existing subpopulation with pfmdr1 amplification either present in a multiclonal infection or at undetectable levels in a monoclonal primary infection. Second, an infection could be newly acquired in the presence of declining concentrations of antimalarial drugs. Third, there could be de novo amplification and subsequent growth of a resistant parasite population. It was not possible to quantify the parasite subpopulations within apparently clonal infections, and, therefore, we are unable to determine the exact frequency or distribution of de novo pfmdr1 amplification from the current data. Amplification could have occurred through unequal crossover at either mitosis or meiosis in the mosquito vector, but the ability to select this in vitro [23, 24], the high frequency of changes in pfmdr1 copy number observed within patients, and recent genetic data analyzing amplicon size, sequence, and break points strongly suggest that, in the human host, amplification events occur relatively frequently and do so during mitotic DNA replication in the blood-stage infection. In the presence of appropriate drug pressure favoring survival of the amplified genotype, increased copy number will, therefore, be readily selected.
Drug pressure is largely determined by the elimination half-life of the antimalarial drug used [25], which, in this study, varied considerably, with average values of 1 h for the active metabolite of artesunate, 16 h for QU, 4 days for lumefantrine, and 14 days for MFQ. Although increased pfmdr1 copy number is associated with reduced susceptibility to ARTs [5], QU, and lumefantrine [8], when recrudescences followed these rapidly eliminated drugs, amplification was uncommon. In fact, if the primary isolate already had increased pfmdr1 copy number, deamplification to a single copy predominated. This suggests that, in the absence of the drug-derived selective pressure, there is a parasite fitness disadvantage to having increased pfmdr1 copy number and that this competitive growth disadvantage is sufficient for these unfit parasites to be outgrown by their fitter rivals in 8–12 asexual cycles (the time between elimination of the drug and recrudescence of the parasite).
In our previous studies, we have demonstrated that increased pfmdr1 copy number is the major determinant of treatment response to MFQ as well as to MAS3. These observations were confirmed in recent laboratory studies, which demonstrated a key role of pfmdr1 copy number and expression in determining MFQ susceptibility in vitro [26]. In the present study, we high-light that gene amplification is selected very commonly in patients treated with either regimen. Overall recrudescence rates after MAS3 have averaged 5%, of which 70% (i.e., net 3.5%) showed amplification [5]. However, this unstable trait conferring a survival advantage in the presence of a drug is readily lost when the selection pressure is removed, even in a single host.
Recent studies from Africa have demonstrated high rates of reversion of SNPs to wild-type pfmdr1 in recurrent isolates after AL therapy [27-29]. This generally represents selection of newly acquired infections in the presence of residual drug, rather than de novo mutations in recrudescent infections. The N86Y and N1042D SNPs of pfmdr1 are associated with decreased susceptibility to chloroquine but increased susceptibility to lumefantrine, MFQ, and artesunate [8, 22]. In Thailand, we observed an increase in the prevalence of the 1042N wild-type allele after MFQ and MAS3 therapy, although this was not apparent for any of the other drugs or SNPs tested. We also observed an increase in the prevalence of pfmdr1 amplification after reinfection following treatment with MAS3 (32%–63%) and AL (59%–88%), but numbers were small, and this increase did not reach significance.
Among field isolates, pfmdr1 amplification occurs almost exclusively in isolates with the wild-type allele at codons 86, 1034, and 1042 [5]. Hence, reversion to 86N and 1042N may represent the first stage of the process leading to pfmdr1 amplification in the field. Once present, clinical resistance is associated with increased gametocyte carriage and, thus, increased transmissibility [30], which, under appropriate drug pressure, translates into a survival advantage for parasites with increased pfmdr1 copy number, fueling the spread of antimalarial drug resistance. The force that drives this selection is determined by the relative treatment failure rate and consequent transmission advantage of the resistant over the sensitive genotype. Although increased pfmdr1 copy number confers a major selective advantage when MFQ alone is used for treatment (cure rates decrease from almost 100% to 50%), when MAS is given, the advantage is lessened as efficacy remains high. This reduces the differential survival advantage and, thus, the force driving the spread of resistance. The greatest increment in MFQ resistance is an increase from 1 to 2 copies, which is associated with a 2.4-fold decrease (95% CI, 1.3–2.5–fold decrease) in in vitro susceptibility [5]. Further increments confer proportionally less resistance but may incur further fitness costs. This probably explains why MFQ resistance has stabilized in this area over the past 10 years [31, 32]. A state of relative equilibrium has been reached in which the survival advantage of pfmdr1 amplification is balanced by its fitness disadvantage.
Acknowledgment
We thank the staff of the Shoklo Malaria Research Unit for their excellent work.
Financial support: Wellcome Trust (Career Development Award [074637] to R.N.P.; grant 066201 for molecular work performed at St. George’s, University of London).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plowe CV, Cortese JF, Djimde A, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–6. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 3.Nosten F, ter Kuile F, Chongsuphajaisiddhi T, et al. Mefloquine-resistant falciparum malaria on the Thai-Burmese border. Lancet. 1991;337:1140–3. doi: 10.1016/0140-6736(91)92798-7. [DOI] [PubMed] [Google Scholar]
- 4.Wilson CM, Volkman SK, Thaithong S, et al. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–60. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- 5.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–9. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 7.Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–3. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price RN, Uhlemann A-C, van Vugt M, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–7. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ter Kuile FO, Dolan G, Nosten F, et al. Halofantrine versus mefloquine in treatment of multidrug-resistant falciparum malaria. Lancet. 1993;341:1044–9. doi: 10.1016/0140-6736(93)92409-m. [DOI] [PubMed] [Google Scholar]
- 10.Nosten F, Luxemburger C, ter Kuile FO, et al. Treatment of multidrug-resistant Plasmodium falciparum malaria with 3-day artesunate-mefloquine combination. J Infect Dis. 1994;170:971–7. doi: 10.1093/infdis/170.4.971. [DOI] [PubMed] [Google Scholar]
- 11.Price RN, Nosten F, Luxemburger C, et al. Artesunate versus artemether in combination with mefloquine for the treatment of multidrug-resistant falciparum malaria. Trans R Soc Trop Med Hyg. 1995;89:523–7. doi: 10.1016/0035-9203(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 12.Price RN, Nosten F, Luxemburger C, et al. Artesunate/mefloquinetreatment of multi-drug resistant falciparum malaria. Trans R Soc Trop Med Hyg. 1997;91:574–7. doi: 10.1016/s0035-9203(97)90032-8. [DOI] [PubMed] [Google Scholar]
- 13.Price R, van Vugt M, Nosten F, et al. Artesunate versus artemether for the treatment of recrudescent multidrug-resistant falciparum malaria. Am J Trop Med Hyg. 1998;59:883–8. doi: 10.4269/ajtmh.1998.59.883. [DOI] [PubMed] [Google Scholar]
- 14.Price R, Luxemburger C, van Vugt M, et al. Artesunate and mefloquine in the treatment of uncomplicated multidrug-resistant hyperparasitaemic falciparum malaria. Trans R Soc Trop Med Hyg. 1998;92:207–11. doi: 10.1016/s0035-9203(98)90750-7. [DOI] [PubMed] [Google Scholar]
- 15.McGready R, Nosten F. The Thai-Burmese border: drug studies of Plasmodium falciparum in pregnancy. Ann Trop Med Parasitol. 1999;93(Suppl 1):S19–23. doi: 10.1080/00034989957709. [DOI] [PubMed] [Google Scholar]
- 16.van Vugt M, Brockman A, Gemperli B, et al. Randomized comparison of artemether-benflumetol and artesunate-mefloquine in treatment of multidrug-resistant falciparum malaria. Antimicrob Agents Chemother. 1998;42:135–9. doi: 10.1128/aac.42.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vugt MV, Wilairatana P, Gemperli B, et al. Efficacy of six doses of artemether-lumefantrine (benflumetol) in multidrug-resistant Plasmodium falciparum malaria. Am J Trop Med Hyg. 1999;60:936–42. doi: 10.4269/ajtmh.1999.60.936. [DOI] [PubMed] [Google Scholar]
- 18.Luxemburger C, Thwai KL, White NJ, et al. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans R Soc Trop Med Hyg. 1996;90:105–11. doi: 10.1016/s0035-9203(96)90102-9. [DOI] [PubMed] [Google Scholar]
- 19.Luxemburger C, Ricci F, Nosten F, Raimond D, Bathet S, White NJ. The epidemiology of severe malaria in an area of low transmission in Thailand. Trans R Soc Trop Med Hyg. 1997;91:256–62. doi: 10.1016/s0035-9203(97)90066-3. [DOI] [PubMed] [Google Scholar]
- 20.Brockman A, Paul RE, Anderson TJ, et al. Application of genetic markers to the identification of recrudescent Plasmodium falciparum infections on the northwestern border of Thailand. Am J Trop Med Hyg. 1999;60:14–21. doi: 10.4269/ajtmh.1999.60.14. [DOI] [PubMed] [Google Scholar]
- 21.Price RN, Cassar C, Brockman A, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–9. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 23.Triglia T, Foote SJ, Kemp DJ, Cowman AF. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol Cell Biol. 1991;11:5244–50. doi: 10.1128/mcb.11.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 1992;11:3067–75. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins WM, Sibley CH, Hastings IM. The search for effective and sustainable treatments for Plasmodium falciparum malaria in Africa: a model of the selection of resistance by antifolate drugs and their combinations. Am J Trop Med Hyg. 2005;72:163–73. [PubMed] [Google Scholar]
- 26.Sidhu ABS, Uhlemann A-C, Valderramos SG, Valderramos J-C, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sisowath C, Strömberg J, Mårtensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–7. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 28.Mårtensson A, Strömberg J, Sisowath C, et al. Efficacy of artesunate plus amodiaquine versus that of artemether-lumefantrine for the treatment of uncomplicated childhood Plasmodium falciparum malaria in Zanzibar, Tanzania. Clin Infect Dis. 2005;41:1079–86. doi: 10.1086/444460. [DOI] [PubMed] [Google Scholar]
- 29.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–5. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price RN, Nosten F, Luxemburger C, et al. Effects of artemisinin derivatives on malaria transmissibility. Lancet. 1996;347:1654–8. doi: 10.1016/s0140-6736(96)91488-9. [DOI] [PubMed] [Google Scholar]
- 31.Nosten F, van Vugt M, Price R, et al. Effects of artesunate-mefloquine combination on incidence of Plasmodium falciparum malaria and mefloquine resistance in western Thailand: a prospective study. Lancet. 2000;356:297–302. doi: 10.1016/s0140-6736(00)02505-8. [DOI] [PubMed] [Google Scholar]
- 32.Carrara VI, Sirilak S, Thonglairuam J, et al. Deployment of early diagnosis and mefloquine-artesunate treatment of falciparum malaria in Thailand: the Tak Malaria Initiative. PLoS Med. 2006;3:e183. doi: 10.1371/journal.pmed.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]