Abstract
Splicing of mRNA precursors can occur cotranscriptionally and it has been proposed that chromatin structure influences splice site recognition and regulation. Here we have systematically explored potential links between nucleosome positioning and alternative splicing regulation upon progesterone stimulation of breast cancer cells. We confirm preferential nucleosome positioning in exons and report four distinct profiles of nucleosome density around alternatively spliced exons, with RNA polymerase II accumulation closely following nucleosome positioning. Hormone stimulation induces switches between profile classes, correlating with a subset of alternative splicing changes. Hormone-induced exon inclusion often correlates with higher nucleosome occupancy at the exon or the preceding intronic region and with higher RNA polymerase II accumulation. In contrast, exons skipped upon hormone stimulation display low nucleosome densities even before hormone treatment, suggesting that chromatin structure primes alternative splicing regulation. Skipped exons frequently harbor binding sites for hnRNP AB, a hormone-induced splicing regulator whose knock down prevents some hormone-induced skipping events. Collectively, our results argue that a variety of chromatin architecture mechanisms can influence alternative splicing decisions.
Keywords: alternative splicing, nucleosome, progesterone, RNA polymerase II
INTRODUCTION
Eukaryotic genomes are tightly packaged inside the nucleus in the form of chromatin nucleoprotein complexes that modulate the access of the DNA to replication, recombination, and transcription factors and therefore play an important role in the regulation of gene duplication, stability, and expression (for review, see Ballaré et al. 2013b; Becker and Workman 2013; Bartholomew 2014). The primary unit of chromatin is the nucleosome, composed of an octamer of four different histone proteins around which ∼147 bp of DNA are wrapped (Luger et al. 1997, 2012). Histones are small, globular proteins characterized by an unstructured tail protruding outside of the nucleosome core, which can be post-transcriptionally modified in a highly dynamic manner (for review, see Kouzarides 2007). The state of these modifications is essential to regulate chromosome compaction, transcription, and gene silencing.
The affinity of DNA for histone octamers is influenced by its primary sequence, which is therefore an important determinant of nucleosome positioning along the genome (Segal et al. 2006; Kaplan et al. 2010). Nevertheless, nucleosome positioning is highly dynamic and its regulation is crucial to control chromatin accessibility and recruitment of chromatin modifiers and transcription factors (Ballaré et al. 2013b; Becker and Workman 2013; Bartholomew 2014). Genome-wide studies correlated chromatin structure with the exon–intron organization of the genes and argued that nucleosomes are preferentially positioned in exons compared with introns in a variety of species, from worms to humans (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009). While the pattern of nucleosome occupancy at the transcription initiation and termination sites (TSS and TTS, respectively) is tightly connected with gene expression levels, nucleosome positioning over internal exons seems independent of transcription changes and levels of expression of the gene (Tilgner et al. 2009). Because exons with weak splice sites display stronger nucleosome densities, it has been argued that nucleosome positioning in exons facilitates exon recognition (Schwartz et al. 2009; Tilgner et al. 2009).
At least 90% of human genes are alternatively spliced (Pan et al. 2008; Wang and Burge 2008), either in a cell type-specific manner or in response to different stimuli. Defects in alternative splicing regulation are linked to different diseases (for review, see Singh and Cooper 2012). Therefore intron removal needs to be both precise and plastic and thus is tightly controlled at multiple levels. In addition to the assembly of core components of the spliceosome on splice site signals, additional factors bind to intronic and exonic sequences of the pre-mRNA to regulate the inclusion of specific exons (for review, see Wang and Burge 2008; Chen and Manley 2009; Braunschweig et al. 2013). The interplay between these sequence elements and their binding factors is the main determinant of alternative splicing outcome (Barash et al. 2010). Nonetheless, intron removal happens in large part cotranscriptionally (Khodor et al. 2011, 2012; Tilgner et al. 2012), and thus chromatin structure and transcription dynamics can also be involved in the regulation of exon inclusion (for review, see Luco et al. 2011; Iannone and Valcárcel 2013; Zhou et al. 2014).
RNA polymerase II kinetics can be a determinant of the regulation of specific alternatively spliced exons. Slow elongation rates can favor inclusion of exons with weak splice sites by providing them with a kinetic advantage over competing downstream sites (de la Mata et al. 2003, 2010). Moreover, the carboxy-terminal-domain (CTD) of RNA polymerase II interacts with different RNA processing factors at transcription loci and can help to recruit them to nascent transcripts (McCracken et al. 1997; Bird et al. 2004). Furthermore, histone modifications can serve as a landing platform for “adaptor” proteins that recruit splicing factors (Luco et al. 2010; Saint-Andre et al. 2011) or indirectly impact splicing outcomes by modulating Pol II elongation rate through changes of level of compaction of chromatin (Schor et al. 2009).
It has been proposed that nucleosome positioning over alternative exons regulates RNA polymerase II processivity and consequently exon inclusion (Schwartz et al. 2009), but the general effect of nucleosome density on alternative splicing regulation has not been investigated systematically. We have explored this issue by analyzing parallel changes in nucleosome positioning and alternative splicing induced by progesterone. Progesterone is a steroid hormone that can traverse the cell membrane and activate its nuclear receptors (PRs), which act as ligand-inducible transcription factors (Beato 1996). A small fraction of PRs is also bound to the plasma membrane, where upon ligand binding activates signaling cascades. This was initially known as the “nongenomic” pathway; however, recent studies revealed an intense cross-talk between kinase cascades and the transcriptional functions of PRs (Lange 2004; Vicent et al. 2006). Upon binding of the hormone to its C-terminal domain, progesterone receptor (PR) gets activated, binds to its DNA target sequences (hormone responsive elements, HREs) and modulates gene expression by recruiting a plethora of coregulators, among which chromatin modifiers that remodel nucleosome organization (Beato and Klug 2000).
As for other transcription factors, PR binding is strongly influenced by nucleosome density and chromatin compaction. PRs are thought to bind preferentially to HREs exposed in nucleosome-depleted chromatin regions, in which “pioneer factors” such as AP1 and FOXA1 are already bound (Carroll et al. 2005; Hurtado et al. 2011; John et al. 2011). A recent genome-wide analysis of global correlations between PR binding and chromatin dynamics induced by hormones in breast cancer cells (Ballaré et al. 2013a) revealed that PR binds preferentially to HREs that are at the same time enriched in nucleosomes prior to hormone induction and marked by DNAse I hypersensitive sites (DHS). Upon PR binding, nucleosomes get remodeled (but not evicted) and activation of gene expression begins (Ballaré et al. 2013a).
Our results confirm the higher density of nucleosomes associated with exons and its general decrease upon hormone stimulation. Four different classes of nucleosome density profiles that correlate with the distribution of GC content and that show a strong correlation with RNA polymerase II accumulation are observed. Particular switches between nucleosome profile classes are correlated with hormone-induced alternative splicing changes: High nucleosome density 5′ of alternative exons is associated with increased exon inclusion, while increased exon skipping is typically associated with low nucleosome densities even before hormone treatment. Exon skipping can be assisted by hormone induction of the splicing regulatory factor hnRNP AB. Our results suggest that changes in nucleosome positioning can influence alternative splicing decisions by a variety of mechanisms not limited to influencing RNA polymerase II dynamics.
RESULTS
Progesterone-induced alternative splicing changes are largely independent of transcriptional changes
T47D-MMTVluc cells are breast cancer cells containing a single copy of the Mouse Mammary Tumor Virus promoter fused to a luciferase reporter. These cells are frequently used to study progesterone-induced transcriptional regulation (Truss et al. 1995). Cells were treated with 10 nM of the synthetic progestin R5020, total RNA was extracted 6 h after treatment (R6) or from untreated cells (T0) and high-throughput Solexa RNA-sequencing was carried out using single-end reads (190–200 million reads/sample, see Materials and Methods). To identify alternative exons regulated by R5020, we analyzed RNA-seq data using the mixture of isoform probabilistic model (MISO) (Katz et al. 2010) (see also Materials and Methods) focusing on internal cassette exons. We identified 247 R5020-regulated alternative exon cassettes that showed changes in percent spliced in index (ΔPSI) >15%, corresponding to 239 genes (Supplemental Table S1). Semiquantitative PCR analysis confirmed the alternative splicing changes in five out of seven events analyzed (Supplemental Fig. S1). Progesterone treatment was associated with a similar number of higher inclusion events (124 exons, henceforth “R5020-included” exons), which on average display a change in PSI from 0.32 to 0.53, and higher skipping events (123 exons, henceforth “R5020-skipped” exons), which on average change PSI from 0.61 to 0.35 (Fig. 1A). Of these, 34 R5020-included exons and 39 R5020-skipped exons showed ΔPSI of >25%.
FIGURE 1.
Progesterone-induced alternative splicing changes in T47D breast cancer cells. (A) Scheme summarizing the experimental procedure and number of alternative splicing changes identified. (B) Fraction of alternative exon changes (increased inclusion or skipping) that occurs in genes without expression changes (no change), up-regulated (up), or down-regulated (down) genes. (C) Fraction of expressed genes at different times after hormone treatment that contain progesterone receptor (PR) binding sites. Different gene categories are represented with different colors, as indicated. (D,E). Gene ontology terms enrichment for progesterone-regulated exons (D) and genes (E). Statistical significance (P-value) of GO terms enrichment is indicated.
It has been previously reported that R5020 treatment modulates transcription of 3835 genes (Ballaré et al. 2013a). However, the majority of R5020-regulated cassette exons occur in genes that are not transcriptionally regulated by the hormone, as determined by global microarray analysis and RNA-seq data (Ballaré et al. 2013a): 74% for genes with included exons, and 73.7% for genes with skipped exons (Fig. 1B). A similar proportion of transcriptional changes was observed for genes that do not display splicing changes, 76% of which do not experience gene expression changes upon hormone induction. Among the genes displaying changes in both gene expression and alternative splicing, a preference was observed for included exons to be more frequent in down-regulated genes and for skipped exons to be more frequent in up-regulated genes, possibly related to differences in transcriptional elongation (Fig. 1B).
Gene ontology analysis using GOrilla software (http://cbl-gorilla.cs.technion.ac.il/) argued that genes displaying alternative splicing changes belong to functional categories distinct from those displaying hormone-induced transcriptional changes (Fig. 1D,E). This is in line with previous observations in a variety of systems suggesting that gene expression and alternative splicing coordinate different layers of cellular function (Pan et al. 2004). Neither RNA processing nor RNA binding proteins were among the GO categories enriched in regulated genes or alternative splicing events.
Previous ChIP-seq results identified over 25,000 DNA progesterone receptor (PR) binding sites in the genome, occupied at different time points after R5020 treatment. Progesterone-modulated genes show enrichment in PR binding sites compared with nonregulated genes, some of which nevertheless also show PR binding upon hormone stimulation. To assess whether PR binding could influence alternative splicing regulation, we searched for PR binding sites around R5020-regulated cassette exons. Less than 3% of the regulated exons had a ChIP-seq signal in a window of 3000 bp flanking the alternative exon (data not shown). Around 40% of genes containing regulated exons display PR binding sites anywhere in the gene body or within 3000 bp upstream of the TSS, at any of the analyzed time points (Fig. 1C). While PR binding site densities increase in genes transcriptionally regulated by R5020, these increases do not correlate with the presence of R5020-regulated exons. We conclude that PR binding is unlikely to substantially contribute to progesterone-induced changes in alternative splicing.
Nucleosome occupancy and RNA polymerase II dynamics in alternatively spliced genomic regions
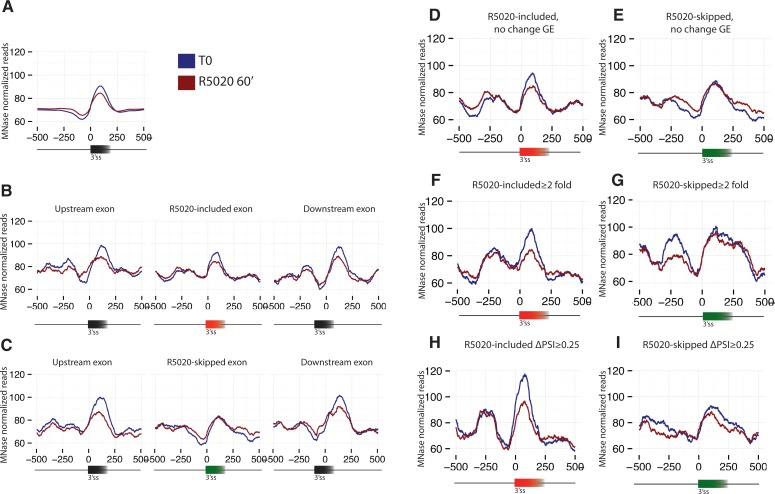
To correlate genome-wide, high-resolution maps of nucleosome density with alternative splicing changes, we analyzed data obtained by sequencing mononucleosomes generated by MNase digestion of chromatin in permeabilized T47D-MMTVluc cells untreated or treated with R5020 for 60 min (Ballaré et al. 2013a). We first analyzed the distribution of nucleosome density over all internal exons. In agreement with previous reports (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009), an obvious peak of nucleosome density centered on the exon and surrounded by regions of lower density in the flanking introns was observed (Fig. 2A). This enrichment was detected both in control cells and in cells treated with R5020, but a clear decrease in the exonic nucleosome peak was observed upon hormone treatment, concomitant with a slight increase in nucleosome density in the flanking intronic regions (Fig. 2A). These patterns of nucleosome occupancy and changes upon hormone induction were generally observed regardless of the extent of inclusion (PSI) of the exons (Supplemental Fig. S2). These results indicate that hormone treatment decreases nucleosome positioning in exons, which could, in principle, contribute to reduced exon recognition and hormone-induced exon skipping.
FIGURE 2.
Progesterone-induced changes in nucleosome positioning in exons and flanking regions. (A) General profile of nucleosome density in exons and flanking sequences in control and hormone-stimulated T47D cells. Chromatin from permeabilized cells treated with MNase was isolated, mononucleosome-associated DNA deep-sequenced and the number of reads covering each nucleotide represented, after normalization by genomic DNA reads, relative to the 3′ splice site of exons. Profiles of nucleosome densities in R5020-included (B) and R5020-skipped (C) exons and their flanking exons and introns in control and hormone-stimulated T47D cells. (D,E) Profiles of nucleosome densities as in B,C, corresponding to regulated alternative exons located in genes that do not display changes in expression upon hormone stimulation. (F–I) Profiles of nucleosome densities as in B,C for alternative exons that experience a change in inclusion or skipping higher than twofold or with a ΔPSI of ≥0.25, as indicated.
To evaluate whether nucleosome occupancy changes can be correlated with alternative splicing regulation, we investigated nucleosome occupancy of R5020-regulated exon cassettes and of their flanking exons in treated and untreated samples (Fig. 2B,C). Both R5020-included and R5020-skipped exon cassettes display higher nucleosome occupancy on exonic regions compared with flanking introns in the absence of hormone treatment, although the peak of R5020-skipped exons is distinctly lower than those of the flanking exons. This indicates that hormone-induced skipped exons have distinctive features (relatively lower nucleosome peaks compared with their flanking introns than the rest of the exons) even before hormone treatment. This suggests that hormone treatment tends to weaken the recognition of exonic regions that do not harbor well-positioned nucleosomes. Upon hormone treatment, nucleosome peaks are generally reduced, except the peak of the alternative skipped exons, which is not reduced further. Similar patterns were observed for exons displaying splicing changes in genes whose expression was not altered by hormone treatment (Fig. 2D,E), and for regulated exons displaying inclusion or skipping changes higher than twofold (33 skipped exons and 42 included exons) (Fig. 2F,G) or displaying ΔPSI >25% (39 skipped exons and 34 included exons) (Fig. 2H,I). Of relevance, exons that display inclusion changes higher than twofold have relatively low inclusion ratios in the absence of hormone (0.16 on average, increasing to an average of 0.4 upon hormone treatment). This indicates that these exons have a clear nucleosome peak despite showing low basal levels of exon inclusion and, indeed, exons with low PSI display high nucleosome peaks (Supplemental Fig. S2). Conversely, strongly R5020-skipped exons, which have an average basal exon inclusion ratio of 0.40 (reduced to 0.15 upon hormone treatment), display low nucleosome occupancy both in the absence or presence of hormone, suggesting that these exons display intrinsic features associated with low nucleosome density. Similar observations apply also to exons whose ΔPSI >0.25. Collectively, the results of Figure 2 suggest that particular profiles of nucleosome occupancy around alternatively spliced exons could influence their regulation.
Transcribing RNA polymerase II pauses at nucleosomes (Churchman and Weissman 2011; Kwak et al. 2013) and at the 3′ end of introns (Churchman and Weissman 2011; Kwak et al. 2013) and its kinetics have been associated with alternative splicing regulation (de la Mata et al. 2003, 2010; Dujardin et al. 2014). Nucleosome positioning has been proposed to represent a barrier for RNA polymerase II progression, leading to reduced elongation rates that can provide more extensive opportunities to facilitate exon recognition by polymerase-associated splicing factors (Schwartz et al. 2009). In contrast, our results reveal decreased nucleosome density upon R5020-induced exon inclusion, and low densities with or without hormone in exons with induced skipping. In fact, skipped exons display a particularly pronounced peak of low nucleosome density in the 3′ splice site region before the exon (Fig. 2C).
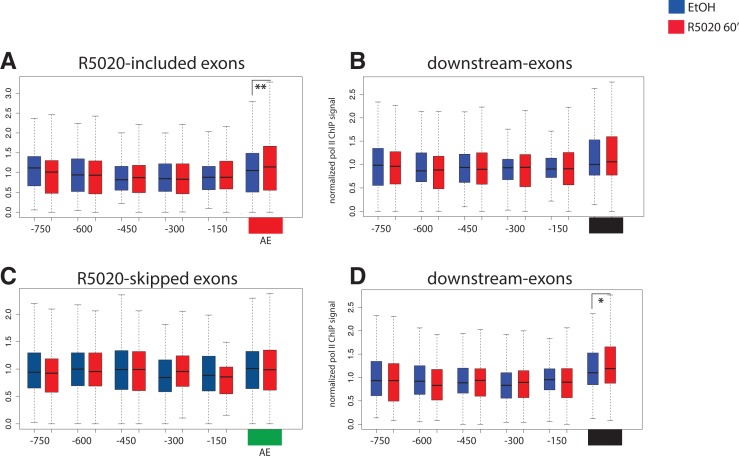
To investigate RNA polymerase II dynamics in response to hormone induction along regulated exons, we carried out total Pol II ChIP-seq in T47D-MMTVluc cells, either treated for 60 min with 10 nM R5020 or mock treated with the ethanol carrier. The meta-profile of the ChIP-seq signal shows, as expected, accumulation of RNA polymerase II on transcription start sites (TSS) and low signals in the gene body in both conditions (Supplemental Fig. S3). We investigated RNA polymerase II accumulation over R5020-regulated exons and their upstream introns partitioned in segments (bins) of 150 bp, and the same regions of the corresponding downstream constitutive exons as control (Supplemental Table S3). While RNA polymerase II signals tend to accumulate in exonic regions compared with upstream intronic regions (Fig. 3) a similar tendency is observed when sequences from the input material are analyzed (Supplemental Fig. S4), consistent with a sequencing bias caused by differences in mappability between exon and intron sequences due to their divergent GC content, as previously reported by Schwartz et al. (2011). Nevertheless, hormone treatment increases RNA polymerase II accumulation in R5020-included exons (P-value = 0.0056) (Fig. 3A), compatible with the possibility that further accumulation of RNA polymerase II upon hormone treatment facilitates recognition and inclusion of these exons. A similar (albeit less significant) increase in RNA polymerase II accumulation is also observed in exons downstream from R5020-skipped exons. The possible functional significance of the latter observation is unclear.
FIGURE 3.
Distribution of RNA polymerase II in exons and upstream intronic regions. Boxplots represent the density of deep sequencing reads of DNA molecules associated with RNA polymerase II ChIP experiments, corresponding to alternative exons and 750 nt upstream, normalized to local average, mock-treated with ethanol (blue) or with R5020 for 60 min (red). Density maps correspond to R5020-included or R5020-skipped exons (A and C, respectively), their constitutive downstream exon (B,D). P-values calculated by paired two-sample t-test are indicated by (*) 0.05 > P > 0.01, (**) 0.01 > P > 0.001.
Classification of alternative exons according to their nucleosome profiles
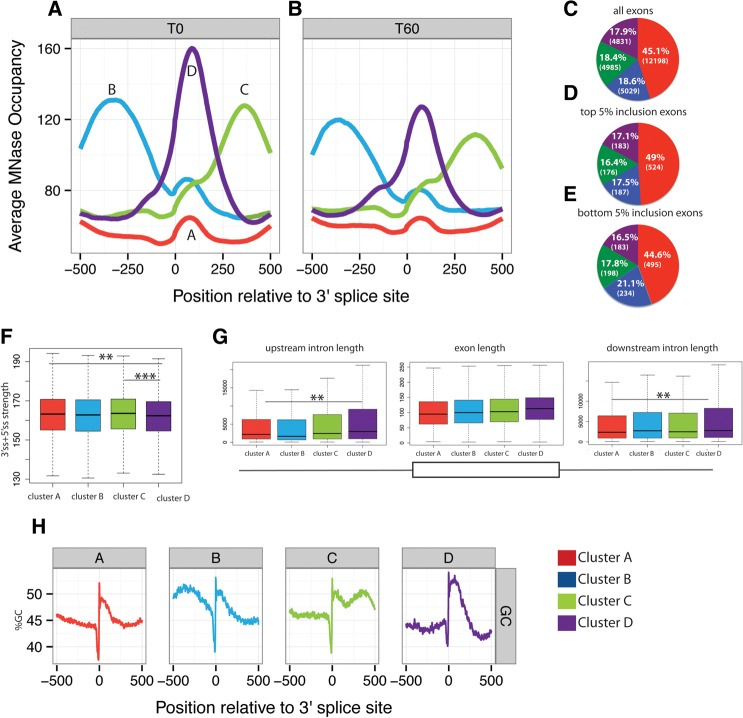
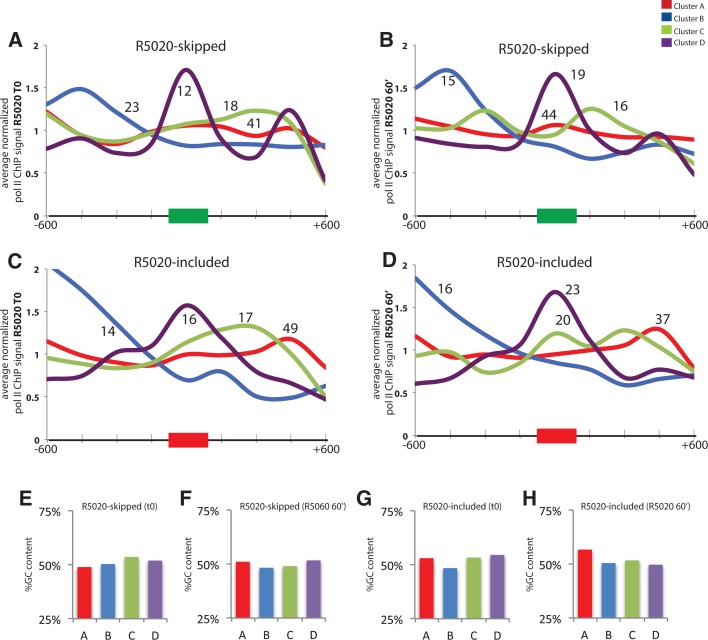
The pattern of nucleosome occupancy over internal exons has been so far described as a single sharp peak standing out on exons compared with the flanking introns (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009). Such pattern is generated by averaging the profile of nucleosomes across all exons, possibly masking distinct profile categories associated with particular subclasses of exons. We analyzed the MNase-seq nucleosome data of T47D-MMTVluc untreated cells corresponding to all cassette exons whose expression was detectable using the MISO annotation. We subsequently classified them into four different categories according to their profile of nucleosome occupancy using K-means clustering (see Materials and Methods) (Fig. 4A). While all the categories display a clear nucleosome peak in the exon, distinct features were also identified (Fig. 4A). Cluster A corresponds to the most populated class (45% of analyzed exons), and is characterized by a relatively low peak of nucleosome density centered on the alternative exon. The remaining exons are similarly distributed among three other categories (B = 18.6%, C = 18.4%, and D = 17.9%) (Fig. 4C). Two clusters show higher nucleosome density either in the upstream intron (cluster B) or downstream intron (cluster C) relative to the cassette alternative exon. Cluster D exons are characterized by a strongly positioned exonic nucleosome.
FIGURE 4.
Clusters of nucleosome profiles around alternatively spliced exons. (A,B) Four classes of nucleosome profiles identified using K-means in T47D cells before (A) and 60 min after (B) progesterone stimulation. Profiles of MNase reads relative to the position of 3′ splice sites (0) are represented as in Figure 2 for four distinct clusters of exons before (T0) (A) and after progesterone treatment (T60) (B). (C–E) Distribution of exons in the four classes of nucleosome profile clusters for all exons (C), exons in the top 5% of exon inclusion range (D), or exons in the bottom 5% of exon inclusion range (E). The colors correspond to each of the clusters defined in panels A–B. Figures between brackets indicate the number of exons in each category. (F) Distribution of splice site strengths associated with exons in the four classes of nucleosome profiles. Combined strength of 5′ and 3′ splice sites was measured by Analyze Splice Tool (http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm). Colors correspond to the clusters defined in A,B. P-values are represented by (*) 0.05 > P > 0.01, (**) 0.01 > P > 0.001, (***) 0.001 > P in the difference between splice site strengths between the indicated clusters, according to two sample t-test. (G) Distribution of alternative exon and flanking intron lengths for the four clusters of nucleosome profiles defined in A,B. P-values are represented by (**) 0.01 > P > 0.001, (***) 0.001 > P in the difference between lengths between the indicated clusters, according to two sample t-test. (H) Distribution of GC content (in percentage) relative to the 3′ splice site of alternative exons in the four classes of nucleosome profiles defined in A,B. Colors correspond to the clusters defined in A,B.
To evaluate the possible impact of these nucleosome profile classes on exon inclusion, we selected the alternative exons with the 5% lowest and highest inclusion level in untreated cells and analyzed their distribution in the four clusters. The distribution of exons in the different categories was essentially maintained, with cluster B (harboring a nucleosome peak before the exon) only slightly more frequent among the least included exons, and the most included exons displaying a slightly increased fraction in cluster A (Fig. 4C–E). Analysis of splice site strengths showed that exons classified under cluster D (strongly positioned nucleosome in the exon) are usually associated with weaker 3′ and 5′ splice sites (Fig. 4F), in agreement with previous reports that correlated higher nucleosome occupancy with weaker splice sites (Tilgner et al. 2009). We found that cluster B exons, characterized by high nucleosome occupancy upstream of the exon, are associated with weaker 3′ splice sites but not with weaker 5′ splice sites, suggesting a role for nucleosome positioning upstream of the exon in 3′ splice site recognition (data not shown). The analogous correlation was not found for cluster C exons (harboring a nucleosome downstream from the exon) and weaker 5′ splice sites (data not shown). While there is no difference in average exon length, cluster D exons (marked by strongly positioned nucleosomes) are associated with longer flanking introns (Fig. 4G). Clusters B and C are not enriched in exons with short introns (Fig. 4G), arguing against the possibility that upstream/downstream nucleosome accumulation was caused by peaks associated with other exons nearby.
DNA sequence is a strong determinant of nucleosome positioning (Kaplan et al. 2010), and exons show a different GC composition compared with introns (Schwartz et al. 2009). We analyzed the average GC content distribution in the four different clusters and found a clear correspondence between peaks of high GC content and nucleosome occupancy. In fact, the distribution of GC content of each cluster mimics the corresponding nucleosome occupancy profiles, pointing to the DNA sequence as a contributor to nucleosome distribution (Fig. 4H).
Changes in nucleosome profiling associated with alternative splicing regulation
Next we analyzed whether hormone treatment was accompanied by changes in nucleosome profile that would represent switches between cluster classes. For this we compared the MNase-seq data obtained from control cells and from R5020-induced T47D-MMTVluc cells and clustered the alternatively spliced exons according to their nucleosome profiles. The same four main patterns of nucleosome occupancy were identified upon hormone treatment (Fig. 4B), although the peaks were less sharp, as expected from the generally less dense nucleosome profiles observed in all the exons (Fig. 2A).
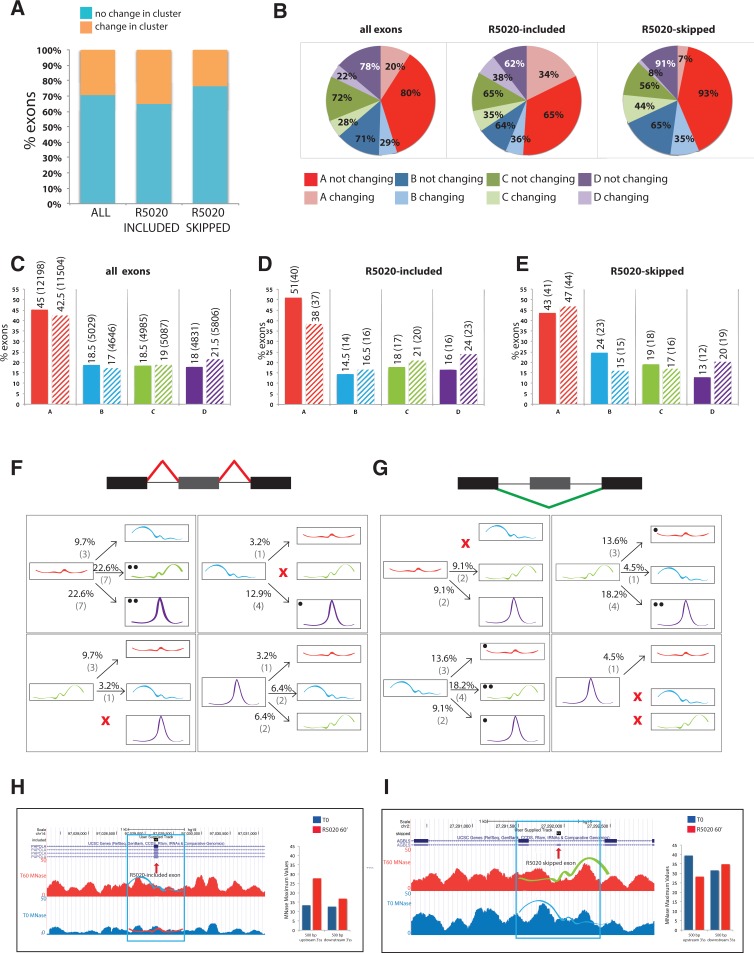
Even though the classification of exons in different nucleosome occupancy patterns appears to be affected by GC content distribution (Fig. 4H), 30% of all the exons analyzed switch the nucleosome profile class upon hormone treatment, indicative of active modulation of nucleosome occupancy (Fig. 5A). The proportion is higher for R5020-included exons than for R5020-skipped exons, suggesting that nucleosome remodeling could play a more important role in the regulation of exon inclusion than of exon skipping. No significant difference in splice site strength was observed between exons that change cluster and those that do not.
FIGURE 5.
Switches in the distribution of nucleosome profile clusters upon hormone stimulation. (A) Fraction of exons (total, R5020-induced, and R5020-skipped with a minimum ΔPSI of 0.15) that switch or not between nucleosome profile clusters upon hormone stimulation. (B) Fraction of exons in each class that switch (light colors) or not (dark colors) between nucleosome profile clusters upon hormone stimulation. Color codes are as in Figure 4. Values are represented for all, R5020-induced, and R5020-skipped exons. (C–E) Distribution of exons in nucleosome profile clusters before (solid bars) and after (striped bars) progesterone treatment. Values are represented for all (C), R5020-induced (D), and R5020-skipped (E) exons. Color codes representing each profile cluster are as in Figure 4. Figures between brackets indicate exon numbers in each category. Distribution of nucleosome profile cluster switches for R5020-included (F) and R5020-skipped (G) exons. Classes are indicated by schemes of the profiles. The fraction and number of exons displaying each switch (out of the total number of exons switching profile) is indicated. Single and double black dots indicate switches between categories which are more frequent in hormone-regulated exons compared with all alternative exons. (H,I) Genome browser visualization of MNase profiles before (blue) and after (red) hormone induction around two R5020-regulated exons. Maximum MNase peak values are also represented. Both exons change in nucleosome cluster type upon hormone induction. R5020-included exon in the gene PAPOLA, switches from cluster A in uninduced (T0) cells to cluster B 60′ after R5020 induction. R5020-skipped exon in the gene AGBL5, switches from cluster B in uninduced (T0) cells to cluster C 60′ after R5020 induction.
The proportion of exons from each class changing their profile ranged between 20% and 29% (Fig. 5B). Globally, the number of exons in the more populated A class are slightly reduced, while the number of exons in clusters C and D increase slightly (Fig. 5C). R5020-included exons generally increase in classes B, C, and D (all of them harboring strongly positioned nucleosomes), while they decrease in class A (harboring low nucleosome peaks) (Fig. 5B,D), once again suggesting that nucleosome positioning could influence hormone-induced exon inclusion. In contrast, R5020-skipped exons increase in A and D and decrease in B and C (Fig. 5B,E).
We next focused our analysis on those exons undergoing nucleosome profile changes upon R5020 treatment, analyzing the frequency of class transitions that correlate with hormone-induced alternative splicing changes. Figure 5, F and G, summarizes the relative frequency of transitions associated with alternatively spliced exons, the class switches particularly enriched in R5020-included or -skipped exons being labeled by black dots. The majority of R5020-included exons switch from cluster A (poorly positioned nucleosomes) to C or D (strongly positioned nucleosome on the exon or downstream from it), suggesting that increased nucleosome positioning could facilitate alternative exon recognition. Figure 5H shows an example of this. Regarding R5020-skipped exons, favored shifts are from C to D/A, and from B to C/A, i.e., from strongly positioned nucleosomes upstream of or downstream from the alternative exon to other categories. Figure 5I shows an example of such transition. Interestingly, R5020-skipped exons were rarely associated with nucleosome positioning upstream of the alternative exon after hormone treatment. This and the results of Figure 2F are consistent with nucleosome positioning upstream of the alternative exon contributing to exon recognition. The picture is however complex, because examples of transitions to a well-positioned exonic nucleosome (cluster D) are observed for all regulated exons, regardless of the inclusion/skipping outcome.
Next we analyzed profiles of RNA polymerase II accumulation along R5020-included and -skipped exons, classified according to their nucleosome occupancy profiles before and after hormone induction. RNA polymerase II accumulation follows quite closely the profiles of nucleosome density, with polymerase peaks in the areas of higher nucleosome occupancy (Fig. 6). The correlation is particularly clear in exons with strongly positioned nucleosomes upstream (cluster B) or on the alternative exon (cluster D). RNA polymerase II accumulation is observed in these cases, however, regardless of the splicing regulation outcome. A peak of RNA polymerase II accumulation is particularly noticeable in R5020-included exons belonging to cluster C (Fig. 6D). As these regions harbor a nucleosome in the region downstream from the exon, upstream accumulation of RNA polymerase II may contribute to higher exon recognition. Of relevance for the previous discussion on exon sequence mappability (Schwartz et al. 2011), no clear correlation was observed between RNA polymerase II accumulation and GC content in different categories of exon clusters (Fig. 6E–H).
FIGURE 6.
Distribution of RNA polymerase II in regulated exons and flanking introns. Average normalized RNA polymerase II ChIP-seq reads are represented relative to the alternative exons and flanking introns in each nucleosome profiling class (color code as given in previous figures), for R5020-skipped (A,B) or R5020-included exons (C,D), 60 min after mock (A,C) or (B,D) hormone treatment. (E–H) Average percentage of GC content of exon clusters in each of the categories represented in A–D.
Possible regulation of hormone-induced exon skipping by hnRNPAB
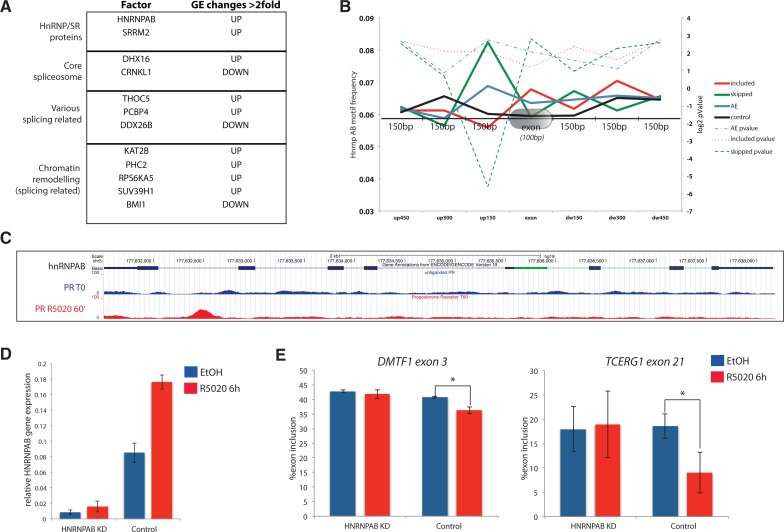
Taken together, our findings suggest that R5020-included exons may be more influenced by hormone-dependent nucleosome remodeling than skipped exons. To better understand the mechanisms behind hormone-induced exon skipping, we analyzed whether hormone induction leads to changes in the expression of splicing regulators. Several genes encoding splicing regulators, core splicing factors as well as splicing-related chromatin remodeling factors were found to be up- or down-regulated upon hormone treatment (Fig. 7A). We were particularly interested in HnRNP AB, whose expression was found increased two- to 2.5-fold 6 h after hormone treatment (Fig. 7D), particularly because sequence motif analysis using the predicted binding sequence for hnRNP AB (Ray et al. 2013) revealed a clear, statistically significant enrichment of the motif in the region 150 bp upstream of R5020-skipped exons (Fig. 7B). Moreover, the gene contained a clear peak of PR binding after hormone treatment (Fig. 7C). These observations, together with the involvement of hnRNP AB in splicing regulation (Fomenkov et al. 2003) (see also Discussion), suggest that hnRNP AB overexpression upon hormone treatment could contribute to hormone-induced exon skipping. Consistent with this idea, knock down of hnRNP AB (Fig. 7D) attenuated two tested hormone-induced exon skipping events corresponding to alternative spliced regions harboring predicted hnRNP AB binding sites (Fig. 7E).
FIGURE 7.
A potential role for hnRNP AB in hormone-induced exon skipping. (A) Hormone-induced gene expression changes of genes encoding proteins involved in splicing and its regulation. The direction of the change (up- or down-regulation) upon hormone treatment is indicated. (B) Frequency of hnRNP AB binding motifs relative to exons that are included or skipped upon hormone treatment, alternative exons (AE), or control exons. Frequencies correspond to segments of 150 bp upstream, downstream, or at the exons. P-values for motif enrichment in each region are also represented. (C) Schematic representation of Genome browser view of the hnRNP AB gene displaying tracks of PR binding before and 60 min after R5020 treatment. (D) RT-qPCR analysis of hnRNP AB mRNA levels 6 h after R5020 treatment or after vehicle treatment, relative to GAPDH mRNA levels in control cells or cells depleted of hnRNP AB by RNAi. (E) Percentage of inclusion of the indicated exons measured by semiquantitative RT-PCR under the same conditions as in D. (*) P-value 0.05 > P > 0.01.
DISCUSSION
In this work, we aimed to test the hypothesis that nucleosome positioning influences alternative exon recognition. We compared detailed maps of nucleosome density before and after progesterone induction with changes in alternative splicing induced by the hormone. This is a relevant system to address this question because progesterone is known to have profound effects on chromatin architecture (Beato and Klug 2000). The key roles that progesterone plays in development and tissue homeostasis, together with the links between elevated progesterone levels and mammary carcinogenesis (Brisken 2013), make a detailed understanding of the molecular mechanisms behind hormone-induced transcription and splicing changes an issue of significant biological and medical interest. We found 247 alternative splicing changes induced by progesterone stimulation, affecting genes with biological functions distinct from those associated with hormone-induced transcriptional changes, thus offering novel information about another layer of gene expression modulation by the hormone.
Regulation of alternative splicing of mRNA precursors relies upon the combinatorial effects of RNA motifs in the pre-mRNA and the activities of their cognate trans-acting factors (Smith and Valcárcel 2000; Wang and Burge 2008; Chen and Manley 2009; Braunschweig et al. 2013). It has also become clear in recent years that chromatin architecture reflects the gene intron–exon structure (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009) and that chromatin structure and transcriptional dynamics play important roles in alternative splicing regulation (for review, see Luco et al. 2011; Iannone and Valcárcel 2013; Zhou et al. 2014). While it has been reported that enrichment of specific histone modifications modulates alternative splicing regulation of some exon cassettes (Luco et al. 2010; Saint-Andre et al. 2011), it remained unclear whether nucleosome positioning can by itself influence exon inclusion. A straightforward model is that nucleosome density regulates exon inclusion by either acting as “speed bump” for elongating RNA polymerase and its associated splicing factors, or by acting as a “recruiting platform” for splicing regulators. In this model, increased nucleosome density at exons is associated with higher levels of exon inclusion, while decreased nucleosome occupancy is generally associated with exon skipping (de la Mata et al. 2003, 2010), although more complex scenarios have been documented (Dujardin et al. 2014). Some of our results are consistent with this standard model, but other data suggest the operation of a more diverse range of effects and mechanisms. Also, it seems unlikely that every hormone-induced alternative splicing event is influenced by chromatin structure. In this regard, expression of a number of splicing factors and regulators, including hnRNP AB, is affected by progesterone stimulation, and this can also contribute to the observed changes in alternative splicing.
We confirmed that internal exons display, on average, higher nucleosome density than their flanking introns. A more detailed analysis revealed four main distinct classes of nucleosome profiles in internal expressed cassette exons, which closely follow the distribution of GC content in these regions. The most frequent class is characterized by a small peak of nucleosome density at exons compared with the flanking introns, a profile shared by another major class displaying much more pronounced exonic peak, which correspond to exons frequently associated with weaker splice sites and longer flanking introns, consistent with previous findings (Tilgner et al. 2009). The remaining two classes show also an exonic peak but display an additional higher density peak upstream or downstream from the exon. An overall correlation was observed in the different classes between nucleosome positioning and RNA polymerase II accumulation at (or 5′ of) the region of higher nucleosome density, which is of potential relevance considering the link between the dynamics of RNA polymerase II transcription and alternative splicing decisions discussed above. Exons whose inclusion is increased upon progesterone treatment undergo more nucleosome changes and seem also to be more influenced by RNA polymerase II dynamics than exons that increase skipping. For example, R5020-included exons tend to increase RNA polymerase II occupancy upon hormone treatment (Figs. 3A, 6D) and the majority of the class transitions associated with these exons involve increased nucleosome density at—or 3′ of—the exon (Fig. 5F), consistent with the standard model. Another clear association is observed between nucleosome positioning upstream of alternative exons and increased exon inclusion. It is conceivable that accumulation of RNA polymerase II in a region 5′ of the alternative exon provides more time for the loading of splicing factors that can facilitate exon recognition once the polymerase is released from the region of higher nucleosome density. Altering RNA polymerase II dynamics could also influence the timing at which exons are processed, affecting the ratio of co- versus post-transcriptional processing and consequently the concentration/range of regulators to which the primary transcripts are exposed (Vargas et al. 2011). Conversely, it is conceivable that changes in cotranscriptional splicing can also influence nucleosome occupancy, similar to the effects of splicing and RNA-binding proteins on epigenetic modifications (de Almeida et al. 2011; Zhou et al. 2011; de Almeida and Carmo-Fonseca 2014).
In contrast, exons that are more skipped upon progesterone treatment display weaker nucleosome densities and lower RNA polymerase II accumulation after—and in some cases even before—hormone treatment. This suggests that (the absence of) chromatin architecture somehow primes these exons for hormone-induced skipping. It is conceivable that lower nucleosome density leads to a state of high RNA polymerase II processivity in which nonchromatin regulatory effects can easily lead to inhibition of splice site recognition. We found that hormone stimulation induces higher expression of the splicing factor hnRNP AB. This protein, also known as APOBEC1-binding protein 1 (ABBP-1) has been involved in RNA editing of Apolipoprotein B mRNA transcripts (Lau et al. 1997) and in alternative splicing regulation of the FGFR2 gene, the latter through interactions with the carboxy-terminal region of p63, of relevance for the ankyloblepharon-ectodermal dysplasia-clefting (AEC) syndrome (Fomenkov et al. 2003). Consistent with a role for this factor in splicing regulation and the frequent function of hnRNP proteins in exon skipping (for review, see Martinez-Contreras et al. 2007), hnRNP AB binding sites are clearly enriched in the region upstream of R5020-skipped exons. Upon hnRNP AB knock down, hormone-induced exon skipping in the genes DMTF1 and TCERG1 is attenuated, consistent with the hypothesis that hormone-induced expression of hnRNP AB contributes to regulation of at least some exons harboring upstream hnRNP AB binding sites.
We propose that hormone-induced changes in nucleosome positioning influence splice site recognition, partly through changes in RNA polymerase II dynamics and contribute, together with other factors and mechanisms, to the regulation of alternative splicing by a steroid hormone.
MATERIALS AND METHODS
Cell culture and hormone treatments
T47D-MTVL human breast cancer cells, carrying one stably integrated copy of the luciferase reporter gene driven by the MMTV promoter (Truss et al. 1995), were routinely grown as described (Beato and Vicent 2011). Cells were plated in RPMI medium without phenol red, supplemented with 10% dextran-coated charcoal-treated fetal bovine serum (FBS). After 48 h, the medium was replaced by fresh medium without serum. After 24 h in serum-free conditions, cells were incubated with 10 nM R5020 or EtOH for different times at 37°C. For RNAi experiments, T47D cells were plated the day before transfection at 250 × 103 cells per well in six-well plates with RPMI medium without phenol red, and transfected with two rounds of 100 nM of scrambled or HnrnpAB siRNA Oligonucleotides (Dharmacon) using Lipofectamine RNAiMAX (Invitrogen). Hormone induction was then performed as previously described.
RT-PCR, capillary electrophoresis, and gel quantification
Total RNA was extracted from T47D-MMTVluc cells untreated or treated as described in Cell Culture and Hormone Treatments, using Maxwell 16 LEV simplyRNA Tissue Kit (Promega) following manufacturer's instructions. Five hundred nanograms of total mRNAs were reverse-transcribed using super script III retro transcriptase (Invitrogen, Life Technologies) following the manufacturer's recommendations. PCR reactions for every splicing event to validate were carried out with GOTaq DNA polymerase kit (GoTaq, Promega) following the manufacturer's recommendations, in a final reaction volume of 30 µL, including 1 µL of cDNA and forward and reverse primers in exonic sequences flanking the alternatively spliced region of interest (0.5 µM final concentration, Sigma). PCR protocols were set up for 96-well plates and performed in a Zephyr compact liquid handling workstation (Caliper, PerkinElmer). Primers used in this study are listed in Supplemental Table S2. High-throughput capillary electrophoresis measurements for the different splicing isoforms were performed in 96-well format in a Labchip GX Caliper workstation (Caliper, PerkinElmer) using a HT DNA High Sensitivity LabChip chip (PerkinElmer). Data values were obtained using the Labchip GX software analysis tool (version 3.0). PCR products were run in 6% acrylamide gels, and relative amount of inclusion and skipping bands were quantified with ImageJ.
RNA-sequencing
RNA-seq paired-end reads were mapped using GEM RNA Mapping Pipeline (v1.7) using last Gencode Annotation version 18, as follows:
Reads were mapped to genome reference index (including chromosome 1 to X) with maximum mismatch ratio of 0.06 and base quality threshold of 26.
Reads were mapped to a transcriptome index generated from previous reference index using the same options as step 1.
De novo transcriptome index were created and the reads mapped again to this index using maximum mismatch ratio of 0.04, minimum split size of 15 bp and maximum of 500,000 bp.
Reads were mapped again to de novo transcriptome index created using the same options as step 1.
The created mappings and pair alignments were merged using base quality threshold of 26 and maximum insertion size of 500,000 bp.
As a result, BAM alignment files were obtained and used to generate genome-wide normalized profiles using RSeQC software. Exon quantifications (summarized per-genes) were used for expression level determination, either as raw read counts or as reads per kilobase per million mapped reads (RPKM) using Flux Capacitor (http://liorpachter.wordpress.com/tag/flux-capacitor/).
Detecting hormone-dependent alternate splicing exons
Isoform expression and differentially spliced cassette exons in the T60′ post-treatment samples versus the untreated (T0) samples were determined with the MISO pipeline (Katz et al. 2010) using the “skipped exon” event type option. Briefly, PSI (“percent-spliced-in”) values were estimated for each of the three timepoints using the MISO-provided reference exon-centric annotation (hg19 v1.0). Next a list of high confidence differentially expressed cassette exons for the comparison (T60′ versus T0) was compiled by applying the following filters:
An absolute difference of PSIs of at least 0.15 (|ΔPSI| > 0.15).
At least three reads supporting exon skipping in one of the two conditions.
At least 10 reads mapped to the flanking exons in both conditions.
A P-value <0.01 for a different mean between the two posterior PSI distributions (two-tailed t-test).
Only strictly internal cassette exons events are retained.
PR binding site analysis
RefSeq coordinates (including 3000 bp 5′ of the TSS) for genes containing R5020-regulated exons and 243 genes not harboring R5020-regulated exons selected at random, displaying or not transcriptional changes upon R5020 treatment, were intersected with PR ChIP-seq tracks (Ballaré et al. 2013a) and the percentage of genes containing PR binding sites was subsequently determined for each category.
Chromatin immunoprecipitation
RNA polymerase II-ChIP was carried out as previously described (Paronetto et al. 2010) with minor modifications of the protocol. T47D-MMTVluc cells (∼2 × 106 cells/sample) were incubated with 1% (vol/vol) formaldehyde in culture medium for 10 min at room temperature. Cells were then washed in cold phosphate-buffered saline (PBS), harvested and lysed to isolate nuclei in a hypotonic buffer containing 5 mM PIPES 8.0, 85 mM KCl, and 0.5% NP-40. Nuclei were then resuspended, lysed in a buffer containing 1% SDS, 10 mM EDTA, and 50 mM Tris/HCl at pH 8.1, and sonicated in 15 mL tubes with Bioruptor UCD-200 Diagenode (ultrasonic wave output power 250 W, 14 × 30 sec) to yield chromatin sizes of 150–400 bp, and 100 μg of DNA/sample were used for immunoprecipitation with 15 μg of anti-RNA polymerase II clone 4H8 antibodies (Millipore 05-623). Coprecipitated DNA and input DNA were purified and subjected to deep sequencing using the Solexa Genome Analyzer (Illumina). The sequence reads were aligned to the human genome reference (assembly hg19), keeping only sequence tags that mapped uniquely to the genome allowing up to two mismatches. To assess accumulation of RNA polymerase II over exons, reads mapped to alternative exons and downstream exons, including 300 bp downstream from the 5′ss and 750 bp upstream of the 3′ss were downloaded from the UCSC browser and averaged in five consecutive bins of 150 bp in a window of 750 bp 5′ of the 3′ss, on the exonic regions and on two consecutive bins of 150 bp 3′ of the 5′ss. These values were then normalized to local averages for every exon and its upstream bins. Two-sample t-test was used to determine the associated P-values.
MNase sequence processing
Single- and paired-end Illumina sequencing data from MNase-digested chromatin was obtained from Ballaré et al. (2013a) for T47D-MTVL breast cancer cells untreated or treated for 60 min with R5020. FASTQ files were aligned to the reference human genome GRCh37/hg19 (Lander et al. 2001) using BWA (Li et al. 2009). At the time of writing, BWA does not fill all BAM fields completely, so “samtools fixmate” was run to fill in information about a paired-end read's mate. Resulting bam files (Li et al. 2009) were then processed in the following way using custom software:
If 40% or more of a read's quality values were demarked by a “#” (Illumina 1.8 Phred quality), i.e., unknown quality value at that base, then the read was flagged with the 0 × 200 bit (the read does not pass quality controls), and tagged with ZL:Z:0.40.
If the read overlaps with a highly duplicated region (HDR) (Pickrell et al. 2011), the read is also flagged with the 0 × 200 bit and tagged with “ZR:Z:HDR.”
If both the read and its mate's coordinates match one or more pair's coordinates, the read is tagged “ZD:Z:x.y,” where in this case x indicates the total number of pairs duplicated, and y represents an index for one of the pairs in question, numbered from 1 to x.
If flags and tags are encountered on a read's mate, then they are added to the read.
The final BAM file is then indexed using “samtools index.” Because no reads are ever removed at any stage, the original FASTQ files can be regenerated from the BAMs.
Nucleosome occupancy signal generation
Reads not flagged as unsuitable were gathered, and single-end reads were extended to be 147 bp long. Using custom software, genome-wide read depth profiles were then generated and subsequently normalized at each base using alternative read depth profiles where the reads were extended from their middle bases 10 kb upstream and downstream. In this way, the profiles are normalized to their 20 kb background, and consequently allow the comparison between samples with different sequencing depths. In principle, the normalization also attenuates issues arising from the aneuploidity of the T47D genome.
Clustering and aggregation of nucleosome signal
Skipped exons were extracted from all models in the MISO annotation to define a general set of internal exons (39,922 total). The coordinates of these exons were then projected (“lifted”) from genome reference hg18 to hg19 using the liftOver tool from the UCSC Genome Browser website. Sixteen exons were discarded because did not lift correctly, leaving 39,206. Further exons were removed in the following order:
One hundred eighty-two were without Ensembl v68 gene IDs (39,024 total).
Forty-five were on chrY (38,979 total).
Of note, 11,748 exons were filtered where 1-kb region flanking the 3′ splice site of the exon and 1 kb region flanking the 3′ splice sites of both flanking exons did not all have mappability above 90% (27,231 total).
Using bwtool (Pohl and Beato 2014), with the -starts option and 500 bases upstream and downstream, clusters of T0 normalized nucleosome occupancy signal were then obtained from the general internal exon set using a k-means clustering parameter of k = 4. The averaged values of nucleosome signal at each base in the 1 kb flanking the 3′ splice site were then calculated (using bwtool) for each cluster, first at T0 and then at T60, of the MNase-seq data.
Splice site scoring and GC content percent
All exons expressed in T47D at T0 were extracted from MISO annotation, as described in Clustering and Aggregation of Nucleosome Signal, and scored using “Analyzer-Splice-Tool” (http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm), attributing to each annotated exon the sum of its acceptor and its donor score. Two-sample t-tests were used to assign P-values.
Fifty base pair mappability and five base-window GC percent data were downloaded from the UCSC Genome Browser.
HnRNP AB motif analysis
The motif WRGWUAG derived from the RNA-compete tool for the RNA binding domain (RBD) of hnRNP AB in Tetraodon nigroviridis (Ray et al. 2013) (http://cisbp-rna.ccbr.utoronto.ca/index.php), which has an identity of 0.814 with the human RBD. Using SFmap (http://sfmap.technion.ac.il/index.html), enrichment of the predicted hnRNP AB motif was assessed in R5020-regulated exons and a subset of 247 random, not R5020-regulated alternative internal exons in the following regions: 100 bp starting from the 3′ss, 450 bp upstream of the 3′ss, and downstream from the 5′ss, divided in bins of 150 bp. Fisher's exact test corrected for multiple testing was used to calculate P-values between enrichment frequencies on regulated and random regions.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our labs and of the Guigó’s laboratory for discussions, support throughout this work, and critical reading of the manuscript. We are grateful to Juan Ramón Tejedor and Luisa Vigevani for technical help and experimental advice. C.I. was supported by a la Caixa predoctoral fellowship. Work in J.V.’s laboratory is supported by grants from Fundación Botín, Ministerio de Economía y Competitividad and Consolider RNAREG.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048843.114.
REFERENCES
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C, Castellano G, Gaveglia L, Althammer S, González-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP, et al. 2013a. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 49: 67–79. [DOI] [PubMed] [Google Scholar]
- Ballaré C, Zaurin R, Vicent GP, Beato M 2013b. More help than hindrance: Nucleosomes aid transcriptional regulation. Nucleus 4: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59. [DOI] [PubMed] [Google Scholar]
- Bartholomew B 2014. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem 83: 671–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M 1996. Chromatin structure and the regulation of gene expression: remodeling at the MMTV promoter. J Mol Med (Berl) 74: 711–724. [DOI] [PubMed] [Google Scholar]
- Beato M, Klug J 2000. Steroid hormone receptors: an update. Hum Reprod Update 6: 225–236. [DOI] [PubMed] [Google Scholar]
- Beato M, Vicent GP 2011. When every minute counts: the enzymatic complexity associated with the activation of hormone-dependent genes. Cell Cycle 10: 2407–2409. [DOI] [PubMed] [Google Scholar]
- Becker PB, Workman JL 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G, Zorio DA, Bentley DL 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol 24: 8963–8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C 2013. Progesterone signalling in breast cancer: a neglected hormone coming into the limelight. Nat Rev Cancer 13: 385–396. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Carmo-Fonseca M 2014. Reciprocal regulatory links between cotranscriptional splicing and chromatin. Semin Cell Dev Biol 32: 2–10. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 9: 977–983. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR 2010. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR 2014. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 54: 683–690. [DOI] [PubMed] [Google Scholar]
- Fomenkov A, Huang YP, Topaloglu O, Brechman A, Osada M, Fomenkova T, Yuriditsky E, Trink B, Sidransky D, Ratovitski E 2003. P63 α Mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J Biol Chem 278: 23906–23914. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannone C, Valcárcel J 2013. Chromatin's thread to alternative splicing regulation. Chromosoma 122: 465–474. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E 2010. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17: 918–920; author reply 920–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, Rosbash M 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M 2012. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA 18: 2174–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lange CA 2004. Making sense of cross-talk between steroid hormone receptors and intracellular signaling pathways: Who will have the last word? Mol Endocrinol 18: 269–278. [DOI] [PubMed] [Google Scholar]
- Lau PP, Zhu HJ, Nakamuta M, Chan L 1997. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem 272: 1452–1455. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ 2012. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS 2009. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle 8: 3420–3424. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Cappellari M, Busa R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C 2010. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res 70: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gaffney DJ, Gilad Y, Pritchard JK 2011. False positive peaks in ChIP-seq and other sequencing-based functional assays caused by unannotated high copy number regions. Bioinformatics 27: 2144–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, Beato M 2014. bwtool: a tool for bigWig files. Bioinformatics 30: 1618–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Andre V, Batsche E, Rachez C, Muchardt C 2011. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol 18: 337–344. [DOI] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Alló M, Kornblihtt AR 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci 106: 4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16: 990–995. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Oren R, Ast G 2011. Detection and removal of biases in the analysis of next-generation sequencing reads. PLoS One 6: e16685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA 2012. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18: 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcárcel J 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25: 381–388. [DOI] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigó R 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M, Candau R, Chávez S, Beato M 1995. Transcriptional control by steroid hormones: the role of chromatin. Ciba Found Symp 191: 7–17; discussion 17–23. [DOI] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S 2011. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell 24: 367–381. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H 2011. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci 108: E627–E635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Luo G, Wise JA, Lou H 2014. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res 42: 701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.