Abstract
The epithelium of the mouse tongue and soft palate consists of at least three distinct epithelial cell populations: basal cells, keratinized cells organized into filiform and fungiform papillae, and taste receptor cells present in tight clusters known as taste buds in the fungiform and circumvallate papillae and soft palate. All three cell types develop from the simple epithelium of the embryonic tongue and palate, and are continually replaced in the adult by cell turnover. Previous studies using pulse-chase tritiated thymidine labeling in the adult mouse provided evidence for a high rate of cell turnover in the keratinocytes (5–7 days) and taste buds (10 days). However, little is known about the localization and phenotype of the long-term stem or progenitor cells that give rise to the mature taste bud cells and surrounding keratinocytes in these gustatory tissues. Here, we make use of a tamoxifen-inducible K14-CreER transgene and the ROSA26 LacZ reporter allele to lineage trace the mature keratinocytes and taste bud cells of the early postnatal and adult mouse tongue and soft palate. Our results support the hypothesis that both the pore keratinocytes and receptor cells of the taste bud are derived from a common K14+K5+Trp63+Sox2+ population of bipotential progenitor cells located outside the taste bud. The results are also compatible with models in which the keratinocytes of the filiform and fungiform papillae are derived from basal progenitor cells localized at the base of these structures.
Keywords: Taste bud, Keratinocytes, Tongue, Progenitor cell, Lineage mapping
Introduction
The tongue and soft palate are gustatory tissues containing numerous taste buds (TBs) to detect taste stimuli. A typical mature TB consists of a tight intragemmal cluster of 50–100 elongated taste receptor/sensory epithelial cells surrounded by an outer layer of flattened keratinocytes. These form the perigemmal layer and taste “pore.” TBs are located in several places in the tongue (fungiform, foliate, and circumvallate papillae) and oral cavity (soft palate). Depending on their location, TBs develop either perinatally in the anterior tongue and soft palate or postnatally in the foliate and circumvallate (CV) papillae. In all cases, they arise during development from the local surface epithelium rather than from neuronal or neural crest cells [1, 2]. Indeed, studies in rodents have shown that both the receptor cells of the TBs and the stratified keratinized epithelium of the fungiform papillae in which they reside, as well as the numerous filiform papillae of the tongue, are all derived from the same simple epithelium present in the embryo [3, 4].
Recent studies have begun to identify genetic pathways controlling the differentiation of embryonic tongue epithelium into either TB precursors or stratified, keratinized epithelium. For example, we have found that the Sry-related transcription factor Sox2 is more highly expressed in developing and mature TBs than in the surrounding epithelial cells [5]. Newborn pups hypomorphic for Sox2 lack mature TBs and fungiform papillae. In contrast, when Sox2 is overexpressed in the embryonic tongue epithelium, keratinocyte (filiform) differentiation is completely blocked, with the absence of trichohyalin (AE15 antigen) expression [5]. These findings suggest that the level of Sox2 in the immature tongue epithelial cells is critical for the cell fate decision between TB cells and keratinocytes. Furthermore, Sox2 expression in regenerating CV TBs is dependent on gustatory innervation [6]. Thus, Sox2 also likely plays an important role in the maintenance of adult TB cells. Pax9 is also known to be an important transcription factor for the proper differentiation of filiform papillae [7]. Trp63 (p63), a member of the p53 transcription factor family, is critical for tongue development because Trp63-deficient embryos have only a thin tongue epithelial layer [8]. Thus, gene-targeting studies suggest that complex transcriptional hierarchies and signaling networks regulate the differentiation and maintenance of tongue epithelial cells.
In the adult tongue, both filiform papillae and TBs undergo continual turnover. In the case of the keratinocytes of the filiform papillae, pulse-chase tritiated thymidine studies support a model in which long-term, self-renewing stem cells reside in the base of the papillae. The transit amplifying and differentiating descendants move up along the basal lamina and exit into the superficial layers to be shed from the surface over the course of about 5–7 days [9]. In the case of the TBs of the fungiform papillae, previous studies have suggested a turnover time of about 10 days in the adult [10, 11]. However, the localization of the long-term progenitor cells of the TBs and whether they reside exclusively inside or outside the bud are still under debate. For example, studies in the rat suggest that CV TB cells are rapidly renewed from cells outside the bud [12, 13]. In order to learn more about the identify of progenitor cells of the TB and the niche in which they reside, we characterized the expression patterns of the epithelial markers keratin 8 (K8), K14, K5, and Trp63 in mouse gustatory tissues. Based on these observations, we used the tamoxifen-inducible K14-CreER/Rosa26R reporter system [14, 15] to carry out cell lineage mapping studies in the tongue and in the soft palate at early postnatal and adult stages. Our results support a model in which K14+ epithelial cells located outside the TBs function as bipotential progenitor cells to continuously generate both mature TB receptor cells and surrounding keratinocytes (perigemmal and taste pore cells). Our findings are also compatible with models in which the keratinocytes of the filiform and fungiform papillae are derived from basal progenitor cells localized to the bottom of these structures.
Materials and Methods
Mice
All mice were maintained under a 12-hour light/dark cycle and handled under Institutional Animal Care and Use Committee-approved protocols. Outbred ICR mice were used as controls at the embryonic and postnatal stages. The K14-CreER transgenic mouse line was kindly provided by Barry Stripp [15] and was maintained as hemizygotes. ROSA26 mice (Gt(ROSA)26Sortm1Sor), in which LacZ is a reporter for Cre recombination, were maintained as homozygous [16]. For genotyping, genomic DNA was extracted from tail tips and assayed using polymerase chain reaction (PCR) primer sets for the Rosa26R allele (5′-AAAGTCGCTCTGAGTTGTTAT-3′, 5′-GCGAAGAGTTTGTCCTCAACC-3′, 5′-GGAGCGGGAGAAATGGATATG-3′) and for the K14-CreER allele (5′-TCGATGCAACGAGTGATGAG-3′, 5′-TTCGGCTATACGTAACAGGG-3′).
Tamoxifen Injection
K14-CreER males were crossed with Rosa26R females. Tamoxifen (TM) dissolved in corn oil (20 mg/ml) was given by i.p. injection at a dose of 0.25 mg/g body weight (one injection at postnatal day 2 (P2) for the mother, and five injections at 1-day intervals for P30 adult mice).
LacZ Staining
For each time point, 4–10 independent tongues and soft palates were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 2 hours after dissection, and then washed in PBS. The tissues were next stained in X-gal solution (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl, 0.02% NP-40, 1 mg/ml X-gal) at 37°C overnight. Tissues were postfixed in 4% PFA, processed for sectioning at 7 µm, and counterstained with eosin.
Histology and Immunohistochemistry
Tongues and soft palates were fixed in 4% PFA in PBS for 2 hours or overnight at 4°C, processed through the appropriate dehydration steps, and embedded in paraffin. The 7-µm thick sections were pretreated by boiling in 10 mM sodium citrate buffer (pH 6.0). For the primary antibody reaction, the sections were incubated with the following antibodies: Trp63 (4A4; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com), Sox2 (AB5603; Chemicon, Temecula, CA, http://www.chemicon.com), K14 (LL002; Chemicon), K8 (TROMA I; Developmental Studies Hybridoma Bank, Iowa City, IA, http://www.uiowa.edu/~dshbwww), K5 (AF138; Covance, Princeton, NJ, http://www.covance.com), trichohyalin (AE15, ab58755; Abcam, Cambridge, U.K., http://www.abcam.com), and phosphohistone H3 (PHHS) (Ser10) (Upstate, Charlottesville, VA, http://www.upstate.com). Signal detection was performed with the following secondary antibodies: AlexaFluor 488, AlexaFluor 546-labeled anti-mouse, anti-rat, and anti-rabbit IgG antibodies (Molecular Probes Inc., Eugene, OR, http://probes.invitrogen.com) and the DAKO EnVision system-HRP (DakoCytomation, Glostrup, Denmark, http://www.dakocytomation.com).
Results
Expression of Epithelial Markers in the Development of the Anterior Tongue
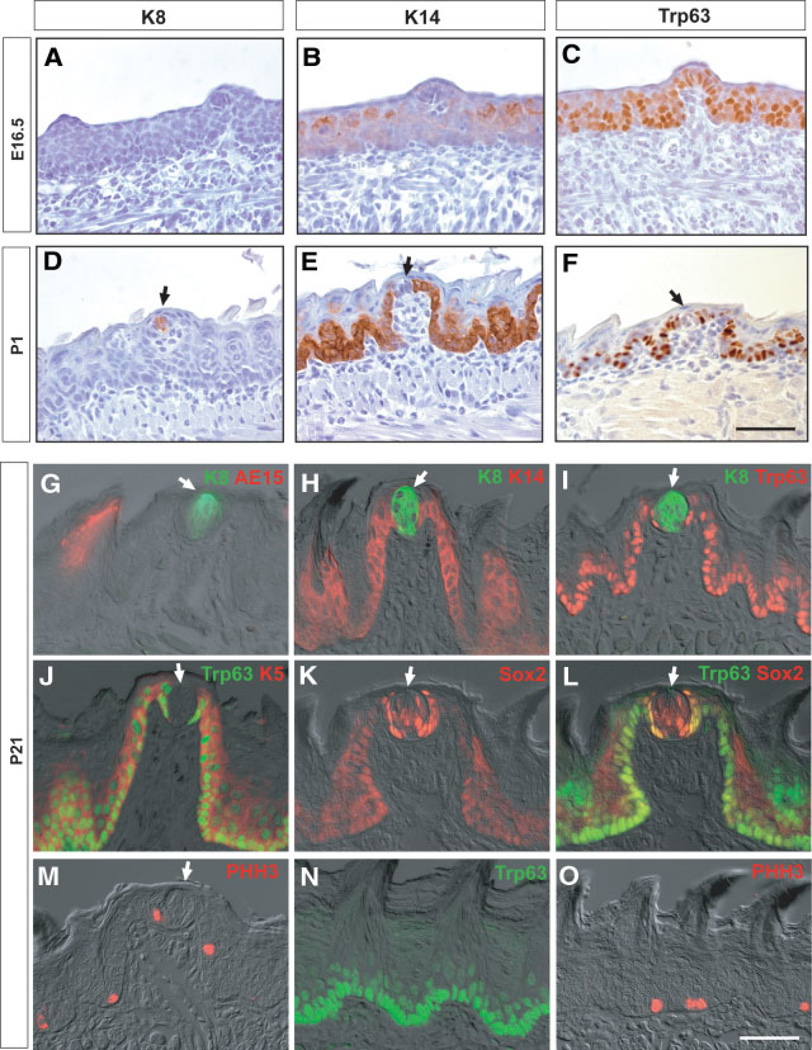
In the mouse, the TBs and keratinized epithelial structures of the tongue and soft palate develop from the embryonic epithelium with slightly different timing, depending on their location. To follow this development in the anterior tongue we performed immunohistochemistry with several markers at different stages (Fig. 1). K8, a marker of mature intragemmal cells [17, 18], is first expressed in the TBs of the fungiform papillae perinatally (P1), and its level gradually increases (Fig. 1A, 1D, 1G). In contrast, K14 expression is absent from intragemmal cells at P1 but clearly expressed in perigemmal cells and basal keratinocytes (Fig. 1B, 1E, 1H). Likewise, the transcription factor Trp63 is initially expressed throughout the basal layer of the epithelium, including the developing fungiform papillae, but is completely absent from the intragemmal cells at P1 and later (Fig. 1C, 1F, 1I, 1J, 1L). K5, which is coexpressed with K14 and Trp63 in basal cells of other epithelia [19], has a similar expression pattern to Trp63 in the adult tongue (Fig. 1J). The mutually exclusive expression of K8 inside the TB, versus Trp63, K14, and K5 in the perigemmal keratinocytes outside the TB and in basal keratinocytes, is clearly shown by double immunohistochemistry in the adult tongue (Fig. 1G–1J). For example, there is a very sharp boundary between K8+ and K14+ cells inside and outside the TB (Fig. 1H) as well as between K8+ and Trp63+ cells (Fig. 1I). The transcription factor Sox2 is clearly expressed at high levels in the mature TB cells, but at lower levels in the basal epithelium of the fungiform and filiform papillae (Fig. 1K). However, unlike K8, it is also present in the nuclei of perigemmal cells, where its expression overlaps with that of Trp63, as shown by double labeling (Fig. 1L). Finally, immunohistochemistry shows that trichohyalin (AE15 antigen), a differentiation marker for keratinocytes of the filiform papillae, is not expressed in fungiform papillae or TB cells (Fig. 1G). To localize mitotic cells in the adult tongue, we used antibodies to PHH3. This shows that, in the anterior tongue, PHH3+ cells are present in the perigemmal layer as well as in the Trp63+ epithelial basal layer of the filiform papillae (Fig. 1M, 1O).
Figure 1.
Expression of epithelial markers in the anterior tongue at different stages. Sections of the anterior tongue show expression of keratin 8 (K8) (A, D), K14 (B, E), and Trp63 (C, F) at E16.5 and P1. (G–L): Fluorescence immunostaining for epithelial and proliferation markers in fungiform papillae at P21. Shown are expression of K8/AE15 (G), K8/K14 (H), K8/Trp63 (I), Trp63/K5 (J), Sox2 (K), Trp63/Sox2 (L), and phosphohistone H3 (PHH3) (M) merged with differential interference contrast images. Note in particular the sharp boundary between K8 inside and K14 and Trp63 outside the taste bud (TB), and the coexpression of Trp63 and Sox2 in the perigemmal cells around the TB (L). (N, O): Sections of filiform papillae showing expression of Trp63 (N) and PHH3 (O). Arrows indicate TBs. Scale bar, 50 µm. Abbreviations: E, embryonic day; P, postnatal day.
Expression of Epithelial Markers in the Development of the CV Papilla and Soft Palate
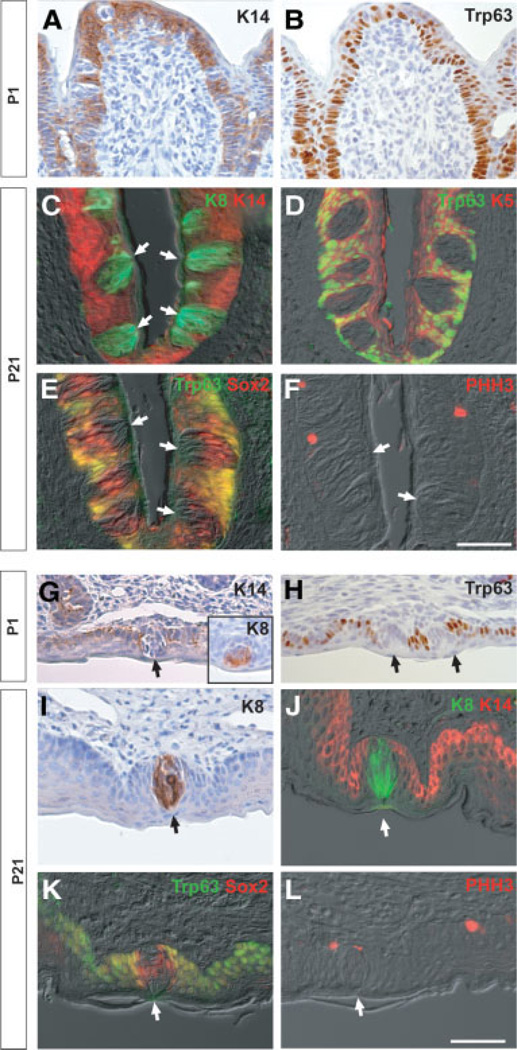
We next examined the expression of epithelial markers in the CV papilla and soft palate, in which the TBs develop perinatally. At P1, K14+Trp63+ CV epithelium is already invaginated into the posterior tongue (Fig. 2A, 2B), but mature TBs have not yet appeared. In contrast, in the perinatal soft palate, distinct K8+ TB cells are already differentiated (Fig. 2G, inset) and the expression of K14 and Trp63 is not observed in TBs (Fig. 2G, 2H). At P21, immunohistochemistry shows that many K8+ intragemmal cells are present in the TBs at both locations. These cells are completely negative for K14, K5, and Trp63, which are coexpressed in the epithelium outside the TBs (Fig. 2C, 2D, 2J). As in the tongue, Sox2 is expressed in the TBs of the CV and soft palate in intragemmal cells and in perigemmal cells that are positive for Trp63 (Fig. 2E, 2K). Finally, PHH3+ mitotic cells are frequently detected in the basal layer of the CV and soft palate epithelium and in perigemmal cells adjacent to TBs (Fig. 2F, 2L). Taken together, these results in all gustatory tissues are compatible with a model in which TB intragemmal cells are derived from a K14+K5+Trp63+ cell population outside the bud.
Figure 2.
Expression of epithelial markers in the circumvallate papilla and soft palate at different stages. (A–F): Sections of circumvallate papilla at P1 (A, B) and P21 (C–F). Shown are the expression of keratin 14 (K14) (A) and Trp63 (B) at P1 and the expression of K8/K14 (C), Trp63/K5 (D), Trp63/Sox2 (E), and phosphohistone H3 (PHH3) (F) merged with differential interference contrast (DIC) images. Note the exclusion of K14, Trp63, and K5 from the interior of the taste bud (TB), which contains K8+ cells with a sharp boundary in perigemmal cells (C, D). Note also the coexpression of Sox2 and Trp63 in cells around the TBs (E). (G–L): Sections of soft palate at P1 (G, H) and P21 (I–L). Shown are expression of K14 (G), K8 (G, inset), and Trp63 (H) at P1 and expression of K8 (I), K8/K14 (J), Trp63/Sox2 (K), and PHH3 (L) merged with DIC images. Note again the sharp boundary between K8+ and K14+ cells and the coexpression of Trp63 and Sox2 in perigemmal cells adjacent to the TBs (J, K). Arrows indicate TBs. Scale bar, 50 µm. Abbreviation: P, postnatal day.
Cell Lineage Mapping in the Postnatal Tongue and Soft Palate
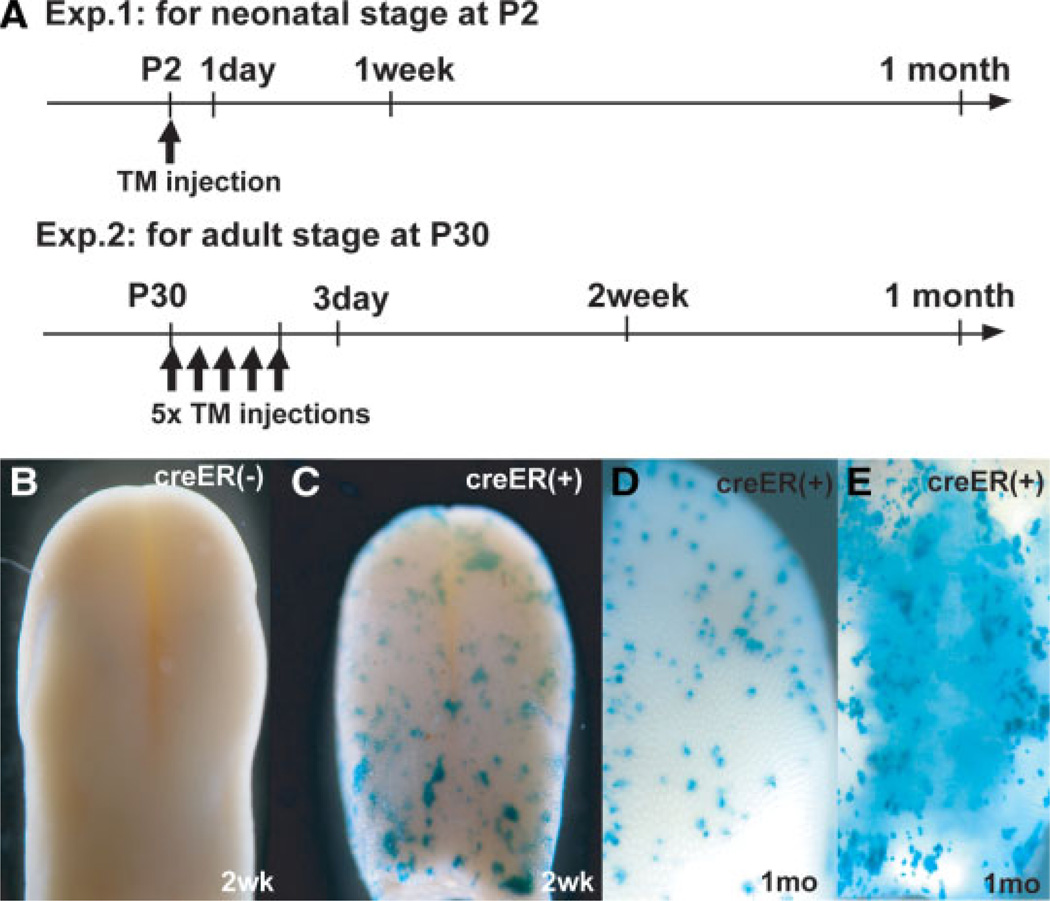
To determine the long-term progenitor cells for the maintenance of mature TB cells and keratinocytes in the postnatal mouse, we performed cell lineage mapping experiments to follow the descendants of cells that originally express the K14-CreER transgene. This experiment was carried out at two different times, namely, at P2 and at P30 (Fig. 3A). Since it is difficult to inject TM into pups soon after birth, a single dose was administered to the lactating mother so that the drug would pass to the pups through the milk. By 2 weeks, whole-mount staining revealed patches of LacZ+ cells throughout the tongue and soft palate of K14-CreER/Rosa26R compound transgenic pups, but not in littermates lacking the CreER transgene (Fig. 3B–3E).
Figure 3.
Experimental design and whole mount LacZ staining. (A): Time course of cell lineage mapping experiments for postnatal and adult stages. TM, tamoxifen (B, D): Typical examples of whole-mount LacZ staining of tongues in CreER−/Rosa26R (B) and CreER+/Rosa26R (D) tongues chased for 2 weeks from P30. (E, F): Anterior tongue surface (E) and soft palate (F) of CreER+/Rosa26R mice chased for 1 month from P30. Abbreviation: P, postnatal day.
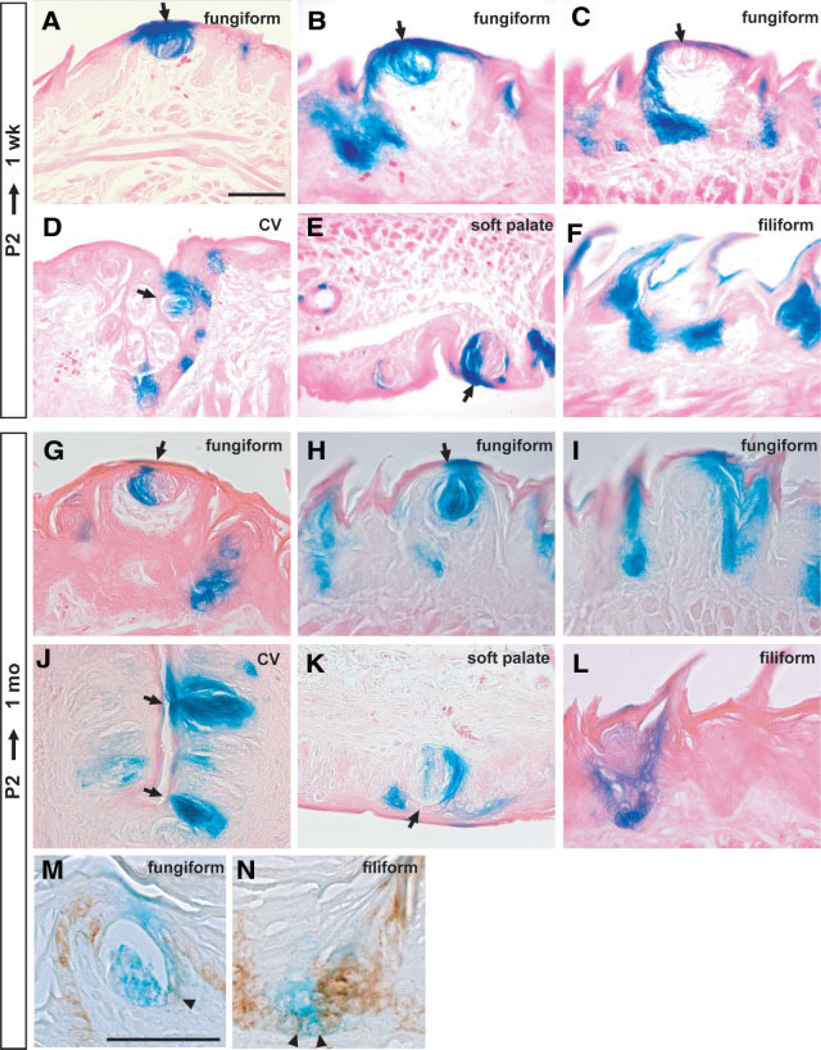
Histological sections of the tongues after 1 week showed that many fungiform TB cells were positive for LacZ activity, even though, as shown in Figure 1, these cells do not themselves express K14 (Fig. 4A, 4B). LacZ staining was also seen in keratinocytes and pore cells immediately surrounding the TBs. The fact that some TBs were completely negative (Fig. 4C) is evidence that the LacZ staining, when present, does not represent endogenous β-galactosidase activity. Similar patterns of labeling of TB cells and surrounding keratinocytes were seen in the CV and soft palate (Fig. 4D, 4E). In the tongue, LacZ+ cells were also present in both basal cells and stratified keratinocytes of many filiform papillae (Fig. 4F).
Figure 4.
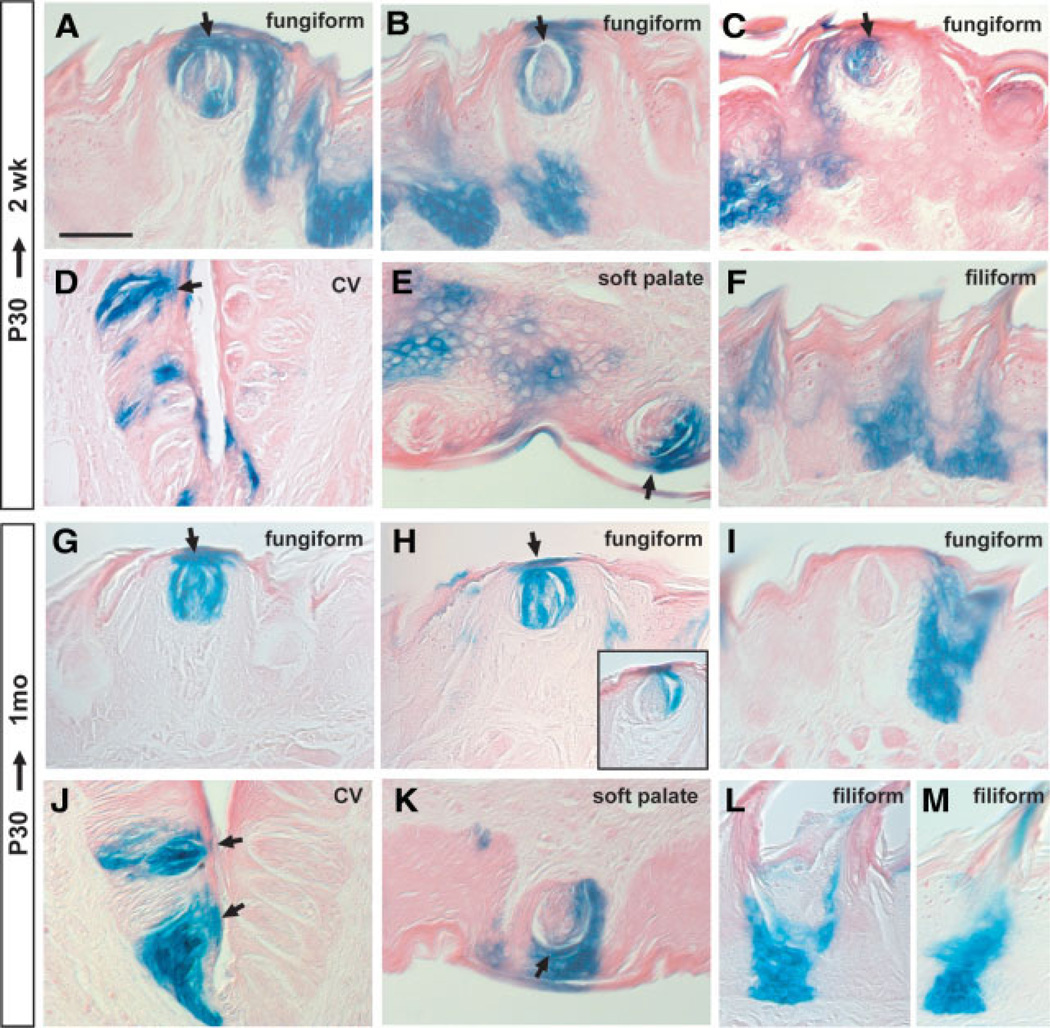
Cell lineage analysis after a 1-week chase from P2. (A–F): LacZ staining for the tongue and soft palate of K14-CreER+/Rosa26R mice dissected at 1 week after tamoxifen administration. (A–C): Different examples of fungiform papillae. LacZ staining is seen in taste bud (TB) cells and in the surrounding keratinocyte epithelial cells (pore cells). In the case of circumvallate (CV) (D) and soft palate TBs (E), examples are shown in which adjacent epithelial cells and about half of the taste receptor cells are labeled. (F): Filiform papillae. (G–L): Cell lineage analysis after a 1-month chase from P2. Shown are typical staining patterns of fungiform papillae (G–I). Note that only a subset of TB cells are stained (G). All TB receptor cells and adjacent epithelial cells are strongly stained (H). The surrounding keratinized epithelial cells are also strongly stained in the absence of TB labeling in (I). (J) TBs in CV; (K) TB in soft palate. (L): Typical labeling of filiform papillae. (M, N): K14/LacZ double staining in fungiform (M) and filiform (N) papillae. Arrows show TBs. Arrowheads show K14/LacZ coexpressing cells. Scale bar, 50 µm. Abbreviation: P, postnatal day.
When the chase period was extended to 1 month, patches of LacZ+ cells were still seen in the tongue and soft palate. Significantly, TB cells were still strongly stained, as well as perigemmal cells and adjacent keratinized epithelial cells (pore cells) (Fig. 4G, 4H). Another typical pattern of LacZ staining is illustrated in Figure 4I, which shows labeling of cells of the fungiform papilla and taste pore cells. TBs in the CV (Fig. 4J) and soft palate (Fig. 4K) are also strongly stained. As with the shorter chase period, labeled cells were present in both basal cells and stratified keratinocytes of the fungiform and filiform papillae (Fig. 4I, 4L). Expression of K14 and LacZ overlapped in some cells adjacent to the TB cells and in the basal layer of the filiform papillae (Fig. 4M, 4N).
Cell Lineage Mapping in the Adult Tongue and Soft Palate
In a second series of experiments, TM was injected five times into individual mice at P30 (Fig. 3A), and the fate of the descendants of K14+ cells in which recombination had taken place was followed after 2 weeks and 1 month from the last TM injection (Fig. 3A). This regimen of injection was adopted because control experiments showed that one or two injections of TM did not produce lineage-labeled cells (data not shown), probably due to the inefficiency of the K14-CreER transgene in tongue and soft palate tissue. Other control experiments showed that, 3 days after the last injection, only single cells or small groups of cells were labeled in the basal cells or perigemmal regions of all gustatory tissues such as the fungiform papillae, CV, and soft palate (supporting information Fig. 1A–1D). Significantly, labeling was never seen inside the TBs. After a 2-week and 1-month chase, whole-mount staining revealed patches of LacZ+ cells in both the tongue and soft palate (Fig. 3C–3E). At both times, no LacZ+ cells were seen in littermates that did not inherit the K14-CreER transgene (Fig. 3B).
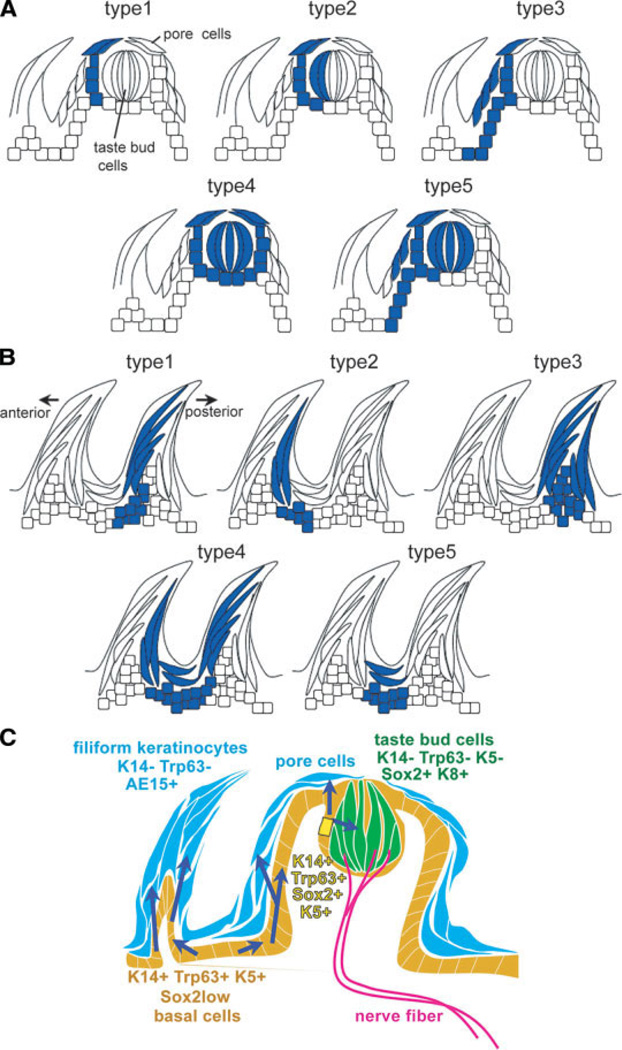
Histological analysis was carried out after both chase periods, and serial sections were examined for patterns of LacZ staining. As shown in Figure 5 and summarized in Figure 6, the patterns were very similar to those seen in the first series of experiments carried out in the early postnatal period. Significantly, even after 1 month, labeled receptor cells were found inside the TBs in all locations (fungiform and CV in the tongue and in the soft palate) (Fig. 5). The different staining patterns in the fungiform papillae and TBs were classified and quantified as shown in Figure 6A and Table 1. Typical patterns for the CV are shown in supporting information Figure 2. These results allowed several conclusions to be drawn. First, in all cases where TB cells were labeled, some positive cells were also seen in the surrounding pore cells, as would be expected if the TB cells are continually derived from K14+ bipotential cells immediately outside the bud. In no case were only the internal TB receptor cells positive and not the immediate outside (perigemmal) cells. Second, not all the internal cells (intragemmal cells) within positive TBs were labeled (Fig. 5C–5E, 5H inset, 5K), suggesting that several progenitor cells contribute descendants to a single TB [2]. Third, in the longer chases, there were few or no patches (1 of 43 examined after 1 month from P2, and 0 of 24 after 1 month from P30) in which both keratinocytes within the fungiform papillae (i.e., nonperigemmal cells) and the TBs were labeled (Fig. 6A, type 5, Table 1).
Figure 5.
Cell lineage analysis after a 2-week chase from P30. (A–F): The tongues and soft palate were dissected 2 weeks after tamoxifen injection and stained for LacZ. Shown are typical examples of labeling of taste bud (TB) cells and adjacent keratinized epithelial cells (A), adjacent epithelial cells but not TBs (B), and a subset of TB cells and adjacent perigemmal cells (C). (D): LacZ expression is seen in the TBs, basal cells, and adjacent keratinized cells in the circumvallate (CV) papilla. (E): Some TB cells are stained in the soft palate. (F): Strong LacZ staining in the base of each filiform papillae. (G–M): Cell lineage analysis after a 1-month chase from P30. Shown are various staining patterns in the fungiform papillae (G–I). LacZ expression is seen in some TB cells and in adjacent epithelial cells (H) (inset shows another example). (J): TBs in the CV papilla; (K): TB in the soft palate. (L, M): Different lineage labeling patterns are seen in the filiform papillae. Arrows show TBs. Scale bar, 50 µm. Abbreviation: P, postnatal day.
Figure 6.
A possible model based on cell lineage analyses. (A, B): Schematic representation of typical LacZ lineage labeling patterns seen in the fungiform (A) and filiform (B) papillae. (A): Type 1 shows staining only in taste pore cells and adjacent epithelial cells. Type 2 shows staining in a subset of taste bud (TB) cells and adjacent pore and epithelial cells. Type 3 shows staining in both the basal cells of the fungiform papilla and the epithelial cells around the TBs. Type 4 involves labeling of TB cells and surrounding pore cells and adjacent epithelial cells. Type 5 is a combination of Type 3 and 4. (B): Different patterns of labeling seen within the filiform papillae. (C): A possible model based on genetic mapping of keratin 14 (K14)+ cells in the tongue epithelium. Adjacent to the TBs, K14+Trp63+Sox2+K5+ cells (yellow) are bipotential progenitor cells that can generate both TB receptor cells (green) and keratinized pore cells (blue). Daughter cells committed to the TB lineage upregulate Sox2 and K8 expression, and downregulate Trp63 and K14. On the other hand, daughter cells going towards the keratinocyte lineage lose both Sox2 and Trp63 expression. In the filiform and fungiform papillae, long-term progenitor cells (K14+Trp63+K5+Sox2Low) are located in the basal layer (orange) and move up and differentiate into K14−Trp63−AE15+ filiform keratinocytes (blue).
Table 1.
Relative frequency of different LacZ staining patterns in the fungiform and filiform papillae
| Fungiform papillae |
Filiform papillae |
|||
|---|---|---|---|---|
| P2 to 1 month |
P30 to 1 month |
P2 to 1 month |
P30 to 1 month |
|
| Type 1 | 4/43 | 3/24 | 26/111 | 29/89 |
| Type 2 | 12/43 | 3/24 | 21/111 | 15/89 |
| Type 3 | 16/43 | 12/24 | 16/111 | 12/89 |
| Type 4 | 10/43 | 6/24 | 19/111 | 7/89 |
| Type 5 | 1/43 | 0/24 | 29/111 | 26/89 |
The relative frequency is the number of each type/total number of LacZ+ stained fungiform or filiform papillae. Abbreviation: P, postnatal day.
Cell Lineage Mapping in the Postnatal and Adult Filiform Papillae
In the case of the filiform papillae, several different LacZ staining patterns were seen (Figs. 4F, 4L, 5F, 5L, 5M). In most cases, the general pattern of labeling was consistent with the progenitor cells residing in the base of each conical papilla and the transit-amplifying cells moving up along the basal lamina of the walls of the filiform papilla and then exiting into the superficial layer. In some cases (Fig. 6B, type 1), the labeled cells only populated the anterior aspect of the papilla, corresponding approximately to the anterior column of cell proposed by Hume and Potten [9]. In other cases, only the posterior aspect of the papilla was labeled (Fig. 6B, type 2). In some cases (Fig. 6B, type 3, 4) the labeled cells populated both the anterior and posterior aspects of the same or adjacent papillae. Finally, examples were seen (Fig. 6B, type 5) in the which the descendants of labeled progenitor cells populated only the keratinocytes of the interpapillary column [9]. The patterns and their relative frequencies are thus generally consistent with a model in which there is little mixing of epithelial cells between the columns or between one papilla and the next.
Discussion
Long-Term Progenitor Cells of the Fungiform Papillae and Taste Buds
In this study, we used an inducible, Cre recombination-based cell lineage labeling system to trace the origin of TB cells in different gustatory tissues of postnatal and adult mice. Our results support a model in which TB cells in all locations (fungiform papillae, CV, and soft palate) and at both stages are continuously derived from K14+ perigemmal cells immediately outside the TB. Moreover, our 1-month chase studies suggest that these K14+ cells give rise to mature TB cells over the long term (~1 month). This is because the TB cells would have turned over three times within the chase period, based on the estimated turnover time of 10 days [10]. Our lineage tracing and PHH3 staining results are consistent with recent findings in the rat suggesting that TBs of the CV papilla are descendants of immediately adjacent K14+ cells that can undergo proliferation as judged by 5-bromo-2′-deoxyuridine incorporation [13]. However, these studies did not use lineage labeling and did not address which specific cell types generate TB cells over the long term. The fact that, in our studies, some TBs contain both labeled and unlabeled cells also supports previous conclusions from mosaic analyses in the mouse that more than one progenitor cell contributes to the maintenance of TBs [1, 2]. Thus, our results provide the first clear evidence that the K14+ cells that gives rise to mature TB cells are bipotential and give rise to descendants that differentiate into either TB receptor cells or surrounding keratinocytes (perigemmal and pore cells). This conclusion is based on the fact that we never saw labeled TB cells in the absence of staining of keratinocytes (pore cells) around the bud (Fig. 6A, Table 1). It is possible that the staining patterns classified as type 2 and type 4 in Figure 6A are the result of the simultaneous labeling of two adjacent progenitor cells with different potentials—one that gives rise exclusively to TB cells and one that generates only local keratinocytes/pore cells. However, two observations argue against the existence of two discrete unipotential populations immediately outside the TB. First, after a short chase period, most of the lineage-labeled cells were single cells, with only a few small clusters (supporting information Fig. 1). Second, according to this model, the type 2 and type 4 lineage-labeling patterns in the fungiform papilla should be relatively rare, while in fact they were seen more frequently than type 1, in which only keratinocytes around the TBs were labeled.
In the case of the keratinocytes making up the bulk of the fungiform papillae, the most frequent lineage-labeling pattern (Fig. 6A, type 3) suggests that these cells are derived from K14+Trp63+ cells that reside at the base of the papillae. If this model is correct, then descendants of a single progenitor cell in the base move up along the basal lamina and give rise to differentiating keratinocytes in the superficial layers. It is possible that some of the descendants of the cells in the base of the papilla also give rise to the bipotential K14+ progenitor cells around the TBs. Indeed, in short-term chase experiments (2 weeks), there are many cases of the lineage-labeling pattern (Fig. 6A, type 5) that would support this model (Figs. 4B, 5A). However, in a longer chase experiment (1 month), this pattern was only seen once, in a total of 67 fungiform papillae examined (Table 1). A more parsimonious interpretation is, therefore, that this pattern did result from the chance labeling of two progenitor cells— one in the base of the papilla and one local to the TB. To resolve this crucial question, it will be necessary to use a more efficient system than the K14-CreER inducible transgene for lineage labeling of single cells in the tongue. Further experiments will also be needed to characterize the mechanisms determining whether daughter cells give rise to receptor cells or keratinocytes. Relevant to this question, our studies on the dynamics of expression of Trp63 and K14 during development suggest that both of these genes are downregulated in the TB cells but remain on in the progenitor cells. This idea is supported by a recent enhancer analysis indicating that K14 is a direct target of Trp63 (ΔNp63) [19, 20]. In addition, we found that Sox2 expression is maintained not only in the mature TB, but also in perigemmal cells immediately outside it that also coexpress Trp63 (Figs. 1, 2). Our previous studies have shown that Sox2 plays a critical role in the development of the TB cell lineage and that when Sox2 falls below a threshold, differentiation of the TB lineage is inhibited and fungiform papilla formation is lost [5]. We therefore suggest a model in which the putative bipotential progenitor cells of the TBs are Sox2+, Trp63+, K14+, and K5+ (Fig. 6C). Daughters of these cell that move into the TB maintain a high level of Sox2 expression and switch on K8, but lose K14 and Trp63 expression as they become committed to the TB cell lineage. In contrast, other daughters that move into the keratinocyte lineage lose both Sox2 and Trp63 expression and upregulate genes associated with being pore cells.
Long-Term Progenitor Cells of the Filiform Papillae
Filiform papillae are highly organized, curved, cone-shaped epithelial structures covering the dorsal surface of the tongue. They are tilted so that the anterior face overlaps the posterior. The keratinocytes in the anterior and posterior regions express different soft or hard keratins, such as Krt1–5, Krt2–16, Krt1–17, and keratins recognized by the AE13 antibody [7, 21], possibly because the two faces of the papillae experience different stresses and pressures during the process of mastication. Furthermore, there is some evidence that the Wnt signaling pathway is differentially regulated between the anterior and posterior compartments of the filiform papillae, with higher levels of Lef1 and BAT-gal canonical Wnt reporter expression anteriorly [5, 21]. According to the model previously proposed by others [9], stem cells located in the base of the anterior compartment (position 1) give rise to transit-amplifying daughters that move up along the basal lamina and exit into the superficial layers to differentiate into keratinocytes. According to this model, there is little mixing between the compartments making up the anterior, posterior, buttress and interpapillary columns. Our lineage mapping results generally support this model and the idea that the stem cells reside in niches at the base of the papilla.
A Possible Model for the Stem Cell System of Gustatory Tissues
In conclusion, we propose a model for the location and niche of long-term stem or progenitor cells with the capacity for self-renewal and multilineage differentiation in the tongue (Fig. 6C). Adjacent to the TBs, K14+Trp63+K5+Sox2+ cells are bipotential progenitor cells that can generate both mature TB receptor cells and keratinocytes (pore cells). Daughter cells committed to the taste bud lineage keep high levels of Sox2 expression and switch on K8 expression, and downregulate basal markers such as Trp63, K14, and K5. On the other hand, daughter cells going towards the keratinocyte lineage (pore cells) lose both Sox2 and Trp63 expression. In the filiform and fungiform papillae, long-term progenitor cells located in the basal layer are characterized as Trp63+K14+K5+Sox2Low, and the daughter cells continuously move up during keratinization (Fig. 6C). We also propose that a similar model is involved in the CV papilla and soft palate. The gustatory tissue of the oral cavity thus provides an accessible experimental system to study in comparison with the stem cells of the hair follicles, epidermis, and intestinal crypt [22, 23]. Further investigations are needed to identify the local cues and transcriptional hierarchy that regulate the proliferation and differentiation capacity of the different cell types in the tongue and oral cavity.
Acknowledgments
The antibody to K8 was obtained from the Developmental Studies Hybridoma Bank. This work was supported by NIH grant HL071303 to B.L.M.H.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Author contributions: T.O.: conception and design, data collection and analysis, manuscript writing; C.C.: data collection and analysis, B.L.M.H.: conception and design, manuscript writing.
See www.StemCells.com for supporting information available online.
References
- 1.Stone LM, Finger TE, Tam PP, et al. Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proc Natl Acad Sci U S A. 1995;92:1916–1920. doi: 10.1073/pnas.92.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stone LM, Tan SS, Tam PP, et al. Analysis of cell lineage relationships in taste buds. J Neurosci. 2002;22:4522–4529. doi: 10.1523/JNEUROSCI.22-11-04522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farbman AI. Electron microscope study of the developing taste bud in rat fungiform papilla. Dev Biol. 1965;11:110–135. doi: 10.1016/0012-1606(65)90040-0. [DOI] [PubMed] [Google Scholar]
- 4.Baratz RS, Farbman AI. Morphogenesis of rat lingual filiform papillae. Am J Anat. 1975;143:283–302. doi: 10.1002/aja.1001430303. [DOI] [PubMed] [Google Scholar]
- 5.Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki Y. Expression of Sox2 in mouse taste buds and its relation to innervation. Cell Tissue Res. 2008;332:393–401. doi: 10.1007/s00441-008-0600-1. [DOI] [PubMed] [Google Scholar]
- 7.Jonker L, Kist R, Aw A, et al. Pax9 is required for filiform papilla development and suppresses skin-specific differentiation of the mammalian tongue epithelium. Mech Dev. 2004;121:1313–1322. doi: 10.1016/j.mod.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Mills AA, Zheng B, Wang XJ, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 9.Hume WJ, Potten CS. The ordered columnar structure of mouse filiform papillae. J Cell Sci. 1976;22:149–160. doi: 10.1242/jcs.22.1.149. [DOI] [PubMed] [Google Scholar]
- 10.Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delay RJ, Kinnamon JC, Roper SD. Ultrastructure of mouse vallate taste buds: II. Cell types and cell lineage. J Comp Neurol. 1986;253:242–252. doi: 10.1002/cne.902530210. [DOI] [PubMed] [Google Scholar]
- 12.Hamamichi R, Asano-Miyoshi M, Emori Y. Taste bud contains both short-lived and long-lived cell populations. Neuroscience. 2006;141:2129–2138. doi: 10.1016/j.neuroscience.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 13.Asano-Miyoshi M, Hamamichi R, Emori Y. Cytokeratin 14 is expressed in immature cells in rat taste buds. J Mol Histol. 2008;39:193–199. doi: 10.1007/s10735-007-9151-0. [DOI] [PubMed] [Google Scholar]
- 14.Vasioukhin V, Degenstein L, Wise B, et al. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong KU, Reynolds SD, Watkins S, et al. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soriano P. Generalized LacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 17.Knapp L, Lawton A, Oakley B, et al. Keratins as markers of differentiated taste cells of the rat. Differentiation. 1995;58:341–349. doi: 10.1046/j.1432-0436.1995.5850341.x. [DOI] [PubMed] [Google Scholar]
- 18.Liebl DJ, Mbiene JP, Parada LF. NT4/5 mutant mice have deficiency in gustatory papillae and taste bud formation. Dev Biol. 1999;213:378–389. doi: 10.1006/dbio.1999.9385. [DOI] [PubMed] [Google Scholar]
- 19.Candi E, Rufini A, Terrinoni A, et al. Differential roles of p63 isoforms in epidermal development: Selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 20.Romano RA, Birkaya B, Sinha S. A functional enhancer of keratin14 is a direct transcriptional target of deltaNp63. J Invest Dermatol. 2007;127:1175–1186. doi: 10.1038/sj.jid.5700652. [DOI] [PubMed] [Google Scholar]
- 21.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs E. Skin stem cells: Rising to the surface. J Cell Biol. 2008;180:273–284. doi: 10.1083/jcb.200708185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]