Abstract
Background
A novel fusion gene of echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma kinase (ALK) has been recently identified in non-small-cell lung cancers (NSCLCs). Patients with the EML4-ALK fusion gene demonstrate unique clinicopathological and physiological characteristics. Here we present a meta-analysis of large-scale studies to evaluate the clinicopathological characteristics of NSCLC patients harboring the EML4-ALK fusion gene.
Methods
Both English and Chinese databases were systematically used to search the materials of the clinicopathological characteristics of patients with NSCLC harboring the EML4-ALK fusion gene. Pooled relative risk (RR) estimates and the 95% confidence intervals (95% CI) were calculated with the fixed or random effect model. Publication bias and chi-square test were also calculated.
Results
27 retrospective studies were included in our meta-analysis. These studies included a total of 6950 patients. The incidence rate of EML4-ALK fusion in NSCLC patients was found to be 6.8% (472/6950). The correlation of the EML4-ALK fusion gene and clinicopathological characteristics of NSCLC patients demonstrated a significant difference in smoking status, histological types, stage, and ethnic characteristics. The positive rate of the EML4-ALK fusion gene expression in females were slightly higher than that in males, but not significantly (P = 0.52). In addition, the EML4-ALK fusion gene was mutually exclusive of the EGFR and KRAS mutation genes (P = 0.00).
Conclusion
Our pooled analysis revealed that the EML4-ALK fusion gene was observed predominantly in adenocarcinoma, non-smoking and NSCLC patients, especially those diagnosed in the advanced clinical stage of NSCLC. Additionally, the EML4-ALK fusion gene was exclusive of the EGFR and KRAS mutation genes. We surmise that IHC assay is a valuable tool for the prescreening of patients with ALK fusion gene in clinical practice, and FISH assay can be performed as a confirmation method. These insights might be helpful in guiding the appropriate molecular target therapy for NSCLC.
Introduction
Lung cancer is the most prevalent cancer and leading cause of cancer-related deaths worldwide [1]. Despite improvements in therapeutic methodology, including surgery, chemotherapy and radiotherapy, the average prognosis of lung carcinoma still remains unsatisfactory and the five year survival rate is merely 15% [2]. Among those lung cancer patients, non-small cell lung cancer (NSCLC) accounted for approximately 80%-85% of lung cancer cases [3]. Conventionally, the clinical pathological stage is the important system for predicting the survival rate in patients [4]; the recent discovery of novel molecular signal alterations also may be involved in defining a therapy, which may turn out to be more effective and with less side effects than conventional treatment.
Increased attention has been garnered in the development and application of drugs that target specific molecules which expressed on NSCLC cells and great success has been reported in NSCLC patient study groups [5,6]. These methods include signaling transduction and angiogenesis inhibitors, such as the epidermal growth factor receptor (EGFR) targeted drugs [7]. Therefore, the identification of the key ontogeny for cancer was a critical step in developing molecular-targeting agents.
In the year 2007, the fusion of echinoderm microtubule-associated protein-like 4 (EML4) genes with anaplastic lymphoma kinase (ALK) was found in lung cancer [8]. The fusion of the N-terminal half of EML4 and the intracellular kinase domain of ALK within chromosome 2p lead to expression of chimeric tyrosine kinase [9]. The EML4-ALK fusion gene possessed potent critical biological activity in vitro and in vivo, such as cell proliferation, apoptosis and metastasis [10], which can be effectively blocked by the ALK kinase inhibitor (Crizotinib) [11], which lends a supporting role for the EML4-ALK fusion gene in lung tumorigenesis. To identify patients likely to benefit from Crizotinib, it is necessary to develop a robust and effective diagnostic algorithm to detect the EML4-ALK fusion gene when screening patients for treatment with Crizotinib. Currently, the following three methodologies are used to detect the EML4-ALK fusion gene, which include: fluorescence in situ hybridization (FISH), reverse transcriptase-polymerase chain reaction (RT-PCR) and immunohistochemical (IHC). However, the best algorithm for screening the EML4-ALK fusion gene in clinical lung cancer populations remains to be determined, since the three methodologies described above have different advantages and disadvantages. To improve the detection efficiency of the three methodologies, we investigated if combining the clinicopathological characteristics of NSCLC with the EML4-ALK fusion gene would yield useful information for the effective pre-screening of patients with the EML4-ALK fusion gene in clinical practice.
Despite a large number of studies on patients harboring the EML4-ALK fusion gene demonstrating unique clinical physiological and pathological characteristics [12], detailed clinicopathological profiles remain unclear because of the small number of cases identified. To correlate the EML4-ALK fusion gene with the NSCLC profile (including smoking status, gender, tumor types, stage and ethnic characteristics) and ascertain the relationship of EML4-ALK with EGFR and KRAS mutations, we performed the present meta-analysis of 6950 patients from 27 studies.
Methods
Search Strategy
Electronic searches were performed until April 2014 and included various sources, such as MEDLINE, Embase Databases, Elsevier Science Direct, ISI Web of Science, China National Knowledge Internet, China Biology Medical Literature Database, and the Database of Chinese Scientific and Technical Periodicals. No language restrictions were applied. The keywords were as follows: “non-small cell lung cancer or NSCLC”, “echinoderm microtubule-associated protein-like 4 or EML4”, “anaplastic lymphoma kinase or ALK”, “fusion gene”, “physiological and pathological characteristics”. We searched the reference lists of relevant reviews, editorials, studies, meeting abstracts and letters. We used the Sciences Citation Index to cross reference for further studies that fulfilled the eligibility criteria.
Study selection
The studies included in this meta-analysis according to our predetermined criteria are as follows: (1) the trials that include the full text of the paper published in peer-reviewed English and Chinese journals or reports of presentations at major oncology meetings; (2) evaluation of the associations between the EML4-ALK fusion gene and clinicopathological characteristics in NSCLC patients; (3) similarity in the patients’baseline characteristics.
Data extraction and quality assessment
Two reviewers (Zhao and Lei) independently collected the data with the standard protocol. The following criteria were set to screen the articles which were eligible for our study: (1) expression of the EML4-ALK fusion gene was evaluated in primary lung cancer tissue as opposed to metastatic tissue; (2) methods used to detect the EML4-ALK fusion gene expression, including IHC, FISH or RT-PCR; (3) the histological type of the tumors was NSCLC; (4) comparison of the risk ratio (RR) and its confidence interval (CI) between patients who harbor the EML4-ALK fusion gene over expression and the counterparts were described or statistically extractable from the data in the article; (5) when multiple articles were published by the same authors or groups, the most informative or newest single article was selected. The studies were evaluated with the Downs and Black quality assessment method[13]; (6) potential disagreements were resolved by discussion and consensus with senior investigator (Xu).
Statistical and Sensitivity Analysis
This meta-analysis was performed in the RevMan 5.2 software. Statistical calculations were used with SPSS (version 17.0 SPSS Inc., IL, USA). The relative risk (RR) and the mean difference with 95% confidence intervals (95% CI) were calculated for the continuous outcomes and dichotomous outcomes, respectively. P<0.05 was considered as a significant difference in the value between the two groups. The I2 statistic was used to investigate the heterogeneity among the studies. The heterogeneity was explored by I2 and χ2, I2<50% indicated a small inconsistency and I2>50% indicated a large inconsistency. When there was a statistical difference in terms of the heterogeneity (I2>50%), the random-effect model was used to pool the data; Otherwise, a fixed-effect model was selected.
Publication bias
For publication bias estimating, we can visually observe any significant statistically symmetrical differences, according to the funnel plot.
Results
Description of the Studies
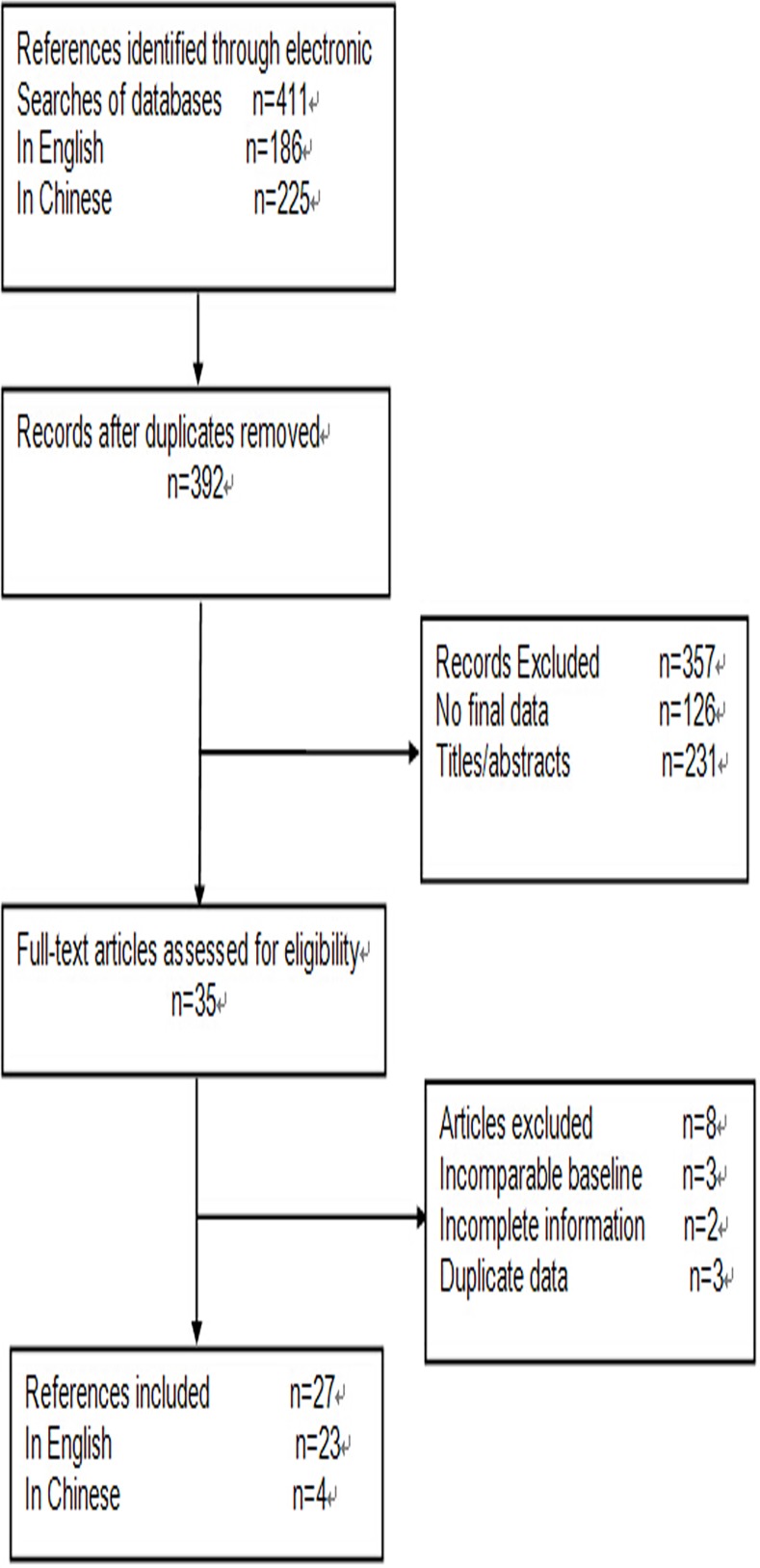
27 retrospective cohort studies coincided with our criteria and are included in this meta-analysis. A total of 6950 patients were included in the 27 studies, among which 24 (5130 cases) estimated the association of fusion of the EML4-ALK gene in NSCLC with a history of smoking; while 17 studies emphasized the association of the EML4-ALK rearrangement to tissue types (3360 cases); 13 papers reflected the relation of the EML4-ALK fusion gene to clinical stages (2876 cases) and 26 researches showed the association of this fusion gene and the gender of patients (5797 cases). To search algorithm, the results of the selection criteria and search strategies are shown in Fig. 1, and the characteristics of patients and detected methods of the EML4-ALK fusion gene are shown in Table 1.
Fig 1. Flow diagram of study selection.
Table 1. Characteristics of included studies regarding patients and detected methods.
| Author(ref.) | Year | Total | Gender | Smoking | Pathological Type | TMN(stage) | methods | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | (-) | (+) | AD | NAD | I | II | III | IV | ||||
| Inamura K [14] | 2008 | 221 | 80 | 69 | 65 | 84 | 149 | 72 | 63 | 85a | - | - | RT-PCR |
| Soda M[15] | 2007 | 75 | 22 | 11 | 9 | 24 | 18 | 15 | - | - | - | - | RT-PCR |
| Shinmura K[16] | 2008 | 77 | 39 | 38 | 41 | 22 | 50 | 27 | - | - | - | - | RT-PCR |
| Kentaro I[17] | 2008 | 253 | 134 | 119 | 142 | 110 | - | - | - | - | - | - | RT-PCR |
| Shaw AT [18] | 2009 | 141 | 48 | 93 | 59 | 82 | 89 | 52 | 25 | 1 | 9 | 96 | FISH |
| Wong DW [19] | 2009 | 266 | 132 | 134 | 141 | 125 | 209 | 57 | 153 | 47 | 60 | 6 | RT-PCR |
| Inamura K [20] | 2009 | 363 | 134 | 119 | 105 | 147 | 253 | 110 | 143 | 110a | - | - | RT-PCR, FISH |
| Martelli MP [21] | 2009 | 120 | 96 | 24 | 16 | 101 | 63 | 57 | 65 | 21 | 22 | 11 | RT-PCR |
| Rodig SJ[22] | 2009 | 358 | 138 | 220 | 85 | 243 | 358 | 0 | 169 | 29 | 67 | 93 | FISH, IHC |
| Jokoji R [23] | 2010 | 254 | 130 | 124 | 51 | 84 | - | - | - | - | - | - | IHC |
| TakahashiT [24] | 2010 | 313 | 111 | 100 | 92 | 119 | 211 | 102 | 141 | 20 | 42 | 8 | RT-PCR |
| Zhang X [25] | 2010 | 103 | 74 | 29 | 52 | 51 | 62 | 41 | 63 | 18 | 20 | 2 | RT-PCR |
| Sanders HR [26] | 2011 | 55 | - | - | - | - | 37 | 18 | - | - | - | - | RT-PCR |
| Shaw AT [27] | 2011 | 411 | 177 | 234 | 175 | 237 | 377 | 35 | - | - | - | - | FISH |
| Sequist LV [28] | 2011 | 546 | 228 | 318 | 128 | 415 | 440 | 106 | 165 | 32 | 105 | 241 | RT-PCR |
| Jin G [29] | 2012 | 167 | 85 | 82 | 73 | 94 | 121 | 46 | 93 | 74 | - | - | RT-PCR |
| Kim HR [30] | 2012 | 229 | 30 | 199 | - | - | 215 | 14 | 43 | 31 | 61 | 94 | FISH, |
| Koivunen JP [31] | 2012 | 305 | 187 | 204 | 69 | 184 | 208 | 97 | 183 | 59 | 50 | 9 | RT-PCR |
| Lin XM [32] | 2012 | 102 | 54 | 48 | 73 | 29 | 73 | 29 | 34 | 17 | 40 | 11 | RT-PCR |
| Han XH[33] | 2013 | 137 | 56 | 81 | 107 | 32 | 135 | 4 | - | - | 27 | 112 | RT-PCR, FISH, IHC |
| Takamochi k [34] | 2013 | 222 | 117 | 105 | 101 | 120 | - | - | 150 | 71a | - | - | RT-PCR, FISH, IHC |
| Zhang YG [35] | 2013 | 473 | 314 | 159 | 180 | 293 | 341 | 132 | 166 | 209b | - | 98 | RT-PCR, FISH, IHC |
| Fang P[36] | 2013 | 60 | 34 | 26 | 35 | 25 | - | - | 16 | 16 | 18 | 10 | FISH |
| Zhong S [37] | 2013 | 268 | 183 | 85 | 118 | 123 | 132 | 79 | - | - | - | - | RT-PCR |
| Wang M [38] | 2013 | 245 | 178 | 67 | 118 | 127 | 114 | 131 | 62 | 113 | 40 | 30 | IHC |
| Li Y[39] | 2013 | 208 | 147 | 61 | 78 | 130 | 95 | 113 | 49 | 43 | 106 | 10 | RT-PCR |
| Yang JJ[40] | 2014 | 977 | 182 | 212 | 308 | 86 | 377 | 17 | 75c | - | 319d | - | RT-PCR, FISH, IHC |
| Total | 6950 | 3238 | 2734 | 2035 | 2871 | 3655 | 1224 | 1734 | 404 | 561 | 821 | - | |
M:male; F:female AD: adenocarcinom; NAD: non-adenocarcinoma; a.Patients of stage II-IV; b. Patients of stage II-IIIA; c. Patients of stage I-II; d. Patients of stage III-IV; RT-PCR: real-time polymerase chain reaction; IHC: immunohistochemestry; FISH: fluorescence in situ hybridization.
Direct meta-analysis and pooled outcomes
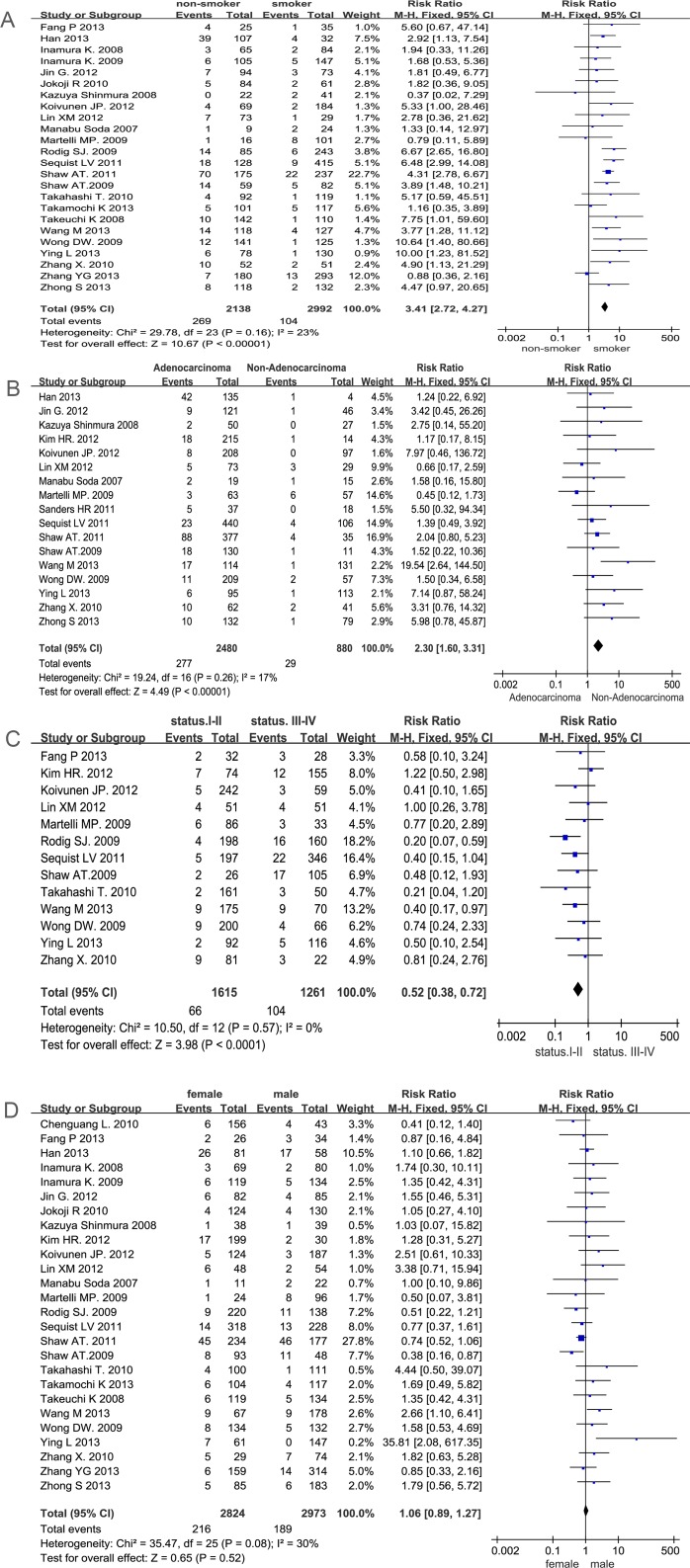
Meta-analysis of the literature revealed 27 publications, which included 6950 NSCLC patients; 472 of these patients (6.8%) harbored the EML4-ALK fusion gene. 24 studies out of 27 documented the correlation between smoking history and the EML4-ALK fusion gene. We detected no significant bias between the two groups (P = 0.16 I2 = 23%) when the fixed effects model was used. The combined result is shown in Fig. 2A. Compared with smoking cases, non-smokers with NSCLC have a statistically significant higher risk in the presence of the EML4-ALK fusion gene (12.6% vs 3.4%, RR = 3.41, 95%CI, 2.72–4.27, P<0.01). 17 studies assessed the EML4-ALK fusion gene in adenocarcinoma and non-adenocarcinoma groups, and heterogeneity was indentified through the 16 reports (P = 0.26 I2 = 17%). Then data were analyzed using a fixed effects model. The results indicated that the EML4-ALK fusion frequency is higher in the adenocarcinoma group than in the non-adenocarcinoma group (11.2% vs 3.3%, RR = 2.30, 95%CI, 1.60–3.31, P<0.01) in Fig. 2B. 13 studies expressed the association between tumor stage and the EML4-ALK fusion gene. There is no significant bias between stage I-II and stage III-IV (P = 0.57 I2 = 0%); therefore, data were analyzed using a fixed effects model. Our results suggest that there was a statistically significant increase in the frequency of EML4-ALK mutations in stage III-IV than in stage I-II (8.2% vs 4.0%, RR = 0.52, 95%CI, 0.38–0.72, P<0.01) in Fig. 2C. In addition, 26 out of these 27 studies documented the EML4-ALK fusion in female and male groups. We detected no significant bias between the two groups (P = 0.08 I2 = 30%) and analyzed the data using a fixed effects model. Our results suggest that there was no significant difference between the male and female groups. (7.6% versus 6.3%, RR = 1.06, 95%CI, 0.89–1.27, p = 0.52) in Fig. 2D.
Fig 2. Meta-analysis of data for EML4-ALK.
(A smokers vs no-smokers; B adenocarcinomas vs non-adenocarcinomas; C stages I-II vs stages III-IV; D male vs female). Forest plot of the Relative Risk (RR) of the clinicopathological characteristics with EML4-ALK fusion gene patients. The RR estimate of each individual trial corresponds to the middle of the squares and the horizontal line gives the 95% CI. On each line, the numbers of events are represented as fractions of the total number; random choices are shown for both treatment groups. For each subgroup, the sum of the statistics, along with the summary RR are represented by the middle of the solid diamonds. A test of heterogeneity between the trials within a subgroup is given below in summary of the statistics.
We also extended our study to perform three methods (FISH, RT-PCR and IHC) for direct comparison of sensitivity and specificity. Among the 3,813 patients included for the FISH study, 336 patients (8.8%,) were found to have the EML4-ALK rearrangement. Among the 5,236 patients, 280 (5.3%) were significantly positive by detection of RT-PCR. A total of 2,688 patients had successful IHC staining detection of the EML4-ALK fusion expression and 191 patients harbored the EML4-ALK fusion gene (7.1%) in Table 2.
Table 2. The detecting methods of the EML4-ALK fusion gen.
| Methods | ELM4-ALK fusion | Total | |
|---|---|---|---|
| positive | negative | ||
| FISH | 336(8.8%) | 3,477(91.2%) | 3,813 |
| RT-PCR | 280(5.3%) | 4,956(94.7%) | 5,236 |
| IHC | 191(7.1%) | 2,477(92.9%) | 2,668 |
The χ2 test indicate there was significant difference among the three diversified methods in the diagnostic detection rate of EML4-ALK fusion gene (χ2 = 21.04, p = 0.000).
In this analysis, 27 studies evaluated the EML4-ALK fusion gene in different ethnicity groups. The difference of fusion rates in Asian and non-Asian population was significant; there were 6,950 patients selected from 27 randomized trials, 4906 in Asian and 2,044 from non-Asian populations. Patients in non-Asian ethnicity groups had a higher mutation rate than those in Asian ethnicity groups (8.5% versus 6.1% χ2 = 12.80 P = 0.00) in Table 3.
Table 3. Comparison of EML4-ALK mutation rate between Asian and non-Asian.
| Group | EML4-ALK fusion | Total | |
|---|---|---|---|
| positive | Negative | ||
| Asian | 299(6.1%) | 4,607(93.9%) | 4,906 |
| non- Asian | 173(8.5%) | 1,871(91.5%) | 2,044 |
| Total | 472(6.8%) | 6,478(93.2%) | 6,950 |
The χ2 test indicated that the rate of ALK mutation in non-Asian group was statistically higher than the Asian group (χ2 = 12.8, p = 0.000).
There were 694 patients with EGFR mutations and 107 patients with KRAS mutations. Additionally, there were 214 patients with the EML4-ALK fusion gene, as summarized in Table 4. 199 patients who harbor the EML4-ALK fusion gene had wild-type EGFR and KRAS. Statistical analysis demonstrated a significant association of the EML4-ALK fusion gene with wild-type EGFR (P = 0.00 McNemar test) and KRAS (P = 0.00 McNemar test). Nevertheless, we identified 15 EML4-ALK fusion NSCLC patients in our study that showed coexistent mutations in EGFR in Table 4.
Table 4. The correlation with ALK fusion mutation and EGFR/KRAS mutation.
| Group | EGFR | Total | KRAS | Total | |||
|---|---|---|---|---|---|---|---|
| (+) | (-) | (+) | (-) | ||||
| EML4-ALK | (+) | 15(2.1%) | 146(97.9%) | 161 | 0(0%) | 53 (100%) | 53 |
| (-) | 679(12.1%) | 1,059(87.9%) | 1,738 | 107(8.3%) | 588(91.7%) | 695 | |
The McNemar test illustrated that there was a statistically significant difference in the case number between EML4-ALK fusion and EGFR mutation (p = 0.000), the outcome of KRAS followed the same pattern (p = 0.000).
Publication Bias
Publication bias can exist when non-significant findings remain unpublished. Begg’s funnel plot was performed to assess the potential publication bias in all literature. As shown in Fig. 3, the symmetric shape of funnel plots does not reveal any evidence of publication bias.
Fig 3. Funnel plot of the outcome of clinicopathological characteristics and the EML4-ALK fusion gene.
(smokers vs non-smokers; B adenocarcinomas vs non-adenocarcinomas; C stages I-II vs stages III-IV; D male vs female).
Discussion
According to tumor-specific biological characteristics, molecularly targeted therapies recently showed a new strategy that demonstrated the importance of small subgroup patients. The EML4-ALK fusion gene represents a new subgroup of NSCLC patients who respond positively to ALK inhibitors [11].
Presently, the most comprehensive meta-analysis regarding the clinical characteristics of the EML4-ALK fusion gene was done in our study. We fully analyzed 6,950 cases from 27 articles. Our findings showed a low incidence (6.8%) of the EML4-ALK translocation among unselected NSCLC patients; this proved consistent with previous reports (1.4%~11.6%) [16,17,20]. Since the incidence of EML4-ALK is low in NSCLC patients, it is necessary to elucidate clinicopathological characteristics of the EML4-ALK fusion gene-positive lung cancer to improve screening efficiency. Our results indicated that the EML4-ALK fusion gene occurred predominantly in non-smoking, adenocarcinoma patients, although no statistical difference was found between male and female patients. The likely interpretation of this phenomenon is that adenocarcinoma account for a major portion of the female patients who seldom smoke. The findings above showed that EML4-ALK fusion gene-related carcinogenesis might be different from chronic inflammation induced by smoking or tuberculosis [41]. Therefore, we believe that clinical characteristics, such as smoking status, and adenocarcinoma, should be used to select patients for EML4-ALK fusion gene screening.
Meanwhile, we found that the incidence of EML4-ALK fusion III-IV patients was slightly higher than that in stage I-II patients. We suggest that the NSCLC patients should be finished EML4-ALK fusion detection before ALK molecular inhibitor treatment. In addition, this unbalanced stage distribution could have been due to the availability of fresh frozen tissues for RT-PCR, which is more likely in operable patients (stage I and III), but difficult for stage II and IV patients. However, the incidences of EML4-ALK rearrangements in stage III patients were higher than in the other stages [25,42,43]. In Takamochi’s study, the proportion of lymph node involvement in EML4-ALK fusion gene-positive adenocarcinoma was significantly more frequent than in the negative counterpart [34]. According to Paik’s report, EML4-ALK fusion gene-positive adenocarcinomas may metastasize to lymph nodes [44], and Vincent reported that EML4-ALK fusion gene-positive tumors tend to have lymph node and brain metastases [45]. All findings stated above were potentially involved in determining why the fusion gene was more frequently seen in the advanced NSCLC.
Currently, several methodologies are used to detect EML4-ALK fusion, including FISH, RT-PCR and IHC. In our meta-analysis, the positive rate of FISH, RT-PCR and IHC was 8.8%, 5.3% and 7.1% respectively. An accurate and reliable method for the detection of EML4-ALK fusion is crucial for selecting NSCLC patients who are candidates for treatment with ALK inhibitors. Although FISH assay has been used to identify patients with the EML4-ALK fusion gene in clinical trials, a gold standard method to determine the EML4-ALK fusion gene has not been established. Our investigation revealed that the popular methods used to detect the EML4-ALK fusion gene are FISH and RT-PCR. Theoretically, RT-PCR and FISH are two approaches for detecting genes fusion; however, both have considerable limitations in clinical practice. RT-PCR required fresh tissue samples for RNA extraction and a reliable FISH assay required a good fluorescence scope and technical expertise. IHC for testing EML4-ALK fusion is a well-established method, particularly since the cost of IHC is much lower than that of FISH. So IHC could be a much more convenient and cost-effective screening method for the EML4-ALK fusion gene in NSCLC patients [41]. However, because RT-PCR methodology may not identify novel rearrangements involving previously uncharacterized EML4-ALK variants or unknown fusion partners and its process may be readily contaminated, its sensitivity and specificity remain to be validated [35]. IHC has the strengths of being widely available, relatively easy to perform and retains morphological information, which allows confident assessment of aberrant genes in tumor cells. Several ALK antibodies, reported in recent studies, shows that IHC has high concordance with FISH. Thus, these results suggested that in routine practice, IHC assay is a tool of value for the prescreening of patients with ALK fusion gene in clinical practice, and FISH assay can be performed as a confirmation method. This is consistent with previous reports [34,35,46].
It should be noted, that for first time in our studies, we determined that the EML4-ALK fusion gene appeared more frequently in non-Asian patients, as opposed to their Asian counterparts; this means that the prevalence of EML4-ALK fusion in the non-Asian population is higher than that in other ethnicities. This result indicates that the EML4-ALK fusion may be linked to non-Asian ethnicity, as opposed to EGFR mutations, which are linked to Asians. Wu recently reported that among NSCLC patients with available date on ethnicity and variant type data for the EML4-ALK fusion gene, variant 3(52.3%) was the most common type in the Chinese population, while variant 1(75.7%) was most common in the Caucasian population [47]. These results further indicated that the prevalence of the EML4-ALK fusion gene may vary amongst different ethnic groups.
Although EML4-ALK fusion and EGFR mutations were previously reported to be mutually exclusive, several studies have shown that EML4-ALK rearrangements can occur concurrently with EGFR mutations [48,49]; however, these may be rare events. Our data demonstrated that there are 15(15/6950) patients who harbored concomitant EML4-ALK fusion and EGFR mutations. In addition, comparison of EML4-ALK rearrangement with the KRAS mutations in the same NSCLC samples revealed that the EML4-ALK fusion gene was mutually exclusive of the KRAS mutations. Therefore, a stepwise mode to select for gene mutations in NSCLC is suggested: first for KRAS, second for EGFR, EML4-ALK translocation, and then for concomitant EML4-ALK fusion and EGFR mutations. If a patient is positive for a KRAS mutation, no further molecular testing will be required. The treatment approach will focus on chemotherapy, as tumors with somatic mutations in KRAS, which encodes a GTPase downstream of EGFR, exhibit greater resistance to the targeted drugs. If the patient is negative for KRAS mutations, it will be necessary to screen for EGFR mutations and the EML4-ALK fusion gene. A positive result for either will indicate molecular treatment using EGFR tyrosine kinase inhibitors (TKI) or ALK inhibitor. When patients harbor concomitant EML4-ALK fusion and EGFR mutations, treatment strategies may be helpful, since this subgroup has a specific genotype with dual therapeutical targets. As Yang’s report [40], testing of the relative phosphorylation levels of EML4-ALK and EGFR might help to guide the selection of ALK inhibitor or EGFR-TKIs in clinical practice.
Conclusion
Our analysis indicated that EML4-ALK-positive NSCLC comprised a unique subgroup of adenocarcinomas with distinct clinicopathological characteristics. We also concluded that EML4-ALK fusion was mutually exclusive of EGFR mutation KRAS mutations. Compared with non-EML4-ALK-positive NSCLC, this group is significantly enriched for non-smoking patients with adenocarcinoma. The positive rate of the EML4-ALK fusion gene expression in females was slightly higher than that in males, but not significantly. These patients typically present in late stages, which is not amenable for surgical resection. Therefore, the molecular target regimens that target the EML4-ALK fusion protein would be an effective, novel therapeutic method for them. Our studies represent EML4-ALK fusion based on the unique clinicopathological characteristics. In addition, HC assay is a tool of value for the prescreening of patients with ALK fusion gene in clinical practice, and FISH assay can be performed as a confirmation method.
Supporting Information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Natural Science Foundation of China (81273814). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 1:69–90. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. (2006) Cancer statistics. Cancer J Clin 56: 106–130 [DOI] [PubMed] [Google Scholar]
- 3. Ettinger DS, Bepler G, Bueno R, Chang JY, Chirieac LR, et al. (2006) Non-small cell lung cancer clinical practice guidelines in oncology. J Natl Compr Canc Netw 4:548–582. [DOI] [PubMed] [Google Scholar]
- 4. Greene FL, Sobin LH (2002) The TNM system: our language for cancer care. J Surg Oncol 80:119–20. [DOI] [PubMed] [Google Scholar]
- 5. Miller VA, Kris MG, Shah N, Patel J, Azzoli C, et al. (2004) Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol 22: 1103–1109. [DOI] [PubMed] [Google Scholar]
- 6. Tiseo M, Gelsomino F, Boggiani D, Bortesi B, Bartolotti M, et al. (2011) EGFR and EML4-ALK gene mutations in NSCLC: a case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer 71: 241–243. 10.1016/j.lungcan.2010.11.014 [DOI] [PubMed] [Google Scholar]
- 7. Giuseppe G (2002) Targeted therapy in non-small cell lung cancer. Lung Cancer 38: S29–S32. [DOI] [PubMed] [Google Scholar]
- 8. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 9. Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G, et al. (2008) The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer 8:11–23. [DOI] [PubMed] [Google Scholar]
- 10. Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, et al. (2008) A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci USA 105:19893–19897. 10.1073/pnas.0805381105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, et al. (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363:1693–1703. 10.1056/NEJMoa1006448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Solomon B, Varella-Garcia M, Camidge DR (2009) ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol 4: 1450–1454. 10.1097/JTO.0b013e3181c4dedb [DOI] [PubMed] [Google Scholar]
- 13. Downs SH, Black N (1998) The feasibility of creating a checklist for the assessment of the methodological quality both of randomized and non-randomised studies of health care interventions. J Epidemiol Community Health 52:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inamura K, Takeuchi K, Togashi Y, Nomura K, Ninomiya H, et al. (2008) EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 3:13–17. 10.1097/JTO.0b013e31815e8b60 [DOI] [PubMed] [Google Scholar]
- 15. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 16. Shinmura K, Kageyama S, Tao H, Bunai T, Suzuki M, et al. (2008) EML4- ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 61: 163–169. 10.1016/j.lungcan.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 17. Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, et al. (2008) Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 14: 6618–6624. 10.1158/1078-0432.CCR-08-1018 [DOI] [PubMed] [Google Scholar]
- 18. Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, et al. (2011) Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 12:1004–1012. 10.1016/S1470-2045(11)70232-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, et al. (2009) The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 115:1723–1733. 10.1002/cncr.24181 [DOI] [PubMed] [Google Scholar]
- 20. Inamura K, Takeuchi K, Togashi Y, Hatano S, Ninomiya H, et al. (2009) EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 22:508–515. 10.1038/modpathol.2009.2 [DOI] [PubMed] [Google Scholar]
- 21. Martelli MP, Sozzi G, Hernandez L, Pettirossi V, Navarro A, et al. (2009) EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol 174:661–670. 10.2353/ajpath.2009.080755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, et al. (2009) Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 15:5216–5223. 10.1158/1078-0432.CCR-09-0802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jokoji R, Yamasaki T, Minami S, Komuta K, Sakamaki Y, et al. (2010) Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol 63: 1066–1070. 10.1136/jcp.2010.081166 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, et al. (2010) Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 17:889–897. 10.1245/s10434-009-0808-7 [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Zhang S, Yang X, Yand J, Zhou Q, et al. (2010) Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 9:1–12. 10.1186/1476-4598-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanders HR, Li HR, Bruey JM, Scheerle JA, Meloni-Ehrig AM, et al. (2011) Exon scanning by reverse transcriptase-polymerase chain reaction for detection of known and novel EML4-ALK fusion variants in non-small cell lung cancer. Cancer Genet 204: 45–52. 10.1016/j.cancergencyto.2010.08.024 [DOI] [PubMed] [Google Scholar]
- 27. Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, et al. (2009) Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27:4247–4253. 10.1200/JCO.2009.22.6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, et al. (2011) Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 22:2616–2624. 10.1093/annonc/mdr489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin G, Jeon HS, Lee EB, Kang HR, Yoo SS, et al. (2012) EML4-ALK fusion gene in Korean non-small cell lung cancer. J Korean Med Sci 27:228–230. 10.3346/jkms.2012.27.2.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, et al. (2012) Distinct clinical features and outcomes in never-smokers with non-small cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer 118:729–739. 10.1002/cncr.26311 [DOI] [PubMed] [Google Scholar]
- 31. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, et al. (2008) EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 14:4275–4283. 10.1158/1078-0432.CCR-08-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin XM, Mo JM, Zou M, Zeng AP, Yu QT, et al. (2012) Detection of EML4-ALK fusion gene and analysis of its clinical features in NSCLC patients with EGFR mutation. Chinese Journal of Pathophysiology 28:1135–1139. [Google Scholar]
- 33. Han XH, Zhang NN, Ma L, Lin DM, Hao XZ, et al. (2013) Immunohistochemistry reliably detects ALK rearrangements in patients with advanced non-small-cell lung cancer. Virchows Arch 463:583–591. 10.1007/s00428-013-1472-7 [DOI] [PubMed] [Google Scholar]
- 34. Takamochi K, Takeuchi K, Hayashi T, Oh S, Suzuki K (2013) A rational diagnostic algorithm for the identification of ALK rearrangement in lung cancer: A comprehensive study of surgically treated Japanese patients. PLoS One 8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang YG, Jin ML, Li L, Zhao HY, Zeng X, et al. (2013) Evaluation of ALK rearrangement in Chinese non-small cell lung cancer using FISH, Immunohistochemistry, and Real-Time Quantitative RT-PCR on paraffin-embedded tissues. PLoS One 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang P, Feng GH, Lin L, Hu B, Wu H, et al. (2013) Expression of EML4-ALK and EGFR mutation in lung ademocarcinoma. J Med Res 42:109–112. [Google Scholar]
- 37. Zhong S, Zhang HP, Zheng J (2013) Detection of EMIA-ALK fusion gene in non-small cell lung cancer and its clinicopathologic correlation. Chin J Pathol 42:252–256. [DOI] [PubMed] [Google Scholar]
- 38. Wang M, Yang JY, Li JC, Chen YX (2013) Expression of EML4-ALK in non-small cell lung cancer and its clinical significance. Chinese Clinical Oncology 18:688–690. [Google Scholar]
- 39. Li Y, Li YG, Yang T, Wei S, Wang J, et al. (2013) Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang JJ, Zhang XC, Su J, Xu CR, Zhou Q, et al. (2014) Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res 20:1383–1392. 10.1158/1078-0432.CCR-13-0699 [DOI] [PubMed] [Google Scholar]
- 41. Nalbandian A, Yan BS, Pichugin A, Bronson RT, Kramnik I (2009) Lung carcinogenesis induced by chronic tuberculosis infection: the experimental model and genetic control. Oncogene 28:1928–1938. 10.1038/onc.2009.32 [DOI] [PubMed] [Google Scholar]
- 42. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, et al. (2008) EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 14: 4275–4283. 10.1158/1078-0432.CCR-08-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, et al. (2009) The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 115: 1723–1733. 10.1002/cncr.24181 [DOI] [PubMed] [Google Scholar]
- 44. Paik JH, Choi CM, Kim H, Jang SJ, Choe G, et al. (2012) Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer 76: 403–409. 10.1016/j.lungcan.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 45. Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, et al. (2013) Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 15:415–453 [DOI] [PubMed] [Google Scholar]
- 46. Fallet V, Cadranel J, Doubre H, Toper C, Monnet I, et al. (2014) Prospective screening for ALK: Clinical features and outcome according to ALK status. Eur J Cancer 50:1239–1246. 10.1016/j.ejca.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 47. Wu YC, Chang IC, Wang CL, Chen TD, Chen YT, et al. (2013) Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS One 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuo YW, Wu SG, Ho CC, Shih JY (2010) Good response to gefitinib in lung adenocarcinoma harboring coexisting EML4-ALK fusion gene and EGFR mutation. J Thorac Oncol 5: 2039–2040. 10.1097/JTO.0b013e3181f43274 [DOI] [PubMed] [Google Scholar]
- 49. Tiseo M, Gelsomino F, Boggiani D, Bortesi B, Bartolotti M, et al. (2011) EGFR and EML4-ALK gene mutations in NSCLC: a case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer 71: 241–243. 10.1016/j.lungcan.2010.11.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.