Abstract
Advances in small RNA sequencing technologies and comparative genomics have fueled comprehensive microRNA (miRNA) gene annotations in humans and model organisms. Although new miRNAs continue to be discovered in recent years, these have universally been lowly expressed, recently evolved, and of debatable endogenous activity, leading to the general assumption that virtually all biologically important miRNAs have been identified. Here, we analyzed small RNAs that emanate from the highly repetitive rDNA arrays of Drosophila. In addition to endo-siRNAs derived from sense and antisense strands of the pre-rRNA sequence, we unexpectedly identified a novel, deeply conserved, noncanonical miRNA. Although this miRNA is widely expressed, this miRNA was not identified by previous studies due to bioinformatics filters removing such repetitive sequences. Deep-sequencing data provide clear evidence for specific processing with precisely defined 5′ and 3′ ends. Furthermore, we demonstrate that the mature miRNA species is incorporated in the effector complexes and has detectable trans regulatory activity. Processing of this miRNA requires Dicer-1, whereas the Drosha–Pasha complex is dispensable. The miRNA hairpin sequence is located in the internal transcribed spacer 1 region of rDNA and is highly conserved among Dipteran species that were separated from their common ancestor ∼100 million years ago. Our results suggest that biologically active miRNA genes may remain unidentified even in well-studied organisms.
Keywords: miRNA, ribosomal RNA, repetitive sequence, noncanonical miRNA processing
INTRODUCTION
Two classes of 21–23 nt small regulatory RNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs), play important roles in gene regulation (Okamura and Lai 2008; Bartel 2009). In Drosophila, these classes of small RNAs are sorted into distinct Argonaute effector complexes, resulting in preferential binding of miRNAs and siRNAs to AGO1 and AGO2, respectively (Czech and Hannon 2011). miRNAs generally down-regulate target gene expression by destabilization and/or translational inhibition of target mRNAs through partial complementarity to target sites in 3′ UTRs (Bartel 2009). On the other hand, endogenous siRNAs usually down-regulate highly complementary targets by guiding the cleavage of target RNA molecules via AGO2 (Okamura and Lai 2008).
The ribosome is a ribonucleoprotein complex that catalyzes protein synthesis, and the eukaryotic ribosome generally contains four RNA components, 5S, 5.8S, 18S, and 28S rRNAs. These are transcribed by RNA polymerase I (Pol I) as a 45S polycistronic precursor transcript, except for 5S rRNA, which is transcribed by RNA polymerase III as a monocistronic transcript (Granneman and Baserga 2004). The 45S polycistronic rRNA precursor is processed into mature rRNAs after the removal of internal and external transcribed spacers (ITSs and ETSs). Transcription and processing of rRNAs occur in a nuclear structure, the nucleolus (Boisvert et al. 2007). In Drosophila, 5.8S and 28S rRNAs have additional cleavage sites and produce two pieces of rRNAs from each (5.8S/2S and 28S-1/-2 RNAs) (Tautz et al. 1988). The DNA sequence encoding 45S pre-rRNA is flanked by intergenic spacers (IGS) (Morgan et al. 1983; Kuhn and Grummt 1987; Paalman et al. 1995). The rDNA is present as tandem direct repeats that are organized in one or several clusters in the genome (Long and Dawid 1980). The copy number of rDNA units per genome shows a great variation ranging from <50 to >10,000 among eukaryotes (Long and Dawid 1980).
Ribosome biogenesis is a regulated process whose dysregulation is associated with various diseases, including cancers and Diamond–Blackfan anemia (Stumpf and Ruggero 2011). In mice, noncoding transcripts from the IGS region regulate chromatin/DNA modification at the promoter region by recruiting the NoRC (nucleolar remodeling complex) (Mayer et al. 2006). Furthermore, transcription of the antisense rDNA strand is often observed in a wide range of organisms and may also regulate rRNA biogenesis (Chekanova et al. 2007; Bierhoff et al. 2010). In Arabidopsis, a nuclear Argonaute AtAGO4 forms foci in nucleoli, and the formation of these foci is dependent on the siRNA pathway and 24-nt siRNAs produced from the pre-rRNA sequence (Li et al. 2006; Pontes et al. 2006).
Although ITS regions are proposed to play roles in rRNA processing by studies in yeast (Musters et al. 1990), ITS sequences are generally considered nonfunctional due to their poor evolutionary conservation. Nevertheless, phylogenetic studies of ITS sequences have uncovered conserved stem–loop sequences in particular taxa (Schlotterer et al. 1994; Armbruster et al. 2000).
Here we show that two distinct classes of small RNAs are produced from the Drosophila rDNA locus. First, we provide clear evidence for siRNA production from the Drosophila rDNA. In addition, we unexpectedly identify a conserved miRNA encoded in the ITS1 region and show that the mature species is highly expressed throughout fly development. Therefore, this study provides a potential explanation for the unusual conservation of the fly ITS hairpin sequence that was identified two decades ago (Schlotterer et al. 1994). Considering the possibility that highly repetitive sequences may not be correctly assembled in reference genome sequences, our identification of a conserved, active miRNA gene from a highly repetitive region raises a possibility that such miRNAs may have escaped attention even in well-studied organisms.
RESULTS
Drosophila rDNA generates siRNAs
Previous studies revealed that rDNAs are transcribed bidirectionally in some organisms (Chekanova et al. 2007; Bierhoff et al. 2010), and that small RNAs can be produced from rDNA (Li et al. 2006; Pontes et al. 2006; Lee et al. 2009; Wei et al. 2013). In flies, the involvement of the siRNA pathway in rDNA regulation was proposed because mutants in core RNAi factors, namely ago2 and dcr-2 mutants, exhibit rDNA phenotypes including aberrant nucleolar morphology, lower H3K9me2 occupancy at the rDNA loci and elevated extrachromosomal circular rDNA levels (Peng and Karpen 2007). However, the production of siRNAs from rDNA in flies has not been explicitly shown.
To address this, we took advantage of small RNA libraries made from AGO1 complexes purified from fly ovaries of wild-type and RNAi-defective genotypes. In principle, bulk siRNAs should be dependent on Dcr-2 and/or segregate to AGO2. However, as the endo-siRNA population is extremely diverse, individual siRNAs are often not well-expressed (thus potentially challenging approaches to assay their depletion in dcr-2 mutants), and immunoprecipitation techniques do not necessarily distinguish intrinsically and peripherally bound species (thus potentially challenging the assessment of all reads in AGO2-IP libraries as bona fide siRNAs). As an alternative strategy, we previously showed that bulk endo-siRNAs are resorted to AGO1 complexes in mutants of the siRNA loading factor R2D2 or the siRNA effector AGO2 (Okamura et al. 2011). Such resorting patterns, in which AGO1-IP small RNA reads exhibit relative abundance of r2d2 or ago2 ≫ wild-type > dcr-2, provide a powerful means for functional categorization of endo-siRNAs.
To improve the detection sensitivity of our previous samples, we resequenced AGO1-IP libraries from wild-type, dcr-2, r2d2, and ago2 mutant ovaries, and obtained 33–35 million reads from each (Supplemental Table S1, Sheet 1); the previous and current data were combined for this study. We normalized read counts by the number of reads mapping to known miRNA hairpins in each library, and expressed values in reads per million miRNA reads (RPM). We believe that this normalization method permits a more appropriate comparison of small RNA abundances, compared with conventional normalization using total numbers of reads perfectly mapping to the genome sequence, for the following reasons. First, the proportions of resorted known siRNAs (from transposons and hairpin RNAs) in the AGO1 complexes substantially varied in the four AGO1-IP libraries (from 0.3% in dcr-2 mutant to 25.1% in r2d2 mutant, Supplemental Table S1, Sheet 2). Second, our previous Northern blotting analysis suggested that the levels of abundant miRNA species were not changed in these mutants (Okamura et al. 2011). According to this normalization scheme, transposon-derived siRNAs were 27.3 and 36.5 times more abundant in the AGO1-IP libraries from r2d2 and ago2 mutant ovaries, respectively, than the AGO1-IP libraries from wild-type (Supplemental Table S1, Sheet 2). In the AGO1-IP library from dcr-2 mutant, these reads were strongly reduced, indicating that the libraries contain pure endo-siRNA populations.
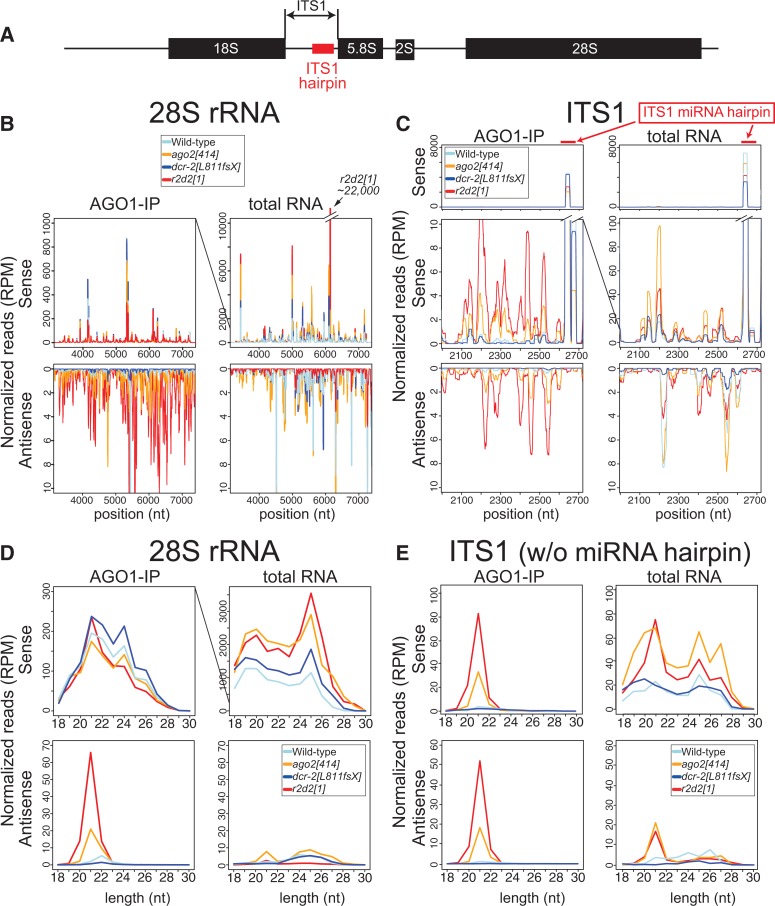
We attempted to detect siRNAs from the Drosophila rDNA locus using these data. However, even with AGO1 immunopurification from r2d2 and ago2 mutants, we failed to detect the siRNA signature in the sense strand of rRNA coding regions (Fig. 1A,B,D, upper panels; Supplemental Fig. S1). The size distribution of small RNAs matching to the sense rRNA sequence did not show a specific peak at any length, suggesting that the majority of these reads may represent degradation products from mature rRNAs. However, it was possible that siRNA reads might be hidden by the abundant background reads.
FIGURE 1.
siRNAs produced from the rDNA locus. (A) Schematic representation of the Drosophila rDNA gene structure. (B,C) Small RNA read densities on the sense (upper panels) and antisense (bottom panels) strands of 28S rRNA (B) and ITS1 (C) regions were plotted. Read counts were normalized by the number of reads mapping to miRNA hairpins. The x-axis shows the nucleotide position on the D. melanogaster rDNA sequence (NCBI M21017.1). Upper panels in C are the same plots as the middle panels with broader y-axis ranges to accommodate the high ITS1 miRNA peaks. (D,E) Size distribution of small RNA reads mapping on the sense (upper panels) and antisense (bottom panels) strands of 28S rRNA (D) and ITS1 (E) regions. In order to compare the different region sizes, values were further normalized to RPM per kilobase region mapped. To obtain the size distribution of ITS1 mapping reads, we removed the mir-10404/mir-ITS1 hairpin sequence that produces a large number of miRNA reads. Results for other regions are shown in Supplemental Figure S1.
To avoid the abundant mature rRNA-derived reads, we decided to analyze the ITS regions and the antisense strand of the coding/noncoding regions. In all these regions, we were able to detect clear siRNA signals (Fig. 1; Supplemental Fig. S1). In the AGO1 complex from r2d2 or ago2 mutant, small RNAs mapping to the antisense strand were observed throughout the rDNA region. These antisense mapping reads were at a very low level in the AGO1 complex from wild-type or dcr-2 mutant. Furthermore, AGO1-IP reads mapping to the antisense rDNA strand showed a sharp peak at 21 nt (Fig. 1D, lower panels). We also observed the siRNA signature on both strands of the ITS regions (Fig. 1C,E; Supplemental Fig. S1A,B). The densities of siRNAs from the sense and antisense strands were similar in the ITS regions, suggesting that rDNA siRNAs derive from dsRNA produced from bidirectional transcription of the pre-rRNA sequence.
Our results indicated that siRNAs are produced from rDNA in Drosophila. It has to be seen if rDNA-derived siRNAs play roles in the nucleolar phenotypes observed in dcr-2 or ago2 mutant (Peng and Karpen 2007). In Neurospora and rice, rDNA-derived small RNAs are involved in DNA damage response (Lee et al. 2009; Chen et al. 2013). Furthermore, a new role for Dicer in suppression of rDNA antisense transcription and copy number maintenance of rDNA repeats was recently uncovered in fission yeast (Castel et al. 2014). It would be interesting to test whether Drosophila rDNA-derived siRNAs have analogous functions.
rDNA ITS1 encodes a conserved miRNA
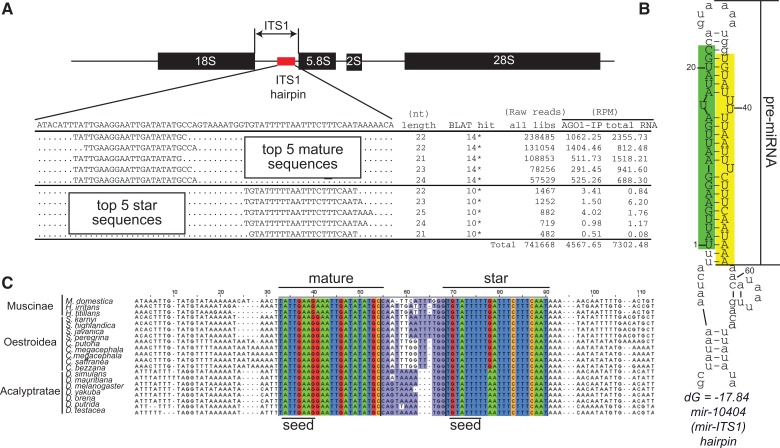
In the course of these analyses, we noticed a large number of small RNA reads mapped to a small region on the sense strand of the ITS1 region (Fig. 1C, red arrow). There are two major clusters of ∼22 nt reads with precisely defined 5′ ends (Fig. 2A), which paired to each other with short 3′ overhangs within an apparent ∼60-nt hairpin precursor structure (Fig. 2B). These reads were present at comparably high levels in total RNA and AGO1-IP libraries in wild-type (Fig. 2A), and we found no evidence for resorting in ago2 or r2d2 mutant background (Supplemental Table S2), indicating their normal preferential sorting to AGO1. Altogether, these observations indicated that the ITS1 hairpin encodes a novel miRNA. Notably, reads mapping to this hairpin in the total RNA libraries are substantially abundant (ranging from 3397.5 RPM in the dcr-2 mutant library to 7249.6 RPM in the wild-type library) and the ITS1 hairpin could be ranked as the 22nd–31st most abundant miRNA among the 238 miRNA genes in these libraries (Supplemental Table S3). Since these features satisfy the criteria for miRNA gene annotation (precise 5′ ends from the duplex structure with short 3′ overhangs) (Kozomara and Griffiths-Jones 2014), the hairpin was named mir-10404/mir-ITS1.
FIGURE 2.
A conserved miRNA in rDNA ITS1. (A) Small RNA reads mapping to the ITS1 hairpin region. Total small RNA read counts were obtained from ∼90 small RNA libraries listed in Supplemental Table S1. For normalized read counts in ovary AGO1-IP and total RNA libraries, we used the number of reads mapping to annotated miRNA hairpins as a normalizer. The asterisk (*) indicates that the BLAT hit numbers shown on the table are the numbers of genomic positions the sequence can be perfectly mapped in the D. melanogaster reference genome. Note that the actual copy number of rDNA is not reflected in the genome assembly. Each of the two rDNA arrays in Drosophila melanogaster is estimated to contain hundreds of rDNA units (Long and Dawid 1980). (B) RNA folding of mir-10404/mir-ITS1 hairpin. The RNA sequence producing reads shown in A was folded by Mfold (Zuker 2003). The most abundant mature (5p) and star (3p) sequences were highlighted by green and yellow, respectively. The numbers beside the hairpin structure indicate relative nucleotide positions counting from the 5′ end of the major 5p species. The pre-mir-10404/mir-ITS1 hairpin sequence was defined as the sequence from the 5′ end of 5p to the 3′ end of 3p. Note that the flanking sequences do not form a stem structure, suggesting a lack of the lower stem structure that is essential for processing by the Microprocessor complex. (C) Sequence alignment of fly mir-10404/mir-ITS1 hairpin. The hairpin sequence is conserved in distant fly species whereas the surrounding sequences are highly diverged. The loop region (purple) shows faster evolution compared with the mature and star regions, exhibiting a typical “saddle-shape” conservation pattern. Note that the nucleotide substitutions occurring in the stem region are outside of the seed sequence.
The rDNA ITSs have generally been considered as nonfunctional sequences that are under a low selective pressure, although their potential roles in rRNA processing have been proposed in yeast (Musters et al. 1990). However, within the 12 sequenced Drosophila genomes, the duplex sequence was perfectly conserved, whereas the flanking sequences were highly diverged (Supplemental Fig. S2). In fact, the hairpin encoding mir-10404/mir-ITS1 was identified in the 1990s as an unusually highly conserved hairpin (Schlotterer et al. 1994). We reanalyzed conservation of the region corresponding to the mir-10404/mir-ITS1 hairpin across a broad set of Dipteran species (Fig. 2C). As previously reported, clearly orthologous hairpins were identified in distant fly species including Musca, which is estimated to have diverged from the Drosophilid ancestor ∼100 million years ago (Beverley and Wilson 1984; Schlotterer et al. 1994). The hairpin appears to have arisen within Schizophoran radiation from other Dipteran species, because a clear counterpart of this hairpin could not be found in the orthologous ITS1 region of Nematoceran species (Anopheles or Culex).
In conserved miRNAs, the stem region generally shows a lower rate of divergence compared with the loop region, likely due to the selective pressure maintaining the target-miRNA complementarity and the hairpin structure (Lai et al. 2003; Berezikov et al. 2005). Close examination of divergence patterns in the ITS1 miRNA hairpin revealed that substitutions occurred primarily in the loop region (8 and 2 variable nucleotides in the loop and stem regions, respectively). The two nucleotide substitutions found in the stem regions were located outside of the seed sequence regions (Fig. 2C). Furthermore, these substitutions were predicted to preserve the pairing state of the hairpin structure (Supplemental Fig. S3). Therefore, these substitutions are not predicted to have major effects on processing and functions of this miRNA. These features strongly argue that the mir-10404/mir-ITS1 hairpin is under selective pressure to maintain function as a miRNA.
Endogenous expression and Argonaute loading of ITS1-miRNA
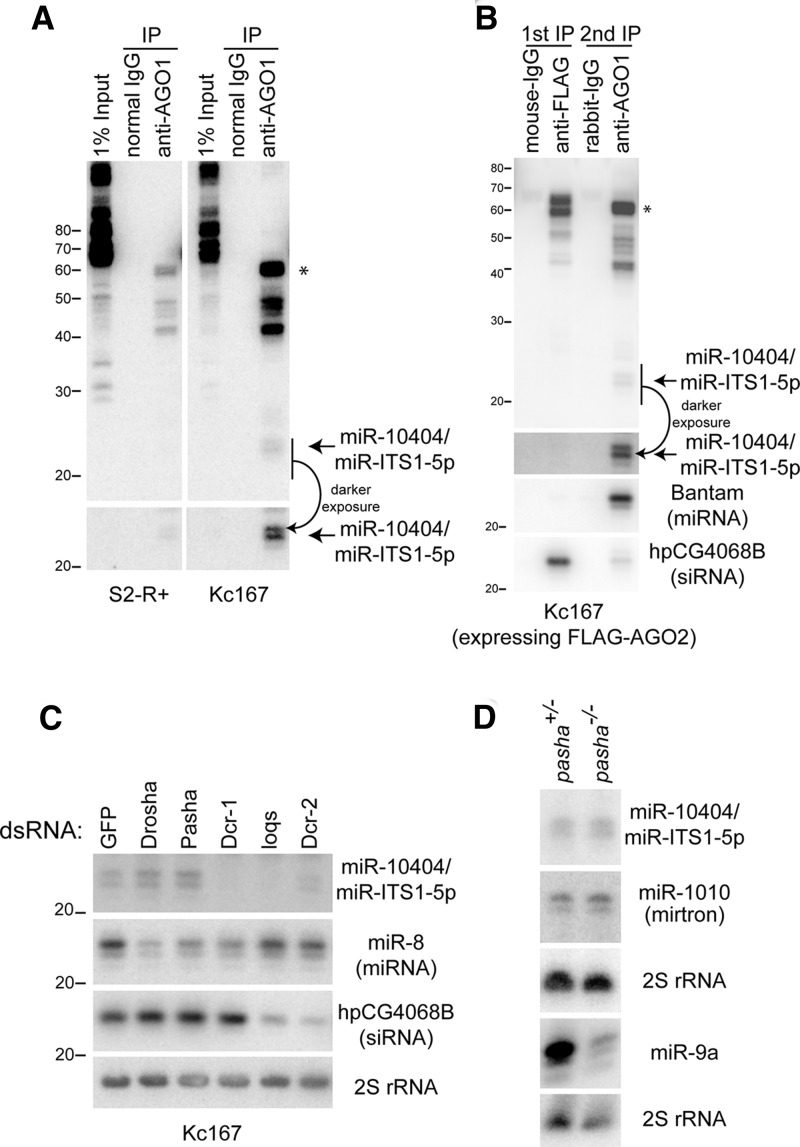
We sought to directly detect mature miR-10404/miR-ITS1 by Northern blotting. To distinguish AGO1-loaded species from other RNAs, we performed AGO1-IP from S2-R+ and Kc167 cell lysates. We observed many bands in the input lanes from both of the cell lines (Fig. 3A, left panels). In Kc167 cells, only a few bands including the 22 nt species (Fig. 3A, arrow) were strongly enriched after AGO1-immunopurification (Fig. 3A, left panels). In S2-R+ cells, the ∼40–60 nt species could be easily detected in the AGO1-complex, but only faint signals were observed in the ∼22 nt region (Fig. 3A, left panels). The ∼60 nt species that was enriched in the AGO1-complex (Fig. 3A, asterisk) may represent the hairpin precursor species loaded to AGO1 before dicing, although we usually observe depletion of pre-miRNA species from Argonaute IP samples with fly materials (Okamura et al. 2013).
FIGURE 3.
Expression and sorting of ITS1 miRNA. (A) Loading of miR-10404/miR-ITS1-5p to AGO1 in S2-R+ cells and Kc167 cells. AGO1-complexes were purified from S2-R+ (left panels) or Kc167 cells (right panels). RNA extracted from ∼1% lysate was used for input analysis. The mature species (∼22 nt, arrow) and the presumable precursor hairpin (60 nt, asterisk) were enriched in the AGO1 complex. (B) Sorting of miR-10404/miR-ITS1-5p to AGO1 in Kc167 cells. FLAG-AGO2 and AGO1 complexes were sequentially purified from Kc167 cells stably expressing FLAG-tagged AGO2 protein. miR-10404/miR-ITS1-5p is enriched in AGO1. Successful IP was verified by probing the same membrane for Bantam miRNA and hp-CG4068B siRNA, which are enriched in the AGO1 and FLAG-AGO2 complexes, respectively. The arrow and the asterisk indicate the mature and hairpin species. (C) Processing of mir-10404/mir-ITS1. RNA samples were prepared from Kc167 cells soaked with indicated dsRNAs. miR-10404/miR-ITS1 production was reduced when Dcr-1 or Loqs was depleted. On the other hand, Drosha knockdown did not reduce expression of miR-10404/miR-ITS1. A canonical miRNA miR-8 is slightly (∼50%) reduced in Drosha knockdown cells. Four micrograms of small RNA-enriched RNA was loaded in each lane. Quantified signal intensities are shown in Supplemental Figure S5A. (D) Production of miR-10404/miR-ITS1 in pasha mutant. RNA was extracted from heterozygous or homozygous pasha mutant third instar larvae. miR-10404/miR-ITS1 expression did not change in pasha mutant. A canonical miRNA (miR-9a), but not a mirtron (miR-1010), was reduced in pasha mutant. 2S rRNA panels are shown as loading control for the miR-10404/miR-ITS1 and miR-1010 panels (middle 2S panel) or for the miR-9a panel (bottom 2S panel). For the three panels from the top (miR-10404/miR-ITS1-5p, miR-1010, and 2S rRNA), 4 µg small RNA-enriched RNA was loaded. For other panels, 20 µg total RNA was loaded in each lane. Signal quantification results are shown in Supplemental Figure S5B.
Next, we were interested to analyze the expression pattern of mir-10404/mir-ITS1. mir-10404/mir-ITS1 reads were found in all of the ∼90 published small RNA libraries analyzed in this study, which included four body parts/organs, eight developmental time points and 14 cell lines (Supplemental Table S2). While broadly expressed, miR-10404/miR-ITS1 exhibited variable levels in different tissues. For example, the read counts from this ITS1 hairpin were 1569.8 and 180.8 RPM in the Kc167 and S2-R+ libraries, respectively (Supplemental Table S3). Beyond D. melanogaster, the evolutionary conservation of the hairpin sequence suggested that mature miR-10404/miR-ITS1 species is produced in other Dipterans (Fig. 2C; Supplemental Fig. S2A). Indeed, a discrete ∼22 nt species was readily detectable by Northern blotting in all five other Drosophilid species tested (Supplemental Fig. S2B). These results further support the conclusion that this conserved hairpin encodes an evolutionarily conserved miRNA gene.
In Drosophila, miRNAs and siRNAs are preferentially sorted to AGO1 and AGO2, respectively (Czech and Hannon 2011). To understand Argonaute sorting of miR-10404/miR-ITS1, we reanalyzed published AGO1- and AGO2-IP libraries prepared from S2-R+ cells (Okamura et al. 2013). The sorting ratio (AGO1 raw read count/AGO2 raw read count) of these miR-10404/miR-ITS1 reads was similar to those of known miRNAs and much higher than those of endo-siRNAs (Supplemental Table S3; Fig. S4A). This suggested that miR-10404/miR-ITS1 is preferentially loaded in AGO1. We verified preferential AGO1-sorting of miR-10404/miR-ITS1 by Northern blotting (Fig. 3B). AGO1- and AGO2-complexes were immunopurified from Kc167 cells stably expressing FLAG-tagged AGO2 (Czech et al. 2008). The ∼22-nt miR-10404/miR-ITS1-5p species was not detectable in the AGO2 complex (Fig. 3B). The major species in the AGO2 complex migrated at ∼55–60 nt. These bands were distinct from major bands detected in the AGO1-IP lane, suggesting that AGO1 and AGO2 preferentially bind distinct species derived from the mir-10404/mir-ITS1 hairpin.
In summary, we were able to verify preferential AGO1-loading of the mature miR-10404/miR-ITS1-5p species and found that miR-10404/miR-ITS1 is expressed throughout fly development in a broad range of tissues.
Biogenesis of ITS1-miRNA
Most miRNAs are processed by a stepwise processing mechanism catalyzed by two RNase III complexes, the Drosha/Pasha and Dcr-1/loqs complexes (Kim et al. 2009). The Drosha/Pasha complex recognizes hairpins of ∼3 helical turns (∼33 nt), and releases miRNA precursor hairpins from primary transcripts, which is then cleaved by the Dcr-1/loqs complex to produce ∼22 nt small RNA duplexes. The mir-10404/mir-ITS1 hairpin structure containing a relatively short (∼25 nt) stem region (Fig. 2B) suggested that the mir-10404/mir-ITS1 hairpin precursor may not be a product of the Drosha/Pasha complex (Han et al. 2006). On the other hand, our reanalysis of published small RNA libraries from Dcr-1 knockdown S2 cells (Zhou et al. 2009; Yang et al. 2014) suggested that the maturation of miR-10404/miR-ITS1 is mediated by Dcr-1 (Supplemental Fig. S4B). Based on these observations, we hypothesized that mir-10404/mir-ITS1 is a Drosha-independent, Dcr-1-dependent miRNA.
We assessed its biogenesis dependencies by using RNAi to knockdown small RNA processing factors in Kc167 cells (Fig. 3C). Mature miR-10404/miR-ITS1-5p was reduced in cells depleted of Dcr-1 or loqs, suggesting that the ITS1 hairpin is cleaved by the Dcr-1/loqs complex similar to canonical miRNAs. In contrast, depletion of Drosha or Pasha did not reduce the ∼22-nt miR-10404/miR-ITS1-5p species although expression of a canonical miRNA miR-8 was reduced by ∼70% following knockdown of Drosha in Kc167 cells (Fig. 3C; Supplemental Fig. S5A). To further verify Drosha/Pasha-independent processing in a more stringent assay, we used a genetic knockout of the pasha gene (Martin et al. 2009). In homozygous pasha mutant third instar larvae, miR-9a expression was significantly reduced (∼50%), whereas expression of a Drosha/Pasha-independent miRNA (mirtron miR-1010) was unchanged (Fig. 3D; Supplemental Fig. S5B). The accumulation of mature miR-10404/miR-ITS1-5p was unaffected in pasha mutant larvae (Fig. 3D; Supplemental Fig. S5B), providing clear support for its status as a Microprocessor-independent miRNA.
These results indicated that ITS1 miRNA was processed by a noncanonical miRNA processing pathway that bypasses cleavage by the Drosha/Pasha complex but requires the Dcr-1/loqs complex for its maturation.
Trans regulatory activity of ITS1-miRNA
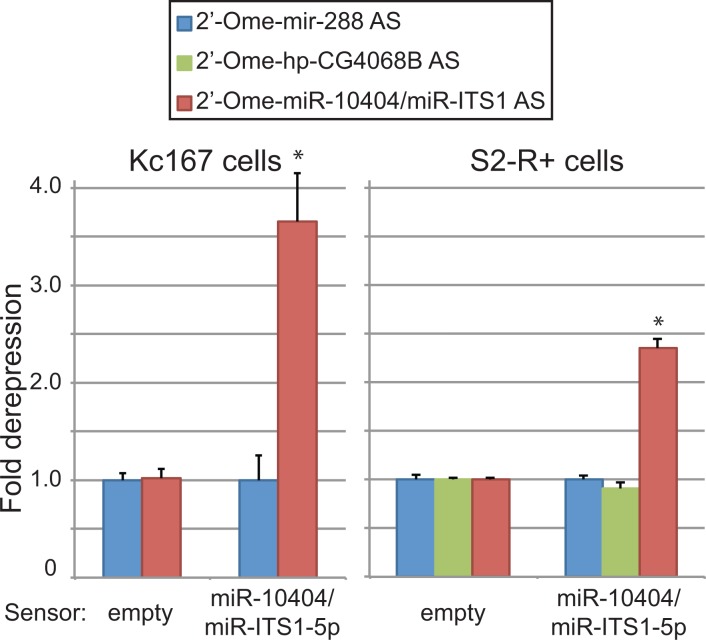
To test whether endogenous small RNAs from the ITS1 miRNA hairpin have detectable activity as trans-regulatory species, we constructed a Renilla luciferase sensor bearing target sequences perfectly complementary to miR-10404/miR-ITS1-5p. This reporter plasmid was cotransfected with a specific antisense oligonucleotide inhibitor against miR-10404/miR-ITS1-5p or unrelated inhibitors. In Kc167 cells, we observed approximately threefold derepression of miR-10404/miR-ITS1-5p reporter expression when the plasmid was cotransfected with the miR-10404/miR-ITS1-5p inhibitor (Fig. 4). We also observed weaker, but clear derepression in S2-R+ cells, consistent with the lower expression level of miR-10404/miR-ITS1-5p in this cell line (Fig. 4; Supplemental Table S3).
FIGURE 4.
Regulatory activity of endogenous miR-10404/miR-ITS1-5p. Luciferase sensors bearing no target site or a two tandem copy of target sequences that is perfectly complementary to miR-10404/miR-ITS1-5p were cotransfected with 2′-O-methylated RNA oligonucleotide inhibitor against mature miR-288, miR-10404/miR-ITS1-5p, or hp-CG4068B. The sensors were derepressed only when the cognate inhibitor was cotransfected. Note that stronger derepression was observed in Kc167 cells consistent with the expression levels of miR-10404/miR-ITS1 in these cell lines. The columns and error bars depict means and standard deviations, respectively (N = 4). Asterisks indicate statistically significant differences compared with the values with the control miR-288 antisense oligonucleotide (P < 0.05, t-test).
These results indicated that endogenous miR-10404/miR-ITS1-5p is an active repressor against their targets. Altogether, we identified a conserved, noncanonical miRNA with demonstrable regulatory activity that is encoded in the repetitive rDNA.
DISCUSSION
Two classes of small RNAs from rDNA
Using AGO1-IP libraries from wild-type and RNAi defective mutant ovaries, we demonstrated that the Drosophila rDNA is a source of two distinct classes of Argonaute-dependent small RNAs. The first class comprises endo-siRNAs produced from pre-rRNAs and its antisense transcripts (Fig. 1). rDNA siRNAs show the typical siRNA signature (AGO1-IP reads from r2d2 or ago2 mutant ≫ wild type > dcr-2 mutant), and provide compelling evidence that the ∼21 nt reads represent genuine siRNA species (Fig. 1). Notably, this approach can be applied to small RNA reads from any locus to test whether the locus produces endo-siRNAs. Even in D. melanogaster, comprehensive annotation of siRNA loci using large sequencing data sets has only been performed using cultured cell lines (Wen et al. 2014), therefore, additional AGO1-IP small RNA libraries from other tissues using the same set of genotypes would facilitate accurate siRNA gene annotation.
The second class is an evolutionarily conserved miRNA (Fig. 2; Supplemental Fig. S2). Endogenous activity of miR-10404/miR-ITS1 was readily detectable by luciferase sensor assays (Fig. 4). Given the important roles of ribosome biogenesis in cell growth and homeostasis, it is conceivable that small RNAs found in the present study are relevant to these processes. Future studies should be aimed to further elucidate biological roles of these small RNAs from rDNA in regulation of ribosome biogenesis and cell growth.
rRNA-derived miRNAs in other organisms
Is pre-rRNA-derived miRNA unique to flies? In mammals, there are miRNA genes located in rDNAs, including human (hsa-mir-663) and mouse (mmu-mir-696, mir-712, mir-714 and mir-715) genes (Kozomara and Griffiths-Jones 2014). mmu-mir-712 was recently reported to play roles in endothelial inflammation and atherosclerosis (Son et al. 2013); however, clear evidence for production of specific species or Argonaute loading has been lacking. We analyzed a number of published mammalian Argonaute IP libraries (Valen et al. 2011; Dueck et al. 2012; Li et al. 2013; Maillard et al. 2013; Polikepahad and Corry 2013), and these data failed to provide evidence for the production of mature miRNAs from these loci (Supplemental Table S4). Recently, a few mouse miRNA genes located in rDNAs (mmu-mir-2182, mir-5102, mir-5105, mir-5109, and mir-5115) were removed from miRBase due to the lack of evidence for specific processing (Castellano and Stebbing 2013). We further identified additional miRBase dead entries that have similarities to the coding (mmu-mir-2134, mmu-mir-2143, hsa-mir-1826, and hsa-mir-6087) and 5′ ETS (hsa-mir-3687) sequences of the mouse or human rDNA. Again, we were not able to verify specific processing of these hairpins by small RNA library analysis (Supplemental Table S4). Therefore, the previously annotated mammalian rDNA miRNAs may be erroneous annotations. This highlights that care must be taken when annotating miRNAs, especially ones that potentially derive from abundant noncoding RNAs.
Nevertheless, as our study provides a clear precedent for pre-rRNA-derived miRNAs, the possibility remains in other organisms. Previous molecular phylogenetics studies using ITS sequences have noted interesting conservation patterns in rDNA ITS regions in particular taxa (Borsuk et al. 1994; Armbruster et al. 2000). The conserved ITS sequences were generally speculated to play roles in rRNA processing in the previous studies, but the importance of the conserved ITS sequences in rRNA processing has not been biochemically tested. Our demonstration that rRNA ITS sequences can acquire gene regulatory roles raises an intriguing possibility that conserved ITS regions may have trans-regulatory activity. It will be interesting to see in future studies whether any of conserved ITS regions produces stable RNA species that regulate gene expression in trans.
Implications for miRNA gene annotation
Our findings provide a notable precedent of a highly conserved, abundant miRNA derived from repetitive DNA. It is standard practice to filter such loci when attempting to annotate miRNAs, especially as the provenance of reads derived from repetitive loci can be difficult to ascertain. In addition, multicopy sequences including rRNA, tRNA, and other noncoding RNA genes are often highly expressed, and their degradation products complicate data analysis. In the case of insects, the presence of abundant endogenous siRNAs from transposons makes interpretation of reads from repetitive sequences even more difficult (Okamura and Lai 2008).
Moreover, repetitive regions are systematically underassembled even in relatively “complete” genomes. For example, the assembled D. melanogaster genome has only a fraction (∼14) of the hundreds of rDNA copies it carries on both the X and Y chromosomes, and at the extreme, there is only a single copy of the rDNA cluster present in the C. elegans genome assembly. One wonders whether other bona fide miRNAs may yet exist in repetitive or even unassembled portions of genomes.
MATERIALS AND METHODS
Sequencing of small RNA libraries and sequencing data analysis
Construction of ovary AGO1-associated small RNA libraries was described in our previous study (Okamura et al. 2011). We resequenced the AGO1-IP libraries on Illumina GAII and sequencing data from two sequencing runs were combined. Small RNA data sets were downloaded from DNA Data Bank of Japan (DDBJ) (listed in Supplemental Table S1). Processing was carried out with Fastx-toolkit v0.0.13.2 (http://hannonlab.cshl.edu/fastx_toolkit), BEDOPS v2.20 (Neph et al. 2012), Bedtools v2.19.0 (Quinlan and Hall 2010), UCSC Kent Source Utilities (Karolchik et al. 2014), and custom shell scripts. Reads with length 18–30 nt were mapped to reference sequences with bowtie v1.0.0 (Langmead et al. 2009) and no mismatches allowed. RepeatMasker was used to identify TE siRNAs (Jurka et al. 2005). Hairpin RNA sequences were defined in previous studies (Czech et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). Reference sequences used in this study are listed in Supplemental Table S1, Sheet 7. Raw read counts were first adjusted by the number of mapped alignment hits for a read sequence within the bowtie index. These were then normalized to per million mapped miRNA stem–loop reads (RPM). Values in the read length distribution plots were further normalized to RPM per kilobase region mapped in order to compare the different region sizes on the rDNA sequence.
Ribosomal DNA sequences (dme, M21017.1; mmu, BK000964; hsa, NR046235) were downloaded from NCBI while genomic sequences were obtained from UCSC Genome Browser (dm3, mm10, hg19) (Benson et al. 2014; Karolchik et al. 2014). miRNA stem–loop sequences were downloaded from miRBase (Release 20) (Kozomara and Griffiths-Jones 2014). For dme, the sequence of dme-mir-10404/mir-ITS1 was added to the bowtie index. In order to find previously annotated miRNA hairpins that matched to rDNA, stem–loop sequences from miRBase Release 14–20 were mapped with bowtie for up to three mismatches to the rDNA sequence. mmu-mir-2182, mir-5102, mir-5105, mir-5109, and mir-5115 were not analyzed because similar analysis was done for these miRNAs in a previous study (Castellano and Stebbing 2013).
ITS1 sequences from the 12 Drosophila species were collected from the trace archives at GenBank, with the exception of D. pseudoobscura ITS1 sequence, which was obtained from GenBank: HQ631785. Clones containing rDNA sequences were identified by searching for the Drosophila melanogaster 5.8S rRNA sequence. The consensus ITS1 hairpin sequence obtained with >40 clones was used as the representative sequence of each fly species.
ITS1 homologs from other fly species were identified using BLAST search of the M. domestica rRNA sequence (GenBank Accession: Z28417.1) against the NCBI nucleotide sequence repository. Phylogenetic relationships between fly species were determined using the NCBI Taxonomy Browser. Sequence alignment of mir-10404/mir-ITS1 was performed using the Fast Statistical Aligner (Bradley et al. 2009), and visualized using Jalview (Waterhouse et al. 2009).
Argonaute-IP and Northern blotting
Kc167 cells were stably transfected with a plasmid containing FLAG-HA tagged AGO2 genomic fragment (Czech et al. 2008). Immunoprecipitation was carried out using anti-FLAG (Wako) or anti-AGO1 (AbCam) antibody in RIPA buffer as described previously (Okamura et al. 2013). For detection of miR-10404/miR-ITS1-5p, a DNA probe (Fig. 3A) or an LNA probe (Fig. 3B,C,D) was used. Small RNA enrichment was performed for the top two panels of Figure 3C and the top three panel of Figure 3D using mirVana miRNA isolation kit (Ambion). Oligo probe sequences are listed in Supplemental Table S5. Northern blotting and preparation of pasha mutant samples were described previously (Okamura et al. 2008; Martin et al. 2009).
Luciferase assay
Sensor construction, transfection and luciferase assays were carried out as described previously with modifications (Okamura et al. 2008). For S2-R+ cells, 150 ng sensor plasmid and 50 pmol antisense 2′-O-me oligonucleotides were transfected in the 24-well format using Effectene (Qiagen). For Kc167 cells, we transfected 600 ng plasmid and 200 pmol in the 6-well format. Sequences of antisense oligonucleotides and oligos used to generate sensors are listed in Supplemental Table S5. Three days after transfection, cells were harvested and lysed in 70 µL Passive lysis buffer (Promega), and the lysate was used for luciferase assays.
RNAi in Kc167 cells
Of note, 2 × 106 Kc167 cells were soaked with 15 µg dsRNA in 1 ml serum free medium for 0.5–1 h. After soaking, 1 mL Schneider's medium containing 20% serum was added and cells were incubated at 25°C. Four days later, dsRNA soaking was repeated to ensure efficient knockdown. RNA samples were prepared 4 d after the second soaking.
DATA DEPOSITION
The small RNA library sequencing data generated in this study are available at NCBI SRA under SRP050320.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cai Yu for Kc167 cells, Joanne Yew for Drosophilid species samples, and Gregory Hannon for the FLAG-tagged AGO2 plasmid. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund and the National Institutes of Health/National Institute of General Medical Sciences (R01-GM083300). Research in K.O.’s group was supported by the National Research Foundation, Prime Minister's Office, Singapore under its NRF Fellowship Programme (NRF2011NRF-NRFF001-042). G.T.-K. was supported by NUS Faculty of Science startup grant R-154-000-536-133. The content is solely the responsibility of the authors and does not necessarily represent the official views of these agencies.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.049098.114.
REFERENCES
- Armbruster GFJ, van Moorsel CHM, Gittenberger E 2000. Conserved sequence patterns in the non-coding ribosomal ITS-1 of distantly related snail taxa. J Molluscan Stud 66: 570–573. [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW 2014. GenBank. Nucleic Acids Res 42: D32–D37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24. [DOI] [PubMed] [Google Scholar]
- Beverley SM, Wilson AC 1984. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol 21: 1–13. [DOI] [PubMed] [Google Scholar]
- Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I 2010. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol 75: 357–364. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585. [DOI] [PubMed] [Google Scholar]
- Borsuk P, Gniadkowski M, Kucharski R, Bisko M, Kanabus M, Stepien PP, Bartnik E 1994. Evolutionary conservation of the transcribed spacer sequences of the rDNA repeat unit in three species of the genus Aspergillus. Acta Biochim Pol 41: 73–77. [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L 2009. Fast statistical alignment. PLoS Comput Biol 5: e1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang A-Y, Sánchez M, Valbuena A, Antequera F, Martienssen RA 2014. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 10.1016/j.cell.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano L, Stebbing J 2013. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res 41: 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. 2007. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Chen H, Kobayashi K, Miyao A, Hirochika H, Yamaoka N, Nishiguchi M 2013. Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. PLoS One 8: e55252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ 2011. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. 2008. An endogenous siRNA pathway in Drosophila. Nature 453: 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G 2012. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 40: 9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ 2004. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC genome browser database: 2014 update. Nucleic Acids Res 42: D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H 2008. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature 453: 793–797. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I 1987. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J 6: 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM 2003. Computational identification of Drosophila microRNA genes. Genome Biol 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106. [DOI] [PubMed] [Google Scholar]
- Li N, You X, Chen T, Mackowiak SD, Friedlander MR, Weigt M, Du H, Gogol-Döring A, Chang Z, Dieterich C, et al. 2013. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic Acids Res 41: 3619–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long EO, Dawid IB 1980. Repeated genes in eukaryotes. Annu Rev Biochem 49: 727–764. [DOI] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O 2013. Antiviral RNA interference in mammalian cells. Science 342: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Smibert P, Yalcin A, Tyler DM, Schäfer U, Tuschl T, Lai EC 2009. A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Mol Cell Biol 29: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361. [DOI] [PubMed] [Google Scholar]
- Morgan GT, Reeder RH, Bakken AH 1983. Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc Natl Acad Sci 80: 6490–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Boon K, van der Sande CA, van Heerikhuizen H, Planta RJ 1990. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J 9: 3989–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28: 1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC 2008. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Robine N, Liu Y, Liu Q, Lai EC 2011. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol Cell Biol 31: 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ladewig E, Zhou L, Lai EC 2013. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev 27: 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paalman MH, Henderson SL, Sollner-Webb B 1995. Stimulation of the mouse rRNA gene promoter by a distal spacer promoter. Mol Cell Biol 15: 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S, Corry DB 2013. Profiling of T helper cell-derived small RNAs reveals unique antisense transcripts and differential association of miRNAs with argonaute proteins 1 and 2. Nucleic Acids Res 41: 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS 2006. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotterer C, Hauser MT, von Haeseler A, Tautz D 1994. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol 11: 513–522. [DOI] [PubMed] [Google Scholar]
- Son DJ, Kumar S, Takabe W, Kim CW, Ni CW, Alberts-Grill N, Jang IH, Kim S, Kim W, Won Kang S, et al. 2013. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat Commun 4: 3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Ruggero D 2011. The cancerous translation apparatus. Curr Opin Genet Dev 21: 474–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA 1988. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol Biol Evol 5: 366–376. [DOI] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH 2011. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol 18: 1075–1082. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Zhou B, Zhang F, Tu Y, Hu Y, Zhang B, Zhai Q 2013. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS One 8: e56842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Mohammed J, Bortolamiol-Becet D, Tsai H, Robine N, Westholm JO, Ladewig E, Dai Q, Okamura K, Flynt AS, et al. 2014. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res 24: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Smibert P, Westholm JO, Jee D, Maurin T, Lai EC 2014. Intertwined pathways for Argonaute-mediated microRNA biogenesis in Drosophila. Nucleic Acids Res 42: 1987–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ 2009. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA 15: 1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.