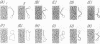Abstract
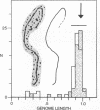
Five clonal isolates of purified polyoma defective DNA [Fried, M. (1974) J. Virol. 13, 939-946] have been examined by electron microscopy. Four isolates (D-92, D-74, D-50, and D-47) are largely homogeneous in sequence, whereas, the fifth isolate (D-80) is somewhat heterogeneous.
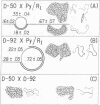
Polyoma nondefective DNA is cleaved in a unique region of the genome by the EcoRI endonuclease. Hybridization of the resulting linear molecules with randomly nicked defective DNA reveals distinguishable types of heteroduplex structures for each of the different defective DNAs. Although the defective DNAs are shorter than polyoma nondefective DNA, the heteroduplex experiments demonstrate that they are not simply deletion mutants containing only a portion of the viral genome. Three isolates (D-92, D-50, and D-47) contain regions of homology to polyoma DNA covalently linked to non-homologous regions. One isolate (D-74) contained no regions of detectable homology to polyoma DNA. Another isolate (D-80) contained a large proportion of molecules with duplicated-inverted regions. Some of these isolates of defective DNA may contain specific host sequences at the site(s) of integration of the polyoma genome during the lytic cycle in mouse cells. A process we term “abortive replication” may explain the formation of different types of defective DNAs.
Keywords: genetic deletions, electron microscopy, restriction endonuclease, DNA replication, episomes
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chow L. T., Davidson N. Electron microscope study of the structures of lambdadv DNAs. J Mol Biol. 1974 Jun 15;86(1):69–89. doi: 10.1016/s0022-2836(74)80008-2. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Syrett C., Wilde A. The replication of polyoma DNA. J Gen Virol. 1973 Dec;21(3):515–521. doi: 10.1099/0022-1317-21-3-515. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Nathans D. Bidirectional replication of Simian Virus 40 DNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3097–3100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareed G. C., Garon G. F., Salzman N. P. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol. 1972 Sep;10(3):484–491. doi: 10.1128/jvi.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M. Isolation and partial characterization of different defective DNA molecules derived from polyoma virus. J Virol. 1974 May;13(5):939–946. doi: 10.1128/jvi.13.5.939-946.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Tsai A. Direction of DNA replication in mammalian cells. J Mol Biol. 1973 Mar 25;75(1):5–12. doi: 10.1016/0022-2836(73)90525-1. [DOI] [PubMed] [Google Scholar]
- Lavi S., Winocour E. Acquisition of sequences homologous to host deoxyribonucleic acid by closed circular simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):309–316. doi: 10.1128/jvi.9.2.309-316.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson D. L., Kasamatsu H., Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A. 1972 Mar;69(3):737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne H. V. Detection of size heterogeneity in the supercoiled fraction of Polyoma virus DNA. J Mol Biol. 1968 Jul 14;35(1):215–226. doi: 10.1016/s0022-2836(68)80049-x. [DOI] [PubMed] [Google Scholar]
- Thorne H. V., Evans J., Warden D. Detection of biologically defective molecules in component I of polyoma virus DNA. Nature. 1968 Aug 17;219(5155):728–730. doi: 10.1038/219728a0. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Yoshiike K. Studies on DNA from low-density particles of SV40. I. Heterogeneous defective virions produced by successive undiluted passages. Virology. 1968 Mar;34(3):391–401. doi: 10.1016/0042-6822(68)90059-7. [DOI] [PubMed] [Google Scholar]
- Yoshike K. Studies on DNA from low-density particles of SV40. II. Noninfectious virions associated with a large-plaque variant. Virology. 1968 Mar;34(3):402–409. doi: 10.1016/0042-6822(68)90060-3. [DOI] [PubMed] [Google Scholar]