Abstract
Ginger is one of the most commonly used herbal medicines for the treatment of numerous ailments and improvement of body functions. It may be used in combination with prescribed drugs. The coadministration of ginger with therapeutic drugs raises a concern of potential deleterious drug interactions via the modulation of the expression and/or activity of drug-metabolizing enzymes and drug transporters, resulting in unfavorable therapeutic outcomes. This study aimed to determine the molecular interactions between 12 main active ginger components (6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol) and human cytochrome P450 (CYP) 1A2, 2C9, 2C19, 2D6, and 3A4 and to predict the absorption, distribution, metabolism, excretion, and toxicity (ADMET) of the 12 ginger components using computational approaches and comprehensive literature search. Docking studies showed that ginger components interacted with a panel of amino acids in the active sites of CYP1A2, 2C9, 2C19, 2D6, and 3A4 mainly through hydrogen bond formation, to a lesser extent, via π–π stacking. The pharmacokinetic simulation studies showed that the [I]/[Ki] value for CYP2C9, 2C19, and 3A4 ranged from 0.0002 to 19.6 and the R value ranged from 1.0002 to 20.6 and that ginger might exhibit a high risk of drug interaction via inhibition of the activity of human CYP2C9 and CYP3A4, but a low risk of drug interaction toward CYP2C19-mediated drug metabolism. Furthermore, it has been evaluated that the 12 ginger components possessed a favorable ADMET profiles with regard to the solubility, absorption, permeability across the blood–brain barrier, interactions with CYP2D6, hepatotoxicity, and plasma protein binding. The validation results showed that there was no remarkable effect of ginger on the metabolism of warfarin in humans, whereas concurrent use of ginger and nifedipine exhibited a synergistic effect on platelet aggregation in humans. Moreover, ginger components showed a rapid half-life and no to low toxicity in humans. Taken together, this study shows that ginger components may regulate the activity and expression of various human CYPs, probably resulting in alterations in drug clearance and response. More studies are warranted to identify and confirm potential ginger–drug interactions and explore possible interactions of ginger with human CYPs and other functionally important proteins, to reduce and avoid side effects induced by unfavorable ginger–drug interactions.
Keywords: CYP, drug metabolism, ginger, drug interaction, docking
Introduction
Herbal medicine is becoming increasingly popular worldwide for improving body functions and managing numerous ailments, despite the lack of evidence for efficacy and long-term toxicological data in humans.1 Ginger is one of the most widely used herbal medicines with a wide range of pharmacological activities, including the reduction of the level of lipid, blood glucose, prostaglandin E2, and thromboxane B2; downregulation of the cyclooxygenase activity; blockage of Ca2+ channel; and inhibition of the production of cytokines and chemokines.2 So far, there are at least 128 compounds identified in ginger.3,4 Ginger is rich in pungent components such as phenylpropanoids and sesquiterpenes and contains approximately 2.5%–3.0% volatile oils.5 Gingerols are the most abundant pungent constituents in fresh ginger, and the content is slightly reduced in dry ginger, with the most abundant gingerol being 6-gingerol. In dry ginger, shogaols are the primary constituents, which are the major gingerol dehydration products, with 6-shogaol being the most abundant. Volatile oil mainly contains (−)-zingiberene, ar-curcumene, β-bisabolene, and β-phellandrene.5,6 The clinical beneficial effects of ginger are attributed to these major components. For example, interactions between 5-hydroxytryptamine receptors and shogaol/gingerol are partially responsible for the antiemetic activity.5 So far, there are a number of studies on the pharmacological activities of ginger extract and its single component;4 however, the data on the effect of ginger on the activity and/or expression of drug-metabolizing enzymes, in particular, cytochrome P450s (CYPs), are sparse.
Human CYPs are the major Phase I drug-metabolizing enzyme family accounting for metabolism of more than 95% drugs and some natural products.7 In particular, CYP1A, 2C9, 2C19, 2D6, and 3A4 are the most important CYPs in the oxidative metabolism, with capability of catalyzing the biotransformation of a large number of endogenous and exogenous compounds.7 CYP-related metabolism is important for the disposition of many herbal compounds. However, herbal compounds can also exert regulatory effects on the activity and expression of CYPs. Alteration on the activity and expression of CYPs may cause changes in CYP-mediated drug metabolism, resulting in pharmacokinetic changes, which will potentially lead to therapeutic failure or even lethal event.8 As such, modulation of CYP-mediated drug metabolism in multidrug therapy including ginger is always a major safety concern from drug development to clinical practice.9 There are a number of studies indicating that 14%–31% of prescription drug users combine herbal products with traditional medicines.10–12 The combination of herbal medicines and therapeutic drugs may cause undesired drug interaction, which is a major cause of therapeutic failure or toxic effect.13 Of note, there are over 100,000 lethal events occurring every year in USA due to drug interactions.14 Thus, it is of great importance to identity potential drug–drug and herb–drug interactions.
Nowadays, in many Western countries including USA, along with the significant increase in the use of ginger to manage various chronic diseases or to promote health, the potential risk of ginger-induced drug interaction involving CYP has drawn increasing attention.15 However, after comprehensive data search using the terms “drug metabolism”, “pharmacokinetic interaction”, “pharmacodynamic interaction”, “drug safety”, and “adverse event” in combination with “ginger” and “CYP or cytochrome P450”, there are scattered studies on the effects of ginger on CYP-mediated drug metabolism and ginger–drug interaction. On the other hand, there are increasing applications of computational approaches to predict absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of compounds, in particular, the applications in the evaluation of ADMET properties for herbal medicines. Compared to the conventional experimental approaches, the computational approach is a fast, efficient, and high-throughput strategy to screen, predict, and identify the ADMET properties of compounds with reduced labor and animal usage. Thus, the aim of this study was to explore the effects of ginger on CYP-mediated drug metabolism and the potential drug interactions with a focus on the estimation of the binding modes and interaction potential of ginger components with human CYP1A2, 2C9, 2C19, 2D6, and 3A4 and evaluation of ADMET properties through molecular docking and pharmacokinetic simulation studies.
Methods
Literature search
The comprehensive data search was performed using the key terms “drug metabolism”, “pharmacokinetics”, “drug response”, “clearance”, “interaction”, “safety”, “side effect”, and “mechanism” in combination with “ginger” and “cytochorme P450 or CYP” in Medline (via PubMed), ScienceDirect, Current Contents Connect (ISI), Cochrance Library, and Embase (all from inception to October 2014).
Molecular docking
In order to determine the molecular interactions between main active components of ginger and human CYPs, 12 compounds including 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol were separately docked into the active sites of CYP1A2, 2C9, 2C19, 2D6, and 3A4, using the Discovery Studio program 3.1 designed by Accelrys Inc. (San Diego, CA, USA) as previously described.16–18 Briefly, the crystal structures of human CYP1A2 (protein data bank [PDB] code 2HI4), 2C9 CYP2C9 (PDB code 1OG2), 2C19 (PDB code 4GQS), 2D6 (PDB code 2F9Q), and 3A4 (PDB code 1W0F) were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/). The protein and ligand were prepared prior to the docking. Human CYP1A2, 2C9, 2C19, 2D6, and 3A4 were cleaned, modified, and prepared for defining and editing the binding sites. For the preparation of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol, the duplicate structures were deleted and ionization change, tautomer or isomer generation, Lipinski filter, and 3D generator were all set true. A harmonic potential with the force constant of 300 kcal/mol was applied outside the grid boundary. Following preparation of CYPs, ginger compounds, and grid setting, the CYP–ginger compound interactions were determined.
Prediction of ginger–drug interactions
The prediction of potential ginger–drug interactions was performed as previously described.19,20 If the following equation is true, a ginger-induced drug interaction involving CYP inhibition in vivo is likely.
| (1) |
Where [I] is the inhibitor concentration and Ki is the inhibition constant. The degree of increase (Rc) in the area under the plasma concentration–time curve (AUC)iv caused by ginger–drug interaction is dependent on the route of administration, hepatic fraction (fh), and other factors. If drugs are administered by intravenous (iv) bolus and the dose or infusion rate is constant, the Rc can be calculated by the following equation:19
| (2) |
Where CLh is the hepatic clearance and CL’ is the clearance in the presence of the inhibitor. In case of high clearance drugs administered by iv bolus, CLh is rate-limited by the flow rate (CLh=Qh). When the altered CLh remains rate-limited by the flow rate (CLh′=Qh), then CLh′=CLh, indicating no change in AUCiv, Rc=1. However, this is not true when the inhibition is extensive that CLh is not limited by the flow rate.
In case of low clearance drugs administered by iv, fh is hepatic fraction and fm is metabolic pathway fraction in hepatic metabolism. Rc is expressed by the following equation:19
| (3) |
where CLint is the intrinsic clearance inhibited by the inhibitor, ′ represents the value after alteration by drug interaction, fh is the hepatic fraction, and fm is the metabolic pathway fraction in hepatic metabolism. In the clinical settings, [S] (concentration of substrate) is often much less than Km, then Rc is calculated by the following equation:19
| (4) |
Prediction of ADMET of main ginger components
The ADMET profiles of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol were predicted using the Discovery Studio program 3.1 designed by Accelrys Inc. Briefly, the chemical structures of these compounds were retrieved from PubChem database (Figure 1). All the structures were introduced into Discovery Studio program and prepared. During the process of structure preparation, the duplicate chemical structures were deleted and ionization change, tautomer or isomer generation, Lipinski filter, and 3D generator were all set true. Following the structure preparation, 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol were subject to ADMET evaluation using the ADMET Predictor module. The parameters predicted included solubility, absorption, permeability across the blood–brain barrier (BBB), interactions with CYP2D6, hepatotoxicity, and plasma protein binding (PPB). There are four different ADMET absorption levels, which are 0, 1, 2, and 3, representing good, moderate, low, or very low absorption, respectively. ADMET aqueous solubility level was classified into extremely low (0), no; very low, but possible (1); yes, low (2); yes, good (3); yes, optimal (4); no, too soluble (5); and warning, molecules with one or more unknown AlogP98 types (6). ADMET BBB permeability has six different levels, including very high (0), high (1), medium (2), low (3), undefined (4), and warning, molecules with one or more unknown AlogP98 calculation (5). There are two predicted classes of ADMET CYP2D6 ligand, noninhibitor (0) and inhibitor (1). ADMET hepatotoxicity is categorized into nontoxic (0) and toxic (1) effects. There are three different ADMET PPB levels, including binding <90% (0), binding ≥90% (1), and binding ≥95% (2).
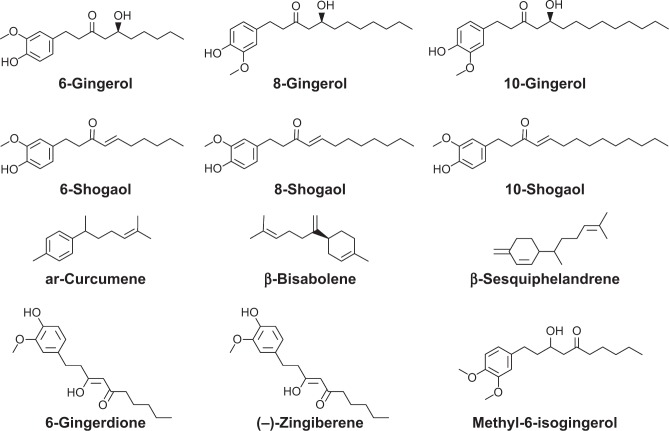
Figure 1.
The chemical structures of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol.
Results
Ginger components bind to the active sites of human CYP1A2, 2C9, 2C19, 2D6, and 3A4 via hydrogen bond formation and π–π stacking
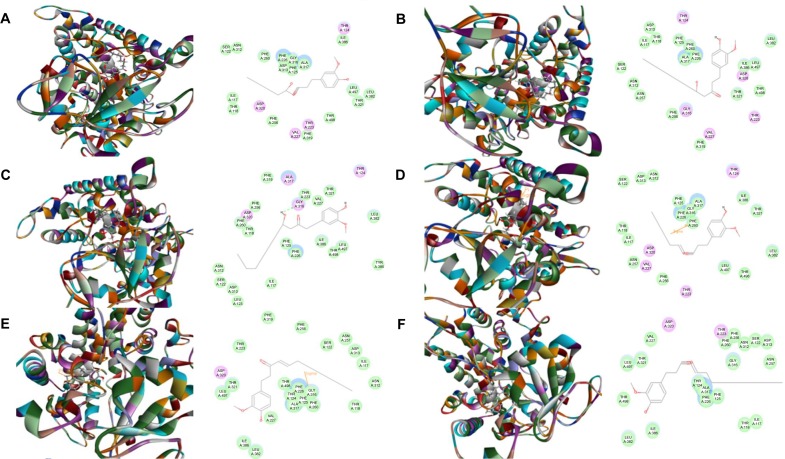
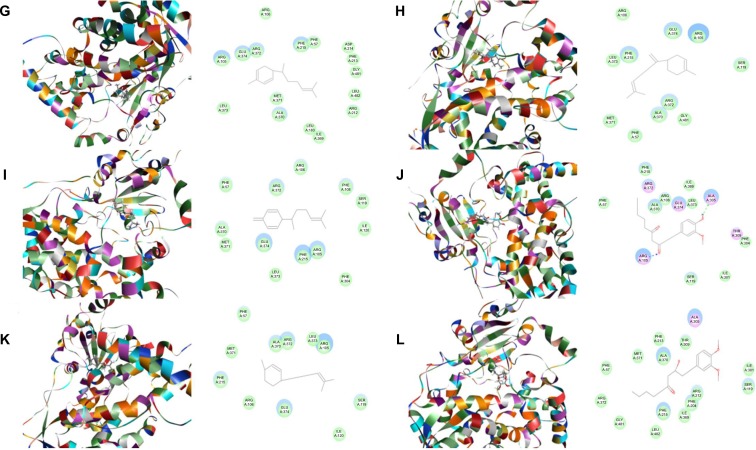
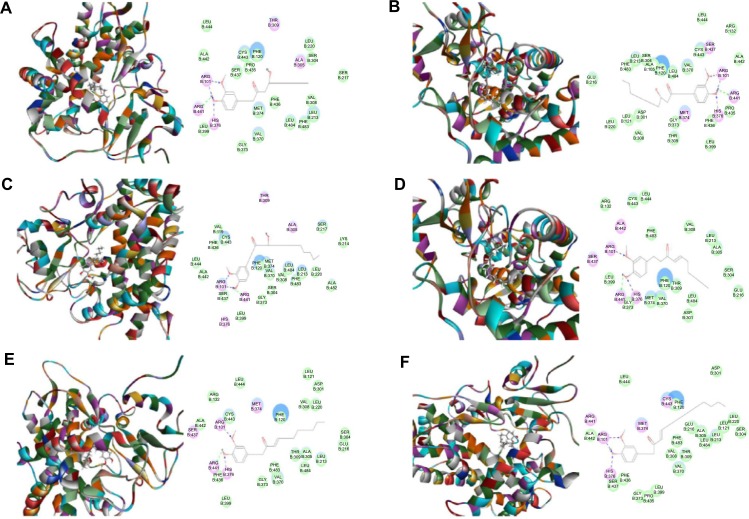
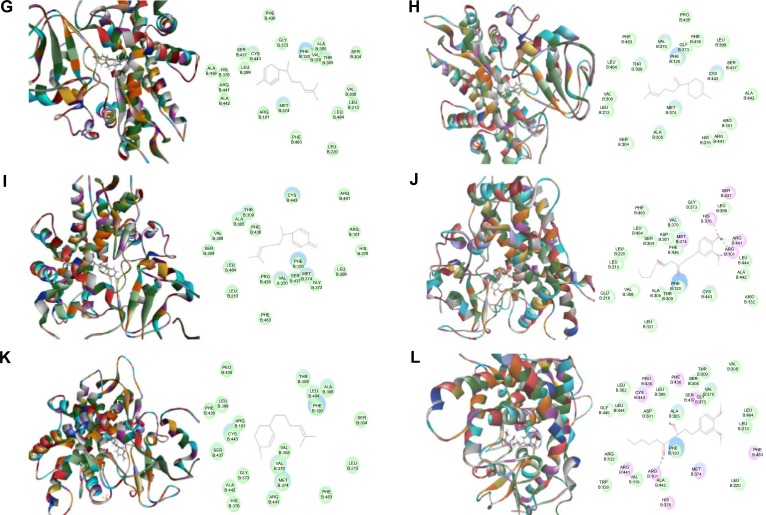
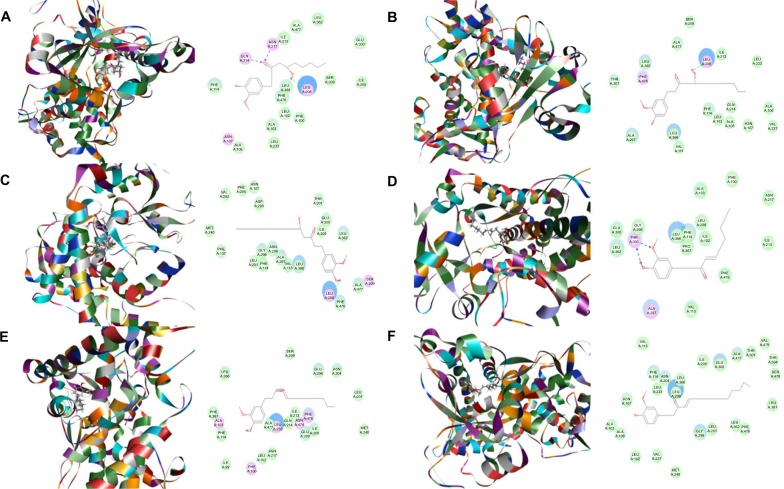
First, we performed a computational docking study of 12 main active ginger components using Discovery Studio 3.1. Twelve ginger components including 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol were docked into the active sites of CYP1A2, 2C9, 2C19, 2D6, and 3A4, respectively. After docking the main active ginger components into the active sites of five human CYPs, 10 positions were generated for each compound–CYP interaction. The CDOCKER interaction energy ranged from 23.0 to 63.1 kcal/mol. Each compound–CYP complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected (Figures 2–6).
Figure 2.
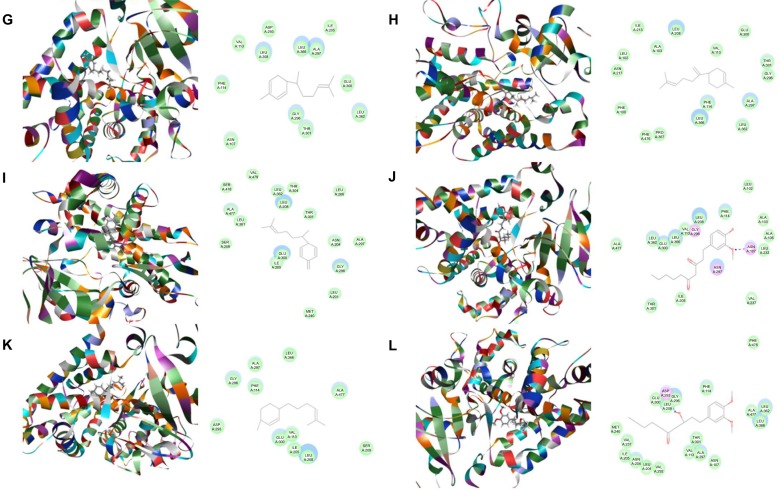
Binding modes of main active ginger components to human CYP1A2 (PDB code 2HI4).
Notes: Each compound–CYP1A2 complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected. (A) 6-Gingerol; (B) 8-gingerol; (C) 10-gingerol; (D) 6-shogaol; (E) 8-shogaol; (F) 10-shogaol; (G) ar-curcumene; (H) β-bisabolene; (I) β-sesquiphelandrene; (J) 6-gingerdione; (K) (−)-zingiberene; and (L) methyl-6-isogingerol.
Abbreviation: CYP, cytochrome P450.
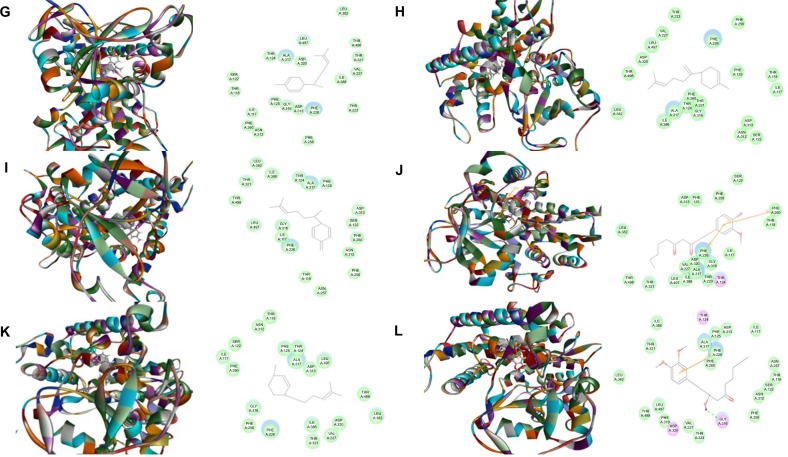
Figure 6.
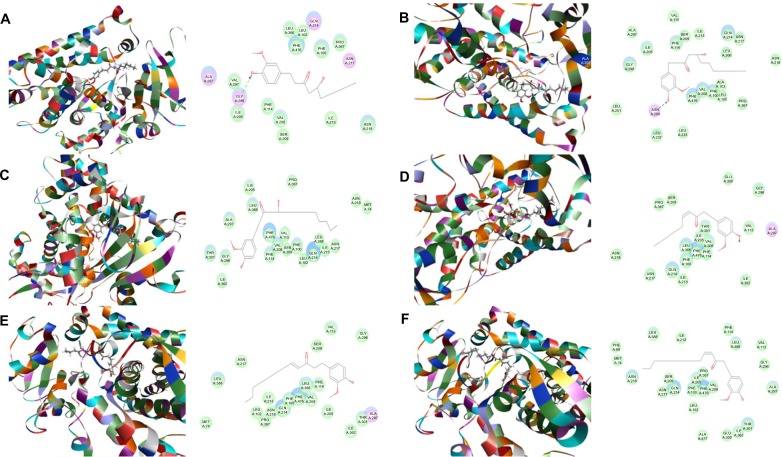
Binding modes of main active components of ginger in human CYP3A4 (PDB code 1W0F).
Notes: Each compound–CYP3A4 complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected. (A) 6-Gingerol; (B) 8-gingerol; (C) 10-gingerol; (D) 6-shogaol; (E) 8-shogaol; (F) 10-shogaol; (G) ar-curcumene; (H) β-bisabolene; (I) β-sesquiphelandrene; (J) 6-gingerdione; (K) (−)-zingiberene; and (L) methyl-6-isogingerol.
Abbreviation: CYP, cytochrome P450.
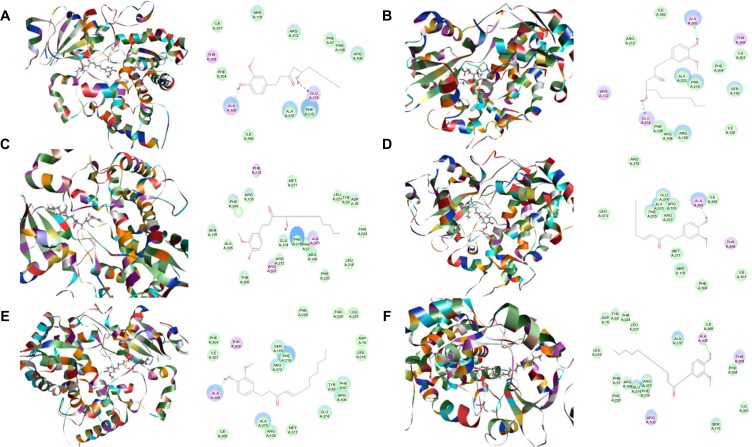
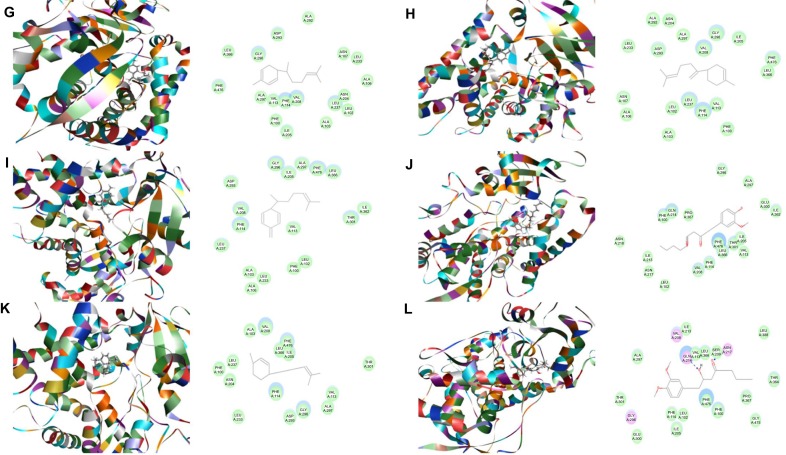
Based on our docking studies, we identified the interactions between amino acid residues in the CYP active sites and ginger components (Table 1). Most interactions occurred in the active site of CYP2D6 (Figure 5A–L), and, to a lesser extent, in the active sites of CYP3A4 (Figure 6A–L), 2C9 (Figure 3A–L), 1A2 (Figure 2A–L), and 2C19 (Figure 4A–L). It suggests that CYP2D6 may be the most favorable enzyme for ginger components. Indeed, CYP2D6 is known to detoxify the toxic xenobiotics in plants.21 H-bond formation was the main interaction between ginger components and CYPs (Table 1). The hydroxyl- and keto-groups were two major reaction sites for the ginger–CYP interactions. The most involved amino acids were Arg and Asn. Arg101, Arg441, and His376 were the most important amino acid residues accounting for the interaction between ginger compounds and CYP2D6 active site. Asn204, Asn217, Asn218, Gly296, and Gln214 were involved in the interaction in the active site of CYPC19. There were only five ginger compounds interacting with the active site of CYP2C19, which is the least among the five CYPs. Gly316, Phe226, and Phe260 were responsible for the interaction between ginger compounds and CYP1A2 active site. Thr301 and Leu208 were the most important residues in the active site of CYP2C9 and Asn107 and Asp293 played a lesser important role. For CYP3A4, Ala305 and Glu374 were the most important residues responsible for the interaction in the active site. π–π stacking only occurred between CYP1A2 and 6-shogaol, 8-shogaol, 6-gingerdione, and methyl-6-isogingerol at Phe226 and Phe260 (Table 1). Taken together, the in silico data show that ginger components interact with human CYPs, suggesting that the 12 ginger components may act as ligands for CYP1A2, 2C9, 2C19, CYP2D6, and 3A4 and that the interactions between ginger components and CYPs may influence the activity of CYPs with a consequence of inhibition in vivo.
Table 1.
Interacting forces of main active components of ginger with human CYP1A2, 2C9, 2D6, 2C19, and 3A4
| Compound | CDCOKER interaction energy (CIE kcal/mol) | H-bond number | Residues involved in H-bond formation | Charge interaction | Residues involved in charge interaction | π–π stacking | Residues involved in π–π stacking |
|---|---|---|---|---|---|---|---|
| CYP1A2 (PDB ID: 2HI4) | |||||||
| 6-Gingerol | 55.9802 | 0 | / | 0 | / | 0 | / |
| 8-Gingerol | 60.0329 | 0 | / | 0 | / | 0 | / |
| 10-Gingerol | 61.3568 | 1 | H-Gly316 | 0 | / | 0 | / |
| 6-Shogaol | 55.9354 | 0 | / | 0 | / | 1 | Phe260 |
| 8-Shogaol | 58.6712 | 0 | / | 0 | / | 1 | Phe260 |
| 10-Shogaol | 56.4091 | 0 | / | 0 | / | 0 | / |
| Ar-Curcumene | 36.2667 | 0 | / | 0 | / | 0 | / |
| β-Bisabolene | 35.991 | 0 | / | 0 | / | 0 | / |
| β-Sesquiphelandrene | 31.8433 | 0 | / | 0 | / | 0 | / |
| 6-Gingerdione | 59.0853 | 0 | / | 0 | / | 2 | Phe226 Phe260 |
| (−)-Zingiberene | 36.363 | 0 | / | 0 | / | 0 | / |
| Methyl-6-isogingerol | 51.232 | 1 | H-Gly316 | 0 | / | 1 | Phe226 |
| CYP2C9 (PDB ID: 1OG2) | |||||||
| 6-Gingerol | 38.3346 | 3 | H-Leu208 O-Gln214 O-Asn217 |
0 | / | 0 | / |
| 8-Gingerol | 41.0866 | 1 | H-Leu208 | 0 | / | 0 | / |
| 10-Gingerol | 45.8869 | 1 | H-Leu208 | 0 | / | 0 | / |
| 6-Shogaol | 38.4989 | 2 | H-Thr301 O-Thr301 |
0 | / | 0 | / |
| 8-Shogaol | 40.3159 | 0 | / | 0 | / | 0 | / |
| 10-Shogaol | 45.2874 | 0 | / | 0 | / | 0 | / |
| Ar-Curcumene | 27.0167 | 0 | / | 0 | / | 0 | / |
| β-Bisabolene | 24.3503 | 0 | / | 0 | / | 0 | / |
| β-Sesquiphelandrene | 24.6193 | 0 | / | 0 | / | 0 | / |
| 6-Gingerdione | 38.5734 | 1 | O-Asn107 | 0 | / | 0 | / |
| (−)-Zingiberene | 25.4757 | 0 | / | 0 | / | 0 | / |
| Methyl-6-isogingerol | 39.2082 | 1 | H-Asp293 | 0 | / | 0 | / |
| CYP2C19 (PDB ID: 4GQS) | |||||||
| 6-Gingerol | 30.5894 | 1 | H-Gly296 | 0 | / | 0 | / |
| 8-Gingerol | 34.7882 | 1 | O-Asn204 | 0 | / | 0 | / |
| 10-Gingerol | 41.7354 | 0 | / | 0 | / | 0 | / |
| 6-Shogaol | 31.298 | 1 | O-Gln214 | 0 | / | 0 | / |
| 8-Shogaol | 37.7374 | 1 | O-Asn218 | 0 | / | 0 | / |
| 10-Shogaol | 42.1221 | 0 | / | 0 | / | 0 | / |
| Ar-Curcumene | 26.7476 | 0 | / | 0 | / | 0 | / |
| β-Bisabolene | 24.9395 | 0 | / | 0 | / | 0 | / |
| β-Sesquiphelandrene | 25.8704 | 0 | / | 0 | / | 0 | / |
| 6-Gingerdione | 33.6531 | 0 | / | 0 | / | 0 | / |
| (−)-Zingiberene | 23.3969 | 0 | / | 0 | / | 0 | / |
| Methyl-6-isogingerol | 30.036 | 2 | H-Gln214 O-Asn217 |
0 | / | 0 | / |
| CYP2D6 (PDB ID: 2F9Q) | |||||||
| 6-Gingerol | 49.7194 | 4 | H-Arg441 O-His376 O-Arg101 O-Arg101 |
0 | / | 0 | / |
| 8-Gingerol | 55.2159 | 4 | H-Arg441 O-Arg101 O-Arg101 O-His376 |
0 | / | 0 | / |
| 10-Gingerol | 57.6051 | 3 | H-Arg441 O-Arg10 O-Arg101 |
0 | / | 0 | / |
| 6-Shogaol | 46.6251 | 3 | H-Arg441 O-Arg 101 O-His376 |
0 | / | 0 | / |
| 8-Shogaol | 52.6567 | 3 | H-Arg441 O-Arg101 O-His376 |
0 | / | 0 | / |
| 10-Shogaol | 56.3996 | 3 | O-His376 O-Arg101 O-Arg101 |
0 | / | 0 | / |
| Ar-Curcumene | 29.9457 | 0 | / | 0 | / | 0 | / |
| β-Bisabolene | 30.5663 | 0 | / | 0 | / | 0 | / |
| β-Sesquiphelandrene | 28.6495 | 0 | / | 0 | / | 0 | / |
| 6-Gingerdione | 50.0314 | 4 | H-Arg441 O-Arg101 O-Arg101 O-His376 |
0 | / | 0 | / |
| (−)-Zingiberene | 29.6369 | 0 | / | 0 | / | 0 | / |
| Methyl-6-isogingerol | 51.8517 | 1 | O-Arg101 | 0 | / | 0 | / |
| CYP3A4 (PDB ID: 1W0F) | |||||||
| 6-Gingerol | 51.1796 | 2 | H-Ala305 H-Glu374 |
0 | / | 0 | / |
| 8-Gingerol | 53.5899 | 2 | H-Ala305 H-Glu374 |
0 | / | 0 | / |
| 10-Gingerol | 54.8047 | 1 | H-Glu374 | 0 | / | 0 | / |
| 6-Shogaol | 49.1685 | 1 | H-Ala305 | 0 | / | 0 | / |
| 8-Shogaol | 53.9844 | 1 | H-Ala305 | 0 | / | 0 | / |
| 10-Shogaol | 63.0781 | 1 | H-Ala305 | 0 | / | 0 | / |
| Ar-Curcumene | 28.8825 | 0 | / | 0 | / | 0 | / |
| β-Bisabolene | 27.4614 | 0 | / | 0 | / | 0 | / |
| β-Sesquiphelandrene | 26.3732 | 0 | / | 0 | / | 0 | / |
| 6-Gingerdione | 52.9237 | 2 | H-Ala305 O-Arg105 |
0 | / | 0 | / |
| (−)-Zingiberene | 27.9257 | 0 | / | 0 | / | 0 | / |
| Methyl-6-isogingerol | 47.7586 | 0 | / | 0 | / | 0 | / |
Abbreviation: CYP, cytochrome P450.
Figure 5.
Binding modes of ginger main active components in human CYP2D6 (PDB code 2F9Q).
Notes: Each compound-CYP2D6 complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected. (A) 6-Gingerol; (B) 8-gingerol; (C) 10-gingerol; (D) 6-shogaol; (E) 8-shogaol; (F) 10-shogaol; (G) ar-curcumene; (H) β-bisabolene; (I) β-sesquiphelandrene; (J) 6-gingerdione; (K) (−)-zingiberene; and (L) methyl-6-isogingerol.
Abbreviation: CYP, cytochrome P450.
Figure 3.
Binding modes of ginger main active components in human CYP2C9 (PDB code 1OG2).
Notes: Each compound–CYP2C9 complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected. (A) 6-Gingerol; (B) 8-gingerol; (C) 10-gingerol; (D) 6-shogaol; (E) 8-shogaol; (F) 10-shogaol; (G) ar-curcumene; (H) β-bisabolene; (I) β-sesquiphelandrene; (J) 6-gingerdione; (K) (−)-zingiberene; and (L) methyl-6-isogingerol.
Abbreviation: CYP, cytochrome P450.
Figure 4.
Binding modes of main active components of ginger in human CYP2C19 (PDB code 4GQS).
Notes: Each compound–CYP2C19 complex with the lowest CDOCKER interaction energy was selected and the 2D and 3D pictures of them were collected. (A) 6-Gingerol; (B) 8-gingerol; (C) 10-gingerol; (D) 6-shogaol; (E) 8-shogaol; (F) 10-shogaol; (G) ar-curcumene; (H) β-bisabolene; (I) β-sesquiphelandrene; (J) 6-gingerdione; (K) (−)-zingiberene; and (L) methyl-6-isogingerol.
Abbreviation: CYP, cytochrome P450.
Validation of the modulating effects of ginger components on human CYP activity
Because we have observed the interactions between ginger components and CYPs from computational studies, we further validated these potential ginger–CYP interactions using previously published data. The regulatory effect of ginger on CYPs is a major cause of pharmacodynamic/pharmacokinetic alteration in the combination of ginger and other conventional therapeutic drugs.22 However, there are only a few studies about the effect of ginger components on the activities of human CYPs (Table 2).2,8,9,23
Table 2.
Effect of ginger components on the activity and expression of human CYPs
| Ginger | Concentration | Test system | Target CYP | Effect on enzyme | IC50 (μg/mL) | Reference |
|---|---|---|---|---|---|---|
| Ginger extract | 100 μg/mL | Human live microsomes | CYP2C9 | Inhibit enzymatic activity | 10.0 | 25 |
| Aqueous ethanolic extract of ginger | 0.05–5 μg/mL | Human liver microsomes | CYP2C19 | Inhibit enzymatic activity | 3.8 | 24 |
| 6-Gingerol | 21.6 μg/mL | Human liver microsomes | CYP3A4 | Inhibit enzymatic activity | 21.6 | 26 |
| Ginger fraction | 0.01–100 μg/mL | Human liver microsomes | CYP3A4 | Inhibit enzymatic activity | 30.5–42.0 | WO2007071708 |
| Ginger extract | 100 μg/mL | Human liver microsomes | CYP3A4 | Inhibit enzymatic activity | 5.1 | 25 |
| Ginger supplement | – | Human colon carcinoma cell line LS180 | CYP1A2 | Increase gene expression | – | 27 |
| Ginger supplement | – | Human colon carcinoma cell line LS180 | CYP3A4 | Increase gene expression | – | 27 |
| Sho-saiko-to (containing ginger extract) | – | Healthy subjects (n=26) | CYP1A2 | Inhibit caffeine metabolism | – | 29 |
Abbreviations: CYP, cytochrome P450; IC50, half maximal inhibitory concentration.
Kim et al24 reported that aqueous ethanolic extract of ginger possessed inhibitory effects on CYP2C19 in a concentration-dependent manner in human liver microsomes. The results showed that 0.05–5 μg/mL of aqueous ethanolic extract of ginger competitively inhibited CYP2C19 with an IC50 (half maximal inhibitory concentration) value of 3.8 μg/mL, whereas the ginger extract had no effect on other CYPs.24 The effect of ginger extract on CYP2C9 and 3A4 was also tested in human liver microsomes.25 The inhibitory effect of ginger extract on CYP2C9 and 3A4 was observed with IC50 values of 10 μg/mL and 5.1 μg/mL, respectively.25 Zhang and Lim26 also examined the effect of 6-gingerol on CYP3A4-mediated midazolam hydroxylation in human liver microsomes. 6-Gingerol showed inhibitory effect on CYP3A4 with an IC50 value of 21.6 μg/mL. In addition, the effect of ginger on CYP gene expression was also examined in vitro. Brandin et al27 observed an inducing effect of ginger on the expression of CYP1A2 and CYP3A4 genes in human LS180 cells, which are colorectal adenocarcinoma epithelial cells. Based on the in vitro inhibitory effect of ginger extract or ginger components, it is likely that ginger extract might affect the pharmacokinetics of drugs and lead to ginger–drug interactions with drugs that are metabolized by CYP1A2, 2C9, 2C19, and 3A4, in particular those with a narrow therapeutic index.28 Notably, Saruwatari et al29 tested the effect of a Chinese herbal medicine Sho-saiko-to containing ginger in 26 healthy subjects. The results showed that CYP1A2-mediated caffeine metabolism was impaired by Sho-saiko-to, indicating that ginger may exert an inhibitory effect on CYP1A2 activity. However, due to multiple composition of Sho-saiko-to, other herb components-induced inhibitory effect on CYPs cannot be excluded in this study.
Estimation of ginger–drug interaction potential by pharmacokinetic simulation approach
Modulation of CYP-mediated drug metabolism by ginger represents a major safety concern in clinic practice.30 It has been shown that ginger extract and/or its components exerted competitive inhibitory effect on CYPs including CYP1A2, 2C9, 2C19, and 3A4 with various IC50 values (Table 3). Due to the lack of clinical data on ginger-induced drug interactions involving CYPs, it is likely to predict ginger–drug interactions using preclinical data to reduce and avoid the risk of potential ginger–drug interactions. The inhibitory effect of ginger on CYPs may fall into mutual competitive inhibition, mechanism-based inhibition, and nonselective inhibition of CYPs.19 Notably, the prediction of mechanism-based inhibitory effect of ginger on CYPs is different from the reversible inhibition and the application of mechanism-based CYP inhibition for predicting drug interactions has been comprehensively described by us previously.20 Herein, we focused on the competitive inhibition. In order to predict in vivo drug– ginger interaction after iv bolus administration, it is crucial to calculate the values of Ki, [I], fh, and fm· fh and fm can be determined from the urinary recovery of the parent molecule and each metabolite; Ki can be estimated by in vitro inhibition studies using human liver microsomes and recombinant systems.
Table 3.
Estimated AUC ratios when drugs are coadministered with ginger extract or ginger components
| Ginger | IC50 (μg/mL) | [I] (μg/mL) | Ki (μg/mL) | Target CYP | Substrate | [I]/[Ki] | Estimated AUC ratio (R) | Risk of drug interaction | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Ginger extract | 10.0 | 100 | 10.0 | CYP2C9 | Diclofenac | 10 | 11 | High | 25 |
| Ginger extract | 3.8 | 0.05–5 | 3.8 | CYP2C19 | Mephenytoin | 0.013–1.32 | 1.013–2.32 | Low-high | 24 |
| 6-Gingerol | 21.6 | 21.6 | 21.6 | CYP3A4 | Midazolam | 1 | 2 | High | 26 |
| Ginger fraction | 30.5–42.0 | 0.01–100 | 30.5–42.0 | CYP3A4 | Erythromycin | 0.0002–3.27 | 1.0002–4.27 | Low-high | WO2007071708 |
| Ginger extract | 5.1 | 100 | 5.1 | CYP3A4 | Testosterone | 19.6 | 20.6 | High | 25 |
Abbreviations: AUC, area under the plasma concentration–time curve; CYP, cytochrome P450; IC50, half maximal inhibitory concentration; I, concentration of inhibitor; Ki, inhibitory constant.
As shown in Table 3, the [I]/Ki value for CYP2C9 was 10 and the estimated R value was 11, suggesting that ginger extract might exhibit a high risk of drug interaction via inhibition of the activity of CYP2C9. For CYP2C19, the [I]/Ki value ranged from 0.013 to 1.32 and the R value ranged from 1.013 to 2.32, suggesting ginger extract might show a low to high risk of drug interaction through the inhibition of the activity of CYP2C19. For CYP3A4, the [I]/Ki value ranged from 0.0002 to 19.6 and the R value ranged from 1.0002 to 20.6, suggesting that ginger extract may have a low to high risk of drug interaction via inhibition of the activity of CYP3A4.
Notably, the pharmacokinetic simulation studies show that the inhibitory effect of ginger extract on the activity of CYP2C9, 2C19, and 3A4 may result in alterations in the pharmacokinetics and pharmacodynamics of coadministered drugs that are mainly metabolized by these CYPs. In particular, alterations in the pharmacokinetics and pharmacodynamics of drugs with narrow therapeutic windows and mainly metabolized by CYP2C9, 2C19, and 3A4 (such as warfarin) may cause clinically important outcomes. The pharmacokinetic simulation studies suggest that concurrent administration of ginger and warfarin or nifedipine may cause drug interactions in the clinical settings through inhibition of the activity of CYP2C9 and 3A4. Subsequently, the potential ginger–warfarin and ginger–nifedipine interactions were validated by previously published data.
Validation of clinical interactions of ginger with CYPs and drugs
The molecular interactions between ginger components and human CYPs in silico and the inhibitory effect of ginger on CYPs in vitro suggest that ginger may cause pharmacodynamic and/or pharmacokinetic interactions in vivo. Alteration in the ADME of a conventional therapeutic drug is the cause of ginger-induced pharmacokinetic interactions. Pharmacodynamic interactions may occur when the ginger component exerts synergistic or antagonistic effect on the same drug target(s) as the coadministered drug.31,32 In particular, a pharmacodynamic interaction may occur when the coadministered drugs have a narrow therapeutic index. Thus, we have evaluated the potential effects of ginger on the pharmacodynamic and/or pharmacokinetic profiles of warfarin and nifedipine.
Ginger–warfarin interaction
Warfarin is the most commonly prescribed anticoagulant with a narrow therapeutic index. S-warfarin has a stronger anticoagulant activity than R-warfarin.33 CYP2C9 is the major metabolizing enzyme responsible for the S-7-hydroxywarfarin formation.34 In addition, CYP1A2 catalyzes the 6- and 8-hydroxylation of R-warfarin, while CYP3A4 catalyzes the 10-hydroxylation of R-warfarin.35 Thus, alteration of the activity and/or expression of CYP1A2, 2C9, and 3A4 will result in changes in the therapeutic effect of warfarin. There is a growing concern on the pharmacokinetic and/or pharmacodynamic interactions between warfarin and herbal medicines including ginger, St John’s wort, garlic, etc.
Jiang et al36 designed a randomized, open-label, crossover study to examine the effect of ginger on the pharmacokinetics and pharmacodynamics of warfarin in 12 healthy human subjects. When administering warfarin alone, the maximum plasma concentration (Cmax) of S- and R-warfarin was 1.7 μg/mL, and the volume of distribution was 0.12 L/kg. The AUC level was 68 μg/mL·hour and 104 μg/mL·hour, and the plasma half-life was 35.8 hours and 50.3 hours for S- and R-warfarin, respectively. The plasma clearance of S- and R-warfarin was 189 mL/hour and 127 mL/hour, respectively. In coadministration of warfarin with ginger, the AUC level was 66 μg/mL·hour and 102.6 μg/mL·hour and the plasma half-life was 35.8 hours and 50.3 hours for S- and R-warfarin, respectively. The plasma clearance was 201 L/hour and 131 L/hour and the volume of distribution was 0.12 L/kg and 0.11 L/kg for S- and R-warfarin, respectively. Moreover, there was no significant effect of ginger on the international normalized ratio (INR) of warfarin. The INRbaseline was 1.12 for both groups, and the AUC0–168 h of INR was 7.5 and 8.1 for warfarin and warfarin with ginger groups, respectively. The result showed that there was no significant effect of ginger on pharmacokinetics and pharmacodynamics of warfarin. Although Jiang et al36 did not observe any remarkable effect of ginger extract on the pharmacokinetics and pharmacodynamics of warfarin, one needs to be cautious to concurrently administer ginger with warfarin to patients and more clinical studies are warranted to explore the risk of ginger–warfarin interactions.
Ginger–nifedipine interaction
Nifedipine is mainly used as an antihypertensive and antianginal drug. It is mainly metabolized by CYP3A4, and dehydronifedipine is the primary metabolite.37 It has been shown that ginger and nifedipine exerted a synergistic effect on antiplatelet aggregation both in healthy humans and patients with hypertension.38 The results have shown that the inhibitory effect of ginger with nifedipine on platelet aggregation was more than two-fold of that of ginger or nifedipine alone in healthy volunteers. In hypertensive patients, compared to nifedipine, there was a three-fold increase in the inhibition of platelet aggregation in combination of ginger with nifedipine. The results indicate that ginger-induced pharmacodynamic interaction occurs in the combination of nifedipine. This will lead to an enhanced therapeutic effect of nifedipine in the site of action. In addition, an inhibitory effect of ginger on arachidonic acid-induced platelet aggregation was also reported in a male volunteer who consumed large, unspecified quantities of ginger marmalade (15% raw ginger).39
It has been shown that coadministration of ginger resulted in a significant decrease in Cmax and AUC0–t of oral cyclosporine by 70.9% and 63.1%, respectively, in rats.40 A stronger inhibitory effect of ginger was observed when the intake of ginger was 2 hours before cyclosporine, which decreased Cmax and AUC0–t by 51.4% and 40.3%, respectively. However, there was no remarkable change in the pharmacokinetics of iv cyclosporine by oral coadministration of ginger juice.40 These data suggest that ginger exerts a significant inhibitory effect on the absorption of cyclosporine, resulting in a significant reduction in oral bioavailability. To avoid the pharmacokinetic interaction, patients should be advised against using ginger products together with cyclosporine to ensure the efficacy of cyclosporine.
Prediction of the ADME properties and liver toxicity of ginger components
The predicted ADMET properties of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol are shown in Table 4. Ar-Curcumene, β-bisabolene, β-sesquiphelandrene, and methyl-6-isogingerol were predicted to have an extremely low aqueous solubility with the aqueous solubility level of 0. 6-shogaol and 8-shogaol showed a low aqueous solubility with an aqueous solubility level of 2. 6-Gingerol, 8-gingerol, 10-gingerol, 6-gingerdione, and (−)-zingiberene showed a good absorption with ADMET solubility level of 3 (Table 4).
Table 4.
Predicted ADMET properties of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, 6-gingerdione, (−)-zingiberene, and methyl-6-isogingerol
| Compound | ADMET solubility level | ADMET BBB level | ADMET EXT CYP2D6 | ADMET EXT CYP2D6 applicability | ADMET EXT hepatotoxicity | ADMET EXT hepatotoxic applicability | ADMET absorptionlevel | ADMET EXT PPB | ADMET EXT PPB applicability | ADMET AlogP98 | ADMET PSA 2D |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6-Gingerol | 3 | 2 | −1.99449 | All properties and OPS components are within expected ranges. | −10.8609 | OPS PC24 out of range. | 0 | 0.68641 | All properties and OPS components are within expected ranges. | 3.638 | 67.861 |
| 8-Gingerol | 3 | 1 | −2.22953 | All properties and OPS components are within expected ranges. | −12.1404 | OPS PC24 out of range. | 0 | 1.15791 | All properties and OPS components are within expected ranges. | 4.551 | 67.861 |
| 10-Gingerol | 3 | 1 | −2.16239 | All properties and OPS components are within expected ranges. | −13.6718 | OPS PC24 out of range. | 0 | 0.75522 | All properties and OPS components are within expected ranges. | 5.463 | 67.861 |
| 6-Shogaol | 2 | 1 | −0.772781 | All properties and OPS components are within expected ranges. | −10.0805 | All properties and OPS components are within expected ranges. | 0 | −0.00983 | All properties and OPS components are within expected ranges. | 4.717 | 47.046 |
| 8-Shogaol | 2 | 0 | −0.138333 | All properties and OPS components are within expected ranges. | −12.3015 | All properties and OPS components are within expected ranges. | 0 | 1.20105 | All properties and OPS components are within expected ranges. | 5.629 | 47.046 |
| 10-Shogaol | 2 | −0.663161 | False | 15.6598 | False | 15.6978 | 0.88111 | True | 15.3938 | 47.046 | |
| ar-Curcumene | 0 | −0.000351 | False | 12.2567 | False | 10.903 | 2.7504 | True | 10.7617 | 0 | |
| β-Bisabolene | 0 | 0.150831 | False | 12.4332 | False | 10.2596 | 2.96409 | True | 11.3322 | 0 | |
| β-Sesquiphelandrene | 0 | −0.470657 | False | 11.459 | False | 10.7802 | 3.03886 | True | 11.3647 | 0 | |
| 6-Gingerdione | 3 | 1 | −2.10543 | All properties and OPS components are within expected ranges. | −8.359 | All properties and OPS components are within expected ranges. | 0 | −1.57444 | All properties and OPS components are within expected ranges. | 4.044 | 67.861 |
| (−)-Zingiberene | 3 | 1 | −2.10543 | All properties and OPS components are within expected ranges. | −8.359 | All properties and OPS components are within expected ranges. | 0 | −1.57444 | All properties and OPS components are within expected ranges. | 4.044 | 67.861 |
| Methyl-6-isogingerol | 0 | −1.32416 | False | 16.6566 | False | 13.1585 | 2.72612 | True | 12.955 | 55.976 |
Abbreviations: ADMET, absorption, distribution, metabolism, excretion, and toxicity; BBB, blood–brain barrier; CYP, cytochrome P450; EXT, extension; OPS, optimal prediction space; PPB, plasma protein binding; PSA, polar surface area.
The 12 ginger components exhibited a medium to very high BBB penetration capability with the level from −1.3 to 2 (Table 4). 8-Shogaol, 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, and methyl-6-isogingerol showed a very high BBB penetration capability. 8-Gingerol, 10-gingerol, 6-shogaol, 6-gingerdione, and (−)-zingiberene exhibited a high BBB penetration capability. 6-Gingerol was predicted to have a medium BBB penetration capability.
Furthermore, all properties and optimal prediction space (OPS) components of 6-gingerol, 8-gingerol, 10-gingerol, 6-shogaol, 8-shogaol, 6-gingerdione, and (−)-zingiberene were within the expected ranges with regard to the CYP2D6 ligand and PPB level. 10-Shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, and methyl-6-isogingerol were predicted not to be a ligand of CYP2D6 (Table 4). For the prediction of hepatotoxicity, the ADMET predictor showed that 6-shogaol, 8-shogaol, 6-gingerdione, and (−)-zingiberene exhibited good hepatotoxic applicability with all properties and OPS components being within the expected range, whereas OPS24 values of 6-gingerol, 8-gingerol, and 10-gingerol were out of range. Moreover, the hepatotoxicity of 10-shogaol, ar-curcumene, β-bisabolene, β-sesquiphelandrene, and methyl-6-isogingerol were predicted to be false (Table 4). Taken together, the results suggest that ginger components exhibit a largely favorable ADMET profile.
Validation of the ADME properties and liver toxicity of ginger
Following the prediction of ADMET profile of 12 main ginger components, we evaluated the clinical pharmacology and toxicology of ginger using published data.
There are two pharmacokinetic studies on 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol in humans (Table 5).41,42 The results showed that the plasma half-lives for the four components and their metabolites were 1–3 hours in humans. The glucuronide and sulfate conjugates were the main detectable metabolites of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol in human plasma.41,42 In a study by Yu et al, there was no free form of 6-gingerol and 8-gingerol detected over 24 hours in the plasma after a single oral dose of 2.0 g ginger extract; only 10-gingerol and 6-shogaol were detected in free form.42 It was considered that 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol are rapidly metabolized in humans. In order to fully confirm the ADMET properties of ginger, more studies are required to investigate the ADMET profile of ginger in humans.
Table 5.
Reported pharmacokinetic parameters of active components of ginger in healthy volunteers
| Component | Dose of ginger extracts (mg) | No of subjects | Administration route | Cmax (μg/mL) | AUC (μg·min/mL) | Tmax (min) | t1/2β (min) |
|---|---|---|---|---|---|---|---|
| Total 6-gingerola | 1,000 | 6 HV | Single po | 0.4±0.2 | 12.6±6.4 | 55.0±7.7 | N/A |
| 1,500 | 3 HV | Single po | 1.69±2.31 | 75.6±110.3 | 60.0±0.0 | N/A | |
| 2,000 | 8 HV | Single po | 0.85±0.43 | 65.6±44.4 | 65.5±22.6 | 110.0±34.9 | |
| Free 6-gingerolb | 2,000 | 9 HV | Single po | N/A | N/A | N/A | N/A |
| 6-Gingerol glucuronidea | 1,000 | 6 HV | Single po | 0.16±0.15 | N/A | N/A | N/A |
| 1,500 | 3 HV | Single po | 0.62±0.62 | N/A | N/A | N/A | |
| 2,000 | 8 HV | Single po | 0.62±0.56 | N/A | N/A | N/A | |
| 6-Gingerol glucuronideb | 2,000 | 9 HV | Single po | 0.45±0.25 | 44.4±33.6 | 61.8±24.6 | 98.4±52.8 |
| 6-Gingerol sulfatea | 1,000 | 6 HV | Single po | 0.02±0.03 | N/A | N/A | N/A |
| 1,500 | 3 HV | Single po | 0.04±0.04 | N/A | N/A | N/A | |
| 2,000 | 8 HV | Single po | 0.33±0.41 | N/A | N/A | N/A | |
| 6-Gingerol sulfateb | 2,000 | 9 HV | Single po | 0.26±0.13 | 25.8±15.6 | 61.8±24.6 | 107.4±59.4 |
| Total 8-gingerola | 1,000 | 6 HV | Single po | 0.1±0.1 | 2.1±2.2 | 52.5±8.7 | N/A |
| 1,500 | 3 HV | Single po | 0.1±0.1 | 2.6±2.0 | 60.0±0.0 | N/A | |
| 2,000 | 8 HV | Single po | 0.23±0.16 | 18.1±20.3 | 73.1±29.4 | 113.5±41.1 | |
| Free 8-gingerolb | 2,000 | 9 HV | Single po | N/A | N/A | N/A | N/A |
| 8-Gingerol glucuronideb | 2,000 | 9 HV | Single po | 0.16±0.11 | 15.6±16.2 | 63.6±24.0 | 61.8±20.4 |
| 8-Gingerol sulfateb | 2,000 | 9 HV | Single po | 0.027±0.015 | 2.16±2.1 | 61.8±22.8 | 75.0±15.6 |
| Total 10-gingerola | 1,000 | 6 HV | Single po | 0.1±0.1 | 2.9±3.2 | 60.0±0.0 | N/A |
| 1,500 | 3 HV | Single po | 0.1±0.02 | 7.7±5.3 | 80.0±34.6 | N/A | |
| 2,000 | 8 HV | Single po | 0.53±0.4 | 50.1±49.3 | 75.0±27.8 | 128.7±38.8 | |
| Free 10-gingerolb | 2,000 | 9 HV | Single po | 0.009±0.002 | 0.48±0.24 | 66.6±24.0 | 107.4±19.2 |
| 10-Gingerol glucuronideb | 2,000 | 9 HV | Single po | 0.41±0.24 | 52.8±36.0 | 71.4±30.6 | 125.4±26.4 |
| 10-Gingerol sulfateb | 2,000 | 9 HV | Single po | 0.017±0.006 | 3.54±0.9 | 68.4±31.2 | 194.4±109.8 |
| Total 6-shogaola | 1,000 | 6 HV | Single po | 0.1±0.1 | 0.8±1.5 | 55.0±8.7 | N/A |
| 1,500 | 3 HV | Single po | 0.04±0.08 | 1.6±2.8 | 60.0±0.0 | N/A | |
| 2,000 | 8 HV | Single po | 0.15±0.12 | 10.9±13.0 | 65.6±22.6 | 120.4±42.0 | |
| Free 6-shogaolb | 2,000 | 9 HV | Single po | 0.011±0.007 | 1.44±0.78 | 60.0±24.6 | 79.2±26.4 |
| 6-Shogaol glucuronideb | 2,000 | 9 HV | Single po | 0.080±0.057 | 6.6±4.8 | 61.8±24.6 | 92.4±39.6 |
| 6-Shogaol sulfateb | 2,000 | 9 HV | Single po | 0.049±0.026 | 4.74±2.64 | 63.6±22.2 | 84.0±21.6 |
Notes:
Data are from the study by Zick et al.41
Data are from the study by Yu et al.42
Abbreviations: AUC, area under the concentration–time curve; Cmax, maximum plasma concentration; HV, healthy volunteers; PSA, polar surface area; NA, not available; t1/2β, elimination half-life; Tmax, time to Cmax.
Furthermore, the previous data show that ginger is generally considered safe and that there is no hazard or moderate-to-severe side effect reported.43 Only some minor adverse effects of ginger are found in humans. In a clinical trial in which 12 healthy volunteers participated, 400 mg ginger taken orally three times per day for 2 weeks caused mild diarrhea, which only occurred during the first 2 days of ginger pretreatment in one subject.2 Another human study has shown that ginger may cause ulcer formation and gastric irritant in a dose higher than 6 g.44 Other mild adverse reactions include heartburn, gas production, and bloating.2 Taken together, ginger is considered to be safe when used alone.
Discussion
Clinically, a number of pharmacodynamic/pharmacokinetic herb–drug interactions have been documented and many of them result in significant effects on therapeutic outcomes.45,46 There is an estimation that 5% of hospital admissions are associated with drug interactions,47,48 and there are over 100,000 deaths per year that may have been caused by drug interactions, including herb–drug interaction.14 Increasing usage of ginger raises the requirement of clarification of its pharmacokinetics and pharmacodynamics in humans. In fact, there is a significant gap between the growing usage and clinical evidence of ginger. Furthermore, when the consumers think that ginger or ginger products are natural products and should be “safe” and there is lack of solid data on ginger–CYP and ginger–drug interactions, there is an increased concern on the potential ginger-induced drug interactions.
Our docking studies indicate that CYP2D6 is the most likely enzyme interacting with ginger components, whereas CYP3A3, 2C9, 1A2, and 2C19 have a lower binding affinity. The hydroxyl- and keto-groups are two major interaction sites for the ginger–CYP interactions. The most involved amino acids are neutral or acidic in CYP3A4, 2C9, 1A2, and 2C19. It suggests that ginger components may compete in the interaction site with other substrates/inhibitors of CYPs. Ginger possesses inhibitory effects on CYPs in vitro; however, the clinical evidence for the inhibitory effect of ginger on CYPs is lacking. Although extrapolating in vitro and animal data to predict potential ginger-induced drug interaction is an option, in vitro inhibitory potencies of ginger do not necessarily translate directly into relative extents of inhibition in humans.
Inducing or inhibiting CYPs is the primary underlying mechanism of altered pharmacodynamics/pharmacokinetics by coadministrated ginger products. Based on the in vitro inhibitory effect of ginger on CYPs, inhibition of CYPs by ginger compounds may have important clinical implications. Thus, it is very important to evaluate the toxicity and drug interaction potential of ginger products. A strategy of risk identification, risk reduction or elimination, and a final evaluation of the adopted risk reduction can be used to assess the risk of ginger–drug interaction.49 However, the clinical ginger–drug interaction is difficult to predict because of multiple factors relating to individual variability and history of use.14 Obviously, the inhibitory effect of ginger extract or component on CYPs varies too much in vitro, and if [I]/Ki>0.2 is true, the risk of a ginger-induced drug interaction involving CYP2C9, 2C19, and 3A4 in vivo is possible. Although Jiang et al36 reported that ginger did not affect the pharmacokinetics of warfarin in humans and ginger-induced drug interaction has been seldom reported in humans, it requires more clinical studies to purge the potential risk of ginger-induced drug interactions.
Due to the growing usage and inadequate evidence of the effect of ginger on CYP-mediated drug interaction, further well-designed studies are warranted to address the underlying mechanism(s) and clinical significance of important ginger–drug interactions and predict the potential ginger–drug interactions. There are numerous ginger products accessible to customers even though there is a lack of clinical evidence on pharmacokinetics and pharmacodynamics.3 As a matter of fact, there is a significant gap between the growing demand in the usage of ginger and low regulatory controls on it. In particular, there is a lack of evidence on ginger–drug interactions with the involvement of CYPs, which may cause therapeutic or toxic effects in the clinical setting. For example, alteration on CYP2C9-mediated drug metabolism with narrow therapeutic index (warfarin) can lead to a serious clinical event.
Computational approach has been considered to be a practical strategy to timely, precisely, and widely predict and identify the potential drug–drug interactions, herb–drug interactions, and herb–herb interactions, avoiding dangerous outcomes in clinic resulting from modification of the activity of drug-metabolizing enzymes. Meanwhile, because the degree of change in AUC of a drug (Rc) is determined by Ki, [I], fh, and fm, but not by Km or [S], using in vitro data of ginger to estimate the risk of drug interactions and predict potential drug interaction is also practical. Therefore, integration of computational approach with in vitro data prediction method may be capable of assessing the risk of ginger–drug interactions and deciphering the potential mechanisms of ginger–drug interactions. Although there are limitations in the extrapolation of data from in vitro to in vivo, this integrated approach also can be extended to predict the drug interaction potential of other commonly used herbal medicines such as St John’s wort, garlic, ginseng, and ginkgo. They may interact with a wide range of therapeutic drugs, including some clinically important therapeutic drugs with a narrow therapeutic index to cause, in some cases, life-threatening and lethal consequences.9
CYP-mediated drug metabolism is the major biotransformation of most drugs and some herb compounds, which is a determinant factor for the pharmacokinetics of the substrates. An increased understanding of CYPs has aided in profiling drug pharmacokinetics in clinical practice. Modulation of CYP-mediated drug metabolism in multidrug therapy is the main cause of drug–drug interaction resulting in alterations in pharmacokinetics and pharmacodynamics, which eventually may cause therapeutic failure and/or toxic effect. Concurrent administration of ginger with a drug often occurs, in particular, with drugs with narrow therapeutic index, which causes a major concern of potential ginger–drug interactions.
In humans, ginger-induced changes in pharmacokinetic can be estimated by the degree of increase (Rc) in AUC. It is dependent on the administration route. Ki, [I], fh, and fm are the determining factors. In general, the potential inhibitory effect of ginger on the activity of CYPs may lower the rate of biotransformation and clearance resulting in changes in pharmacokinetics and pharmacodynamics in humans. Thus, a combination use of ginger with therapeutic drugs should be avoided in clinical practice. In particular, ginger should be advised against concurrent administration with drugs with narrow therapeutic index, such as warfarin. In addition, although ginger only has mild adverse effects, one needs to be cautious while consuming a large dose of ginger.
In summary, the integration of computational and experimental approaches can facilitate the rapid and precise prediction and validation of the potential ginger–drug interaction and the ADMET properties of ginger components to reduce and avoid the potential ginger-induced side effects with the involvement of CYPs. This study may help to bridge the gap between the increasing use of herbal medicines and the safety concerns. However, due to the lack of evidence on ADMET profile, clinical efficacy and molecular targets, and ginger-induced drug interactions, more well-designed studies are certainly needed to address these important issues.
Acknowledgments
The authors appreciate the Startup fund from College of Pharmacy, University of South Florida, Tampa, FL, USA. Dr Zhi-Wei Zhou is a holder of a postdoctoral scholarship from College of Pharmacy, University of South Florida, Tampa, FL, USA.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.De Smet PA. Herbal remedies. N Engl J Med. 2002;347(25):2046–2056. doi: 10.1056/NEJMra020398. [DOI] [PubMed] [Google Scholar]
- 2.Chrubasik S, Pittler MH, Roufogalis BD. Zingiberis rhizoma: a comprehensive review on the ginger effect and efficacy profiles. Phytomedicine. 2005;12(9):684–701. doi: 10.1016/j.phymed.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Kubra IR, Jaganmohanrao L. An overview on inventions related to ginger processing and products for food and pharmaceutical applications. Recent Patent Food, Nutr Agric. 2012;4(1):31–49. [PubMed] [Google Scholar]
- 4.Kubra IR, Rao LJ. An impression on current developments in the technology, chemistry, and biological activities of ginger (Zingiber officinale Roscoe) Crit Rev Food Sci Nutr. 2012;52(8):651–688. doi: 10.1080/10408398.2010.505689. [DOI] [PubMed] [Google Scholar]
- 5.Ali BH, Blunden G, Tanira MO, Nemmar A. Some phytochemical, pharmacological and toxicological properties of ginger (Zingiber officinale Roscoe): a review of recent research. Food Chem Toxicol. 2008;46(2):409–420. doi: 10.1016/j.fct.2007.09.085. [DOI] [PubMed] [Google Scholar]
- 6.Jolad SD, Lantz RC, Chen GJ, Bates RB, Timmermann BN. Commercially processed dry ginger (Zingiber officinale): composition and effects on LPS-stimulated PGE2 production. Phytochemistry. 2005;66(13):1614–1635. doi: 10.1016/j.phytochem.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Coon MJ. Cytochrome P450: nature’s most versatile biological catalyst. Annu Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 8.Zhou SF, Xue CC, Yu XQ, Wang G. Metabolic activation of herbal and dietary constituents and its clinical and toxicological implications: an update. Curr Drug Metab. 2007;8(6):526–553. doi: 10.2174/138920007781368863. [DOI] [PubMed] [Google Scholar]
- 9.Zhou SF, Zhou ZW, Li CG, et al. Identification of drugs that interact with herbs in drug development. Drug Discov Today. 2007;12(15–16):664–673. doi: 10.1016/j.drudis.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg DM, Davis RB, Ettner SL, et al. Trends in alternative medicine use in the United States, 1990–1997: results of a follow-up national survey. JAMA. 1998;280(18):1569–1575. doi: 10.1001/jama.280.18.1569. [DOI] [PubMed] [Google Scholar]
- 11.Ang-Lee MK, Moss J, Yuan CS. Herbal medicines and perioperative care. JAMA. 2001;286(2):208–216. doi: 10.1001/jama.286.2.208. [DOI] [PubMed] [Google Scholar]
- 12.Kaufman DW, Kelly JP, Rosenberg L, Anderson TE, Mitchell AA. Recent patterns of medication use in the ambulatory adult population of the United States: the Slone survey. JAMA. 2002;287(3):337–344. doi: 10.1001/jama.287.3.337. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Yang X, Ho PC, et al. Herb-drug interactions: a literature review. Drugs. 2005;65(9):1239–1282. doi: 10.2165/00003495-200565090-00005. [DOI] [PubMed] [Google Scholar]
- 14.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279(15):1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 15.Bent S. Herbal medicine in the United States: review of efficacy, safety, and regulation: grand rounds at University of California, San Francisco Medical Center. J Gen Intern Med. 2008;23(6):854–859. doi: 10.1007/s11606-008-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang ZX, Sun J, Howell CE, et al. Prediction of the likelihood of drug interactions with kinase inhibitors based on in vitro and computational studies. Fundam Clin Pharmacol. 2014;28(5):551–582. doi: 10.1111/fcp.12069. [DOI] [PubMed] [Google Scholar]
- 17.Qiu JX, Zhou ZW, He ZX, et al. Plumbagin elicits differential proteomic responses mainly involving cell cycle, apoptosis, autophagy, and epithelial to mesenchymal transition pathways in human prostate cancer PC-3 and DU145 cells. Drug Des Devel Ther. 2015;9:349–417. doi: 10.2147/DDDT.S71677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin JJ, Sharma S, Shumyak SP, et al. Synthesis and biological evaluation of novel folic acid receptor-targeted, beta-cyclodextrin-based drug complexes for cancer treatment. PLoS One. 2013;8(5):e62289. doi: 10.1371/journal.pone.0062289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito K, Iwatsubo T, Kanamitsu S, et al. Prediction of pharmacokinetic alterations caused by drug-drug interactions: metabolic interaction in the liver. Pharmacol Rev. 1998;50(3):387–412. [PubMed] [Google Scholar]
- 20.Zhou ZW, Zhou SF. Application of mechanism-based CYP inhibition for predicting drug-drug interactions. Expert Opin Drug Metab Toxicol. 2009;5(6):579–605. doi: 10.1517/17425250902926099. [DOI] [PubMed] [Google Scholar]
- 21.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58(5–6):737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JH. CYP induction-mediated drug interactions: in vitro assessment and clinical implications. Pharm Res. 2006;23(6):1089–1116. doi: 10.1007/s11095-006-0277-7. [DOI] [PubMed] [Google Scholar]
- 23.Colalto C. Herbal interactions on absorption of drugs: mechanisms of action and clinical risk assessment. Pharmacol Res. 2010;62(3):207–227. doi: 10.1016/j.phrs.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Kim IS, Kim SY, Yoo HH. Effects of an aqueous-ethanolic extract of ginger on cytochrome P450 enzyme-mediated drug metabolism. Pharmazie. 2012;67(12):1007–1009. [PubMed] [Google Scholar]
- 25.Kimura Y, Ito H, Hatano T. Effects of mace and nutmeg on human cytochrome P450 3A4 and 2C9 activity. Biol Pharm Bull. 2010;33(12):1977–1982. doi: 10.1248/bpb.33.1977. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Lim LY. Effects of spice constituents on P-glycoprotein-mediated transport and CYP3A4-mediated metabolism in vitro. Drug Metab Dispos. 2008;36(7):1283–1290. doi: 10.1124/dmd.107.019737. [DOI] [PubMed] [Google Scholar]
- 27.Brandin H, Viitanen E, Myrberg O, Arvidsson AK. Effects of herbal medicinal products and food supplements on induction of CYP1A2, CYP3A4 and MDR1 in the human colon carcinoma cell line LS180. Phytother Res. 2007;21(3):239–244. doi: 10.1002/ptr.2057. [DOI] [PubMed] [Google Scholar]
- 28.Chan E, Tan M, Xin J, Sudarsanam S, Johnson DE. Interactions between traditional Chinese medicines and Western therapeutics. Curr Opin Drug Discov Devel. 2010;13(1):50–65. [PubMed] [Google Scholar]
- 29.Saruwatari J, Nakagawa K, Shindo J, Nachi S, Echizen H, Ishizaki T. The in vivo effects of sho-saiko-to, a traditional Chinese herbal medicine, on two cytochrome P450 enzymes (1A2 and 3A) and xanthine oxidase in man. J Pharm Pharmacol. 2003;55(11):1553–1559. doi: 10.1211/0022357022061. [DOI] [PubMed] [Google Scholar]
- 30.Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2(6):875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 31.Chavez ML, Jordan MA, Chavez PI. Evidence-based drug herbal interactions. Life Sci. 2006;78(18):2146–2157. doi: 10.1016/j.lfs.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Singh D, Gupta R, Saraf SA. Herbs-are they safe enough? An overview. Crit Rev Food Sci Nutr. 2012;52(10):876–898. doi: 10.1080/10408398.2010.512426. [DOI] [PubMed] [Google Scholar]
- 33.Chan E, McLachlan A, O’Reilly R, Rowland M. Stereochemical aspects of warfarin drug interactions: use of a combined pharmacokinetic-pharmacodynamic model. Clin Pharmacol Ther. 1994;56(3):286–294. doi: 10.1038/clpt.1994.139. [DOI] [PubMed] [Google Scholar]
- 34.Kim JS, Nafziger AN, Gaedigk A, Dickmann LJ, Rettie AE, Bertino JS., Jr Effects of oral vitamin K on S- and R-warfarin pharmacokinetics and pharmacodynamics: enhanced safety of warfarin as a CYP2C9 probe. J Clin Pharmacol. 2001;41(7):715–722. doi: 10.1177/00912700122010618. [DOI] [PubMed] [Google Scholar]
- 35.Chan E, McLachlan AJ, Pegg M, MacKay AD, Cole RB, Rowland M. Disposition of warfarin enantiomers and metabolites in patients during multiple dosing with rac-warfarin. Br J Clin Pharmacol. 1994;37(6):563–569. doi: 10.1111/j.1365-2125.1994.tb04305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Williams KM, Liauw WS, et al. Effect of ginkgo and ginger on the pharmacokinetics and pharmacodynamics of warfarin in healthy subjects. Br J Clin Pharmacol. 2005;59(4):425–432. doi: 10.1111/j.1365-2125.2005.02322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funaki T, Soons PA, Guengerich FP, Breimer DD. In vivo oxidative cleavage of a pyridine-carboxylic acid ester metabolite of nifedipine. Biochem Pharmacol. 1989;38(23):4213–4216. doi: 10.1016/0006-2952(89)90517-0. [DOI] [PubMed] [Google Scholar]
- 38.Young HY, Liao JC, Chang YS, Luo YL, Lu MC, Peng WH. Synergistic effect of ginger and nifedipine on human platelet aggregation: a study in hypertensive patients and normal volunteers. Am J Chin Med. 2006;34(4):545–551. doi: 10.1142/S0192415X06004089. [DOI] [PubMed] [Google Scholar]
- 39.Dorso CR, Levin RI, Eldor A, Jaffe EA, Weksler BB. Chinese food and platelets. N Engl J Med. 1980;303(13):756–757. [PubMed] [Google Scholar]
- 40.Chiang HM, Chao PD, Hsiu SL, Wen KC, Tsai SY, Hou YC. Ginger significantly decreased the oral bioavailability of cyclosporine in rats. Am J Chin Med. 2006;34(5):845–855. doi: 10.1142/S0192415X06004338. [DOI] [PubMed] [Google Scholar]
- 41.Zick SM, Djuric Z, Ruffin MT, et al. Pharmacokinetics of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol and conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prev. 2008;17(8):1930–1936. doi: 10.1158/1055-9965.EPI-07-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y, Zick S, Li X, Zou P, Wright B, Sun D. Examination of the pharmacokinetics of active ingredients of ginger in humans. AAPS J. 2011;13(3):417–426. doi: 10.1208/s12248-011-9286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White B. Ginger: an overview. Am Fam Physician. 2007;75(11):1689–1691. [PubMed] [Google Scholar]
- 44.Desai HG, Kalro RH, Choksi AP. Effect of ginger and garlic on DNA content of gastric aspirate. Indian J Med Res. 1990;92:139–141. [PubMed] [Google Scholar]
- 45.Izzo AA, Ernst E. Interactions between herbal medicines and prescribed drugs: an updated systematic review. Drugs. 2009;69(13):1777–1798. doi: 10.2165/11317010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 46.Izzo AA, Ernst E. Interactions between herbal medicines and prescribed drugs: a systematic review. Drugs. 2001;61(15):2163–2175. doi: 10.2165/00003495-200161150-00002. [DOI] [PubMed] [Google Scholar]
- 47.Kongkaew C, Noyce PR, Ashcroft DM. Hospital admissions associated with adverse drug reactions: a systematic review of prospective observational studies. Ann Pharmacother. 2008;42(7):1017–1025. doi: 10.1345/aph.1L037. [DOI] [PubMed] [Google Scholar]
- 48.Becker ML, Caspers PW, Kallewaard M, et al. Determinants of potential drug-drug interaction associated dispensing in community pharmacies in the Netherlands. Pharm World Sci. 2007;29(2):51–57. doi: 10.1007/s11096-006-9061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Smet PA. Clinical risk management of herb-drug interactions. Br J Clin Pharmacol. 2007;63(3):258–267. doi: 10.1111/j.1365-2125.2006.02797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]