Abstract
Objective
The aim of this study was to review the current knowledge about involvement of microRNAs in breast cancer, and their potential in the clinic, published in scientific journals searched in Pubmed/Medline database until March 2014.
Results
MicroRNAs (miRNAs) are a family of 21–25 nucleotide small RNAs molecules. Currently, it is well known that miRNA plays a key role in all cellular processes of the organism including tumour initiation and progression. Many studies have shown that circulating miRNAs are attractive, easily detectable tumour biomarkers. Breast cancer is one of the most common cancers in the world. It is clinically established that different subtypes may respond differently to therapies, give metastases and present drug resistance. MicroRNAs have a potential as diagnostic, prognostic and therapeutic tools in breast cancer.
Conclusion
Molecular knowledge is crucial for choosing the most effective therapy for individual patients. MicroRNAs holds a great potential in anticancer therapy.
Keywords: Breast cancer, miRNA, Tumourigenesis, Biomarkers
1. Breast cancer – why is it so hard to cure?
Breast cancer is one of the most common cancers in the world with more than 1,300,000 cases worldwide.1 This very heterogeneous disease is clinically divided by histological types based on expression of specific receptors: estrogen receptor (ER) positive (the most numerous and diverse), progesterone receptor (PR) and HER2 (ERBB2) receptor or the absence of all of them, named triple negative breast cancer (TNBC).1–3,5,26 Another classification of breast cancer distinguished luminal A, luminal B, basal and HER2 enriched groups.4 It is clinically established that different subtypes may respond differently to therapies, give metastases and present drug resistance.1,6 For example, TNBC is associated with high risk of recurrence and distant metastases to the brain, compared to other, receptor positive tumours,7 and responses only to chemotherapy.1 Molecular knowledge is crucial for choosing the most suitable therapy for individual patients combined with cost-effectiveness of the treatment.98 Conventional treatment for breast cancer includes wide local excision, sentinel lymph node biopsy or axillary lymph node dissection, adjuvant medical treatment and radiotherapy to the whole breast.25,99
2. miRNA – briefly about biogenesis, mechanism and function
MicroRNAs (miRNAs) are a family of 21–25 nucleotide small RNAs molecules.8,9 When in 1993 Victor Ambros et al. found out that gene lin-4 did not code a protein,10 no one could predict that this small molecule plays such an important role in gene regulation. Currently, it is well known that miRNA plays a key role in all cellular processes of the organism, including among others: development, proliferation, apoptosis, differentiation, organogenesis.8–12 Therefore, it is not surprising that currently miRNA plays a role in tumour initiation and progression. The number of reports associating miRNA with cancer has risen from 0.002% of total cancer reports in 2002 to 2% at present.11 The biogenesis of miRNAs has been extensively examined.13 miRNA are intergenic, intronic or exonic (in exons of coding or non-coding genes) and can be transcribed as a single miRNA from its own promoter (approximately 50%) or several miRNAs as a cluster from a shared promoter.8,14,15 The transcription of miRNA is driven for the largest part by RNA polymerase II. The long primary transcript (pri-miRNA) can measure more than 1 kb8,16 and contains a 5′-cap structure and a 3′-poly(A) tail. A region with not perfectly complementary sequence creates a secondary hairpin structure recognized and processed by a complex of RNAses. Pri-miRNA is converted into a pre-form by ribonuclease DROSHA and the DGCR8 cofactor which is a hairpin structure of ∼70–80 nucleotides.16 These stem-loop shaped pre-miRNAs are translocated from nucleus into cytosol via exportin 5.17 The RNase III endonuclease DICER1 cleaves the pre-form generating a double-stranded miRNA of 18–25 nucleotides in length.16 The double-strand is unwound and the single strands are incorporated into RNA-Induced Silencing Complex – RISC (Fig. 1). Recent reports show that not only 5′-strand is functional as it was believed in the past, 3′-strand is not always degraded but also can give mature, functional miRNA.18 As a part of this complex, microRNA is able to regulate gene expression at a post-transcriptional level, bind partial complementary target mRNA, mainly leading to mRNA degradation or translation inhibition.19 The interactions between miRNA and mRNA are usually located near 5′ terminus miRNA molecule. These ∼6–8 nt sequence is highly conserved.20 It is well known that 3′-UTR mRNA may contain multiple miRNA-binding sites for different miRNAs, and a single miRNA may bind multiple targets.21,22 The “many targets” hypothesis suggests that ∼60% of the mRNAs have one or more evolutionarily conserved sequences and that they are able to interact with miRNA.21 Computational approaches are used to predict mRNA–miRNA interactions and a number of algorithms have been currently developed and tested for accuracy and precision (using both computational and laboratory techniques).23,24 Currently, 2024 mature miRNAs have been discovered in humans [http://www.mirbase.org/] and it is estimated that around 30% of human genes are regulated by miRNA.8
Fig. 1.
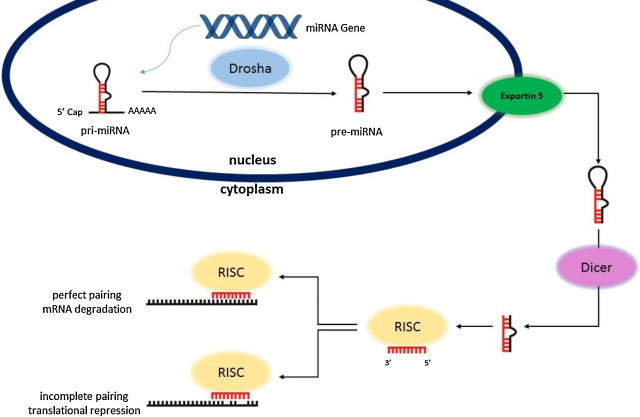
Model for microRNA biogenesis.
3. miRNA in tumourogenesis
The first evidence of the involvement of microRNAs in human cancer was described in 2002 by Calin and et al.27 Studies on chronic lymphocytic leukaemia (CLL) have shown the knock down or knock out of miR-15a and miR-16–1 in approximately 69% of CLLs (deletion of chromosome 13q14). These results pushed the investigators further and they mapped all known microRNA genes.28 Many of them are located in chromosomal loci prone to deletions or amplifications. In the case when microRNAs involved in the negative regulation of a transcript encoding a known tumour suppressor gene are amplified, this amplification results in the increased expression of the miRNA and consequent silencing of the tumour suppressor gene. In opposite, miRNAs repressing oncogenes are often located in fragile foci, where deletions or mutations can occur and result in reduced miRNA levels causing overexpression of the target oncogene. Therefore, in human cancer, alterations of microRNA expression represent a rule rather than being exceptional or incidental.
In 2000 Hanahan and Weinberg described the hallmarks of cancer.63 Below, we present a few examples of how microRNA is involved in the cancerogenesis process.
Uncontrolled proliferation is a special feature of cancer cells. It is well established that Notch signalling maintains between cell proliferation, differentiation and apoptosis, thus, alterations in this signalling pathway are associated with tumourigenesis.65 Several studies have shown that the miR-34 participates in p53 and Notch pathways regulation in consistency with tumour suppressor activity. Ji et al. reported that human gastric cancer cells with miR-34 restoration reduced the expression of target gene Notch66 and that Notch-1 and Notch-2 are downstream genes of miR-34 in pancreatic cancer cells.67 It is worth to mention that they also found that pancreatic cancer stem cells are enriched with tumour-initiating cells or cancer stem cells with high levels of Notch-1/2 and loss of miR-34 which suggests the engagement of miR-34 in pancreatic stem cell self-renewal via modulation of target Notch.
Other miRNA involved in Notch pathway regulation is miR-199. Inhibition of Hes-1 (transcriptional factor) by miR-199b-5p negatively modulated the medulloblastoma (MB) cell growth. Moreover, over-expression of miR-199b-5p causes decreasing of cells with stem-like cells phenotype (CD133+) and blocks expression of several cancer stem-cell genes.68 It is important that the level of miR-199b-5p in the non-metastatic cases was significantly higher than in the metastatic. Patients from first group (high level) have shown a better overall survival.
MiR-290 cluster can directly target the Retinoblastoma-like 2 protein (Rbl2), affect telomere integrity and telomere-length homeostasis.69 Small, non-coding RNA plays also an important role in angiogenesis. For example let7-f, miR-27b and miR-130a have been identified as pro-angiogenic players in vitro.70,71 Lee et al. identified the miR-378 function as an oncogene to enhance tumour cell survival and blood vessel expansion through repression of the expression of tumour suppressors Sufu and Fus-1.72
The most common cause of mortality in human cancers are metastasis. It is a series of complicated processes regulated by multiple factors and it allows the outgrowth of metastatic tumours in the new microenvironment.73 Currently, there is strong evidence that miRNAs coordinate and play important roles in tumour invasion and metastasis.74,75 Si et al. have found that miR-21 was highly overexpressed in breast tumours compared to the matched normal breast tissues among 157 human miRNAs analyzed. They did transfection of breast cancer MCF-7 cells with anti-miR-21 oligonucleotides and found that anti-miR-21 suppressed both in vitro cell growth and tumour growth in the xenograft mouse model.76 They also made a two-dimensional differentiation in gel electrophoresis of tumours treated with anti-mir-21 and identified the tumour suppressor tropomyosin 1 (TPM1) as a potential mir-21 target, which can at least in part explain the notion that mir-21 functions as an oncogene.77 Tumour-suppressor Pdcd4 inhibits transformation and invasion and it is down regulated in cancers. Asangani et al. have shown that miR-21 downregulates Pdcd4 protein and upregulates tumour cell invasion in cultured colon cancer cells. They demonstrated an inverse correlation between miR-21 and Pdcd4-protein.78
4. Methods of detection of miRNA in clinical practice
In the clinical practice, it is necessary to use biomarkers for detection and early diagnosis. miRNAs are stable due to their small size, which makes them a better tool for diagnosis. This molecule can be detected and isolated from frozen and paraffin-embedded tissues, blood,37,38 and different biologic fluids like urine,39 sputum40 or saliva.41 Many studies have shown that circulating miRNAs are attractive, easily detectable tumour biomarkers.42–45 It is necessary to explore the miRNA topic more deeply because of its potential for being used in tumour diagnosis, prognosis and cure.29 Many studies have been developed on the global expression of miRNA genes in normal and diseased tissues30–34,61. In contrast to mRNA global profiling, miRNA expression signatures (miRNome) allowed to discriminate with high accuracy different types of cancer.32,35 It is worth to notice that miRNA expression profile is tissue unique, which has foreseeable benefits in aiding clinical diagnosis and treatment.36
Currently, there is a possibility to use different techniques and methods to detect miRNA.24,46 The most important issue is to determine which miRNAs are changing in a specific disease and if the changes are representative to it, and if only to it.46 Methods and strategies for profiling miRNA expression are described by Kong et al. in the review.24 When we have a based knowledge about the expression profile that is necessary for results interpretation, we can create and develop new methods to act. Some of miRNAs have already been used in clinical practise, for example, a miRNA-based diagnostic assay approved by FDA.47 The most popular methods to detect miRNA that can be easily employed in a routine clinic diagnostic are: quantitative-reverse transcription PCR (qRT-PCR), hybridization-based methods and next generation sequencing [review 47]. Wei et al. systematically screened miRNA expression profile using the Solexa deep-sequencing technology.62 They have created two libraries from T-47D breast cancer cells treated with or without prolactin, identified a number of miRNAs significantly differentially expressed between these two panels, and also detected several new miRNAs associated with prolactin receptor signalling pathway in breast cancer.
Also new technology-based methods have been recommended for the exploration and examination of miRNA, for example electrochemical genosensor that can easily detect miRNA in the serum or other biological samples48 or nanopore sensor based on the α-haemolysin protein.49 Unfortunately, there are still considerable limitations to use miRNA expression as routine in the clinic hence more clinical studies are required.
5. miRNA in breast cancer – problem during effective treatment?
Altered miRNA expression in human breast cancer was first described by Iorio et al. in 2005.50 They analyzed 76 breast cancers, 10 normal breast samples and 14 breast cancer cell lines to identify miRNA whose expression is significantly deregulated in cancer versus normal breast tissues. In this panel there were 29 miRNAs identified with aberrant expression by microarray and Northern blot analyses. MiR-10b, miR-125b, miR-145 (all of them down-regulated), miR-21 and miR-155 (all of them up-regulated) were the most consistently deregulated in breast cancer. This finding suggests that they may act as tumour suppressor (down-regulated) or oncogene (up-regulated) in breast cancer tumourogenesis. It is known that expression of miR-125b is high in differentiated cells and tissues51 so the decreased level of its expression in breast cancer suggests the impairment of differentiation capabilities of cancer cells.50 In 2007, Sempere et al. published a paper about miRNas’ distribution in breast tumour tissue versus normal tissue from more than 100 patients using hybridization in situ.52
As it was mentioned above, miRNAs can inhibit tumourogenesis by repressing oncogenes. Members of the ErbB family play a very important role in development, cellular proliferation and survival in human epithelial malignancies and they are frequently amplified or overexpressed in breast cancer (20–30%); moreover, they are significantly associated with a worse prognosis.53,54 Scott et al. have examined miR-125a and miR-125b overexpression in SKBR3 cells which decreased ErbB2 protein level of approximately 40–65% and ErbB3 level of around 60–80%.54 SKBR3 cells with overexpression of miR-125a and miR-125b were impaired in their malignant cell phenotype (reduced migration and invasion capacities). Wang et al. first providing experimental data to demonstrate that miR-125a, miR-15b and miR-205 act in concert to regulate the expression of ErbB2/ErbB3 in breast cancer cells.55 Because of its role in tumourogenesis, ErbB family is an excellent target for selective anticancer therapies. ErbB-targeted therapies currently used in clinic are divided into two strategies: blocking antibodies (for example transtuzumab targeting ErbB2) and tyrosine kinase inhibitor (for example aslapatinib against EGFR and ErbB2). Because of the lack or low kinase activity ErbB3 receptor can be blocked only using antibody56 and it has been currently clinically studied (http://www.clinical-trials.gov/). ErbB2 required ErbB3 to promote breast cancer cell proliferation57 and ErbB3 has an important role in ErbB2-altered breast cancers.58 The same group reported that ErbB3 contributes to ErbB2-mediated therapeutic resistance to tamoxifen59 and paclitaxel,60 thus they proposed a novel approach to target ErbB2/ErbB3 by reducing their protein levels by microRNA rather than inhibiting only the signalling pathways.55
Biagioni et al. identified 22 differentially expressed miRNAs in HER2 tumours, 31 in basal-like tumours and 33 in luminal tumours.64 Two miRNAs: miR-10b* and miR-139-5p were down-regulated and three: miR-425, miR-454 and miR-301a were up-regulated for all three subtypes. The most important finding from this analyses is that the miR-10b* is a master regulator of breast cancer cell proliferation (lower levels of miR-10b* correspond to higher tumour size, which was also confirmed by soft agar colony formation assay in MCF7 breast cancer cells). Further in vivo examination in a xenograft model confirmed a pivotal role of miR-10b* in breast cancer cell proliferation.64 This experiment can be crucial for exploring the therapeutic potential of miR-10b* in breast cancer.
Zhong et al. demonstrated that an altered miRNA expression pattern is involved in acquiring resistance to adriamycin and docetaxel which are two chemotherapeutic agents commonly used in the treatment of breast cancer. This regulation could be in part via targeting PTEN.79
It was mentioned above that miR-21 plays an important role in tumourogenesis. Wang et al. investigated the association of miR-21 expression with the sensitivity of breast cancer cells to doxorubicin. Using TaqMan RT-PCR and Western blot assay to detect the expression of mature miR-21 and tumour suppressor gene (PTEN) protein they found that dysregulation of miR-21 plays a critical role in the doxorubicin resistance of breast cancer.80
Breast cancer drug resistance is also combined with deregulation of miRNA-200c. Examination of the miRNA-200c expression in tumour specimens obtained from thirty-nine breast cancer patients who had received neoadjuvant chemotherapy showed that miRNA-200c was down-regulated in non-responders as compared to responders.81 Another paper reports that miRNA-30c played a pivotal role in chemoresistance via direct targeting of the actin-binding protein Twinfilin 1 which is responsible for the promoting of epithelial-to-mesenchymal transition.82 Study of miRNA-19 expression levels in three multidrug resistance (MDR) cell lines in comparison with their parent cell line, MCF-7, using a miRNA microarray showed that miRNA-19 was overexpressed in all three MDR cell lines and modulate chemoresistance directly targeting PTEN. Inhibition of miR-19 sensitized MDR cells to chemotherapeutic agents in vitro and in vivo.83
Radiotherapy is an effective and well-established cancer treatment, however, currently, little is known of how microRNA may regulate radiation resistance [revived in 85] in breast cancer. Liu et al. showed that miR-95 promotes radiation resistance and development of an aggressive phenotype on prostate and breast cancer.85 Interestingly, examinations have been performed on different cell line varied with TP53 status, and the results suggest that miR-95 promotes radiation resistance independently of the TP53 function. Xenograft tumour experiments revealed that miR-95 overexpressing tumours have less necrosis and increase proliferation even after irradiation. Also miRNA-21 plays a role in radioresistance acting as a radioresistant miRNA.86 miRNA-21 expression in breast cancer cells contributes to radiation resistance by compromising cell cycle progression (radiation-induced G2/M arrest). Chen et al. showed that antisense targeting miR-155 could increase the sensitivity of breast cancer cells to irradiation.87 Liang et al. also suggest that miRNA-302 could be a potential sensitizer to radiotherapy. miRNA-302 sensitizes resistant breast cancer cells to irradiation in vitro and in vivo.100
MetastamiRs are microRNAs crucial in metastatic spreading. It was proved in migration, invasion and poor prognosis phenotype. Cloonan et al. found that miR-139-5p is de-regulated in human triple negative breast cancer samples and it is able to target several invasive pathways and migratory phenotypes of cancer cells.88 Overexpression of miR-205 inhibits MCF7 cell migration and invasiveness and reduces the growth and colony-formation capacity of MCF7 cells by inducing apoptosis.89 Li et al. demonstrated that decreased expression of miR-720 was correlated with lymph node metastasis and manifests antimetastatic activity by down-regulating TWIST1.90 Studies have shown that ectopic overexpression of miR-301a promote breast cancer cell migration, invasion and metastasis both in vitro and in vivo via constitutively activated Wnt/β-catenin signalled by direct targeting PTEN.91 Also miRNA-155 plays a critical role in breast cancer progression and metastasis. High miR-155 expression was closely correlated with higher tumour grade, advanced tumour stage and lymph node metastasis and it is worth mentioning that relative expression of miR-155 in tumours with lymph node metastasis was significantly higher than that in tumours with no lymph node metastasis.87 Petrović et al. showed that miR-21 expression levels in invasive with non-invasive component and pure invasive cancers were significantly increased compared with normal tissue. The highest difference between non-invasive and pure invasive cancer samples than in other compared group pairs indicates that miR-21 is a strong specific factor to the process of invasion.92
Trastuzumab resistance is a major issue in therapy for HER2+ breast cancers. Gong et al. screened for miRNAs that were differentially expressed in the trastuzumab-resistant breast cancer cells, and identified that miRNA-21 was up-regulated among the reported PTEN-targeting miRNAs. They showed that blocking the action of miR-21 with antisense oligonucleotides re-sensitized the resistant cells to the therapeutic activities of trastuzumab by inducing growth arrest, proliferation inhibition, and G1-S cell cycle checked in the presence of the antibody.93
Mei et al. showed that combined taxol chemotherapy and miR-21 inhibitor treatment via polyamidoamine (PAMAM) dendrimers vector to evaluate the effects of the combination therapy reduced cell viability and invasiveness, thus it might represent a promising novel therapeutic approach for the treatment of breast malignancies.94
Another clinical problem in HER2 positive breast cancer treatment is resistance to tamoxifen (the selective oestrogen receptor modulator). Cittelly et al. analyzed multiple cell models of tamoxifen resistance derived from MCF-7 cells to examine the influence of microRNAs (miRNAs) on tamoxifen resistance.95 They suggest that miR-342 regulates tamoxifen response in breast tumour cell lines via the regulation of expression genes involved and in tumour cell apoptosis and cell cycle progression. Thus, restoring miR-342 expression may be a novel therapeutic approach to sensitizing and suppressing the growth of tamoxifen refractory breast tumours. Another group also demonstrated that tamoxifen resistance in breast cancer is associated with MiR-221/222.96
The data published by Kim in 2011, strongly supported that overexpression of miRNA-145 reduces the levels of cancer cell survival factors and inhibits cancer cell growth and metastasis.97 They demonstrated that miR-145 delivered using an adenoviral vector system showed significant inhibition of tumour growth in breast tumour bearing mice.
U.S. National Institutes of Health approved clinical trial NCT01612871 focuses on the role of miRNAs in resistance and sensitivity in breast cancer. The study concerns women with metastatic invasive breast cancer or locally advanced breast cancer and for whom treatment with tamoxifen or anti aromatase is indicated. This study is currently recruiting participants [http://www.clinicaltrials.gov]. Description of other clinical trials are available online.
As indicated above, deregulation of miRNAs is involved in the development of cancer disease, invasiveness, metastasis and treatment failure. Aberrant miRNA expression has been observed in various types of human tumours playing a role as tumour-suppressor gene or oncogene. miRNA has a great potential in the therapeutic design because of its multiple function in cell homeostasis and one hit-multiple target pathway.
Cited literature indicates that miRNA could be a potential therapeutic tool and promising biomarker in personalized treatment.
Conflict of interest
None declared.
Financial disclosure
Greater Poland Cancer Centre’s internal grant no. 3/2013(54).
Acknowledgements
To Dr. Wiktoria Maria Suchorska and Prof. Dr. hab. Julian Malicki for helpful discussions and motivation.
References
- 1.The Cancer Genome Atlas Network. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed]
- 2.van’t Veer L.J., Dai H., van der Vijver M.J. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad A. Pathways to breast cancer recurrence. ISRN Oncol. 2013;2013 doi: 10.1155/2013/290568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubert K.E., Carey L.A., Wazer D.E. Breast cancer in molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence and response to therapy. Semin Radiat Oncol. 2009;19(6):204–210. doi: 10.1016/j.semradonc.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Gatza M.L., Lucas J.E., Barry W.T. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiser L.M., Sadanandam A., Kuo W.L. Subtype and pathways specific response to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chacón R.D., Costanzo M.V. Triple-negative breast cancer. Breast Cancer Res. 2010;12(2):S3. doi: 10.1186/bcr2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.He L., Hannon G. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Soriano A., Jubierre L., Almazan-Moga A. microRNA as pharmacological targets in cancer. Pharmacol Res. 2013;75:3–14. doi: 10.1016/j.phrs.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophilia. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 13.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 14.Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;3(22):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Saini H.K., Griffiths-Jones S., Enright A.J. Genomics analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y., Kim H., Han J., Yeom K.H., Lee S., Beak S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi R., Quin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Gene Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamura K., Phillips M.D., Tyler D.M., Duan H., Chou Y.T., Lai E.C. The regulatory activity of microRNA species has substantial influence on microRNA and 3′UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the DICER complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org source: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel D.P. MicroRNA: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A., Wong A.K., Tizardi M.L., Moore R.J., Lefevre C. miRNA_targets: a database for miRNA target predictions in coding and non-coding regions of miRNAs. Genomics. 2012;100:352–356. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Kong W., Zhao J.J., He L., Cheng J.Q. Strategies for profiling microRNA expression. J Cell Physiol. 2009;218:22–25. doi: 10.1002/jcp.21577. [DOI] [PubMed] [Google Scholar]
- 25.Bitterman A., Kessner R., Goldman I., Shiloni E., Steiner M. Intraoperative radiotherapy for breast cancer. IMAJ. 2012;14:256–259. [PubMed] [Google Scholar]
- 26.Recchia F., Candeloro G., Desideri G. Triple-negative breast cancer: multipronged approach, single-arm pilot phase II study. Cancer Med. 2012;1(August (1)):89–95. doi: 10.1002/cam4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin G.A., Dumitru C.D., Shimizu M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calin G.A., Sevignani C., Dumitru C.D. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 30.Calin G.A., Ferracin M., Cimmino A. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 31.Liu C.G., Calin G.A., Meloon B. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J., Getz G., Miska E.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 33.Creighton C.J., Reid J.G., Gunarante P.H. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farazi T.A., Horlings H.M., Ten Hoeve J.J. MicroRNA sequence and expression analysis in breast tumours by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volinia S., Calin G.A., Liu S.G. A microRNA expression signature of human solid tumours defines cancer genome targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenfeld N., Aharonov R., Meiri E. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell P.S., Parkin R.K., Kroh E.M. Circulating microRNA as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 39.Hanke M., Hoefig K., Merz H. A robust methodology to study urine microRNA as tumour marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 40.Yu L., Todd N.W., Xing L. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park N.J., Zhou H., Elashoff D. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrie C.H., Gal S., Dunlop H.M. Detection of elevated levels of tumour-associated microRNA in serum of patients with diffuse large B-cell lymphoma. Br J Hematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 43.Henegham H.M., Miller N., Kelly R., Newell J., Kerin M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Z., Huang D., Ni S., Peng Z., Sheng W., Du X. Plasma microRNA are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 45.Xing L., Todd N.W., Yu L., Fang H., Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 46.de Planell-Saguer M., Rodicio M.C. Detection methods for microRNAs in clinic practice. Clin Biochem. 2013;46:869–878. doi: 10.1016/j.clinbiochem.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 47.Szafrańska-Schwarzbach A.E., Adai A.T., Lee L.S., Conwell D.L., Andruss B.F. Development of a miRNA-based diagnostic assay for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn. 2011;11(3):249–257. doi: 10.1586/erm.11.10. [DOI] [PubMed] [Google Scholar]
- 48.Lusi E.A., Passamano M., Guarascio P., Scarpa A., Schiavo L. Innovative electrochemical approach for an early detection of microRNAs. Anal Chem. 2009;81:2819–2822. doi: 10.1021/ac8026788. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Zheng D., Tan Q., Wang M.X., Gu L.Q. Nanopore-based detection of circulating microRNA in lung cancer patients. Nat Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iorio M.V., Ferracin M., Liu C.G. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y.S., Kim H.K., Chung S., Kim K.S., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 52.Sempere L.F., Christensen M., Silahtaroglu A. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 53.Yarden Y., Sliwkowski M.X. Untangling the ERB signaling network. Nat Rev. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 54.Scott G.K., Goga A., Bhaumik D., Berger C.E., Sullivan C.S., Benz C.C. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of microRNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 55.Wang S., Huang J., Lyu H. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013;4:e556. doi: 10.1038/cddis.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoeberl B., Faber A.C., Li D. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer Res. 2010;70:2485–2494. doi: 10.1158/0008-5472.CAN-09-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holbro T., Beerli R.R., Maurer F., Koziczak M., Barbas C.F., 3rd, Hynes N.E. The ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee-Hoeflich S.T., Crocker L., Yao E. A central role for HER3 in HER2-amplified breast cancer: implications for target therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 59.Liu B., Ordonez-Ercan D., Fan Z., Edgerton S.M., Yang X., Thor A.D. Downregulation of ErbB3 abrogates ErbB2-mediated tamoxifen resistance in breast cancer cells. Int J Cancer. 2007;120:1874–1882. doi: 10.1002/ijc.22423. [DOI] [PubMed] [Google Scholar]
- 60.Wang S., Huang X., Lee C.K., Liu B. Elevated expression of ErbB3 confers paclitaxel resistance in ErbB2-overexpressing breast cancer cells via upregulation of survivin. Oncogene. 2010;29:4225–4236. doi: 10.1038/onc.2010.180. [DOI] [PubMed] [Google Scholar]
- 61.van Schooneveld E., Wouters M.C., Van der Auwera I. Expression profiling of cancerous and normal breast cancer tissues identifies microRNA that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;4:R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei Q., He W., Yao J., Guo L., Lu Y., Cao X. Identification and characterization of microRNAs expressed in human breast cancer T-47D cells in response to prolactin treatment by Solexa deep-sequencing technology. Biochem Biophys Res Commun. 2013;432:480–487. doi: 10.1016/j.bbrc.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 63.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 64.Biagioni F., Bossel Ben-Moshe N., Fotemaggi G. Mir-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–1229. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., Li Y., Banerjee S., Sarkar F.H. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279:8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ji Q., Hao X., Meng Y. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ji Q., Hao X., Zhang M. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PloS One. 2009;4:e46816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garzia L., Andolfo I., Cusanelli E. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PloS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benetti R., Gonzalo S., Jaco I. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuehbacher A., Urbich C., Zeiher A.M., Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y., Gorski D.H. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(February (3)):1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee D.Y., Deng Z., Wang C.H., Yang B.B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104(December (51)):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(August (8)):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 74.Steeg P.S. Cancer: micromanagement of metastasis. Nature. 2007;449(7163):671–673. doi: 10.1038/449671a. [DOI] [PubMed] [Google Scholar]
- 75.Bracken C.P., Gregory P.A., Khew-Goodall Y., Goodall G.J. The role of microRNAs in metastasis and epithelial-mesenchymal transition. Cell Mol Life Sci. 2009;66(10):1682–1699. doi: 10.1007/s00018-009-8750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 77.Zhu S., Si M.L., Wu H., Mo Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 78.Asangani I.A., Rasheed S.A., Nikolova D.A. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 79.Zhong S., Li W., Chen Z., Xu J., Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531(November (1)):8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 80.Wang Z.X., Lu B.B., Wang H., Cheng Z.X., Yin Y.M. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res. 2011;42(May (4)):281–290. doi: 10.1016/j.arcmed.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 81.Chen J., Tian W., Cai H., He H., Deng Y. Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol. 2012;29(4):2527–2534. doi: 10.1007/s12032-011-0117-4. [DOI] [PubMed] [Google Scholar]
- 82.Bockhorn J., Dalton R., Nwachukwu C. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang Z., Li Y., Huang K., Wagar N., Shim H. Regulation of miR-19 to breast cancer chemoresistance through targeting PTEN. Pharm Res. 2011;28(December (12)):3091–3100. doi: 10.1007/s11095-011-0570-y. [DOI] [PubMed] [Google Scholar]
- 84.Matheetrairut C., Slack F.J. MicroRNA in the ionizing radiation response and radiotherapy. Curr Opin Genet Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang X., Taeb S., Jahangiri S. miRNA-95 mediates radioresistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer Res. 2013;73(December (23)):6972–6986. doi: 10.1158/0008-5472.CAN-13-1657. [DOI] [PubMed] [Google Scholar]
- 86.Anastasov N., Hofig I., Vasconcellos I.G. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol. 2012;7(1):206. doi: 10.1186/1748-717X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen J., Wang B.C., Tang J.H. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106(3):260–266. doi: 10.1002/jso.22153. [DOI] [PubMed] [Google Scholar]
- 88.Krishnan K., Steptoe A.L., Martin H.C. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA. 2013;19(December (12)):1767–1780. doi: 10.1261/rna.042143.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Z., Liao H., Deng Z. miRNA-205 affects infiltration and metastasis of breast cancer. Biochem Biophys Res Commun. 2013;441(November (1)):139–143. doi: 10.1016/j.bbrc.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 90.Li L.Z., Zhang C.Z., Liu L.L. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35(February (2)):469–478. doi: 10.1093/carcin/bgt330. [DOI] [PubMed] [Google Scholar]
- 91.Ma F., Zhang J., Zhong L. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene. 2014;535(2):191–197. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 92.Petrović N., Mandusić V., Stanojević B. The difference in miR-21 expression levels between invasive and non-invasive breast cancers emphasizes its role in breast cancer invasion. Med Oncol. 2014;31(March (3)):867. doi: 10.1007/s12032-014-0867-x. [DOI] [PubMed] [Google Scholar]
- 93.Gong C., Yao Y., Wang Y. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286(May (21)):19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mei M., Ren Y., Zhou X. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010;9(February (1)):77–86. doi: 10.1177/153303461000900109. [DOI] [PubMed] [Google Scholar]
- 95.Cittelly D.M., Das P.M., Spoelstra N.S. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9(December):317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller T.E., Ghoshal K., Ramaswamy B. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(October (44)):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim S.J., Oh J.S., Shin J.Y. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155(November (3)):427–434. doi: 10.1016/j.jconrel.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 98.Malicki J., Litoborski M., Bogusz-Czerniewicz M., Świeżewski A. Cost-effectiveness of the modifications in the quality assurance system in radiotherapy in the example of in-vivo dosimetry. Phys Med. 2009;25(December (4)):201–206. doi: 10.1016/j.ejmp.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 99.Murawa P., Murawa D., Adamczyk B., Połom K. Breast cancer: actual methods of treatment and future trends. Rep Pract Oncol Radiother. 2014;19(January (3)):165–172. doi: 10.1016/j.rpor.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang Z., Ahn J., Guo D., Votaw J.R., Shim H. MicroRNA-302 replacement therapy sensitizes breast cancer cells to ionizing radiation. Pharm Res. 2013;30(April (4)):1008–1016. doi: 10.1007/s11095-012-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


