Abstract
Background
MicroRNA-21 (miRNA-21 or miR-21) may act as a prognostic biomarker of cancer. However, the available evidence is controversial. Therefore, the present meta-analysis summarizes this evidence and evaluates the prognostic role of this gene in breast cancer.
Methods
The meta-analysis was conducted by searching the databases of PubMed, EMBASE, Web of Science and Chinese database-China National Knowledge Infrastructure (CNKI). Data were extracted from studies that investigated the association between miR-21 expression and survival outcomes in breast cancer patients. With respect to survival outcomes, the pooled hazard ratios (HRs) of miR-21 were calculated given a 95% confidence interval (CI).
Results
Our meta-analysis identified a total of 10 studies involving 1,439 cases. Further investigation demonstrated that a high miR-21 expression can predict poor overall survival (OS) (HR = 2.57, 95% CI: 1.37—4.81, P = 0.003) and shortened disease-free/recurrence-free survival (DFS/RFS) (HR = 1.45, 95% CI: 1.16—1.82, P = 0.001) in breast cancer patients. Moreover, high miR-21 expression was significantly correlated with lowered OS in the Asian group (HR = 5.07, 95% CI: 2.89—8.92, P < 0.001), but not in the Caucasian cohort (HR = 1.44, 95% CI: 0.99—2.10, P = 0.058). Furthermore, odds ratios (ORs) showed that up-regulated miR-21 levels were associated with multiple clinical characteristics.
Conclusion
Our results indicated that miR-21 can predict unfavorable prognoses in breast cancer patients, especially in Asians.
Introduction
Breast cancer is the most frequently diagnosed malignancy and is the leading cause of cancer death among females. It accounts for 23% of all cancer cases and 14% of cancer deaths worldwide [1]. Approximately 232,670 women in the United States are estimated to be diagnosed with invasive breast cancer in 2014, and 40,000 women will die from it [2]. At present, mortality rates are declining in several Western countries because of the increased implementation of mammographic screening and adjuvant systemic therapies for newly diagnosed cases. However, the long-term survival rate and prognosis of advanced-stage patients remain poor, especially in developing countries [2,3]. Currently, both tissue- and serum-based tumor biomarkers are widely used to screen early-stage breast cancer and to predict either its progression or recurrence in advance. These biomarkers include human epidermal growth factor 2 receptor (Her2), estrogen receptor (ER), progesterone receptor (PR), and p53 [4]. Breast cancer is quite complex and heterogeneous in its development, progress, and response to treatment; therefore, researchers are encouraged to identify novel biomarkers for the optimization of breast cancer management.
MiRNAs are a class of 18–25 nucleotide, non-coding RNAs that are significant in the regulation of post-transcriptional gene expression [5]. Studies have shown that miRNAs are involved in virtually all biological processes, including cell proliferation, differentiation, and apoptosis [5,6]. Moreover, miRNAs act as critical regulators of tumorigenesis and are believed to be involved in tumor development and progression [7]. Calin et al. were the first to provide direct evidence of regarding the function of miRNA in human cancer. They reported that aberrant expressions of both miR-15 and miR-16 are related to chronic lymphocytic leukemia [8]. Many subsequent studies identified several miRNAs that are correlated with various human cancers [9,10]. In addition, clinical and quantitative studies determined that the expression levels of some miRNAs are associated with either the stages of diseases or with cancer survival outcomes [11,12]; thus, miRNAs may serve as potential diagnostic or prognostic biomarkers of cancer.
MiR-21 acts as an oncogene and is among the most frequently observed cancer-related miRNAs. It is consistently dysregulated in many types of cancer [13] and is a key factor in tumorigenesis and tumor suppression because it targets tumor suppressor genes such as tropomyosin 1, programmed cell death 4, and phosphatase and tensin homolog [14–17]. Recent accumulated evidence supports the concept of miR-21 as a potential diagnostic or prognostic biomarker in various types of cancer, such as colon cancer, pancreatic cancer, and lung cancer [18–20]. Nonetheless, some of these studies fail to confirm the association between miR-21 and cancer survival outcomes [21,22]; as a result, their results remain controversial. Moreover, the prognostic value of miR-21 varies in different clinical studies with respect to breast cancer [23,24]. Therefore, we conduct this meta-analysis to determine whether miR-21 can predict poor survival rates in breast cancer patients.
Materials and Methods
Search strategy and study selection
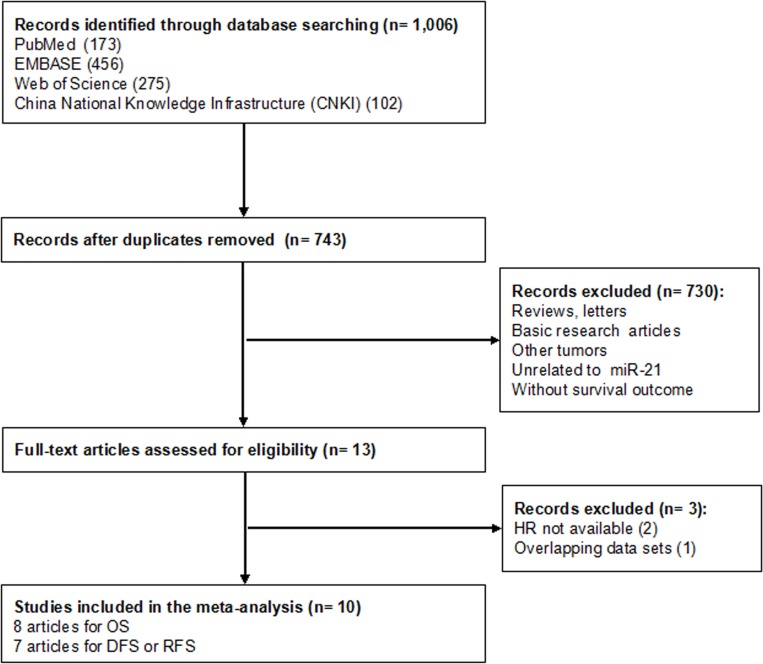
We searched the international databases (PubMed, EMBASE and Web of Science) and Chinese database-China National Knowledge Infrastructure (CNKI) carefully for relevant articles (last update: 24 November 2014). The following key words or text words were used: “breast or mammary”, “cancer or carcinoma”, “tumor or tumour”, and “microRNA-21, miR-21, or miRNA-21”. Studies were considered eligible if they met the following criteria: (i) They diagnosed breast cancer based on histopathological confirmation; (ii) they measured the miR-21 expression in either tumor tissue or serum; and (iii) they investigated the association between the expression level of miR-21 and survival outcomes. Articles were excluded based on any of the following criteria: (i) Reviews, comments, conference abstracts, letters, or basic research articles; (ii) neither English nor Chinese articles; and (iii) those that lacked key information, such as HR, 95% CI, and P value, or useful data for the calculation developed by Parmar, Williamson, and Tierney [25–27]. When multiple publications of a study were identified, we selected the most detailed version for meta-analysis. Two reviewers (YYW and YJZ) identified the qualified studies independently in accordance to the eligibility criteria. Discrepancies were adjudicated by a third reviewer (FXM) until a consensus was reached. A flow diagram of the study selection process is presented in Fig. 1.
Fig 1. Flow diagram of the study selection process.
Quality assessment
Based on a critical review checklist of the Dutch Cochrane Centre proposed by MOOSE, we systematically assessed the quality of all the studies included [28]. The key points of a qualified study include: (i) Detailed information regarding the study population and country of origin; (ii) a clear description of the study design; (iii) a clear description of outcome assessment; (iv) an adequate description of miR-21 measurement; (v) a clear description of the cut-off value of miR-21; and (vi) a sufficient follow-up period. We excluded the studies that did not meet all six of these points.
Data extraction
Two reviewers (YYW and YJZ) extracted the required information from all eligible studies independently. The extracted data included the following: (i) Publication information (name of the author cited first and the publication year); (ii) patients characteristics (age, ethnicity, sample site, stage of disease, histological grade, lymphoid node status, subtypes, Her2/ER/PR status, and follow-up); (iii) miR-21 measurement and cut-off value; and (iv) the HRs of the elevated miR-21 for overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS), as well as their 95% CIs and P values. If miR-21 expression was divided into several categories in a particular study, we combined the corresponding HR estimates using the method proposed by Hamling et al. [29]. If HRs were not provided, we calculated these values based on the total numbers of observed deaths or cancer recurrences and the numbers of samples in each group. If the Kaplan–Meier curves alone were available, we extracted data from the graphical survival plots to estimate the HRs [27]. We also e-mailed the authors of the selected articles to request for additional information and for copies of the original data required for the meta-analysis.
Statistical analysis
The heterogeneity of the combined HRs was evaluated with Cochran’s Q test and the Higgins I-squared statistic. The HRs were considered statistically heterogeneous if they displayed P < 0.05 and/or I2 > 50% [30]. Significant heterogeneities among the studies were resolved with the random-effects model (DerSimonian-Laird method) [31]. Otherwise, the fixed-effects model (Mantel-Haenszel method) was applied [32]. Subgroup analysis was conducted to determine the source of existing heterogeneity. HR values > 1 indicated significant associations with poor prognosis. We also examined the correlation between miR-21 expression and the clinical variables in breast cancer through odds ratio (OR) [33]. Publication bias was analyzed using the funnel plot in combination with Egger's test (P > 0.05 suggested a lack of publication bias). Finally, the influence of a single study on overall HR was assessed for sensitivity analysis. All analyses were conducted with “Stata: Data Analysis and Statistical Software” V12.0 (Stata Corporation, College Station, TX, U.S.).
Results
Summary of the included studies
As shown in Fig. 1, a total of 1,006 published records were initially retrieved in the databases of PubMed, EMBASE, Web of Science and CNKI, and 263 of them were excluded due to duplication. According to the exclusion criteria, 730 studies were further removed based on manual screening of the titles and abstracts. Of the remaining 13 candidate articles, two articles did not provide the essential data for the extraction of HRs and 95% CI [34,35], one study reported duplicate data that had been published previously [36]. A final total of 10 studies, 8 for OS, 7 for DFS/RFS, respectively, were considered in the meta-analysis [23,24,37–44].
The main characteristics and results of the eligible studies are summarized in Table 1 and Table 2. These studies investigated a total of 1,439 cases from China, Italy, Korea, Japan, Greece, the United States, and Germany. The patients were classified as either Asian or Caucasian according to their ethnic background. Radojicic et al. and Dong et al. only selected triple-negative breast cancer cases [24,42], Müller et al. enrolled Her2+ breast cancer patients [44], whereas other studies included mixed type of breast cancer patients. MiR-21 expression was detected by quantitative real-time polymerase chain reaction assay in all studies with 8 in cancerous tissue, 1 in bone marrow and 1 in serum. Furthermore, the cut-off values of miR-21 varied in each study. Median and mean values were extracted from eight studies, and 5.84-fold or 1.5-fold values were considered in the remaining studies. Five of these studies focused on both OS and DFS [24,37,39,40,43]; three studies mainly emphasized OS [23,41,44]; and two investigated DFS or RFS [38,42]. Notably, Müller et al. examined the serum concentrations of circulating miR-21 in breast cancer patients before and after therapy [44], thus we got 2 HR estimates for OS in one study.
Table 1. Main characteristics of enrolled studies.
| Author's name | Year | Country | Ethnicity | N | Stage | Type | Sample | Follow-up, months | HR | Results |
|---|---|---|---|---|---|---|---|---|---|---|
| Yan | 2008 | China | Asian | 113 | I-III | Mixed | FFPE | 66.2(10.4–81) | Reported | OS |
| Qian | 2009 | Italy | Caucasian | 344 | I-IV | Mixed | Frozen tissue | 86.2(8–108) | Reported | OS/DFS |
| Sun | 2009 | China | Asian | 57 | I-III | Mixed | FFPE | 56.3(6–209.8) | Reported | DFS |
| Lee | 2011 | Korea | Asian | 109 | I-III | Mixed | FFPE | 100 | Reported | OS/DFS |
| OTA | 2011 | Japan | Asian | 291 | I-IV | Mixed | Bone marrow | 61 | Reported | OS/DFS |
| Radojicic | 2011 | Greece | Caucasian | 49 | I-IV | Mixed | FFPE | 120 | AP/SC | OS/DFS |
| Walter | 2011 | USA | Caucasian | 25 | NM | TNBC | FFPE | median 35.5 | SC | OS |
| Dong | 2014 | China | Asian | 86 | I-IV | TNBC | Frozen tissue | 95 | Reported | RFS * |
| Markou | 2014 | Greece | Caucasian | 112 | Early stage | Mixed | FFPE | 148.8 | Reported | OS/DFS |
| Müller-b | 2014 | Germany | Caucasian | 127 | I-IV | Her2+ | Serum | 62.15(5.56–66.28) | SC | OS |
| Müller-a | 2014 | Germany | Caucasian | 126 | I-IV | Her2+ | Serum | 62.15(5.56–66.28) | SC | OS |
Abbreviations: FFPE, formalin-fixed paraffin-embedded; AP, author provided; SC, survival curve; NM, not mentioned; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; TNBC, triple-negative breast cancer; Müller-b, results before therapy; Müller-a, results after therapy.
*The extracted OS data is obviously not consistent with the survival curve, so only provided RFS is used for analysis.
Table 2. Main results of enrolled studies.
| Author’s name | Method | Cut-off | Survival analysis | Conclusion | HR(95% CI) for OS | P value | HR(95% CI) for DFS/RFS | P value |
|---|---|---|---|---|---|---|---|---|
| Yan | qRT-PCR | Mean: 1.741 | M | Positive | 4.13(1.80–9.50) | 0.001 | ||
| Qian | qRT-PCR | Median | M | Negative | 0.99(0.56–1.73) | NM | 1.15(0.69–1.93) | NM |
| Sun | qRT-PCR | Mean:5.04 | U | Negative | 1.81(0.96–3.41) | 0.066 | ||
| Lee | qRT-PCR | Mean: 5.92 | M | Positive | 14.21(1.34–15.10) | 0.028 | 0.88(0.09–8.41) | 0.862 |
| OTA | qRT-PCR | 5.84 | M | Positive | 3.40(1.26–9.18) | 0.035 | 1.04(0.71–1.48) | 0.853 |
| Radojicic | qRT-PCR | Median: 0.74 | KM | Negative | 0.85(0.09–8.29) | 0.337 | 2.49(0.72–8.58) | 0.379 |
| Walter | qRT-PCR | Median:5 | KM | Negative | 0.49(0.06–3.71) | 0.99 | ||
| Dong | qRT-PCR | 1.5 | U | Positive | 2.32(1.24–4.12) | 0.033 | ||
| Markou | qRT-PCR | Median | U | Positive | 1.48(0.73–2.98) | 0.274 | 2.49(1.30–4.80) | 0.006 |
| Müller-b | qRT-PCR | Median | KM | Positive | 5.24(1.58–17.35) | 0.009 | ||
| Müller-a | qRT-PCR | Median | KM | Positive | 3.40(1.01–11.38) | 0.037 |
Abbreviations: qRT-PCR, quantitative real-time PCR; M, multivariate analysis; U, Univariate analysis; KM, Kaplan–Meier analysis; NM, not mentioned; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; Müller-b, results before therapy; Müller-a, results after therapy.
Correlation between miR-21 expression and survival outcome
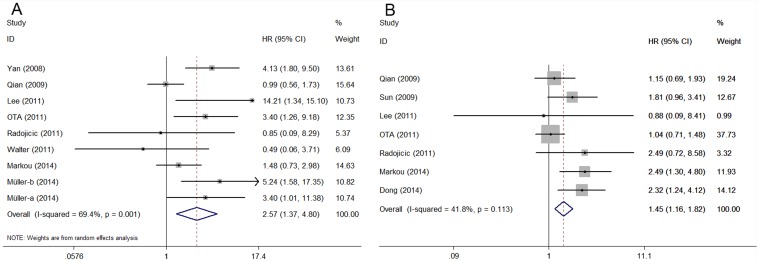
Nine OS-related data displayed heterogeneity (I2 = 69.4%, P = 0.001). Hence, we applied a random model to calculate the pooled HR and its corresponding 95% CI. A high miR-21 expression was significantly associated with poor OS, unlike low miR-21 expression (HR = 2.57, 95% CI: 1.37–4.81, P = 0.003) (Fig. 2A). To reduce the influence of heterogeneity, we conducted subgroup analysis according to ethnicity, cut-off value and types of breast cancer, respectively. In the ethnicity subgroup analysis, heterogeneity was considerably dissolved between the Asian (I2 = 45%, P = 0.162) and Caucasian groups (I2 = 46.6%, P = 0.096). Moreover, elevated miR-21 reduced the OS of Asian cancer patients (HR = 5.07, 95% CI: 2.89–8.92, P < 0.001), but not that of Caucasian ones (HR = 1.44, 95% CI: 0.99–2.10, P = 0.058). In the cut-off subgroup analysis, mean cut-off value was significantly associated with OS (HR = 7.06, 95% CI: 2.13–23.45, P = 0.001) under a random-effects model because the considerable heterogeneity among the pooled studies (I2 = 63.1%, P = 0.1). OS was not significantly lowered in the median cut-off group. In the type subgroup analysis, over-expression of miR-21 was predictive of worse OS (HR = 2.41, 95% CI: 1.09–5.32, P = 0.029) in mixed type of breast cancer patients by random model with prominent heterogeneity among studies (I2 = 77.8%, P < 0.001). Similar result was found in Her2+ group but not triple-negative group (Table 3).
Fig 2. Forest plots of the relationship between elevated miR-21 level and OS (A) and DFS/RFS (B).
Table 3. Subgroup analyses of pooled hazard ratios for overall survival with elevated miR-21 expression.
| Subgroup analysis | Data sets (number) | Number of patients | Model | HR(95% CI) | P value | Heterogeneity (I2, P-value) |
|---|---|---|---|---|---|---|
| All | 9 | 1,296 | Random | 2.57(1.37–4.81) | 0.003 | 69.4%, 0.001 |
| Ethnicity | ||||||
| Asian | 3 | 513 | Fixed | 5.07(2.89–8.92) | <0.001 | 45%, 0.162 |
| Caucasian | 6 | 783 | Fixed | 1.44(0.99–2.10) | 0.058 | 46.6%, 0.096 |
| Cut-off | ||||||
| Mean | 2 | 222 | Random | 7.06(2.13–23.45) | 0.001 | 63.1%, 0.1 |
| Median | 6 | 783 | Fixed | 1.44(0.99–2.10) | 0.058 | 46.6%, 0.096 |
| Type | ||||||
| Mixed | 6 | 994 | Random | 2.41(1.09–5.32) | 0.029 | 77.8%, <0.001 |
| Her2+ | 2 | 253 | Fixed | 4.227(1.81–9.90) | 0.001 | 0%, 0.618 |
| Triple-negative | 1 | 49 | 0.85(0.09–8.29) | 0.337 |
A fixed model was used to examine the seven studies that evaluated DFS/RFS given the absence of heterogeneity among the studies (I2 = 41.8%, P = 0.113). Similarly, miR-21 over-expression significantly predicted poor DFS/RFS in breast cancer (HR = 1.45, 95% CI: 1.16–1.82, P = 0.001) (Fig. 2B).
Correlation between miR-21 expression and clinicopathologic parameters
Based on the ORs derived from each available study, we also evaluated the correlation between miR-21 expression and some clinical characteristics, including TNM stage, lymph node metastasis, histological grade, Her2, ER, and PR status (Table 4). The association between miR-21 expression level and TNM stage was statistically significant (OR = 3.41, 95% CI: 1.92–6.05, P < 0.001). This expression level was also similarly correlated with histological grade (OR = 3.02, 95% CI: 1.95–4.70, P < 0.001. However, it was not significantly linked to lymph node metastasis (P = 0.337). Her2 status was significantly associated with high miR-21 levels (OR = 3.34, 95% CI: 1.97–5.67, P < 0.001) and ER status was negatively related to miR-21 expression (OR = 0.53, 95% CI: 0.35–0.80, P = 0.002), as well as PR status (OR = 0.49, 95% CI: 0.32–0.74, P = 0.001). Detailed information for ORs calculation was summarized in Supporting Information S1 Table.
Table 4. Meta-analyses of miR-21 Expression Classified by clinicopathologic parameters.
| Variables | Number of studies | Number of patients | Model | OR(95% CI) | P value | Heterogeneity(I2, P-value) |
|---|---|---|---|---|---|---|
| TNM stage (III/IV vs. I/II) | 3 [23,39,42] | 294 | Fixed | 3.41(1.92–6.05) | <0.001 | 0.0%, 0.469 |
| Lymph node metastasis (positive vs. negative) | 5 [23,39–42] | 608 | Random | 1.54(0.64–3.73) | 0.337 | 78.2%, 0.001 |
| Histological grad (III vs. I/II) | 4 [39–42] | 497 | Fixed | 3.02(1.95–4.70) | <0.001 | 0.0%, 0.580 |
| Her2 status (positive vs. negative) | 3 [23,39,40] | 391 | Fixed | 3.34(1.97–5.67) | <0.001 | 13.6%, 0.314 |
| ER status (positive vs. negative) | 4 [23,39–41] | 529 | Fixed | 0.53(0.35–0.80) | 0.002 | 48.0%, 0.123 |
| PR status (positive vs. negative) | 4 [23,39–41] | 524 | Fixed | 0.49(0.32–0.74) | 0.001 | 0.0%, 0.464 |
Publication bias and sensitivity analysis
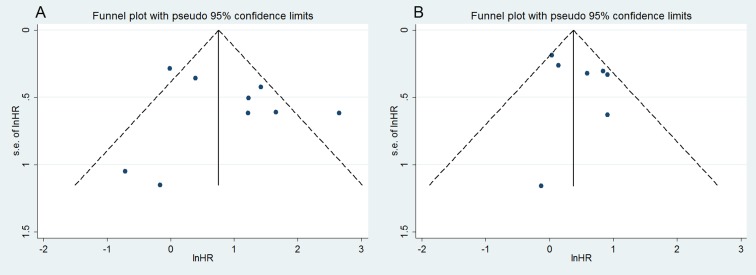
Finally, the publication bias of the included studies was evaluated through funnel plots and Egger's tests. As shown in Fig. 3, the funnel plots were almost symmetric in OS studies, as well as in DFS/RFS studies. The corresponding P values of the Egger's regression intercepts were 0.859 and 0.312, thereby indicating that the meta-analysis did not display publication bias. Meanwhile, one study was omitted to measure its effects on the pooled HR for the OS or DFS/RFS in the sensitivity analysis. No individual study dominantly influenced overall HR, as presented in Fig. 4.
Fig 3. Funnel plot for publication bias analysis: (A) OS; (B) DFS/RFS.
Fig 4. Sensitivity analysis for OS (A) and DFS/RFS (B).
Discussion
As mentioned previously, miR-21 is among the most significantly up-regulated miRNAs in many human cancers, including breast, colon, lung, prostate, ovarian, and stomach cancers [13,45]. The biological characteristics of miR-21 may explain the relationship between its expression and cancer outcome. The miR-21 gene is located in chromosome 17q23.2 within the common fragile site FRA17B. It is often amplified in numerous malignancies. MiR-21 can promote tumor development by down-regulating several tumor suppressor genes. Its identified direct targets include programmed cell death 4, tropomyosin 1, reversion-inducing cysteine-rich protein with kazal, and mapsin [14]. Si et al. showed that suppression or knock-down miR-21 in breast cancer MCF-7 cells can inhibit cell growth and induce apoptosis by downregulating the anti-apoptotic Bcl-2 [46].
Recently, a series of quantitative analyses have been carried out to identify the prognostic role of miR-21 in various cancers. Fu et al. demonstrated that high-level miR-21 predicts unfavorable overall survival in general carcinomas (HR = 1.69, 95% CI: 1.33–2.16, P < 0.001), especially head and neck squamous cell and digestive system carcinomas, moderately well on the basis of 17 studies [47]. Similar results were obtained in Zhou and Zhu’s reports, with pooled HR for OS 1.91 (95%CI: 1.66–2.19, P < 0.001) and 1.903 (95% CI: 1.713–2.113, P < 0.001), respectively [48,49]. In colorectal cancer, Xia et al. reported that higher miR-21 expression also indicates poorer survival with the combined HR to be 1.76 (95% CI: 1.34–2.32, P < 0.001) for OS [50]. However, insignificant or opposite results were also observed in some studies. In a meta-analysis of 1,163 non-small cell lung cancer (NSCLC) cases, Ma et al. showed the HR for OS is 2.19 (95% CI: 0.76–6.30, P = 0.15), which implies the miR-21 expression has limited prognostic significance on NSCLC in spite of the relationship between miR-21 expression and DFS/RFS (HR = 2.31, 95% CI: 1.52–3.49, P < 0.001) [51]. Overall, controversy concerning the prognostic role of miR-21 in cancers still exists. Only one study pooled the HR for OS in breast cancer and the dominant across-study heterogeneity, as well as small number of studies, made their results comparatively weak [48]. Here we performed a meta-analysis including 10 articles to comprehensively evaluate the risk of elevated miR-21 for survival in breast cancer patients.
Elevated miR-21 is correlated with poor survival rates (both OS and DFS) in breast cancer patients in this comprehensive meta-analysis of 1,439 cases from 10 cohorts. Nevertheless, the analysis results of DFS/RFS studies must be considered because they are only weakly significant based on their HR value of 1.45. A prognostic factor with RR > 2 is presumably useful in practice as per Hayes [52]. In addition, the results of the ethnicity subgroup analysis indicated that increased miR-21 expression significantly predicted low OS in Asians (HR = 5.07, 95% CI: 2.89–8.92, P < 0.001), whereas the prediction was not statistically significant in Caucasians (HR = 1.44, 95% CI: 0.99–2.10, P = 0.058). MiRNAs display different expression levels and predictive values across various ethnic groups [51,53]. In this sense, miR-21 is a suitable biomarker for breast cancer prognosis in Asians. Heterogeneity was not eliminated in the mean group during the cut-off subgroup analysis of OS. Moreover, only two studies used mean as the cut-off value and they adopted inconsistent mean values. Therefore, it was not appropriate to conclude that the up-regulated miR-21 significantly associated breast cancer prognosis in the mean cut-off group despite HR >2. As we all know, breast cancer is a kind of heterogeneous disease and its prognosis is closely determined by biological subtypes [54]. Hence, we performed subgroup analysis stratified by types of breast cancer. Positive result was obtained in mixed as well as Her2+ type of breast cancer patients. However, due to limited number of triple-negative and Her2+ studies, the results must be interpreted with caution.
ORs for TNM stage and histological grade were statistically significant in the correlation study of miR-21 expression on the clinical characteristics of patients, whereas the OR of lymph node metastasis was insignificant. This negative result is reasonable given the small population of each study and the diverse enrollment criteria although previous studies reported a connection between miR-21 and lymphoid infiltration in other cancers [51,55]. Furthermore, the process of metastasis was regulated by complicated molecular networks and not miR-21 alone. Currently, hormone receptors and Her2 status are important in the classification and management of breast cancer. The prognoses for Her2+ and triple-negative (Her2-/ER-/PR-) breast cancer types are usually poor because of the unique biology of the tumor itself and the lack of targeting therapy [4,56]. Our data indicated that miR-21 is positively associated with Her2 status but is negatively correlated to hormone receptors. This finding explains the low survival outcome of breast cancer patients with high miR-21 expression to some extent. However, it is noteworthy that the development of breast cancer can be influenced by the composition of tumor subtypes. In our meta-analysis, all studies for ORs calculation did not set limitations on the subtypes except Dong’s research. But no eminent difference was observed if we removed Dong’s study (Data not shown), which suggested the results were not remarkably affected by the type of breast cancer. Of importance, the association between clinical parameters and miR-21 expression need further investigation due to small number of studies and the sample size.
Although the meta-analysis determined the predictive effect of miR-21, especially in the Asian group, this study is limited in several ways. First, the marked heterogeneity of the subjects in the OS group is caused by the differences in the baseline characteristics of patients (age, tumor stage, race, or country), sample sites, the cut-off values of miR-21, the follow-up durations, and treatment strategies. A random-effect model was used in an attempt to minimize the effect of these differences because they may confound the study results. Simultaneously, we conducted subgroup analyses according to ethnic background, miR-21 cut-off value and types of breast cancer. Heterogeneity diminished significantly in the ethnicity subgroup but not in cut-off and type subgroup. These factors should be scrutinized when the conclusions concerned are considered. Second, we calculated several HRs based on data extracted from the survival curve; thus, these values may be slightly erroneous. Meanwhile, three studies reported only the unadjusted HR estimate for survival outcomes [38,42,43]; as a result, HR may have been overestimated. Third, the number of studies regarding specifically miR-21 and the prognosis of breast cancer was small, especially for ORs calculation and subgroup analysis, which was relatively insufficient to confirm the conclusion although no significant publication bias was detected in the meta-analysis. Moreover, we should carefully consider the disregard of other language articles and the preference for publications with significant results. Finally, we failed to specifically define miR-21 over-expression given the lack of uniform cut-off value although most studies defined the median as the cut-off of elevated miR-21 expression.
Furthermore, several concerns should be addressed to practically apply the prognostic value of miR-21. First, only one study detected the miR-21 expressions in serum, while others used tumor tissue or bone marrow samples. As demonstrated in other studies [57], a simultaneous detection of miRNA in serum may conveniently provide additional information regarding host response and prognosis. Hence, researchers should investigate the prognostic value of miR-21 circulation in breast cancer. Second, the lack of a golden standard for the miR-21 cut-off value complicated the exploration of its clinical application. Therefore, the cut-off value of miR-21 level should be clearly defined based on the global population. Third, the examination of a panel of miRNAs may yield more sensitive and specific results than that of a single miRNA given the complex oncogenesis process. For example, Liu et al. showed that a five-miRNA signature (miR-1, miR-20a, miR-27a, miR-34, and miR-423-5p) can serve as a better biomarker for gastric cancer screening than either a carcinoembryonic antigen or a carbohydrate antigen19-9 alone [58].
In conclusion, the present meta-analysis provided statistical evidence that miR-21 up-regulation can predict unfavorable breast cancer prognosis, especially in Asians. Our data also indicated a correlation between miR-21 expression and clinical parameters. Nonetheless, large and well-designed prospective studies should be conducted to confirm these findings before miR-21 can be implemented into routine clinical management.
Supporting Information
(DOC)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from SZZ’s National Natural Science Foundation of China (No. 91229104), www.nsfc.gov.cn. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61: 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Ma JM, Zou ZH, Jemal A. Cancer Statistics, 2014. Ca-a Cancer Journal for Clinicians. 2014; 64: 9–29. 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 3. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010; 127: 2893–2917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 4. Sturgeon CM, Duffy MJ, Stenman UH, Lilja H, Brunner N, Chan DW, et al. National Academy of Clinical Biochemistry Laboratory Medicine Practice Guidelines for Use of Tumor Markers in Testicular, Prostate, Colorectal, Breast, and Ovarian Cancers. Clinical Chemistry. 2008; 54: E11–E79. 10.1373/clinchem.2008.105601 [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 6. Ambros V. The functions of animal microRNAs. Nature. 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 7. Zhang BH, Pan XP, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Developmental Biology. 2007; 302: 1–12. [DOI] [PubMed] [Google Scholar]
- 8. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michael MZ, O'Connor SM, Pellekaan NGV, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Molecular Cancer Research. 2003; 1: 882–891. [PubMed] [Google Scholar]
- 10. Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and Biophysical Research Communications. 2005; 334: 1351–1358. [DOI] [PubMed] [Google Scholar]
- 11. Zhong H, Xu L, Zhong JH, Xiao F, Liu Q, Huang HH, et al. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Experimental and Therapeutic Medicine. 2012; 3: 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J, Zhao JQ, Shi MJ, Ding Y, Sun HQ, Yuan FH, et al. Elevated Expression of miR-210 Predicts Poor Survival of Cancer Patients: A Systematic Review and Meta-Analysis. Plos One. 2014; 9: e89223 10.1371/journal.pone.0089223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006; 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krichevsky AM, Gabriely G. MiR-21: a small multi-faceted RNA. Journal of Cellular and Molecular Medicine. 2009; 13: x–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu SM, Si ML, Wu HL, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). Journal of Biological Chemistry. 2007; 282: 14328–14336. [DOI] [PubMed] [Google Scholar]
- 16. Roy S, Yu Y, Padhye SB, Sarkar FH, Majumdar APN. Difluorinated-Curcumin (CDF) Restores PTEN Expression in Colon Cancer Cells by Down-Regulating miR-21. Plos One. 2013; 8: e68543 10.1371/journal.pone.0068543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. Journal of Biological Chemistry. 2008; 283: 1026–1033. [DOI] [PubMed] [Google Scholar]
- 18. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Jama-Journal of the American Medical Association. 2008; 299: 425–436. 10.1001/jama.299.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is Overexpressed in Pancreatic Cancer and a Potential Predictor of Survival. Journal of Gastrointestinal Surgery. 2008; 12: 2171–2176. 10.1007/s11605-008-0584-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clinical Chemistry. 2008; 54: 1696–1704. 10.1373/clinchem.2007.101741 [DOI] [PubMed] [Google Scholar]
- 21. Menendez P, Padilla D, Villarejo P, Palomino T, Nieto P, Menendez JM, et al. Prognostic Implications of Serum microRNA-21 in Colorectal Cancer. Journal of Surgical Oncology. 2013; 108: 369–373. 10.1002/jso.23415 [DOI] [PubMed] [Google Scholar]
- 22. Busacca S, Germano S, De Cecco L, Rinaldi M, Comoglio F, Faverot F, et al. MicroRNA Signature of Malignant Mesothelioma with Potential Diagnostic and Prognostic Implications. American Journal of Respiratory Cell and Molecular Biology. 2010; 42: 312–319. 10.1165/rcmb.2009-0060OC [DOI] [PubMed] [Google Scholar]
- 23. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna-a Publication of the Rna Society. 2008; 14: 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011; 10: 507–517. [DOI] [PubMed] [Google Scholar]
- 25. Parmar MKB, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine. 1998; 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 26. Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Statistics in Medicine. 2002; 21: 3337–3351. [DOI] [PubMed] [Google Scholar]
- 27. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007; 8: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology—A proposal for reporting. Jama-Journal of the American Medical Association. 2000; 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 29. Hamling J, Lee P, Weitkunat R, Ambuhl M. Facilitating meta-analyses by deriving relative effect and precision estimates for alternative comparisons from a set of estimates presented by exposure level or disease category. Statistics in Medicine. 2008; 27: 954–970. [DOI] [PubMed] [Google Scholar]
- 30. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003; 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986; 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 32. Mantel N, Haenszel W. Statistical Aspects of the Analysis of Data from Retrospective Studies of Disease. Biometrics. 1959; 15: 639–640. [PubMed] [Google Scholar]
- 33. Cornfield J. A Method of Estimating Comparative Rates from Clinical Data—Applications to Cancer of the Lung, Breast, and Cervix. Journal of the National Cancer Institute. 1951; 11: 1269–1275. [PubMed] [Google Scholar]
- 34. Anfossi S, Giordano A, Gao H, Cohen EN, Tin S, Wu Q, et al. High Serum miR-19a Levels Are Associated with Inflammatory Breast Cancer and Are Predictive of Favorable Clinical Outcome in Patients with Metastatic HER2(+) Inflammatory Breast Cancer. Plos One. 2014; 9: e83113 10.1371/journal.pone.0083113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ozgun A, Karagoz B, Bilgi O, Tuncel T, Baloglu H, Kandemir EG. MicroRNA-21 as an Indicator of Aggressive Phenotype in Breast Cancer. Onkologie. 2013; 36: 115–118. 10.1159/000348678 [DOI] [PubMed] [Google Scholar]
- 36. Yan LX, Huang MY, Wu QL, Shao JY. Abnormalities of miR-21 expression in human breast cancer is associated with distinctive pathologic features and shortened postoperative survival. Chinese Journal of Pathophysiology. 2009; 25: 676–681. [Google Scholar]
- 37. Qian BY, Katsaros D, Lu LG, Preti M, Durando A, Arisio R, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta 1. Breast Cancer Research and Treatment. 2009; 117: 131–140. 10.1007/s10549-008-0219-7 [DOI] [PubMed] [Google Scholar]
- 38.Sun B. The evaluation of curative effect of neoadjuvant chemotherapy in breast cancer patients and gene expression profiling for breast cancer prognosis. D.Ph. Thesis, The Academy of Military Medical Sciences, PLA China. 2009. Available: http://www.cnki.net/KCMS/detail/detail.aspx?QueryID=1&CurRec=1&recid=&filename=2009124589.nh&dbname=CDFD0911&dbcode=CDFD&pr=&urlid=&yx=&uid=WEEvREcwSlJHSldTTGJhYlRBMW5tM1phSGNCNW0rWk9ENXJkYVpTQ3YwdExHdkMrTE9zQzZKaTRTd1hFa2xBOUlkdz0=$9A4hF_YAuvQ5obgVAqNKPCYcEjKensW4IQMovwHtwkF4VYPoHbKxJw!!&v=MTE0NzhSOGVYMUx1eFlTN0RoMVQzcVRyV00xRnJDVVJMNmVadVZ2Rnl6aFU3ekFWMTI3RjdLNkd0VEVwcEViUEk=.
- 39. Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast. Journal of Breast Cancer. 2011; 14: 269–275. 10.4048/jbc.2011.14.4.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, Masuda N, et al. Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. International Journal of Oncology. 2011; 38: 955–962. 10.3892/ijo.2011.926 [DOI] [PubMed] [Google Scholar]
- 41. Walter BA, Gomez-Macias G, Valera VA, Sobel M, Merino MJ. miR-21 Expression in Pregnancy-Associated Breast Cancer: A Possible Marker of Poor Prognosis. Journal of Cancer. 2011; 2: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dong GZ, Liang XL, Wang DG, Gao HQ, Wang L, Wang LL, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Medical Oncology. 2014; 31: 57 10.1007/s12032-014-0057-x [DOI] [PubMed] [Google Scholar]
- 43. Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic Significance of Metastasis-Related MicroRNAs in Early Breast Cancer Patients with a Long Follow-up. Clinical Chemistry. 2014; 60: 197–205. 10.1373/clinchem.2013.210542 [DOI] [PubMed] [Google Scholar]
- 44. Muller V, Gade S, Steinbach B, Loibl S, von Minckwitz G, Untch M, et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Research and Treatment. 2014; 147: 61–68. 10.1007/s10549-014-3079-3 [DOI] [PubMed] [Google Scholar]
- 45. Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih IM, Zhang YQ, et al. MicroRNA Expression and Identification of Putative miRNA Targets in Ovarian Cancer. Plos One. 2008; 3: E2436 10.1371/journal.pone.0002436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007; 26: 2799–2803. [DOI] [PubMed] [Google Scholar]
- 47. Fu XN, Han YJ, Wu Y, Zhu XL, Lu X, Mao F, et al. Prognostic role of microRNA-21 in various carcinomas: a systematic review andmeta-analysis. European Journal of Clinical Investigation. 2011; 41: 1245–1253. 10.1111/j.1365-2362.2011.02535.x [DOI] [PubMed] [Google Scholar]
- 48. Zhou X, Wang XP, Huang ZB, Wang J, Zhu W, Shu YQ, et al. Prognostic Value of miR-21 in Various Cancers: An Updating Meta-Analysis. Plos One. 2014; 9: e102413 10.1371/journal.pone.0102413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu W, Xu B. MicroRNA-21 Identified as Predictor of Cancer Outcome: A Meta-Analysis. PloS one. 2014; 9: e103373 10.1371/journal.pone.0103373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xia XC, Yang BX, Zhai XG, Liu XY, Shen K, Wu ZJ, et al. Prognostic Role of microRNA-21 in Colorectal Cancer: a Meta-Analysis. Plos One. 2013; 8: e80426 10.1371/journal.pone.0080426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ma XL, Liu L, Liu XX, Li Y, Deng L, Xiao ZL, et al. Prognostic Role of MicroRNA-21 in Non-small Cell Lung Cancer: a Meta-analysis. Asian Pacific Journal of Cancer Prevention. 2012; 13: 2329–2334. [DOI] [PubMed] [Google Scholar]
- 52. Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: Current and new predictors of metastasis. Journal of Mammary Gland Biology and Neoplasia. 2001; 6: 375–392. [DOI] [PubMed] [Google Scholar]
- 53. Huang RS, Gamazon ER, Ziliak D, Wen YJ, Im HK, Zhang W, et al. Population differences in microRNA expression and biological implications. Rna Biology. 2011; 8: 692–701. 10.4161/rna.8.4.16029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006; 295: 2492–2502. [DOI] [PubMed] [Google Scholar]
- 55. Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007; 72: 397–402. 10.1159/000113489 [DOI] [PubMed] [Google Scholar]
- 56. Duffy MJ. Predictive markers in breast and other cancers: A review. Clinical Chemistry. 2005; 51: 494–503. [DOI] [PubMed] [Google Scholar]
- 57. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Science. 2010; 101: 2087–2092. 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu R, Zhang CN, Hu ZB, Li G, Wang C, Yang CH, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. European Journal of Cancer. 2011; 47: 784–791. 10.1016/j.ejca.2010.10.025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.