Abstract
Splicing factor proline- and glutamine-rich (SFPQ) also commonly known as polypyrimidine tract-binding protein-associated-splicing factor (PSF) and its binding partner non-POU domain-containing octamer-binding protein (NONO/p54nrb), are highly abundant, multifunctional nuclear proteins. However, the exact role of this complex is yet to be determined. Following purification of the endogeneous SFPQ/NONO complex, mass spectrometry analysis identified a wide range of interacting proteins, including those involved in RNA processing, RNA splicing, and transcriptional regulation, consistent with a multifunctional role for SFPQ/NONO. In addition, we have identified several sites of arginine methylation in SFPQ/PSF using mass spectrometry and found that several arginines in the N-terminal domain of SFPQ/PSF are asymmetrically dimethylated. Furthermore, we find that the protein arginine N-methyltransferase, PRMT1, catalyzes this methylation in vitro and that this is antagonized by citrullination of SFPQ. Arginine methylation and citrullination of SFPQ/PSF does not affect complex formation with NONO. However, arginine methylation was shown to increase the association with mRNA in mRNP complexes in mammalian cells. Finally we show that the biochemical properties of the endogenous complex from cell lysates are significantly influenced by the ionic strength during purification. At low ionic strength, the SFPQ/NONO complex forms large heterogeneous protein assemblies or aggregates, preventing the purification of the SFPQ/NONO complex. The ability of the SFPQ/NONO complex to form varying protein assemblies, in conjunction with the effect of post-translational modifications of SFPQ modulating mRNA binding, suggests key roles affecting mRNP dynamics within the cell.
Keywords: polypyrimidine tract-binding protein-associated-splicing factor, splicing factor proline- and glutamine-rich, non-POU domain-containing octamer-binding protein, arginine methylation, citrullination, mRNP binding, mass spectrometry
INTRODUCTION
Splicing factor, proline- and glutamine-rich (SFPQ) also commonly known as polypyrimidine tract-binding protein-associated-splicing factor (PSF) and its binding partner non-POU domain-containing octamer-binding protein (NONO/p54nrb) are highly abundant, multifunctional nuclear proteins (Shav-Tal and Zipori 2002). SFPQ/PSF comprises an N-terminal glycine rich domain (containing three RGG repeats), a proline/glutamine-rich domain (P/Q), two RNA recognition motifs (RRMs), and a C-terminal region with two nuclear localization signals (Patton et al. 1991). NONO/p54nrb is homologous to the C-terminus of SFPQ/PSF (Shav-Tal and Zipori 2002) and has been shown to associate with SFPQ/PSF (Zhang et al. 1993; Straub et al. 1998; Peng et al. 2002; Shav-Tal and Zipori 2002; Hanna et al. 2006).
SFPQ and NONO have been identified in proteomic studies of the spliceosome and in splicing-related complexes (Rappsilber et al. 2002; Peng et al. 2006). Furthermore, in the absence of SFPQ/PSF, spliceosome formation (complexes A and B) is severely impaired (Patton et al. 1993). SFPQ/PSF binds PTB/hnRNPI which interacts with the polypyrimidine tract of intronic mRNA, a region important in the definition of the 3′-splice site in mammalian cells (Patton et al. 1993). In contrast, NONO is not required for spliceosome assembly or splicing (Zhang and Wu 1996). However, it has been shown to interact with important components of the spliceosome such as U5 snRNA and the 5′-splice site (Peng et al. 2002; Kameoka et al. 2004). NONO was also shown to increase the efficiency of splicing in vitro (Zhang and Wu 1996). Consistent with the coupling of transcription and splicing (Rosonina et al. 2005) this complex additionally interacts with the C-terminal domain of RNAPII (Hirose and Manley 2000; Emili et al. 2002). Moreover, NONO binds to the 5′-splice sites in a complex with both splicing and transcription factors (Emili et al. 2002; Kameoka et al. 2004).
Further to their spliceosome-related functions, SFPQ/NONO has been implicated in a wide variety of regulatory roles including the selective nuclear retention of defective mRNAs (Zhang and Carmichael 2001). More recently, SFPQ/NONO were shown to be stimulatory factors for U snRNA export (Izumi et al. 2014) and DNA topoisomerase I (Straub et al. 1998, 2000) together with a role in the nonhomologous end joining response to DNA double strand breaks (Bladen et al. 2005). SFPQ also acts as a transcriptional regulator recruiting Sin3a, which in turn recruits histone deacetylase (HDAC) thus repressing transcription (Mathur et al. 2001; Sewer et al. 2002; Dong et al. 2007). Similarly a PER complex that rhythmically associates with DNA bound CLOCK-BMAL1 at the Per1 promoter represses transcription by virtue of its constituent SFPQ which recruits the SIN3-HDAC complex to deacetylate histones 3 and 4 (Duong et al. 2011). Conversely, in the case of androgen receptor responsive genes, transcriptional activation by SFPQ/NONO has been demonstrated (Ishitani et al. 2003; Kuwahara et al. 2006). Further examples of the functional diversity of SFPQ and NONO are their involvement in developmentally linked gene regulation, specifically, in relation to neuronal tissue development (Chanas-Sacré et al. 1999; Ju et al. 2004) and cell cycle control (Stier et al. 2005).
Protein arginine N-methyltransferases (PRMTs) catalyze the post-translational transfer of a methyl group from the donor S-adensoyl-L-methioinine (AdoMet) to arginine residues (Bedford and Richard 2005; Bedford and Clark 2009). Three forms of arginine methylation have been described NG monomethylarginine, NGNG dimethylarginine (asymmetric dimethylarginine aDMA), and NGN′G dimethylarginine (symmetric dimethylarginine sDMA). Protein arginine methylation is known to play a role in regulating transcription, RNA splicing, and in RNA/protein import/export from the nucleus. Arginine residues are also targeted for an additional type of post-translational modification, which results in conversion to the noncoded amino acid citrulline (Vossenaar et al. 2003). This deimination of arginine is catalyzed by a family of peptidyl arginine deiminases (PADIs or PADs) and results in the loss of a positive charge. This conversion is antagonistic with respect to arginine methylation since deiminated arginine residues cannot be methylated and arginine methylation blocks its own citrullination. Currently, the best understood biological role for this antagonism is in the regulation of chromatin structure. For example, PADI4 catalyzed deimination of histone H3 at Arg17 and histone H4 at Arg 3 has been linked to transcriptional repression (Cuthbert et al. 2004; Wang et al. 2004). Conversely, citrullination of histone H3R26 by PADI2 has been shown to facilitate transcriptional activation at target genes (Zhang et al. 2012). More recently, Christophorou et al. (2014) have demonstrated that citrullination of histone H1 leads to chromatin decondensation during pluripotency.
Many proteins involved in mRNP biogenesis have been shown to be arginine methylated (Liu and Dreyfuss 1995; Yu et al. 2011). The post-translational modification of mRNP components is likely to modulate either the rate or extent of their interactions necessary for the ordered assembly of mRNPs and the associated coupling of transcription with mRNA processing. However, in many cases, it remains to be determined how methylated arginines influence the molecular activities of RNA-binding proteins. Here, we show in mammalian cells that arginine methylation of SFPQ/PSF does not affect the binding of its interacting partner NONO. However, arginine methylation is shown to enhance the interaction between SFPQ/PSF and mRNA.
RESULTS AND DISCUSSION
Mass spectrometry analysis of the SFPQ/NONO complex
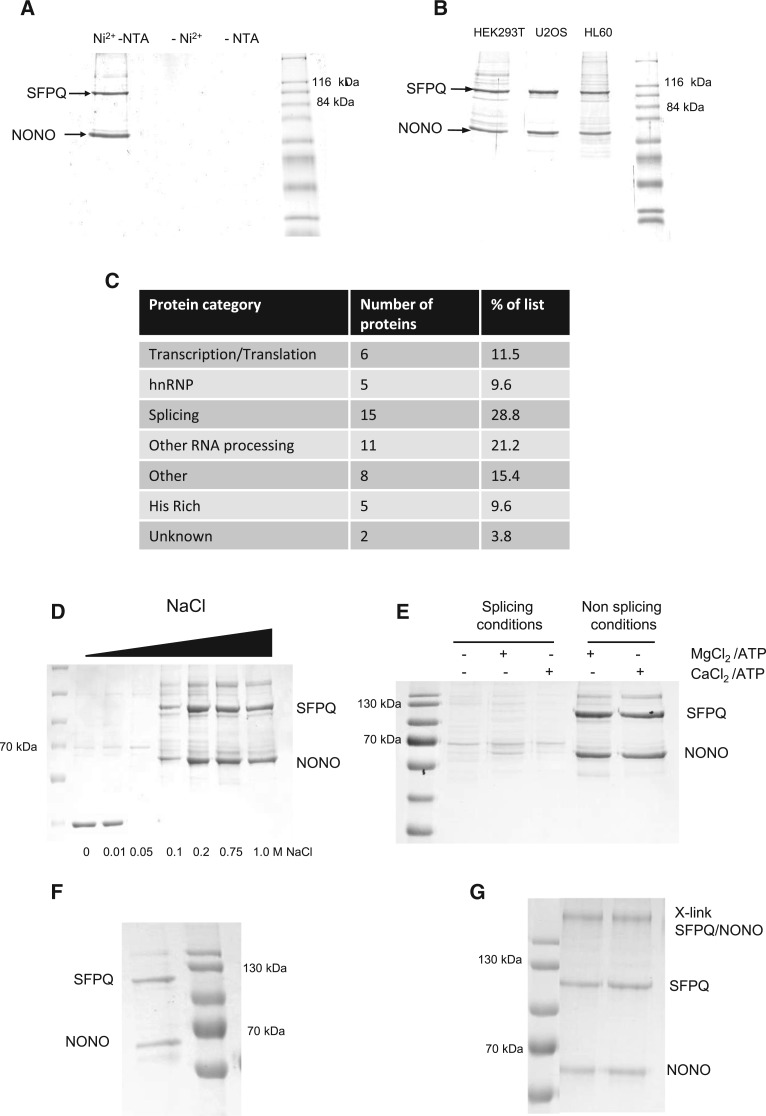
NONO contains an N-terminal histidine rich sequence, enabling purification of the SFPQ/NONO complex from HeLa nuclear extracts (see Fig. 1A) and additional cell lines (see Fig. 1B) using immobilized metal ion affinity chromatography (IMAC) by Ni-NTA open tube capillaries (Ni-NTA PhyCap OTCs) (Hanna et al. 2006; Qureshi et al. 2013). SDS-PAGE analysis of eluted material shows two major proteins captured from the affinity enrichment, that were unambiguously identified as SFPQ and NONO using mass spectrometry (MS). No enrichment of the complex was observed in the absence of Ni2+ and NTA (see Fig. 1A) demonstrating specificity of the IMAC purification. Subsequent MS analysis identified ∼50 coeluting proteins from the Ni-NTA PhyCap OTCs which are listed in Supplemental Table S1, reflecting the multifunctional role of the SFPQ/NONO complex within the cell. An overview of the categorization of proteins based on their functional annotation is shown in Figure 1C and highlights an enrichment for proteins thought to be involved in RNA processing, RNA splicing and transcriptional regulation. Proteins with His-rich motifs were also screened, as they may copurify on the Ni-NTA PhyCap OTC (see Fig. 1C; Supplemental Table S1).
FIGURE 1.
Purification and characterization of the SFPQ/NONO complex. (A) Enrichment of the SFPQ/NONO complex from HeLa nuclear cell extracts using Ni-NTA PhyCap OTC including the absence of Ni2+ and NTA. (B) Ni-NTA PhyCap OTC enrichment of the SFPQ/NONO complex from different cell extracts. Purified complexes were analyzed on SDS-PAGE stained with Coomassie blue. (C) Functional categorization of proteins identified using mass spectrometry analysis of the SFPQ/NONO complex purified using Ni-NTA PhyCap OTCs. (D) Effect of salt concentration on the SFPQ/NONO interaction. Ni-NTA PhyCap OTC enrichment of the SFPQ/NONO complex was performed over a range of NaCl concentrations highlighted. (E) Effect of splicing conditions on the SFPQ/NONO interaction. Ni-NTA PhyCap OTC enrichment of the SFPQ/NONO complex was performed under both splicing and nonsplicing conditions as shown. (F) Repurification of the SFPQ/NONO complex under low salt conditions (6 mM NaCl). (G) Chemical crosslinking of the SFPQ/NONO complex. Following chemical crosslinking the complex was analyzed using SDS-PAGE stained with Coomassie blue.
SFPQ and NONO are known members of the spliceosome and have previously been identified in a large scale antibody-based proteomic analysis (Rappsilber et al. 2002; Peng et al. 2006). A comparison of the proteins identified in these studies with data presented here, reveals a significant corroboration (see Supplemental Table S1). In addition, a number of proteins were identified that were unique to this study including paraspeckle protein 1 (PSPC1), symplekin, FIP1, and nucleophosmin. PSPC1 is a component of a novel subnuclear compartment named paraspeckle and was previously shown to interact with NONO via its coiled-coil domain (Fox et al. 2005). PSPC1 and SFPQ and have also been shown to be coexpressed and interact with Sertoli cells (Kuwahara et al. 2006). Structural studies have shown that PSPC1 and NONO form an extensively intertwined dimer (Passon et al. 2012).
Symplekin and FIP1 are both involved in polyadenylation (Takagaki and Manley 2000) and FIP1 also interacts with the polypyrimidine tract binding protein (PTB), a known binding partner of SFPQ (Patton et al. 1993; Zhao et al. 2005). pre-mRNA cleavage factor 68 kDa was also identified and has previously been identified with partial paraspeckle localization (Dettwiler et al. 2004). Another protein, a 68-kDa pre-mRNA cleavage factor previously identified with partial paraspeckle localization (Dettwiler et al. 2004) was also detected in this analysis. Nucleophosmin is a ubiquitous and multifunctional protein involved in ribosome biogenesis, centrosome duplication, and cell signaling. Our identification of a Ser/Thr phosphatase regulatory subunit, an inhibitor of the α- and γ-isoforms of protein phosphatase (Kreivi et al. 1997) is relevant since another protein phosphatase (1-δ) is known to interact with SFPQ (Hirano et al. 1996). Also relevant to regulation by phosphorylation/dephosphorylation, a component of the ubiquitous cAMP-dependent protein kinase was detected.
Ionic strength alters the SFPQ/NONO complex
The influence of ionic strength upon the purification of the SFPQ/NONO complex from HeLa cell lysates was performed by incorporating NaCl concentrations between 0 and 1 M in the binding, washing, and elution buffers during the Ni-NTA PhyCap OTC enrichment procedure (Fig. 1D). The results show that the SFPQ/NONO complex can be readily purified over a range of ionic strength conditions (0.1–1 M NaCl), indicating that the formation of the SFPQ/NONO complex relies on hydrophobic interactions. Under conditions of low ionic strength (<100 mM NaCl) a significant reduction in the yield of the SFPQ/NONO complex is observed. Interestingly, an alternative protein was purified under these conditions, which was identified via MS as glyceraldehyde 3-phosphate dehydrogenase (GAPDH). These data also demonstrate the relative high stability of the SFPQ/NONO complex, even at elevated ionic strength conditions (1 M NaCl). At low ionic strength (below 100 mM salt) purification of the SFPQ/NONO complex is compromised.
The enrichment of the SFPQ/NONO complex using Ni-NTA OTCs was also performed under typical splicing conditions (60 mM KCl in the presence of additional divalent cations, ATP, and phosphocreatinine) (Peng et al. 2006). Under such conditions, Ni-NTA PhyCap OTC enrichments show a reduction in the yield of purified SFPQ/NONO complex (see Fig. 1E) as expected from the previous results under low salt conditions. The splicing buffer was further altered to test whether the presence of magnesium ions or ATP played a significant role in the enrichment profile using the Ni-NTA PhyCap OTCs. The results shown in Figure 1E demonstrate that the presence of magnesium ions and ATP had negligible impact on enrichment profiles in either splicing buffer or PBS. These findings are consistent with those of Peng et al. (2006) were under in vitro splicing conditions, SFPQ associates with all five splicing snRNPs as well as many other splicing factors regardless of the presence of exogenous pre-mRNA.
It is proposed that the SFPQ/NONO complex exists as a heterodimer previously observed at high salt concentrations (typical intracellular salt concentrations are 150 mM NaCl). Alternatively, at low salt conditions such as those typically used for in vitro splicing, the SFPQ/NONO complex exists as part of larger splicing particles or more heterogeneous protein assemblies. Under these conditions, additional protein interactions including the association of the five snRNPs are facilitated (Peng et al. 2006). The formation of these large heterogeneous protein assemblies precludes the binding of NONO to the Ni-NTA surface of the open tube capillaries. To confirm that the association of SFPQ/NONO as part of the large heterogeneous splicing complex prevents the interaction with the Ni-NTA surface rather than a direct effect of different salt conditions influencing the interaction of NONO with the Ni-NTA PhyCap OTC, the SFPQ/NONO complex enriched under standard conditions was subsequently diluted to significantly reduce the salt concentration (to ∼6 mM NaCl) and repurified under salt-free conditions (see Fig. 1F). These results show that the formation of the SFPQ/NONO complex is enriched in low salt conditions in the absence of additional splicing factors and associated proteins, where formation of the larger splicing complex is prevented.
Chemical crosslinking of the SFPQ/NONO complex
Based on the intensities of each band following Coomassie staining of SDS-PAGE analysis of the SFPQ/NONO complex, we conclude that a stoichiometry of 1:1 fits the observations. To further analyze the size and stoichiometry of the complex enriched using Ni-NTA PhyCap OTCs, the SFPQ/NONO complex was chemically crosslinked using sulfosuccinimidyl suberate. SDS-PAGE analysis of the crosslinked complex is shown in Figure 1G, where a single high MW band is observed, demonstrating the formation of a covalent complex with a calculated mass of ∼250 kDa. These results are consistent with previous gel filtration studies of the SFPQ/NONO complex which suggested a mass of 280 kDa, suggesting that the protein is a tetramer under physiological conditions (Zhang et al. 1993). A number of possible multimeric crosslinked species are consistent with this MW including homotetrameric SFPQ complex or a possible heterotetramer of SFPQ (although SFPQ migrates anomalously as a 100 kDa protein during SDS-PAGE) and NONO (54 kDa). MS analysis of the high MW crosslinked complex identified the presence of both SFPQ and NONO, therefore supporting the formation of the SFPQ/NONO heterotetramer.
These data provide a robust evaluation of the stoichiometry of the interactions between SFPQ and NONO. The application of OTCs in conjunction with MS for the analysis of the complexes, greatly facilitates the experimental investigation of the SFPQ/NONO complex, taken together with the cross linking data, we have developed a platform for the analysis of regulatory processes, which are likely to be an important feature in our dissection of the biological role of these molecules.
Identification and characterization of arginine methylation of SFPQ/PSF by MS
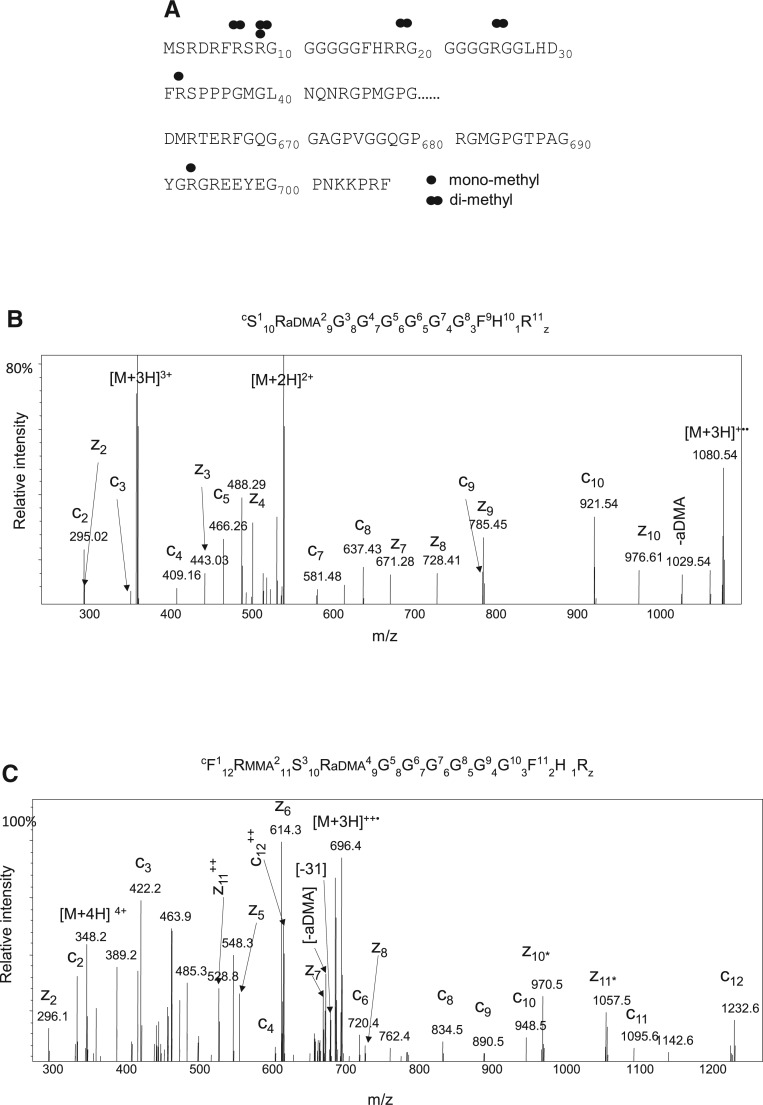
A range of MS approaches were undertaken to comprehensively characterize and identify sites of arginine methylation in SFPQ including the use of both CID and ETD in conjunction with MS analysis (Snijders et al. 2010). In addition, heavy methyl SILAC was also used to verify the sites of arginine methylation in SFPQ. Cells were grown in medium containing heavy methionine (CD3-L-methionine), which is converted in vivo to heavy AdoMet, which serves as a universal methyl-group donor in methyl transfer reactions including arginine methylation (Ong et al. 2004). The MS analysis of SFPQ identified the occurrence of both mono and dimethylation of arginine. A summary of the sites identified is shown in Figure 2A. MS analysis also enabled us to differentiate between the type of arginine dimethylation present by virtue of the characteristic neutral losses. The tandem MS spectra of two arginine methylated peptides SR(aDMA)GGGGGGFHR (heavy methyl labeled) and FR(MMA)SR(aDMA)GGGGGGFHR are shown in Figure 2B,C, respectively. The neutral loss of dimethylamine is highlighted in the spectra, confirming asymmetric dimethylation in each case. Furthermore, arginine methylation was observed in SFPQ purified from a wide range of cell lines including HEK293, U2OS, CHO, HL60, and HeLa cells. A list of all modified peptides identified by MS is shown in Supplemental Table S2.
FIGURE 2.
Identification of arginine methylation of SFPQ/PSF. (A) Dimethylation (R) and both mono or dimethylation (R) sites are mapped on the primary sequence of SFPQ. (B) ETD MS/MS fragmentation spectra of the heavy methylated peptide SR(aDMA)GGGGGGFHR [M+3H]3+. (C) ETD MS/MS fragmentation spectra of the heavy methylated peptide FR(MMA)SR(aDMA)GGGGGGFHR [M+3H]3+. The prominent c, z ions and the characteristic neutral losses associated with asymmetric dimethylation are highlighted (aDMA) dimethylamine, (MMA) monomethylarginine.
PRMT1 methylates SFPQ/PSF in vitro
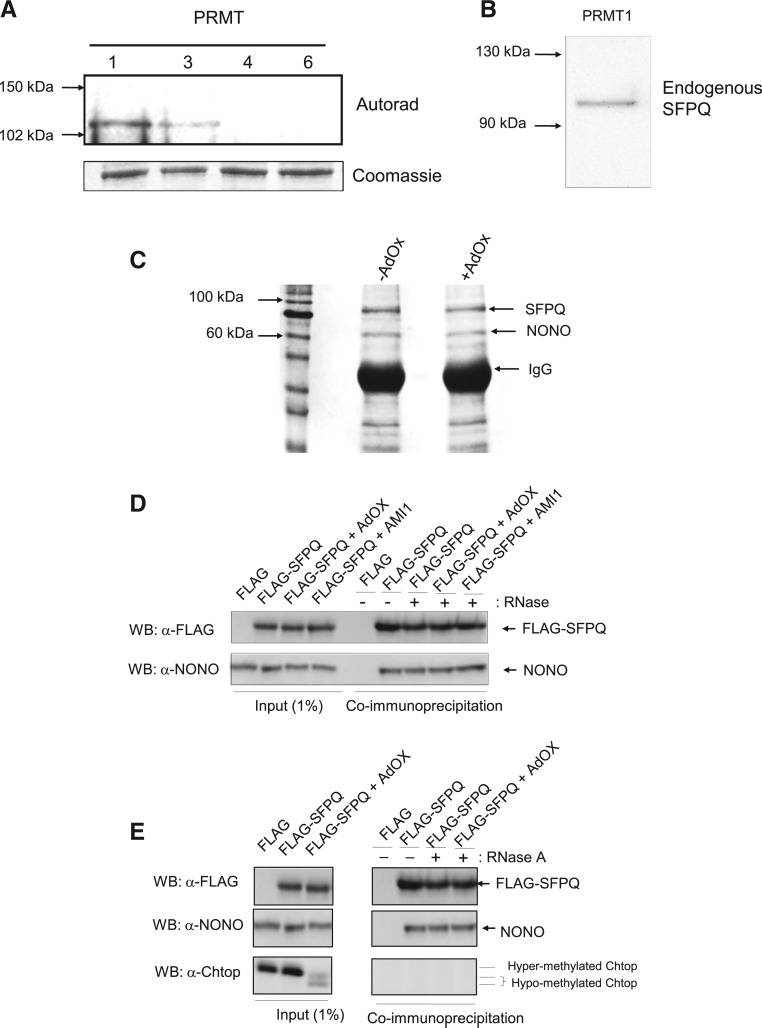
To further characterize the arginine methylation of SFPQ, in vitro methylation reactions were performed on endogenous and recombinant SFPQ using a range of protein arginine methyltransferases (PRMTs 1, 3, 5, and 6), in the presence of tritiated S-adenosyl-L-methionine ([3H] AdoMet). The results are shown in Figure 3A,B and demonstrate that only PRMT1 (and to a lesser extent PRMT3) arginine methylate SFPQ in vitro. Since PRMT3 is cytoplasmic and PRMT1 predominantly nuclear, it is likely that PRMT1 is responsible for the methylation of SFPQ within the nucleus of mammalian cells. Following in vitro methylation by PRMT1, further MS analysis was performed to identify the sites and type of arginine methylation of SFPQ in vitro. A number of sites identified in vivo were also methylated by PRMT1 in vitro (see Supplemental Table S2). Furthermore, methylation of SFPQ by PRMT1 is also consistent with the identification of asymmetric dimethylation in vivo, since PRMT1 is a type I methyltransferase which results in asymmetric dimethylation, in contrast to type II methyltransferases such as PRMT5 which results in symmetric methylation (Wolf 2009).
FIGURE 3.
Arginine methylation of SFPQ/PSF does not affect binding to NONO. (A,B) Methylation assays were performed in conjunction with the addition of tritiated S-adenosyl-L-methionine ([3H] AdoMet) to detect methylation of (A) recombinant SFPQ using PRMT 1, 3, 4, and 6 and (B) endogenous SFPQ using PRMT1. (C) Antibody enrichment of the SFPQ/NONO complex using total extracts from +/− AdOx treated cells. Eluted proteins were analyzed by SDS-PAGE stained with Coomassie blue. (D) Co-IP of FLAG-SFPQ in the presence of RNAse A from 293T cells treated with AdOx/AMI-1 as indicated. Total extract (1% of input) and eluted proteins were analyzed by Western blotting with anti-FLAG and anti-NONO. (E) Co-IP of FLAG-SFPQ in the presence of RNAse A from 293T cells treated with AdOx as indicated. Total extract (1% of input) and eluted proteins were analyzed by Western blotting with anti-FLAG, anti-NONO, and anti-Chtop.
Arginine methylation does not affect the interaction of SFPQ/PSF with NONO/p54nrb
To study the functional effects of arginine methylation on the SFPQ/NONO interaction, extracts were prepared from 293T cells grown in both the presence and absence of the methylation inhibitor adenosine dialdehyde (AdOx). Immunoprecipitation was performed using an antibody specific to SFPQ prior to SDS-PAGE analysis (see Fig. 3C). The results show that methylation of SFPQ does not significantly affect the interaction with NONO. We further investigated the SFPQ/NONO interaction using coimmunoprecipitation (Co-IP) assays. Extracts were prepared from 293T cells transfected with a FLAG-SFPQ expression vector, which had been incubated both with and without the global methylation inhibitor AdOx and a selective inhibitor of protein arginine methyltransferases (AMI-1). This analysis also showed that Co-IP of FLAG-SFPQ and NONO was not affected by arginine methylation (see Fig. 3D). Inhibition of methylation was confirmed by analysis of total extracts which revealed a distinct electrophoretic mobility shift for a known arginine methylated protein Chtop (see Fig. 3E; Chang et al. 2013). Furthermore, inhibition of methylation was also confirmed by MS analysis of purified SFPQ, the MS being consistent with an overall reduction in the level of dimethylated arginine.
Arginine methylation of SFPQ/PSF affects mRNA binding in mammalian cells
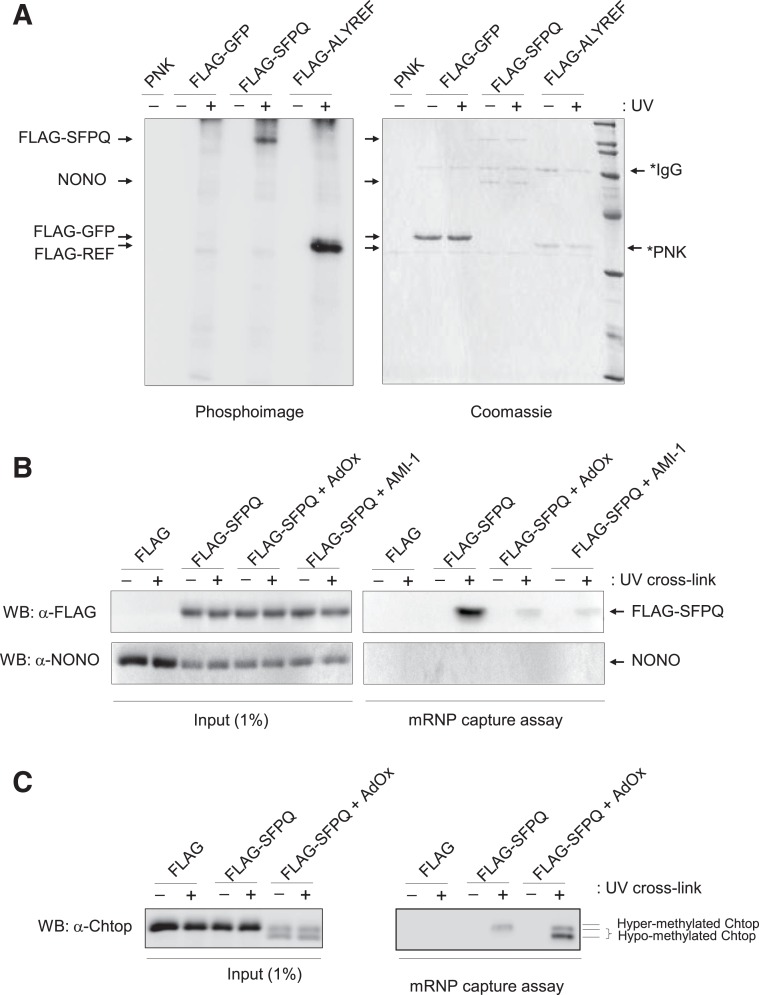
UV crosslinking studies were performed to demonstrate the binding of RNA to SFPQ in mammalian cells. Following overexpression of FLAG-SFPQ and UV crosslinking, immunopurified FLAG-SFPQ under high ionic strength conditions (1 M NaCl) was assayed for binding RNA using partial RNase digestion and end-labeling with [γ−32P]ATP and polynucleotide kinase (see Fig. 4A). The results demonstrate that following crosslinking and purification, RNA was specifically bound to SFPQ, while no RNA was detected in association with NONO. FLAG-tagged ALYREF was used as a positive control showing specific binding to RNA only following covalent cross-linking by UV. To further examine the mRNA binding activity of SFPQ in mammalian cells, we carried out an mRNP capture assay in the presence and absence of AdOx/AMI-1 (Fig. 4B). Whole-cell extracts from UV cross-linked cells treated or not with AdOx/AMI-1 were subjected to oligo(dT) pull down assays under denaturing conditions in order to investigate direct binding of poly(A)+ RNA to SFPQ in mammalian cells. This analysis revealed that the presence of both AdOx/AMI-1 led to a significant reduction in SFPQ bound to mRNA than was found in a methylated untreated control. The mRNA capture was specific since covalent crosslinks were not observed in the absence of UV exposure. Furthermore, the results demonstrate no direct binding of NONO to mRNA, consistent with previous UV crosslinking experiments (see Fig. 4A). Control experiments verifying the reduction of methylation of Chtop and the effect on mRNP binding are shown in Figure 4C, consistent with previous observations (Chang et al. 2013).
FIGURE 4.
Arginine methylation of SFPQ affects mRNA binding. (A) 293T cells cotransfected with a FLAG control, FLAG-SFPQ, and FLAG-ALYREF were UV crosslinked and purified using FLAG affinity beads. Eluted proteins were RNase treated and radiolabeled (32P) using PNK. Resulting complexes were analyzed by SDS-PAGE stained with Coomassie blue (right panel) and Phosphoimage (left panel). (B,C) mRNP capture assay. Poly(A)+ RNA from 293T cells +/− AdOx/AMI-1 transfected with FLAG or FLAG-SFPQ was purified on oligo(dT) in denaturing conditions after UV cross-linking (+) or not (−). Total extract (1% of input) and eluted proteins were analyzed by Western blotting (WB) with a FLAG antibody, NONO antibody, or Chtop antibody.
Citrullination of SFPQ/PSF blocks in vitro methylation
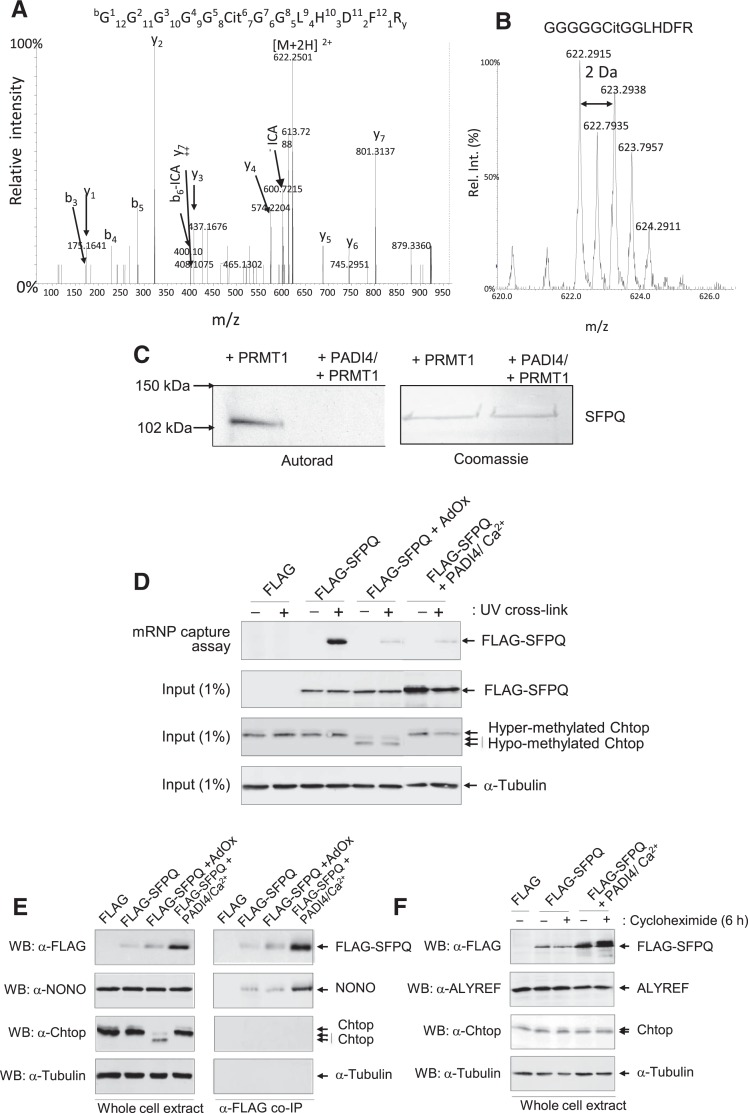
To investigate whether SFPQ is a substrate for PADI4 in vitro, recombinant SFPQ was citrullinated using overexpressed PADI4 in the presence of calcium and analyzed by MS. The MS analysis revealed a number of sites of citrullination in the N-terminus of SFPQ (Supplemental Table S2). The tandem MS spectra of the citrullinated peptide GGGGGCit GGLHDFR is shown in Figure 5A. The characteristic loss of isocyanic acid (−43 Da) from the doubly charged precursor ion and b6 ion was used as a further diagnostic for the identification of citrullination in this peptide (Hao et al. 2009). Owing to the difficulties associated with the MS based identification of citrullination and the discrimination of deamidation of glutamine and asparagine residues (corresponding 1 Da gain), citrullination reactions were also performed in the presence of 50% H218O. This approach enables confident assignment of citrullination peptides by virtue of the isotopic distribution of the citrullinated peptides (Kubota et al. 2005). The MS spectra of the peptide GGGGGCit GGLHDFR citrullinated in the presence of 50% H218O in Figure 5B show incorporation of the stable isotope from the heavy water following the reaction with SFPQ with PADI4, thereby confirming enzymatic citrullination of this peptide. Verification of all citrullinated peptides identified in this study was achieved using both the characteristic neutral loss of isocyanic acid and the isotopic distribution following incorporation of 50% H218O (see Supplemental data). The combined MS results enabled the identification of a number of sites of citrullination in the N-terminus of SFPQ by PADI4, and are summarized in Supplemental Table S2.
FIGURE 5.
Citrullination blocks arginine methylation of SFPQ/PSF in vitro. (A) CID MS/MS fragmentation spectra of the in vitro citrullinated peptide GGGGGCitGGLHDFR [M+2H]2+ from SFPQ. The prominent y, b ions and the characteristic neutral losses associated with isocyanic acid are highlighted. (B) MS spectra highlighting the isotopic distribution due to the incorporation of H218O during the enzymatic citrullination of SFPQ using PADI4. (C) Methylation assays were performed in conjunction with the addition of tritiated S-adenosyl-L-methionine ([3H] AdoMet) to detect methylation of recombinant SFPQ using PRMT before and after citrullination using PADI4. Left panel shows the autorad, right panel shows the Coomassie stained SDS-PAGE. (D) Citrullination of SFPQ affects mRNA binding. Poly(A)+ RNA from 293T cells +/− AdOx transfected with FLAG, FLAG-SFPQ, FLAG-SFPQ + HA PADI4/Ca2+ ionophore was purified on oligo(dT) in denaturing conditions after UV cross-linking (+) or not (−). Total extract (1% of input) and eluted proteins were analyzed by Western blotting (WB) with α-FLAG antibody, α-ALYREF, or Chtop antibody. (E) Co-IP of FLAG control, FLAG-SFPQ in conjunction with PADI4 in the presence of Ca2+ ionophore. Total extract (1% of input) and eluted proteins were analyzed by Western immunoblotting using α-FLAG, α-NONO, α-Chtop, or α-Tubulin antibodies. (F) PADI4/Ca2+ increases SFPQ/PSF abundance in mammalian cells. 293T cells were cotransfected with either a FLAG control, FLAG-SFPQ in conjunction with PADI4 in the presence of Ca2+ ionophore and 5 mM CaCl2. Of note, 100 mM cycloheximide was added for 6 h when indicated (+) in order to block de novo protein synthesis. Twelve percent SDS-PAGE was analyzed using Western immunoblotting using α-FLAG, α-ALYREF, α-Chtop, or α-Tubulin antibodies.
It is interesting to note that all the identified sites of citrullination were also identified as dimethlyated in endogenous SFPQ. No other arginine residues were identified as citrullinated in SFPQ, highlighting the specific nature by which PADI4 citrullinates SFPQ in vitro. These findings offer the tantalizing prospect that citrullination may block arginine methylation of SFPQ and thus regulate function. To determine whether arginine methylation of SFPQ can be blocked by citrullination in vitro, recombinant SFPQ was first citrullinated with PADI4 in the presence of calcium ions. Following citrullination of SFPQ (confirmed by MS analysis), the citrullinated SFPQ was used as a substrate for PRMT1 in the presence of tritiated AdoMet and analyzed using autoradiography. As shown in Figure 5C citrullination does indeed prevent subsequent methylation by PRMT1. Therefore, citrullination has the potential to block methylation of SFPQ in vivo and thus antagonize the effects of arginine methylation. Demethylimination of arginine has also been reported, with the majority of in vitro demethylimination studies demonstrating that dimethylated arginines are poor substrates for PADI4. The ability to demethyliminate endogenous methylated SFPQ was not possible as constitutive methylation of SFPQ is unlikely (verified by the additional methylation of endogenous SFPQ by PRMT1 in vitro). Therefore, the ability of PADI4 to demethyliminate the N-terminal asymmetric dimethylated arginine residues was analyzed using an N-terminal peptide substrate containing the sites of aDMA identified on SFPQ. The MS analysis revealed no demethylimination upon the addition of PADI4 (data not shown, consistent with previous observations, Raijmakers et al. 2007).
Citrullination of SFPQ/PSF affects mRNA binding in mammalian cells
To examine the effects of SFPQ citrullination on mRNA binding, we carried out an mRNP capture assay following the coexpression of both FLAG-SFPQ and PADI4 in the presence of a calcium ionophore (Fig. 5D). This analysis revealed that in the presence of PADI4/Ca2+ a significant reduction in SFPQ bound to mRNA in mammalian cells was observed compared with the methylated control. The mRNA capture was specific since covalent crosslinks were not observed in the absence of UV exposure. Verification of the citrullination of SFPQ was performed using MS, and a number of sites of citrullination were identified including those previously identified as arginine methylated as summarized in Supplemental Table S2. We further investigated the SFPQ/NONO interaction using coimmunoprecipitation (Co-IP) assays. Extracts were prepared from 293T cells transfected with a FLAG-SFPQ and PADI4 in the presence of a calcium ionophore. The results showed that the Co-IP of FLAG-SFPQ and NONO was not affected by the presence of PADI4/Ca2+ and citrullination of SFPQ (see Fig. 5E).
PADI4 increases SFPQ/PSF abundance in mammalian cells
Data from the Co-IP and mRNA capture assays suggested increased expression of SFPQ in the presence of PADI4/Ca2+ ionophore (see Fig. 5D,E). To further investigate the stability of SFPQ in the presence of PADI4, HEK 293T cells were cotransfected with FLAG or FLAG-SFPQ in conjunction with PADI4 in the presence of Ca2+ ionophore. Cycloheximide was added to block de novo protein synthesis. Western blotting was used to detect steady-state levels of SFPQ and control ALYREF or Chtop proteins (see Fig. 5F). The results show that in the presence of PADI4/Ca2+, increased levels of SFPQ were observed in both the presence and absence of cycloheximide treatment. These results indicate that the protein stability is not affected by citrullination, raising the possibility of a mechanism based on increased transcription and/or translation of SFPQ.
CONCLUSIONS
SFPQ and NONO are multifunctional nuclear proteins that appear to play a role in a diverse range of key processes within the cell including RNA processing, RNA splicing, and transcriptional regulation. Here we show that a number of N-terminal arginine residues in SFPQ are asymmetrically dimethylated in mammalian cells, a reaction that can be catalyzed by PRMT1. Arginine methylation of SFPQ leads to an increase in its association with mRNA either via a direct effect of methylation on RNA binding or an indirect effect on protein–protein interactions that alters SFPQ RNA binding specificity and/or affinity. In addition to controlling protein–protein interactions between mRNP components, arginine methylation modulates the ability of certain RNA-binding proteins to target their nucleic acid substrate. Methylation of arginine has the potential to reduce hydrogen bonding networks with RNA or it may sterically hinder the association between the RNA and protein. Alternatively, it has been proposed that methylation enhances the association with RNA by rendering the arginine more hydrophobic, thereby facilitating stacking with the RNA bases (Bedford and Richard 2005).
Arginine methylation has been reported to modulate RNA binding activity in a wide range of RNA binding proteins including hnRNPA1 and Sam68 (Yu 2011). Methylation of the mammalian mRNA export factor ALYREF reduces its RNA-binding capacity to ensure that the message can be efficiently displaced by a second mRNA export factor NXF1 (Hung et al. 2010). However, arginine methylation of FMRP causes mRNA substrate-specific changes in RNA binding activity (Dolzhanskaya et al. 2006). Moreover, arginine methylation was shown to decrease the capacity of RBP16 to associate with gRNA. This is not a general effect on RBP16 RNA binding, however, since methylation conversely increases the association of the protein with mRNA (Goulah and Read 2007). In the case of SFPQ/PSF, it is clear that arginine methylation increases its association with mRNA.
Citrullination of SFPQ/PSF in vitro was demonstrated to block arginine methylation in vitro. Moreover, citrullination of SFPQ/PSF by PADI4 in mammalian cells results in decreased association with poly(A)+/mRNA. It has previously been shown that the ribosomal protein S2 (RPS2) is citrullinated by PADI4 at an RG repeat region of RPS2, which is also methylated by protein arginine methyltransferase 3 (PRMT3) (Guo et al. 2011). Therefore, it is likely that the crosstalk between citrullination and methylation is a widespread phenomenon in RNA binding proteins (RGG repeats). This crosstalk may act as a cellular mechanism for post-translational regulation due to the antagonistic nature of the modifications and play an important role in mRNP dynamics.
MATERIALS AND METHODS
Chemicals
Acetonitrile (LC MS grade), water (HPLC grade), formic acid (FA, HPLC grade) trifluoroacetic acid (TFA, HPLC grade), and ammonium formate (MS grade) were obtained from Thermo Scientific.
HeLa cell nuclear extracts
HeLa cell nuclear extract (5.0 × 109, Cat. no. CC-01-20-50) was obtained from Cilbiotech. This extract was treated with complete protease inhibitor cocktail (Roche), divided into 200 µL aliquots and stored at −80°C. Fifty microliters (≈250 µg) of the HeLa cell nuclear extract was routinely added to 550 µL PBS and passed through the open tube capillary as described below.
Open tube capillary (OTC) enrichment
The OTC enrichments were performed essentially as previously described (Hanna et al. 2006; Qureshi et al. 2013). Briefly, the sample was loaded onto a PhyCap IMAC open-tube capillary (PhyNexus), and the open end submerged in 500 μL wash buffer (PBS + 10 mM imidazole, pH 7.4). This buffer was drawn into and expelled from a 1 mL disposable syringe at a rate of 150 μL/min, for one draw/expel cycle. For elution 2–15 μL of desorption buffer (PBS + 250 mM imidazole, pH 7.4) was used to the entire elution volume to be maintained as a single liquid segment the elution segment was then pushed back to the bottom of the column at 30 μL/min and was not allowed to be expelled through the column opening. Upon completion of the elution cycling, the liquid segment was expelled from the capillary and collected for analyses.
FLAG-SFPQ purification
One 15-cm plate of 293T cells was transfected using calcium phosphate with p3X-FLAG-SFPQ and lysed after 48 h in 500 mL lysis buffer (PBS, 1% Triton X-100) supplemented with 2 mM PMSF (Sigma) and complete protease inhibitor (Roche). Cell debris was removed by centrifugation (16.1 rcf, 10 min, 4°C) and the supernatant incubated for 3 h at 4°C with 30 μL α-FLAG M2-agarose slurry (Sigma) preequilibrated overnight in lysis buffer containing 1% BSA (Sigma). Beads were then washed (3× bead volume) with PBS, 1 M NaCl, 0.5% Triton X-100 and TBS (50 mM Tris, pH 7.4, 150 mM NaCl). Purified protein was eluted in 50 μL 1 × TBS containing 100 mg/mL 3X-FLAG peptide (Sigma) for 30 min at 4°C, before analysis by SDS-PAGE (stained with Coomassie blue).
Coexpression of FLAG-SFPQ and HA-PADI4
One 15-cm plate of 293T cells was transfected using calcium phosphate with p3X-FLAG-SFPQ and pcDNA3.1-HA PADI4 and lysed after 48 h in 500 mL lysis buffer (1 PBS, 1% Triton X-100) supplemented with 2 mM PMSF (Sigma) and complete protease inhibitor cocktail (Roche). FLAG-SFPQ purification was performed as described above.
In vitro methylation reaction
Histidine and GST tagged SFPQ was purified from Escherichia coli. Recombinant GST-PRMT enzymes were overexpressed from pGEX6P constructs at 30°C in LB media by addition of 0.1 mM IPTG and purified using glutathione agarose resin (Pierce). Enzymes were eluted using 20 mM reduced glutathione then dialyzed against 10% glycerol in 50 mM Tris–HCl, pH 8.0. Arginine methylation reactions using recombinant SFPQ, endogenous SFPQ or SFPQ that had been citrullinated in vitro (see below) as substrates, were carried out with bead-bound PRMTs in 50 mM Tris–HCl, pH 9.0, 5 mM MgCl2, 4 mM dithiothreitol and complete protease inhibitor cocktail (Roche) by the addition of 2 μL 3H-AdoMet (PerkinElmer, 67 Ci/mmol) at 30°C for 1 h. Recombinant GST-PRMT enzymes were assayed for activity using GAR as a positive control substrate (data not shown). Reaction products were denatured and resolved on 8% SDS-PAGE and electro-transferred to PVDF membrane. Autoradiography was then performed overnight at −80°C using a Kodak intensifying screen (Thermo Scientific).
In vitro deimination reaction
Purification of recombinant GST-PADI4 and in vitro deimination assays were carried out essentially as described (Cuthbert et al. 2004). Briefly, GST-PADI4 was overexpressed from a pGEX6P construct at 30°C in 2TY broth by the addition of 0.1 mM IPTG and purified using glutathione agarose resin (Thermo Scientific). Deimination reactions were carried out using bead-bound PADI4 in 50 mM Tris, pH 7.5, 2 mM DTT and complete protease inhibitor cocktail (Roche), in the presence or absence of 2 mM CaCl2 at 30°C for 1 h.
Coimmunoprecipitation assays
2 × 10-cm HEK 293T plates were used for each condition and were treated when indicated with 20 µM AdOx or 100 µM AMI-1 for 15 h prior and during transfections (48 h) or with calcium ionophore for 15 min by adding 4 µM Ca2+ ionophore (Sigma A23187) and 5 mM CaCl2 to the culture medium. Fifteen microgram plasmid(s) were transfected per plate with either 15 µg FLAG or 7.5 µg FLAG-SFPQ + 7.5 µg, FLAG or 7.5 µg FLAG-SFPQ + 7.5 µg PADI4. PBS-washed cells were lysed in 1 mL IP lysis buffer per plate supplemented with 2 mM PMSF and complete protease inhibitor cocktail (Roche). Total protein extracts (1.5 mg) were subjected to α-FLAG immunoprecipitation in the presence or absence of 10 µg RNase A with 30 µL FLAG-M2 beads (Sigma) for 1.5 h at 4°C. Beads were then washed three times with 900 µL IP lysis buffer and protein complexes were eluted from the beads in 60 µL IP lysis buffer containing 100 μg/mL 3× FLAG peptide for 30 min at 4°C. Twelve microliter input (∼25 µg) or eluted binding reactions were analyzed by 10% SDS-PAGE. Whole-cell soluble extracts were subjected to α-FLAG immunoprecipitation and analyzed by 12% SDS-PAGE and Western immunoblotting using α-FLAG (1/5000), α-ALYREF (1/3000), α-Chtop (KT64—1/2000), or α-Tubulin (1/5000) antibodies.
RNA binding assay
2 × 6-cm 293T plates transfected with either 6 mg of p3X-FLAG, p3X-FLAG-SFPQ, or p3X-FLAG-ALYREF for each condition were cross-linked (+UV 0.3 J/cm2) or not (−UV) in 500 mL PBS. Cells from 2 × 6-cm plates were lysed in 1 mL IP lysis buffer (50 mM HEPES, pH 7.5/100 mM NaCl/1 mM EDTA/0.5% Triton X-100/10% glycerol) and cleared total extracts were treated with 200 mL 2 M NaCl to disrupt transient protein–protein interactions. Purified complexes bound to beads were treated with 10 μg RNAse A for 15 min at 37°C prior to elution under native conditions using FLAG peptides. Eluted protein/nucleic acids complexes were labeled with γ32P-ATP and polynucleotide kinase (PNK) and subsequently analyzed on SDS-PAGE (stained with Coomassie blue) and phosphoroImaging.
mRNP capture assay
PBS-washed transfected 293T cells were UV-irradiated or not on ice with 0.120 J/cm2 and mRNP capture assays were performed under denaturing conditions as described in Hung et al. (2010). Complexes were directly eluted in 50 μL elution buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 0.4 mg/mL RNase A). Captured mRNA-binding protein complexes were analyzed by SDS-PAGE and Western blotting.
Increased expression of FLAG-SFPQ following citrullination
1 × 6-cm HEK 293T plate transfected with p3X-FLAG/p3X-FLAG-SFPQ, and pcDNA3.1-HA PADI4 were treated or not with Ca2+ ionophore for 15 min by adding 4 µM Ca2+ ionophore (Sigma A23187) and 5 mM CaCl2 to the culture medium. Of note, 100 μM cycloheximide was added for 6 h to block de novo protein synthesis. PBS-washed cells were lysed in 200 μL IP lysis buffer per plate supplemented with 2 mM PMSF and complete protease inhibitor cocktail (Sigma). Whole-cell extracts were analyzed by 12% SDS-PAGE and Western immunoblotting using α-FLAG (1/5000), α-ALYREF (1/3000), α-Chtop (KT64—1/2000), or α-Tubulin (1/5000) antibodies.
Protein digestions
Following purification, SFPQ/NONO complex was digested with trypsin (Sigma, proteomics grade, 0.1–200 ng) in 100 mM ammonium bicarbonate, 20% acetonitrile at 37°C for 1–6 h. The reactions were quenched by the addition of 0.1% TFA. The samples were subsequently dried under vacuum and resuspended in 0.1% final concentration of TFA. Six microliters were analyzed by LC-MS/MS analysis.
LC-MS/MS analysis
Tryptic peptides were identified and characterized by using nanoflow liquid chromatography coupled to mass spectrometry. The systems used were a Dionex Ultimate 3000 LC system (Thermo Scientific) with both HCT Ultra PTM Discovery and Maxis UHR-TOF mass spectrometers (Bruker Daltonics), and a Dionex Ultimate 3000 RSLCnano LC system (Thermo Scientific) with an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). The peptides were separated on a PepMap C18 reverse phase column (75 μm × 15 cm, Thermo Scientific) by a linear gradient from 95% solvent A (0.1% formic acid) to 40% solvent B (0.1% formic acid in 95% acetonitrile) over 40 min at 300 nL/min. Mass spectra were acquired with automated precursor ion selection, using collisionally induced and electron transfer dissociation for MS/MS, either separately or in alternating scans as appropriate for the instrument. Data sets were converted to Mascot Generic Files using scripts supplied by Bruker and searched against the Swiss-Prot database (Release 10.5, 20 April 2010) with Mascot Server 2.2 (Matrix Science). For methylation experiments, methionine oxidation, arginine methylation, and arginine dimethylation were set as variable modifications while for citrullination, methionine oxidation, and asparagine/glutamine/arginine deamidation were selected.
Chemical crosslinking
Samples were prepared using PhyCap open-tube capillary methods described above. Eluted proteins were microdialyzed against PBS for 20 min to remove imidazole. Crosslinking reagents bis(sulfosuccinimidyl) suberate (BS3, Pierce) was dissolved in DMSO and working dilutions were made using ddH2O. A crosslinker was added at a final concentration of 1 mM and allowed to react for 30 min at 37°C, before quenching with Tris–HCl, pH 8.0 at a final concentration of 10 mM. For crosslinking using deuterated reagents, bis(sulfosuccinimidyl) suberate BS3-d0 and BS3-d4 were dissolved in DMSO and mixed in a 1:1 molar ratio. Working dilutions were made and the reaction was carried out as described above. Crosslinked samples were analyzed by 7.5% SDS-PAGE and excised for MS analysis.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council UK [BB/D011795/1]. Chtop antibody was provided to S.A.W. by T.B. van Dijk and S. Philipsen. We thank Vicky Porteous for technical assistance. S.A.W. acknowledges support from the BBSRC and Wellcome Trust.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045138.114.
Freely available online through the RNA Open Access option.
REFERENCES
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S 2005. Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272. [DOI] [PubMed] [Google Scholar]
- Bladen CL, Udayakumar D, Takeda Y, Dynan WS 2005. Identification of the polypyrimidine tract binding protein-associated splicing factor⋅p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem 280: 5205–5210. [DOI] [PubMed] [Google Scholar]
- Chanas-Sacré G, Mazy-Servais C, Wattiez R, Pirard S, Rogister B, Patton JG, Belachew S, Malgrange B, Moonen G, Leprince P 1999. Identification of PSF, the polypyrimidine tract-binding protein-associated splicing factor, as a developmentally regulated neuronal protein. J Neurosci Res 57: 62–73. [DOI] [PubMed] [Google Scholar]
- Chang CT, Hautbergue GM, Walsh MJ, Viphakone N, van Dijk TB, Philipsen S, Wilson SA 2013. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J 32: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, et al. 2014. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. 2004. Histone deimination antagonizes arginine methylation. Cell 118: 545–553. [DOI] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Immediate RNA binding, protein–protein interactions, and subcellular localization. J Biol Chem 279: 35788–35797. [DOI] [PubMed] [Google Scholar]
- Dolzhanskaya N, Merz G, Aletta JM, Denman RB 2006. Methylation regulates the intracellular protein–protein and protein–RNA interactions of FMRP. J Cell Sci 119: 1933–1946. [DOI] [PubMed] [Google Scholar]
- Dong X, Sweet J, Challis JR, Brown T, Lye SJ 2007. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol 13: 4863–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, Weitz CJ 2011. A molecular mechanism for circadian clock negative feedback. Science 332: 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI 2005. p54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulah CC, Read LK 2007. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J Biol Chem 282: 7181–7190. [DOI] [PubMed] [Google Scholar]
- Guo Q, Bedford MT, Fast W 2011. Discovery of peptidylarginine deiminase-4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2. Mol Biosyst 7: 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna C, Gjerde D, Nguyen L, Dickman M, Brown P, Hornby D 2006. Micro-scale open-tube capillary separations of functional proteins. Anal Biochem 350: 128–137. [DOI] [PubMed] [Google Scholar]
- Hao G, Wang D, Gu J, Shen Q, Gross SS, Wang Y 2009. Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J Am Soc Mass Spectrom 4: 723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Erdödi F, Patton JG, Hartshorne DJ 1996. Interaction of protein phosphatase type 1 with a splicing factor. FEBS Lett 389: 191–194. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429. [PubMed] [Google Scholar]
- Hung ML, Hautbergue GM, Snijders AP, Dickman MJ, Wilson SA 2010. Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res 38: 3351–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani K, Yoshida T, Kitagawa H, Ohta H, Nozawa S, Kato S 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem Biophys Res Commun 306: 660–665. [DOI] [PubMed] [Google Scholar]
- Izumi H, McCloskey A, Shinmyozu K, Ohno M 2014. p54nrb/NonO and PSF promote U snRNA nuclear export by accelerating its export complex assembly. Nucleic Acids Res 42: 3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG 2004. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 119: 815–829. [DOI] [PubMed] [Google Scholar]
- Kameoka S, Duque P, Konarska MM 2004. p54nrb associates with the 5′ splice site within large transcription/splicing complexes. EMBO J 23: 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreivi JP, Trinkle-Mulcahy L, Lyon CE, Morrice NA, Cohen P, Lamond AI 1997. Purification and characterisation of p99, a nuclear modulator of protein phosphatase 1 activity. FEBS Lett 420: 57–62. [DOI] [PubMed] [Google Scholar]
- Kubota K, Yoneyama-Takazawa T, Ichikawa K 2005. Determination of sites citrullinated by peptidylarginine deiminase using 18O stable isotope labeling and mass spectrometry. Rapid Commun Mass Spectrom 5: 683–688. [DOI] [PubMed] [Google Scholar]
- Kuwahara S, Ikei A, Taguchi Y, Tabuchi Y, Fujimoto N, Obinata M, Uesugi S, Kurihara Y 2006. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol Reprod 75: 352–359. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol 15: 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur M, Tucker PW, Samuels HH 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol Cell Biol 21: 2298–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mittler G, Mann M 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods 1: 119–126. [DOI] [PubMed] [Google Scholar]
- Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS 2012. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci 109: 4846–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JG, Mayer SA, Tempst P, Nadal-Ginard B 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev 5: 1237–1251. [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev 7: 393–406. [DOI] [PubMed] [Google Scholar]
- Peng R, Dye BT, Pérez I, Barnard DC, Thompson AB, Patton JG 2002. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Hawkins I, Link AJ, Patton JG 2006. The splicing factor PSF is part of a large complex that assembles in the absence of pre-mRNA and contains all five snRNPs. RNA Biol 3: 69–76. [DOI] [PubMed] [Google Scholar]
- Qureshi MS, Sheikh QI, Hill R, Brown PE, Dickman MJ, Tzokov SB, Rice DW, Gjerde DT, Hornby DP 2013. Affinity filtration coupled with capillary-based affinity purification for the isolation of protein complexes. Anal Biochem 439: 47–49. [DOI] [PubMed] [Google Scholar]
- Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ 2007. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol 367: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewer MB, Nguyen VQ, Huang CJ, Tucker PW, Kagawa N, Waterman MR 2002. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54nrb/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology 143: 1280–1290. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D 2002. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett 531: 109–114. [DOI] [PubMed] [Google Scholar]
- Snijders AP, Hung ML, Wilson SA, Dickman MJ 2010. Analysis of arginine and lysine methylation utilizing peptide separations at neutral pH and electron transfer dissociation mass spectrometry. J Am Soc Mass Spectrom 21: 88–96. [DOI] [PubMed] [Google Scholar]
- Stier S, Totzke G, Gruewald E, Neuhaus T, Fronhoffs S, Schoneborn S, Vetter H, Ko Y 2005. Identification of p54(nrb) and the 14-3-3 protein HS1 as TNF-α-inducible genes related to cell cycle control and apoptosis in human arterial endothelial cells. J Biochem Mol Biol 38: 447–456. [DOI] [PubMed] [Google Scholar]
- Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F 1998. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem 273: 26261–26264. [DOI] [PubMed] [Google Scholar]
- Straub T, Knudsen BR, Boege F 2000. PSF/p54nrb stimulates “jumping” of DNA topoisomerase I between separate DNA helices. Biochemistry 39: 7552–7558. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ 2003. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 25: 1106–1118. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306: 279–283. [DOI] [PubMed] [Google Scholar]
- Wolf SS 2009. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci 66: 2109–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MC 2011. The role of protein arginine methylation in mRNP dynamics. Mol Biol Int 2011: 163827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Oberg D, Rush M, Fay J, Lambkin H, Schwartz S 2005. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein. J Virol 79: 4270–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG 2001. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Wu JY 1996. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol 16: 5400–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Zhang LX, Busch RK, Farrés J, Busch H 1993. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem J 290: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, et al. 2012. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc Natl Acad Sci 109: 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.