Abstract
While the human transcriptome contains a large number of circular RNAs (circRNAs), the functions of most circRNAs remain unclear. Sequence annotation suggests that most circRNAs are generated from splicing in reversed orders across exons. However, the mechanisms of this backsplicing are largely unknown. Here we constructed a single exon minigene containing split GFP, and found that the pre-mRNA indeed produces circRNA through efficient backsplicing in human and Drosophila cells. The backsplicing is enhanced by complementary introns that form double-stranded RNA structure to bring splice sites in proximity, but such structure is not required. Moreover, backsplicing is regulated by general splicing factors and cis-elements, but with regulatory rules distinct from canonical splicing. The resulting circRNA can be translated to generate functional proteins. Unlike linear mRNA, poly-adenosine or poly-thymidine in 3′ UTR can inhibit circular mRNA translation. This study revealed that backsplicing can occur efficiently in diverse eukaryotes to generate circular mRNAs.
Keywords: circular RNA, alternative splicing, backsplicing, splicing factors, translation
INTRODUCTION
Although circular RNAs (circRNAs) in higher eukaryotes were discovered more than 20 years ago (Nigro et al. 1991; Cocquerelle et al. 1992), they were often considered as splicing byproducts with low abundance (Nigro et al. 1991; Hentze and Preiss 2013). This paradigm has recently been changed by several reports of a large number of circRNAs in the mammalian transcriptome (Hansen et al. 2013; Jeck et al. 2013; Memczak et al. 2013; Salzman et al. 2013), among which some are even more abundant than their linear counterparts (Burd et al. 2010; Salzman et al. 2012) and others have novel activity in regulating microRNA function (Hansen et al. 2013; Memczak et al. 2013). Sequence annotation suggests that most circRNAs are resulted from pre-mRNA splicing at a reversed order (a.k.a. backsplicing) in which an upstream 3′ splicing site (ss) is joined with a downstream 5′ ss. In addition, circRNA can also be generated through 3′ exonucleolytic degradation of lariat RNAs that are splicing byproducts, resulting an intronic circRNA with 2′,5′-phosphodiester bond (Zhang et al. 2013).
Canonical splicing (i.e., linear splicing) of most metazoan introns requires a stepwise assembly of spliceosomal components in a cotranscriptional fashion (Kornblihtt et al. 2013; Matera and Wang 2014), starting with a process called exon definition in which the 5′ ss and 3′ ss are recognized in pairs across exons through the interaction of U1 and U2 snRNPs (Fox-Walsh et al. 2005; Xiao et al. 2007). In a subsequent transition, U1 and U2 snRNPs undergo rearrangements to form an intron-spanning interaction known as intron definition complex (De Conti et al. 2013). This transition is required for spliceosomal activation and is a key regulatory step of splicing (Sharma et al. 2008). However, as a variation from linear splicing, backsplicing requires spliceosome assembly/activation across exons and thus this transition is either absent or happens in a different configuration. Therefore different mechanisms may be needed to regulate backsplicing.
The general functions of most circRNA remain unclear, although some circRNAs can serve as microRNA sponge to regulate gene expression (Hansen et al. 2013; Memczak et al. 2013). An interesting possibility is that circRNAs may serve as mRNAs to direct protein synthesis, as eukaryotic ribosomes were found to be able to initiate in vitro translation on synthesized circRNAs (Chen and Sarnow 1995). Nevertheless, in vivo protein translation from circular mRNA has not been observed, and exonic circRNAs were not detected in the ribosome-bound fraction with ribosome profiling (Jeck et al. 2013), indicating that the translation from endogenous circRNA is still debatable. A possible explanation for this apparent discrepancy is that translation can be initiated on circRNA but the efficient elongation and termination require auxiliary factors that bind to mRNA ends, as both the 5′ end and poly(A) tail of mRNA are known to mediate various steps of translation (Jackson et al. 2010).
Here we report that a single exon can be efficiently backspliced to generate circRNA. The resulting circRNA can be translated from an IRES to generate a functional GFP protein. The backsplicing is enhanced by intronic complementary sequences that form double-stranded RNA (dsRNA) structure, but such structure is not required. In addition, backsplicing is extensively regulated by canonical splicing regulatory cis-elements and cognate factors. This study provides direct evidence of backsplicing and proves that circRNA can indeed function as mRNA to drive translation inside cells.
RESULTS AND DISCUSSION
Efficient backsplicing produces circular RNA
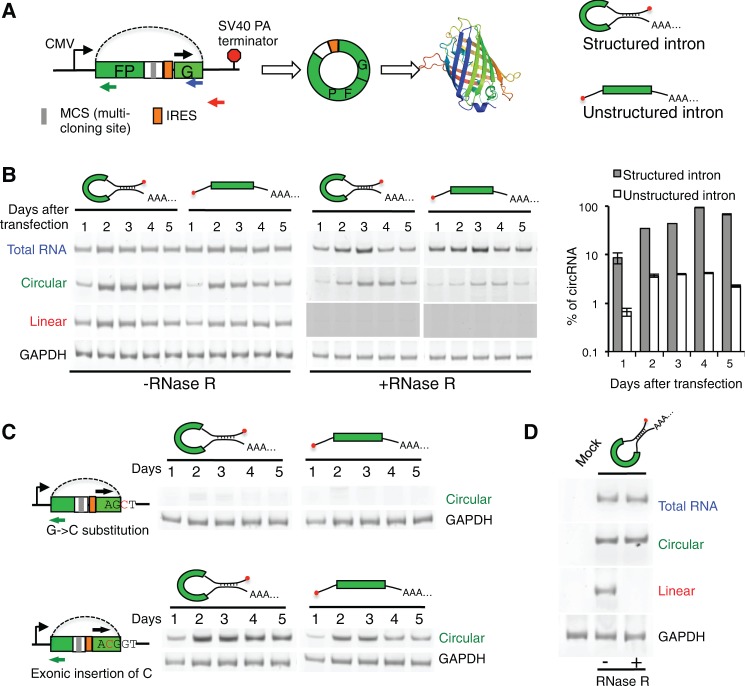
To study circRNA biogenesis, we engineered a minigene reporter that contains a single exon encoding two GFP fragments in a reversed order (Fig. 1A). An internal ribosome entry site (IRES) was inserted upstream of the start codon of GFP, which can drive cap-independent protein synthesis. We also engineered an exonic multicloning site that can be used to study how backsplicing is regulated by exonic sequences. This exon is flanked by two introns with typical 3′ and 5′ splice sites, which can be joined into a circular RNA through backsplicing to generate an intact open reading frame of GFP.
FIGURE 1.
circRNA generated from backsplicing. (A) Schematic diagram of minigene with split GFP in a reverse order. The transcription of minigene is driven by a CMV promoter and terminated by SV40 polyadenylation signal. The exon is represented by box, and the backsplicing is indicated with dotted line. Different pairs of primers were used to detect total, linear, or circular RNAs in RT-PCR. Two versions of the minigene, with structured or unstructured introns, were generated. (B) Different RNAs produced from minigenes with structured or unstructured introns. Total RNAs were purified at day 1 to 5 after transfection, treated with or without RNase R, and analyzed by semiquantitative RT-PCRs (22 cycles) using various primer sets. To ensure no linear product is detected in RNase R treated samples, we used a longer exposure time for the corresponding lanes. The amounts of different RNAs were also quantified by real-time RT-PCR, and the percent of circular RNA is plotted in the right panel. (C) Production of circular RNAs in minigenes with splice site mutation or exonic insertion. The circRNAs were detected at different days after transfection. (D) Production of circRNA in S2 cells. The minigene with structured intron was transfected to S2 cells, and different RNAs were detected as described in panel B.
Previously it was reported that many circRNAs are flanked by ALU sequences that may be paired to each other to form dsRNA across circularizing exons (Jeck et al. 2013), leading to the speculation that intron pairing may promote circRNA production. To directly test this hypothesis, we generated two versions of the minigene that contain the same upstream intron (intron 12 of IGF2BP1) but different downstream introns. One downstream intron includes a reverse complement fragment that forms a dsRNA region of 233 nt with the upstream intron, whereas the other does not form any strong secondary structure (Fig. 1A; Supplemental Fig. S1).
We transfected both minigenes into 293T cells and examined total, linear, and circular RNAs using semiquantitative RT-PCR with three different pairs of primers (Fig. 1A). We detected circRNAs 1 d after transfection of the minigenes with either structured or unstructured introns (Fig. 1B). The production of circRNAs reached the peak at around days 3–4, and was further confirmed with RNase R treatment. The dsRNA structure in flanking introns strongly promoted circRNA production with a 10–30-fold increase in circRNA level as compared with those from unstructured intron (Fig. 1B, right panel). However such dsRNA structure is not required for backsplicing, as the minigene with an unstructured intron also generated detectable circRNA. The formation of circRNA from unstructured introns is especially interesting, as these introns have not been found to be backspliced. To directly examine if the circRNA is indeed generated from backsplicing, we mutated the consensus 5′ ss of the downstream intron by replacing guanine with a cytosine. A similar mutation was generated by inserting a cytosine inside the coding exon (Fig. 1C). We found that the mutation in the consensus 5′ ss disrupted circRNA production, whereas a similar mutation inside the exon restored circRNA production (Fig. 1C), suggesting that a normal 5′ ss is required for backsplicing.
It has been reported that circRNAs are conserved across the eukaryotic tree, suggesting circRNAs may play important biological roles across multiple species (Wang et al. 2014a). When transfecting the minigene into Drosophila S2 cells, we found that the minigene containing human introns can also be efficiently backspliced to produce cricRNA (Fig. 1D), suggesting that the machinery for RNA backsplicing is conserved between invertebrates and vertebrates.
We further measured the cellular level of circular and linear RNAs from the same minigene with real-time RT-PCR, and found that circRNAs accumulated up to 10-fold at day 4 after transfection (Fig. 2A). In contrast, the level of linear RNAs produced from the same minigene was reduced to ∼70% at day 4 and to 55% at day 5. Such accumulation is probably a result of the high stability or a slow production of circRNA through backsplicing, or both. To directly measure RNA stability, we inhibited RNA transcription with actinomycin D, and followed the degradation of different RNA species. We found that the circRNAs generated from two minigenes are extremely stable, with no detectable reduction after 6 h upon actinomycin D treatment. In contrast, the linear RNAs encoded by the same minigenes were degraded in a rate comparable with a typical endogenous mRNA (Fig. 2B).
FIGURE 2.
Stability of circRNAs. (A) The RNA levels relative to day 1 of transfection were measured by qRT-PCR using primer sets for linear or circRNAs. The transfections were conducted with minigene containing structured intron, and all experiments are performed in triplicates with the means and SD of RNA levels plotted. In all panels, the RNA level of GAPDH was used to calibrate all other RNA levels. (B) Change of RNA levels after transcription inhibition by actinomycin D or DMSO control. Two minigenes containing structured or unstructured intron were transfected into 293T cells, and the levels of linear or circular RNAs were determined at different times after drug treatment. The level of a typical endogenous mRNA, SRSF1, was also measured as a control.
Circular RNA function as mRNA for protein translation
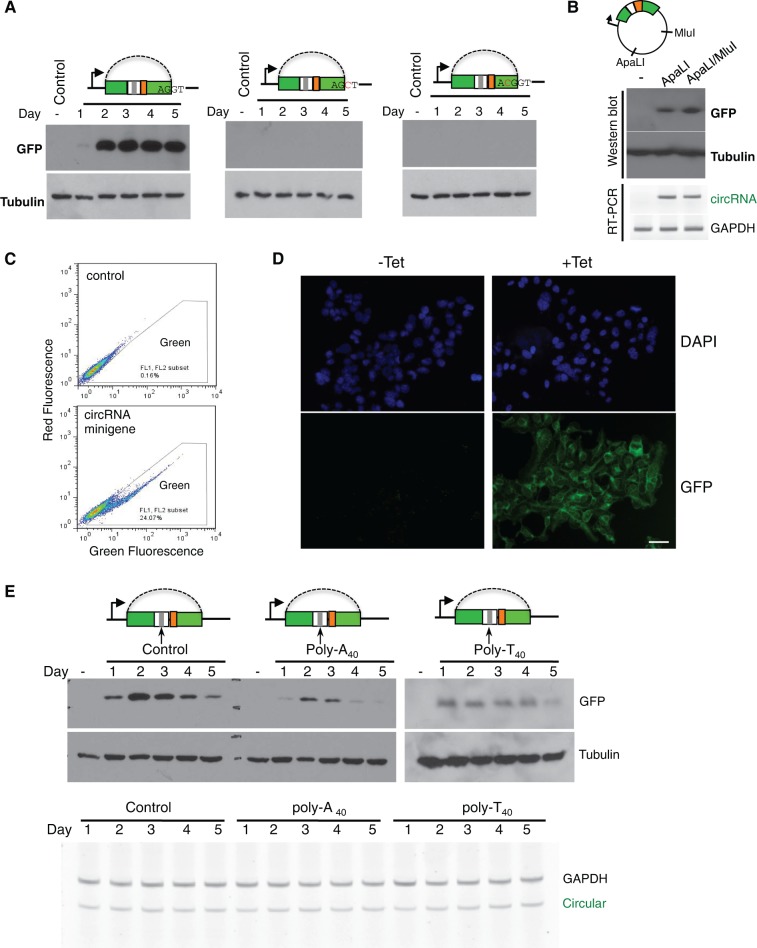
Although some circRNAs were found to function as miRNA sponges (Hansen et al. 2013; Memczak et al. 2013), the in vivo function of most circRNA remains unknown. To examine if circRNA can function as mRNA to direct protein synthesis, we engineered an IRES in our circRNA minigene and detected potential protein production with Western blot (Fig. 3A). We found a robust protein production in minigenes containing structured or unstructured introns (Fig. 3A; Supplemental Fig. S2), suggesting the circRNA can indeed function as mRNA to direct protein synthesis. This result is also consistent with the observation that most circRNA can be transported into cytoplasm (Jeck et al. 2013). In addition, we found the production of a functional GFP was abolished by either the mutation on the 5′ ss that disrupts backsplicing, or by an insertion in the coding region that causes frame shift (same mutations used in Fig. 1C).
FIGURE 3.
In vivo translation directed by circular mRNA. (A) Cells were transfected with wild-type or mutated minigenes containing structured introns, and total proteins were purified at different days after transfection to detect production of GFP. One mutated minigenes has a disrupted 5′ splice site (second panel), while the other contains a C residue inserted into exon to disrupt GFP reading frame (third panel). (B) The circRNA minigene vector has single ApaLI and MluI cut sites in the plasmid backbone. We linearized the wild-type minigene with ApaLI or ApaL1/MluI digestion, and transfected the linearized DNA into 293 cells to detect GFP expression at 2 d after transfection. The production of circRNA was also measured by RT-PCR as described in Figure 1. (C) Cells transiently transfected with backsplicing minigene are assayed by flow cytometry at 2 d after transfection. (D) 293 FlpIn T-REX cells were stably transfected with backsplicing minigene containing structured intron, and the production of GFP was induced by addition of tetracycline at a final concentration of 1 μg/mL. The cells were assayed by fluorescence microscopy 48 h after induction. Scale bar = 50 μm. (E) Production of GFPs from minigenes inserted with poly(A40), poly(T40), or nonrepetitive control sequence downstream from the stop codon was detected with Western blot. The tubulin antibody was used as a loading control. The Western blot at the right panel (poly-T40) was overexposed to show protein production. Bottom, the corresponding RNA levels from each transfected sample were determined with semiquantitative RT-PCR.
There is a small possibility that transcription from minigene may bypass the poly(A) site at the end to produce a long RNA product from entire plasmid that contains concatenated GFP fragments. This long RNA could then be spliced to form a linear RNA containing intact ORF to drive GFP translation. To confirm the translation from circRNA, we digested the plasmid with restriction enzymes that make single cut on the backbone (ApaL1 and MluI). The linearized DNA was gel-purified and transfected into 293 cells. In this case, the transcription cannot go around the entire plasmid. As expected, we detected production of both circRNA and GFP protein (Fig. 3B), proving that the GFP protein can indeed be translated from circular mRNA. The production of GFP was further confirmed by flow cytometry, where we found ∼25% of cells are green at day 2 of transfection (Fig. 3C). The small fraction of cells expressing GFP may be due to low transfection efficiency, slow production of circRNAs, slow translation from IRES, or combination of all factors. In a stably transfected cell line with inducible expression of circRNA, we found close to 100% of cells are green (Fig. 3D).
Translation from linear mRNA is enhanced by the poly(A) tails at the 3′ end of mRNA. This enhancement is mediated through an RNA loop bridged by interactions between poly(A) binding protein (PABP) and eIF4G that recognizes the 5′ end cap of mRNA (Gallie 1998). To study if the poly(A) sequence has similar activities in stimulating translation from circular mRNA, we inserted a 40-nt poly(A) fragment after the stop codon of GFP. Surprisingly, rather than enhancing protein translation, poly(A) sequence actually reduced the protein production as compared with the circular mRNA without poly(A). Since the level of circular mRNA did not change by inserting poly(A) sequences (Fig. 3E, bottom), this effect is likely mediated through translation inhibition. This observation also supports the view that the translation enhancement of linear mRNA by poly(A) is mainly due to RNA looping via PABP and eIF4G rather than direct activation by PABP. Interestingly, we found that insertion of another repetitive sequence, poly(T), also produced similar (or even stronger) inhibition of protein production (Fig. 3E, right panel, same GFP antibody was used in an overexposed blot). We speculate that, when inserted upstream of IRES, these repetitive sequences may somehow interfere with IRES-mediated translation.
Backsplicing of circRNA is regulated by splicing factors
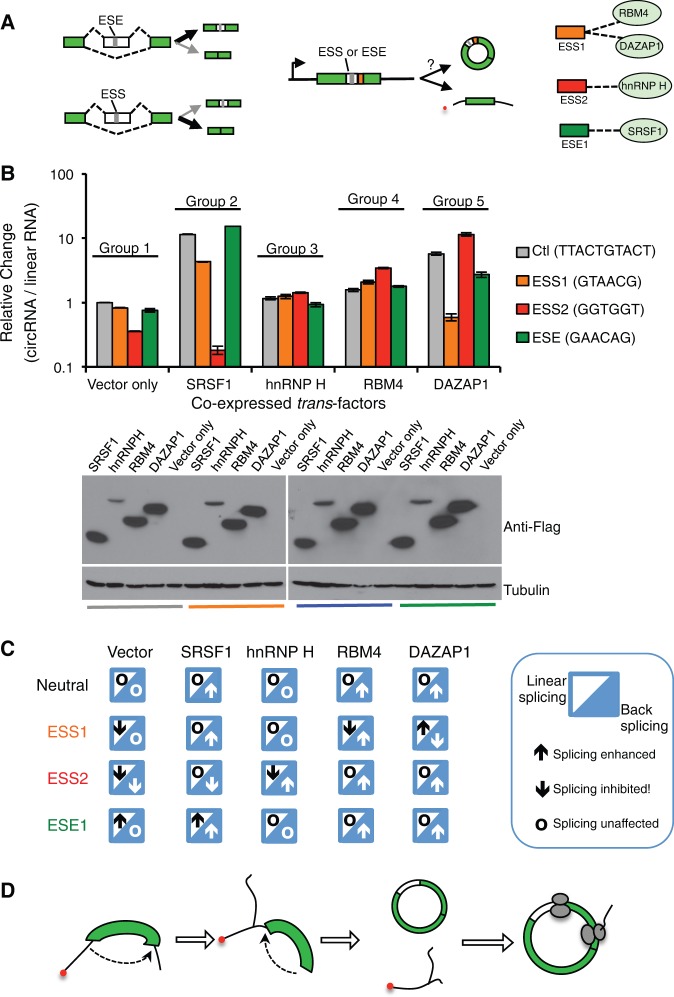
The pairing of splice site is controlled by various splicing factors that enhance or inhibit splicing through binding to cis-acting splicing regulatory elements (SREs) in pre-mRNAs (Wang and Burge 2008). The cis-elements can function as exonic splicing enhancers or silencers (ESEs or ESSs) to regulate exon inclusion. The general rule of regulation is that, by recruiting cognate factors, the ESE promotes exon inclusion to produce a longer isoform and the ESS inhibits splicing to generate a shorter isoform (Fig. 4A). However it is unclear if the same regulatory rules can affect backsplicing. To examine if backsplicing is controlled in a similar fashion, we coexpressed four splicing factors with the minigene containing several known SREs (one ESE, two ESSs, and a control sequence that does not affect linear splicing) (Fig. 4A, right), and measured how the exon circularization is affected by quantitative RT-PCR. We found that the ESS2 substantially inhibited circRNA production through backsplicing, whereas the ESE1 and ESS1 had no obvious effect (Fig. 4B, the first group). This effect is different from that observed in canonical splicing, as the three SREs all affected exon inclusion in a linear splicing reporter (Wang et al. 2012, 2013).
FIGURE 4.
Regulation of backsplicing by cis-elements and splicing factors. (A) Schematic diagram of splicing regulation by SREs and splicing factors in linear and backsplicing. Linear splicing can be promoted or inhibited by ESEs or ESSs, respectively, which is mediated by their cognate splicing factors (left panel). The same ESSs or ESEs were inserted into the backspliced exons, and the resulting minigenes were coexpressed with various splicing factors to measure levels of circular or linear RNA using quantitative RT-PCR. The interactions between SREs and cognate splicing factors were also shown. (B) Backsplicing minigenes inserted with various ESSs or ESE were coexpressed with four different splicing factors (SRSF1, hnRNP H, RBM4, and DAZAP1). The relative levels of circular RNA were compared with the control vector inserted with a neutral sequence. The expression of splicing factors was confirmed by Western blots. (C) Summary of regulatory rules for linear and backsplicing by various SREs and splicing factors. In each case, the enhancement or inhibition of linear splicing and backsplicing was indicated by an upward or downward arrow. The circles indicate the case where splicing was not affected. The activities of each combination of the SRE and splicing factor are based on data from panel B and previous results (Wang et al. 2012, 2013). (D) A model of backsplicing and translation from circular mRNA.
The splicing factors that are specifically bound by these SREs showed distinct effects on circRNA production, however these effects were more complicated than what would be predicted by their activities in controlling linear splicing. For example, SRSF1 appeared to promote circRNA production from most minigenes, with the exception of the minigene containing ESS2 (Fig. 4B, the second group). In contrast, hnRNP H was found to promote backsplicing of the exon inserted with ESS2 but had no effect on other minigenes (cf. group 1 and 3 in Fig. 4B). Both RBM4 and DAZAP1 were found to recognize ESS1 but had opposite activities in controlling exon inclusion in canonical splicing (Choudhury et al. 2014; Wang et al. 2014b). Consistently they showed opposite effects on the backsplicing of circRNA from ESS1-containing exon, although they also affected backsplicing of other minigenes in a sequence independent fashion (Fig. 4B, the fourth and fifth groups).
Using a “mosaic” code, we summarized the regulation rules for linear and backsplicing in different combination of SREs and splicing factors (Fig. 4C). Like linear splicing, the backsplicing process appears to be extensively regulated by canonical SREs and splicing factors, which can noticeably affect circRNA production. Nevertheless, the regulation of backsplicing has fundamentally different rules: In most cases, the same combinations of SREs and splicing factors have distinct or even opposite activity in controlling backsplicing versus linear splicing (Fig. 4C). These intriguing observations suggest that cells may use different mechanisms in controlling backsplicing, and that revealing the backsplicing code will be critical for understanding of the biogenesis for this important type of noncoding RNAs.
In summary, we demonstrated that exons can be efficiently backspliced to generate circular mRNA (Fig. 4D). Previously exon circularization has been observed in vitro at a low frequency, however its efficiency was greatly reduced with increase of exon length (Pasman et al. 1996). In contrast, we found that backsplicing happens efficiently with a relatively long exon (>1.3 kb) in human or insect cells. The complementary sequences in adjacent introns can promote backsplicing, but such dsRNA structure is not required.
We further demonstrated that circRNA can direct translation of an entire functional protein, and the protein production is prolonged for several days after transient transfection. Unlike translation from linear mRNA, poly(A) sequences reduced translation from circular mRNA, and such inhibition is also observed with one other repetitive sequence poly(U). We found that canonical splicing factors that control linear splicing can also regulate backsplicing, however the regulation of backsplicing follows a different set of rules. This simple study presents several key conclusions in circRNA biogenesis and activity, and also provides a unique platform to study regulation of backsplicing. In addition, the stable circular mRNA produced from this system may become a useful tool for long-term protein expression and for examination of the roles of RNA ends in translation control.
MATERIALS AND METHODS
See Supplemental Methods for more details.
Circular RNA reporter constructs
All of the circular RNA reporters were generated from the same backbone, pEGFP-C1, where the transcription of inserted minigene is driven by the CMV promoter and terminated by the SV40 polyadenylation signal. The minigene was inserted with an IRES fragment that was amplified from pIRES2-EGFP vector. A multicloning sites were inserted between the GFP stop codon and the IRES site, resulting a large exon (1333 nt) flanked with canonical introns that only been found in linear splicing (intron 12 of IGF2BP1). This new vector, pCircGFP, was further modified to include intronic complementary pairs or exonic insertions. See Supplemental Figure S1 and Supplemental Methods for more details and the sequences of all primers.
Cell culture and transfection
293T cells were cultured with DMEM medium containing 10% of FBS as described earlier (Wang and Wang 2014). Drosophila S2 cells were cultured with Express Five SFM medium containing 10% L-glutamine. To transiently express circular RNA minigenes, 293 cells were plated into 24-well plates 1 d before transfection. Of note, 0.2 μg of the plasmids containing circular RNA minigene were transfected using lipofectamine 2000 according to the manual. Transfected cells were collected every day for 5 d for further RNA and protein analysis. For transfection of linearized plasmid, the vector was digested with ApaLI or double digested with ApaLI/MluI in matching NEB buffer for 3 h, and the linearized DNA fragments are separated on a 0.8% long agarose gel and purified. The transfection of linearized DNA was essentially same as the plasmid transfection excepting that 0.8 μg linearized DNA was used to compensate the lower transfection efficiency of linear DNA.
To overexpress different splicing factors, cells were plated into 24-well plates 1 d before transfection. To determine the effect of overexpression of splicing factors on circular RNA generation, 0.2 μg of minigene reporters were cotransfected with 0.4 μg of splicing factors distinctly using lipofectamine 2000 according to the manual. After 48 h, cells were collected for further analysis of RNA and protein levels.
To generate a stable cell line expressing the circular RNA minigene upon tetracycline induction, we used pCDNA5 FRT/TO vector and 293 FlpIn/T-Rex cells (Invitrogen). The circular RNA minigene was cloned into the pCDNA5 FRT/TO vector using EcoRV/XhoI site, and transfected with pOG44 in 1:9 ratio. The stably integrated cells were selected with 100 μg/mL hygromycin. One day before the induction, the cells were transferred to hygromycin-free medium. The inductions were carried out by adding tetracycline to a final concentration of 1 μg/mL. The induced cells were applied to immunofluorescence assay 48 h after induction.
Assay of circular and linear RNA with semiquantitative RT-PCR or real-time PCR
The total RNAs were isolated from cells transfected with pCircGFP constructs, and treated with DNase I (37°C, 1 h, followed by heat inactivation of DNase). Total RNA (2 µg) was then reverse-transcribed with SuperScript III (Invitrogen), and one-tenth of the RT product was used for PCR amplification (22 cycles of amplification, with trace amount of Cy5-dCTP in addition to nonfluorescent dNTPs). The products were separated on 10% PAGE gels, scanned with a Typhoon 9400 scanner, and quantified with ImageQuant 5.2.
The real-time PCR was performed with same templates using the Maxima SYBR Green qPCR Master Mix (Thermo Scientific) and a 7500 real-time PCR system (Life Technologies) according to manufacturer's instructions. The expression level of circular RNA, linear RNA was normalized to the endogenous expression of GAPDH.
Protein detection with Western blot and immunofluorescence
Cells were lysed in buffer containing 50 mM HEPES, 150 mN NaCl (4.38 g), 1 mM EDTA, 1% (w/v) CHAPS and Sigma protease inhibitor cocktail. Subsequently the cell lysates were resolved by10% SDS-PAGE and transferred to nitrocellulose membrane. The following antibodies were used: GFP antibody (632381) was purchased from Cell Signaling Technology, Bcl-x antibody (610211) was purchased from Clontech. α-Tubulin antibodies (T5168) were purchased from Sigma-Aldrich. Bound antibodies were visualized with the ECL kit (GE Healthcare).
Two hundred ninety-three cells stably transfected with circular RNA expression vector upon were seeded onto poly-lysine coated glass coverslips in a 6-well plate, and induced with 1 μg/mL tetracycline. At 48 h after induction, the cells were fixed with 4% formaldehyde in 1× PBS for 20 min, washed three times with 1× PBS, and then permeabilized with 0.2% Triton X-100. The cover slips were then mounted with mounting medium (Vector shield's mounting medium with DAPI). Cells were visualized using an Olympus fluorescence microscope, and photographs were generated using a Kodak digital camera.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Daniel Dominguez and Mauro Calabrese for critical reading of the manuscript. This work is supported by National Institutes of Health (NIH) grant R01-CA158283 and the Jefferson Pilot award to Z.W. Y.W. is funded by the Young Thousand Talents Program and the National Natural Science Foundation of China (grants 31471235 and 81422038).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048272.114.
REFERENCES
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE 2010. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6:e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Sarnow P 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417. [DOI] [PubMed] [Google Scholar]
- Choudhury R, Roy SG, Tsai YS, Tripathy A, Graves LM, Wang Z 2014. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun 5:3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B 1992. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L, Baralle M, Buratti E 2013. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA 4:49–60. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci 102:16176–16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR 1998. A tale of two termini: A functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1–11. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Preiss T 2013. Circular RNAs: splicing's enigma variations. EMBO J 32:923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14:153–165. [DOI] [PubMed] [Google Scholar]
- Matera AG, Wang Z 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B 1991. Scrambled exons. Cell 64:607–613. [DOI] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia-Blanco MA 1996. Exon circularization in mammalian nuclear extracts. RNA 2:603–610. [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 15:183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z 2014. Systematical identification of splicing regulatory cis-elements and cognate trans-factors. Methods 65:350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ma M, Xiao X, Wang Z 2012. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z 2013. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol 20:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J 2014a. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9:e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q, Dominguez D, Wang Z 2014b. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 26:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Burge CB 2007. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci 104:18583–18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL 2013. Circular intronic long noncoding RNAs. Mol Cell 51:792–806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.