Abstract
MicroRNAs (miRNAs) play essential roles in a vast array of biological processes, including growth and development, defense against viral infection, and responses to environmental changes in plant. Wheat hybrid necrosis is an interesting genetic phenomenon observed frequency and it is lethal or semi lethal, resulting in gradual death or loss of productivity. However, the molecular basis and mechanisms associated with hybrid necrosis in wheat are still not well understood. Here, we report the population and expression profiles of miRNAs in wheat hybrid necrosis. We identified a total of 57 conserved miRNA families as well as 182 putative novel miRNAs. Expression profiling revealed that expression of 49 known miRNAs and 165 novel miRNAs was changed in hybrid necrosis. And the expression levels of some miRNAs and their predicated targets have been confirmed by qRT-PCR. These results indicate that these miRNAs, especially miR159, miR166, miR167 and miR5072 could be involved in the extensive regulation of gene expression in response to hybrid necrosis.
Introduction
MicroRNAs (miRNAs) are endogenous single-stranded noncoding RNAs (~21 nucleotides in length) that are key posttranscriptional regulator in eukaryotic gene expression by targeting mRNAs for cleavage or translational repression. More and more evidence show that miRNAs play central roles in a vast array of biological processes, including growth and development, defense against viral infection, and responses to environmental changes in eukaryotes [1–4]. In plants, the biogenesis of plant miRNAs is a complex multi-step enzymatic process [5–7]. miRNAs are transcribed by RNA polymerase II to primary miRNAs (pri-miRNAs) which are partially self-complementary and possess the fold-back hairpin structure, The pri-miRNAs are then processed to generate precursor miRNAs (pre-miRNAs) by the protein complex consisting of the Dicer-like 1 (DCL1), the C2H2-zinc finger protein SERRATE 11 (SE), and the double-stranded RNA-binding protein HYPONAS-TIC LEAVES1 (HYL1) in the cell nucleus[8,9]. The Pre-miRNAs or mature miRNAs (miRNA/miRNA* duplexes) are then exported to the cytoplasm mediated by the protein HASTY. After methyl groups are added to the ends of the duplexes catalyzed by the protein HEN1, one strand of the duplexes is selectively incorporated into the RNA-induced silencing complex (RISC) to form the mature miRNAs, whereas the other strand, designated miRNA*, is typically degraded[8, 9]. The miRNA strand is ultimately loaded into the Argonaute (AGO) protein of RNA-induced silencing complex (RISC) to carry out its function [8, 9]. Since the identification of the first miRNA from Caenorhabditis elegans through genetic screens for aberrant development [10, 11], Recently, thousands of miRNAs have been identified in various multicellular eukaryotes and are released in the miRBase database (http://www.mirbase.org/, Release 21, June 2014) [12–14]. Although hundreds of plant miRNAs and their targets have been identified, the majority of studies are still focused on model plant species, such as Arabidopsis thaliana, Oryza sativa, Zea mays and Nicotiana tabacum [12, 13, 15, 16]. To further understand the function of plant miRNAs, more efforts should be made to include plant species with specific developmental and genetic features, which might contain miRNAs that are specific to these features [17]. With the rapid development of high throughput DNA deep sequencing technology, non-conserved miRNAs from divergent plant species, including wheat [18, 19], rose [20], peanut [21], grape [22], barley [23], cucumber [24], olive [25], tomato [26], apple [27], and peach [28] have been identified.
Wheat (Triticum aestivum, AABBDD, 2n = 42) is a globally important crop, occupying 17% of all the cultivated land and accounting for 20 percent of the calories consumed by humans [29, 30], and is, therefore, of great economic importance. Hybrid necrosis has been frequently observed in F1 hybrids between genotypes of common wheat [31]. Hybrid necrosis is usually lethal or semi-lethal, resulting in gradual death or loss of productivity [32–34]. Therefore, hybrid necrosis is a serious barrier either for combining desirable traits from different genotypes of common wheat or for transferring genes from related species to commercial cultivars [32, 35]. The literature presented the hypothesis that hybrid necrosis can result from autoimmunity, perhaps as a pleiotropic effect of evolution of genes that are involved in pathogen response [36]. However, details regarding the molecular basis and mechanisms associated with hybrid necrosis in wheat are still not well understood. Recently, some miRNAs have been isolated and identified from wheat[18], [19], [37–39], but, compared with the number of miRNAs that have been identified in model plants such as Arabidopsis thaliana, Oryza sativa and Nicotiana tabacum, more miRNAs should be identified from wheat to understand the development, genetic phenomenon and stress response in plants. In the present research, we report small RNA profiling in wheat hybrid necrosis to identify new miRNAs and discover hybrid necrosis regulated miRNAs through high-throughput sequencing approach.
Results
Construction and Sequencing of Small RNA Libraries
The F1 hybrids between common wheat Neimai8 (N8) and II469 show hybrid dwarfness belonging to necrosis (S1 Fig.). To obtain a comprehensive survey of miRNAs in hybrid necrosis wheat, we constructed and sequenced two small RNA libraries from wheat F1 hybrids and its parents (mixed tissues of N8 and II469), respectively.
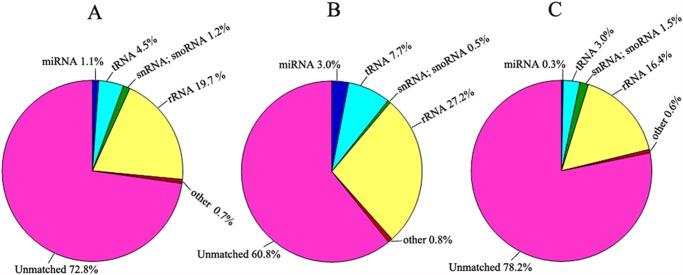
In all, 9,901,074 and 20,872,856 reads were respective obtained from the Illumina Hiseq2000 sequencing machine for the small RNA libraries of the F1 hybrids and its parents. After removing the adaptor/acceptor sequences, filtering the low-quality tags and cleaning up the contamination formed by the adaptor-adaptor ligation, total 15,344,192 clean reads were obtained, representing 1,125,608 unique sequences. Among the total reads, 166,482 were found to be similar to miRNAs. The rest of the sequences were found to be other types of RNA, including non-coding RNA, tRNA, rRNA, snRNA or snoRNA. The proportions of different categories of small RNAs are given in Fig. 1A–C.
Fig 1. Distribution of small RNAs among different categories.
(A) Total small RNAs from the libraries of F1 hybrids and their parents (N8 and II469). (B) Small RNAs from F1 hybrids. (C) Small RNAs from N8 and II469.
In addition, total 11,336,796 clean reads were mapped to genome (S1 Table). The majority of the reads were distributed on contig214090, contig87233 and contig5021661, and accounted for 5958705 reads (52% of the total reads), 3130665 reads (27.56% of the total reads) and 104166 reads (0.92% of the total reads), respectively. 519199 unique sequences from F1 sample and 470596 unique sequences from parents sample were respective mapped to genome (S1 Table).
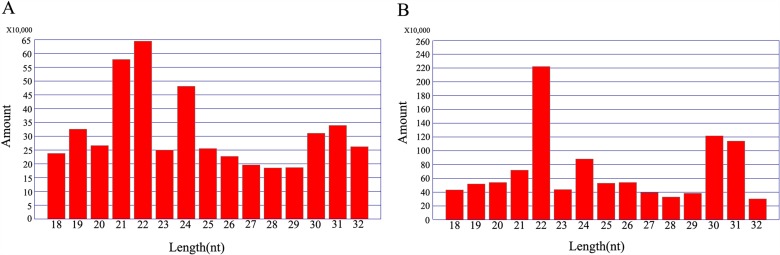
The composition of different categories of small RNAs often reflects the roles in a particular tissue or species and associated biogenetic machines. The length distribution of the small RNAs ranged from 18 to 32 nt was examined and shown in Fig. 2A–B. In F1 sample, 22-nt, 21-nt and 24-nt small RNAs were the major population, while 22-nt, 24-nt, 30-nt and 31-nt small RNAs were the major population in its parents sample. All the small RNA sequences have been deposited into NCBI SRA database under accession number: SRX500281.
Fig 2. Size distribution of small RNA (sRNA) sequences from wheat.
(A) Small RNAs from F1 hybrids. (B) Small RNAs from N8 and II469.
Identification of Conserved miRNAs
There are 24,521 hairpin precursor miRNAs and 30,424 mature miRNAs including thousands of plant miRNAs from more than 200 species deposited in the miRBase database (http://www.mirbase.org/, Release 21, June 2014)[12–14]. Here, we explore the miRNAs in our sequencing data to systematically identify both conserved and species-specific miRNAs in wheat. To identify conserved miRNAs in wheat, we aligned the small RNA sequences against the known plant mature miRNAs registered in the miRBase (Release 21: June 2014), and their corresponding precursor sequences were checked to insure the miRNAs have their expected secondary structures. On the basis of sequence similarity, our analysis revealed that a total of 57 known miRNA families were identified and listed in S2 Table. Among them, 29 families were known and well-conserved, including miR156, miR158, miR159, miR160, miR164, miR165, miR166, miR167, miR168, miR169, miR170, miR171, miR172, miR319, miR393, miR394, miR396, miR397, miR398, miR399, miR444, miR530, miR827, miR1120, miR1128, miR1432, miR1436, miR1511, miR2911. Twenty-eight families were known but not well-conserved, including miR1125, miR1127, miR1136, miR5139, miR6478, miR894, miR1318, miR5071, miR5072, miR5073, miR5077, miR5082, miR5083, miR5538, miR818, miR5049, miR5048, miR6191, miR6203, miR3630, miR4995, miR5368, miR6300, miR5054, miR5062, miR5064, miR5200, miR5203.
Interestingly, wheat has several less-conserved miRNAs which were reported previously only in monocots, such as miR5054, miR5062, miR5064, miR5200, miR5203 in Brachypodium distachyon [40], miR5049, miR5048, miR6191, miR6203 in Hordeum vulgare [23, 41], miR1318, miR5071, miR5072, miR5073, miR5077, miR5082, miR5083, miR5538, miR818 in Oryza sativa [42], miR1125, miR1127, miR1136 in wheat [18].
Identification of Novel miRNAs in wheat
The genome sequences (http://mips.helmholtz-muenchen.de/plant/wheat/uk454survey/index.jsp) [29] and EST sequences (http://wheat.pw.usda.gov/GG2/) of wheat were also used to predict the potential novel miRNAs in wheat. A total of 182 novel miRNAs were obtained and named tae-miR2131a to tae-miR2256 (S3 Table). Those miRNAs which were located different contigs and have the consensus mature sequence were characterized the same family. So these novel miRNAs belong to 126 miRNA families. 31 out of the 126 miRNA families have more than one member. For example, tae-2132 family has seven members.
In addition, the corresponding miRNA* and precursor sequence were identified for these novel miRNAs, further supporting their existence as miRNAs (S3 Table). Furthermore, the stem-loop structures of these miRNAs were also predicted (S3 Table; S2 Fig.).
miRNAs expression patterns in wheat hybrid necrosis
Knowledge of the expression patterns of miRNAs could provide clues about their functions. It has been reported that high-throughput sequencing can be used as a tool for miRNA expression profiling [39, 43]. Therefore, in this study expression profiling of some known and new identified miRNAs was also determined based on illumina sequencing data (Fig. 3A–B; S2 and S3 Tables). Compared to parents (N8 and II469), the expression profiling of both known and new miRNAs in F1 hybrids was different (Fig. 3A–B). In addition, the most normalization abundant three, miR166, miR165, miR168 reached to 98607, 79571, and 30573, respectively (S2 Table). The consensus results were also obtained from rice and wheat in previous study [39, 44, 45].
Fig 3. Distribution of miRNAs expression.
Different expression of known miRNAs (A) and novel miRNAs (B) from F1 hybrids and their parents (N8 and II469).
Compared with the conserved miRNAs, most of the novel miRNAs were relatively in low abundance (S3 Table). Among the 182 novel miRNAs, only three (Tae-2131a, Tae-2131b and Tae-2131c) had 18190 transcripts. Moreover, in order to determine the response or cause of wheat miRNAs to hybrid necrosis, we looked for the miRNAs that were up- or down- regulated in F1 hybrids compared to its parents (S3 Table). We can observed that 49 conserved miRNAs and 165 novel miRNAs were identified to be significantly relative to hybrid necrosis from S2 and S3 Tables, indicating that miRNAs could be involved in the extensive regulation of gene expression in response to hybrid necrosis.
Target predictions for wheat miRNAs
In order to better understand possible biological functions of the newly identified miRNAs as well as the known miRNAs in wheat hybrid necrosis, we searched for putative target genes using the Miranda program (http://www.miRNA.org/miRNA/home.do) with default parameters according to the wheat EST sequences database (http://wheat.pw.usda.gov/GG2/ and http://www.ncbi.nlm.nih.gov/) and the Triticeae Full-Length CDS Database, TriFLDB (http://trifldb.psc.riken.jp/v3/index.pl). Putative targets of 57 known miRNA families and 182 novel miRNAs were predicted (S2, S3, S4, S5 Tables). Among the known miRNAs, total 523 ESTs and 18 full-length cDNAs as target genes were found for miR164 while only one full-length cDNA (RFL_Contig3586) as target was predicted for miR5077 (S2 and S4 Tables). And more than 500 ESTs as target genes were respectively obtained for miR-tae2159 families, miR-tae2192 families and miR-tae2203 families while there was no EST as target for miR-tae2253 (S3 and S5 Tables). There were no more ideal or deep analysis results but the above preliminary target predictions for these miRNAs were obtained, which could be due to the limited of wheat EST sequences available in the databases and the absent of wheat genome annotation and more detailed gene annotation of the full length cDNA library.
miRNA and predicated targets validation
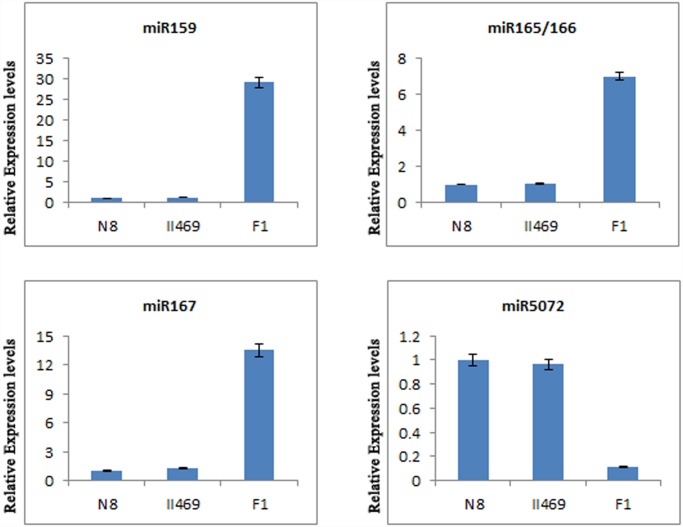
To verify the existence and expression change of the identified wheat miRNAs and predicated targets, RNA preparation subjected to quantitative RT-PCR (qRT-PCR) was the same as those used in the Illumina sequencing assay. In this study, 5 miRNAs (tae-miR159, tae-miR165/166, tae-miR167 and tae-miR5072) were validated and measured using qRT-PCR (Fig. 4). As shown in the Fig. 4, the expression changes of these miRNAs in F1 and its parents are similar to the results of Illumina sequencing. These results suggest that miRNAs had been successfully and accurately discovered from wheat hybrid necrosis with Illumina sequencing.
Fig 4. The expression levels of miR159, miR165/166, miR167 and miR5072.
The expression levels were assayed by qRT-PCR. Bars show standard error.
Moreover, 7 predicated targets (AK248335 and AK249930 for miR165/166, AJ748348_1 and AK248211 for miR159, AK248413 and RFL_Contig2908 for miR167, AK253010 for miR5072) were chosen randomly in TriFLDB (S2 Table) and validated using qRT-PCR (S3 Fig.). As shown in S3 Fig., except AK249930, there is not obvious different for the expression levels of the predicated targets in F1 compared to its parents, indicating that the expression levels of miR159, miR167, miR5072 and their predicated respective targets did not show an obvious negative correlation. There are two reasons for this situation, one is many miRNAs regulate their target(s) by translation inhibition but not slicing activity [9], and another is these predicated targets are not real targets for the miRNA.
Discussion
Hybrid necrosis was belonging to postzygotic hybrid incompatibilities which involve epistatic interactions as predicted by the Bateson-Dobzhansky-Muller (BDM) model [46]. And a further study showed hybrid incompatibility depended on activation of the salicylic acid (SA) stress signaling pathway [47]. Moreover, the hypothesis that hybrid necrosis can result from autoimmunity was also presented [36]. However, Dalal and Khanna-Chopra et al. reported that hybrid necrosis in wheat leaves was associated with oxidative stress without a well-coordinated antioxidant defense system [48, 49]. Although hybrid necrosis in wheat was first reported in the 1940s [50] and a series of classical researches revealed that this phenomenon was genetically controlled by two complementary dominant genes Ne1 and Ne2 located on chromosome arms 5BL and 2BS, respectively [34, 51–54], the molecular basis and mechanisms associated with hybrid necrosis in wheat are still not well understood.
MiRNAs have fundamental functions in regulating almost all aspects of plant development and in response to stress. Although miRNAs have been studied extensively in the past several years, only a few documents have been reported for wheat, one of the most important crops cultivated worldwide. Recently, some miRNAs have been isolated and identified from wheat [18, 19, 37–39], but the identity and function of most wheat miRNAs are still largely unknown. In the present study, we performed small RNA sequencing and determined the expression profiles of miRNAs in wheat hybrid necrosis. We identified 57 known miRNA families and 182 novel miRNAs, as well as their corresponding precursors and prediction targets. Among of them, 49 conserved miRNAs and 165 novel miRNAs were appeared to be differentially expressed during hybrid necrosis (S2 and S3 Tables), indicating that miRNAs could be involved in the extensive regulation of gene expression in response to hybrid necrosis. For conserved miRNAs, except for 15 miRNAs (highlighted in red letter in S2 Table) have been reported in wheat [18, 19, 37–39], 34 miRNAs were identified in wheat by us.
The most interesting and striking miRNAs with high changes during hybrid necrosis were miR165, miR166, miR159 and miR167 (S2 Table). Previous work showed that miR165 and miR166 were different in sequence by only a single nucleotide [55] and their same targets were thought to be the class III homeodomain leucine-zipper (HD-ZIP III) genes [56]. Over expressions of miR165 caused organ polarity alternations and defects in development of vascular tissues and inter fascicular fibers [57]. And a majority of miR166-overexpressing transformants also demonstrated a diverse array of phenotypic alternations such as downward curled leaves and stunted growth; some eventually died after the appearance of a few pairs of rosette leaves [58]. In the present study, the dramatic increase of expressions of miR165 and miR166 in F1 hybrids were observed (S2 Table, Fig. 4), suggesting that miR165 and miR166 should be responsible for wheat hybrid necrosis. Moreover, previously reports showed and confirmed that miR159 regulated the expression of a family of seven transcription factors that includes the two redundant GAMYB-like genes, MYB33and MYB65 as positive regulators of ABA responses though overexpression of miR159 [59, 60] and the loss-of-function mutations (T-DNA insertional mutants) in miR159 [61]. Overexpression of miR159a delayed the flowering of short day grown plants, as well as male sterility due to disruption of anther development [59, 60]. MiR159 over-expression also rendered plants hyposensitive to ABA [62]. And the mir159ab double mutant has pleiotropic morphological defects, including altered growth habit, curled leaves, small siliques, and small seeds [61]. Compared to parents, miR159 presented over expression in F1 hybrids (S2 Table, Fig. 4) in this study. it was reasonable for us to believe that miR159 was relative to hybrid necrosis. In addition, Yang et al. [63] and Wu et al. [64] reported that miR167 controlled patterns of the auxin responsive factor 6 (ARF6) and auxin responsive factor 8 (ARF8), and regulated both female and male reproduction in Arabidopsis and rice. Overexpressing miR167 mimicked the double mutant arf6arf8 phenotypes, such as short hypocotyls, short internode, reduced stem elongation, plant height dwarf [64]. In our study, the same phenotypes and the high increase of miR167 expression were observed in wheat F1 hybrids (S2 Table, Fig. 4), indicating that miR167 maybe play important role in wheat hybrid necrosis. Furthermore, the expression patters of many abiotic/biotic stress relative miRNAs have been sharply changed in this paper (S2 Table), such as miR169 [65], miR444 [66], miR1511 [67], miR5139 [20] and miR5368 [68] which were related to nitrogen-starvation responses, dehydration stress response, microbes response, ethylene, water deficit and rust-stress responses, respectively. Considering those above, we can propose that F1 hybrids between some different genotypes of common wheat give rise to hybrid incompatibility which triggered autoimmunity. In order to response to autoimmunity, many miRNAs expression patterns were changed to adjust their targets. Of these miRNAs were ones that regulated the expression genes which were relative to hormones, development, growth and abiotic/biotic stress. Especially, the markedly increase of expressions of miR165/miR166, miR159 and miR167 were the major cause of hybrid necrosis.
Materials and Methods
Plant materials
Hexaploid wheat (Triticum aestivum L.) cultivar Neimai8 (N8), line II469 and their (cross and reciprocal cross) F1 hybrids were grown in a growth chamber at a relative humidity of 75% and 26/20°C day and night temperature. Seven days later, these seedling plants were transferred into the growth chamber with 4°C temperature because of F1 hybrids presented much more obvious dwarfness in low temperature (4°C) than in higher temperature (above 20°C). After seven days, the whole seedling plants including leaves and roots were frozen immediately in the liquid nitrogen, and stored at -80°C for further use. The same treatment was replicated twice.
Small RNA library development and sequencing
Total RNA was isolated using TRIzol (Invitrogen, USA) and then was purified. Small RNA libraries were prepared and sequenced according to the manufacturer’s instructions and sequenced on an Illumina HiSeq2000 system (Majorbio BioTech, China).
Small RNA analysis and miRNAs Prediction
The bioinformatics analysis of small RNAs was performed as described previously with minor modification [20]. Briefly, the raw sequencing data were processed to trim the adapter sequences and remove low quality sequences, and rRNA, tRNA, snRNA, snoRNA and degradation fragments of mRNAs sequences were also removed. The cleaned small RNA sequences were aligned to the wheat genome http://mips.helmholtz-muenchen.de/plant/wheat/uk454survey/index.jsp) [29] and the wheat transcriptome database sequences (http://wheat.pw.usda.gov/GG2/) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) with perfect matches. Only sRNAs with no more than 20 hits were kept and their flanking sequences on the genome or transcriptome (200 bp on each side) were extracted and then folded in silico using the RNAfold program RNAfold (http://www.tbi.univie.ac.at/RNA/). Resulting folded structures were checked with miRdeep2 (http://www.mdc-berlin.de/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/miRDeep/index.html) with default parameters. Candidate miRNAs whose precursors passed miRdeep2 were then aligned to the miRNA database, miRBase 18.0, using Bowtie. The miRNAs shared homology with known miRNAs were identified as conserved miRNA candidates. Then, they were further confirmed by checking their corresponding precursor structures. Only the candidates with expected structures were identified as conserved miRNAs.
After identifying all candidate miRNAs, those which did not share homology to all known sequences in miRBase were regarded as novel miRNA candidates. And the novel miRNAs’ precursor structures were further analyzed by miRdeep2. Potential miRNA star sequences were identified from the sRNA data set to provide additional evidence supporting miRNA predictions.
miRNAs expression analysis
The different expression patters of the conserved miRNAs and novel miRNAs in sample F1 hybrids and its parents was carried out by DEGseq program (http://www.bioconductor.org/packages/release/bioc/html/DEGseq.html).
Prediction of miRNA targets
All of the conserved and novel rose miRNAs were aligned against wheat transcriptome dataset (http://wheat.pw.usda.gov/GG2/) using Miranda program with default parameters (http://www.miRNA.org/miRNA/home.do) and the Triticeae Full-Length CDS Database, TriFLDB(http://trifldb.psc.riken.jp/v3/index.pl).
miRNA and predicated targets validation
The identified wheat miRNAs were validated by using quantitative real time PCR (qRT-PCR). In this study, 5 miRNAs (tae-miR159, tae-miR165/166, tae-miR167 and tae-miR5072) and 7 predicated targets (AK248335, AK249930, AJ748348_1, AK248211, AK248413, RFL_Contig2908, and AK253010) were validated. The primers were listed in S6 Table.
Total RNA was extracted using TRIzol (Invitrogen, USA). Purified RNA was first treated with DNase I to remove any potential genomic DNA contamination, and then used for cDNA Synthesis.
For examination of the levels of miRNA, miRNA cDNA Synthesis was carried out using miRcute miRNA cDNA kit (Tiangen, China) according to the manufacturer’s instructions. Quantitative real time PCR was performed using the miRcute miRNA qPCR detection kit (Tiangen, China). U6 RNA was used as an internal control.
For examination of the levels of predicated target genes, reverse transcription reaction (RT) was carried out with a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA), and qRT-PCR was performed with an SuperReal PreMix Plus (SYBR Green) PCR master mix kit (Tiangen, China) according to the manufacturer’s instructions. Actin mRNA was used as an internal control.
Values were obtained by normalizing to U6 or Actin and then comparing the normalized values to those of control plants. The relative levels of gene expression were calculated using the 2-△△ cycle threshold method. Three biological replicates were examined to ensure reproducibility.
Supporting Information
(DOCX)
Red colored letter: mature miRNA sequence; yellow colored letter: loop sequence; blue colored letter: miRNA* sequence.
(ZIP)
(DOCX)
(ZIP)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31271420, No.31330017 and No. 30800685), the National Transgenic Major Project (No. 2014ZX0801003B-002) and the Applied Fundamental Research Fund of Sichuan Province (No. 2014JY0006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46: 243–259. [DOI] [PubMed] [Google Scholar]
- 2. Li T, Li H, Zhang YX, Liu JY (2011) Identification and analysis of seven H(2)O(2)-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 39:2821–2833. 10.1093/nar/gkq1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, et al. (2009) genome-wide characterization of microRNA genes in maize. PLoS Genet. 5:e10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sunkar R, Zhu JK (2004) Novel and Stress-Regulated MicroRNAs and Other Small RNAs from Arabidopsis. The Plant Cell 16(8): 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 6. Chen X (2008) MicroRNA metabolism in plants. Curr. Top. Microbiol. Immunol. 320: 117–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramachandran V, Chen X (2008) Small RNA metabolism in Arabidopsis.Trends Plant Sci. 13: 368–374. 10.1016/j.tplants.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44. 10.1146/annurev.cellbio.042308.113417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- 10. Lee RC, Feinbaum RL, Ambros V(1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complemen-tarity to lin-14. Cell 75:843–854. [DOI] [PubMed] [Google Scholar]
- 11. Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862. [DOI] [PubMed] [Google Scholar]
- 12. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39:D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res. 36:D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang S, Wang Y, Li Z, Gui Y, Xiao B, et al. (2012) Identification of wounding and topping responsive small RNAs in tobacco (Nicotiana tabacum) BMC Plant Biol. 12:28 10.1186/1471-2229-12-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li SB, Liu L, Zhuang XH, Yu Y, Liu XG, et al. (2013) MicroRNAs Inhibit the Translation of Target mRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell 153:562–574. 10.1016/j.cell.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, et al. (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18: 1602–1609. 10.1101/gr.080127.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao YY, Guo GG, Ni ZF, Sunkar R, Du JK, et al. (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biology 8: R96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dryanova A, Zakharov A, Gulick PJ (2008) Data mining for miRNAs and their targets in the Triticeae.. Genome 51(6):433–43. 10.1139/G08-025 [DOI] [PubMed] [Google Scholar]
- 20. Pei HX, Ma N, Chen JW, Zheng Y, Tian J, et al. (2013) Integrative Analysis of miRNA and mRNA Profiles in Response to Ethylene in Rose Petals during Flower Opening. PLoS One 8(5): e64290 10.1371/journal.pone.0064290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chi XY, Yang QL, Chen XP, Wang JY, Pan LJ, et al. (2011) Identification and Characterization of microRNAs from Peanut (Arachis hypogaea L.) by High-Throughput Sequencing. Plos One 6 (11): e27530 10.1371/journal.pone.0027530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mica E, Piccolo V, Delledonne M, Ferrarini A, Pezzotti M, et al. (2009) High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genomics 10: 558 10.1186/1471-2164-10-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, et al. (2013) Boron stress responsive microRNAs and their target transcripts in barley. Plose One 8(3): e59543 10.1371/journal.pone.0059543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martinez G, Forment J, Llave C, Pallas V, Gomez G (2011) High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS One 6: e19523 10.1371/journal.pone.0019523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yanik H, Turktas M, Hernandez P, Dorado G, Dundar E, et al. (2013) Genome-wide identification of alternate bearing-associated miRNA in the olive tree (Olea europaea). BMC Plant Biology 13:10 10.1186/1471-2229-13-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zuo JH, Zhu BZ, Fu DQ, Zhu Y, Ma YZ, et al. (2012) Sculpting the maturation, softening and ethylene pathway: The influences of microRNAs on tomato fruits. BMC Genomics 13: 7 10.1186/1471-2164-13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia R, Zhu H, An YQ, Beers EP, Liu ZR (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13: R47 10.1186/gb-2012-13-6-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu H, Xia R, Zhao BY, An YQ, Dardick CD, et al. (2012) Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12: 149 10.1186/1471-2229-12-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, et al. (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710. 10.1038/nature11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill BS, Appels R, Botha-Oberho lster AM, Buell CR, Bennetzen JL, et al. (2004) Aworkshop report on wheat genome sequencing: Interna-tional Genome Research on Wheat Consortium. Genetics 168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsunewaki K (1992) Aneuploid analysis of hybrid necrosis and hybrid chlorosis in tetraploid wheats using the D-genome chromosome substitution lines of durum wheat. Genome 35:594–601. [Google Scholar]
- 32. Tomar SMS, Kochumadhavan M, Nambisan PNN (1991) Hybrid weakness inTriticum dicoccum Schubl. Wheat Inf. Serv. 72:9–11. [Google Scholar]
- 33. Tomar SMS, Singh B (1998) Hybrid chlorosis in wheat X ryecrosses. Euphytica 99:1–4. [Google Scholar]
- 34. Chu CG, Faris JD, Friesen TL, Xu SS (2006) Molecular mapping of hybrid necrosis genesNe1 and Ne2 in hexaploid wheat using microsatellite markers. Theor. Appl. Genet. 112: 1374–1381. [DOI] [PubMed] [Google Scholar]
- 35. Bizimungu B, Collin J, Comeau A, St-Pierre CA (1998) Hybrid necrosis as a barrier to gene transfer in hexaploid winter wheat · triticale crosses. Can J Plant Sci. 78:239–244. [Google Scholar]
- 36. Bomblies K, Weigel D (2007) Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8(5):382–93. [DOI] [PubMed] [Google Scholar]
- 37. Wang Y, Sun FL, Cao H, Peng HR, Ni ZF, et al. (2012) TamiR159 Directed Wheat TaGAMYB Cleavage and Its Involvement in Anther Development and Heat Response. PLoS One 7(11): e48445 10.1371/journal.pone.0048445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao YY, Ni ZF, Peng HR, Sun FL, Xin MM, et al. (2010) Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.).Funct Integr Genomics 10(2): 187–190. 10.1007/s10142-010-0163-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xin MM, Wang Y, Yao YY, Xie J, Peng HR, et al. (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat(Triticum aestivum L.). BMC Plant Biology 10:123 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang JY, Xu YY, Huan Q, Chong K (2009) Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449 10.1186/1471-2164-10-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lv SZ, Nie XJ, Wang L, Du XH, Biradar SS, et al. (2012) Identification and Characterization of MicroRNAs from Barley (Hordeum vulgare L.) by High-Throughput Sequencing. Int J Mol Sci. 13:2973–2984. 10.3390/ijms13032973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei LQ, Yan LF, Wang T (2011) Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 12:R53 10.1186/gb-2011-12-6-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2:e219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V (2007) CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res 35: D829–D833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, et al. (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18:1456–1465. 10.1101/gr.075572.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Orr HA (1996) Dobzhansky, Bateson, and the genetics of speciation. Genetics 144, 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alcázar R, García AV, Parker JE, Reymond M (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106(1):334–339. 10.1073/pnas.0811734106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dalal M, Khanna-Chopra R (1999) Lipid peroxidation is an early event in necrosis of wheat hybrid. Biochem Biophys Res Commun 262:109–112. [DOI] [PubMed] [Google Scholar]
- 49. Dalal M, Khanna-Chopra R (2001) Differential response of antioxidant enzymes in leaves of necrotic wheat hybrids and their parents. Physiol Planta 111:297–304. [DOI] [PubMed] [Google Scholar]
- 50. Caldwell RM, Compton LE (1943) Complementary lethal genes in wheat causing a progressive lethal necrosis of seedlings. J Hered 34:67–70. [Google Scholar]
- 51. Hermsen JGTh (1967) Hybrid dwarfness in wheat. Euphytica 16:134–162. [Google Scholar]
- 52. Nishikawa K, Mori T, Takami N, Furuta Y (1974) Mapping of progressive necrosis gene Ne1 and Ne2 of common wheat by the telocentric method. Japan J Breed 24:277–281. [Google Scholar]
- 53. Singh S, Chaudhary HK, Sethi GS (2000) Distribution and allelic expressivity of genes for hybrid necrosis in some elite winter and spring wheat ecotypes. Euphytica 112:95–100. [Google Scholar]
- 54. Zeven AC (1972) Determination of the chromosome and its arm carrying theNe1-locus ofTriticum aestivum L., Chinese Spring and theNe1-expressivity. Wheat Inf Serv 33–34:4–6. [Google Scholar]
- 55. Liu QL, Yao XZ, Pi LM, Wang H, Cui XF, et al. (2009) The ARGONAUTE 10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 58:27–40. 10.1111/j.1365-313X.2008.03757.x [DOI] [PubMed] [Google Scholar]
- 56. Jung JH, Park CM (2007) MIR166/165genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 225: 1327–1338. [DOI] [PubMed] [Google Scholar]
- 57. Zhou GK, Kubo M, Zhong RQ, Demura T, Ye ZH (2007) Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development inArabidopsis. Plant Cell Physiol. 48: 391–404. [DOI] [PubMed] [Google Scholar]
- 58. Zhu HL, Hu FQ, Wang RH, Zhou X, Sze SH, et al. (2011) Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell 145: 242–256. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365. [DOI] [PubMed] [Google Scholar]
- 60. Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, et al. (2005) Specific Effects of MicroRNAs on the Plant Transcriptome. Developmental cell 8(4): 517–527. [DOI] [PubMed] [Google Scholar]
- 61. Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, et al. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104(41):16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reyes JL, Chua NH (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 49(4):592–606. [DOI] [PubMed] [Google Scholar]
- 63. Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34(6):1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8expression, and regulates both female and male reproduction. Development 133:4211–4218. [DOI] [PubMed] [Google Scholar]
- 65. Zhao M, Ding H, Zhu JK, Zhang FS, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190(4):906–915. 10.1111/j.1469-8137.2011.03647.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kantar M, Unver T, Budak H (2010) Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics 10:493–507. 10.1007/s10142-010-0181-4 [DOI] [PubMed] [Google Scholar]
- 67. Zhai JX, Jeong DH, De Paoli E, Park S, Rosen BD, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25:2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kulcheski FR, de Oliveira LF, Molina LG, Almerão MP, Rodrigues FA, et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12:307 10.1186/1471-2164-12-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Red colored letter: mature miRNA sequence; yellow colored letter: loop sequence; blue colored letter: miRNA* sequence.
(ZIP)
(DOCX)
(ZIP)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.